This article presents evidence, obtained in Arabidopsis thaliana, that two central DNA repair proteins, the recombinases DMC1 and RAD51, are spatially and functionally separated, respectively, during meiotic DNA repair and shows that a well-known DNA damage response factor, the ATR kinase, is involved in regulating DMC1 deposition.
Abstract
Meiosis ensures the reduction of the genome before the formation of generative cells and promotes the exchange of genetic information between homologous chromosomes by recombination. Essential for these events are programmed DNA double strand breaks (DSBs) providing single-stranded DNA overhangs after their processing. These overhangs, together with the RADiation sensitive51 (RAD51) and DMC1 Disrupted Meiotic cDNA1 (DMC1) recombinases, mediate the search for homologous sequences. Current models propose that the two ends flanking a meiotic DSB have different fates during DNA repair, but the molecular details remained elusive. Here we present evidence, obtained in the model plant Arabidopsis thaliana, that the two recombinases, RAD51 and DMC1, localize to opposite sides of a meiotic DSB. We further demonstrate that the ATR kinase is involved in regulating DMC1 deposition at meiotic DSB sites, and that its elimination allows DMC1-mediated meiotic DSB repair even in the absence of RAD51. DMC1’s ability to promote interhomolog DSB repair is not a property of the protein itself but the consequence of an ASYNAPTIC1 (Hop1)-mediated impediment for intersister repair. Taken together, these results demonstrate that DMC1 functions independently and spatially separated from RAD51 during meiosis and that ATR is an integral part of the regular meiotic program.
INTRODUCTION
Reshuffling of genetic information during meiosis depends on the timely and spatially coordinated formation of DNA double strand breaks (DSBs) and their repair. In different organisms, various members of DSB-forming complexes have been identified (reviewed in Keeney, 2001; Edlinger and Schlögelhofer, 2011), with the conserved topoisomerase-related SPO11 protein as the catalytically active factor (Keeney et al., 1997). Two SPO11 proteins work in concert at a given DSB site and become covalently linked to the 5′ ends of DNA. Work mostly performed in Saccharomyces cerevisiae and Schizosaccharomyces pombe demonstrated that the SPO11 proteins are subsequently released together with a short DNA oligonucleotide by an endonucleolytic cleavage adjacent to the break site, mediated by the Mre11/Rad50/Xrs2-Nbs1 (MRX/N) complex in conjunction with Sae2/Com1 (Neale et al., 2005; Hartung et al., 2007; Uanschou et al., 2007; Milman et al., 2009). The DNA break sites therefore have, as demonstrated for S. cerevisiae and S. pombe, protruding 3′ termini, which are further extended by resection of the 5′ termini depending on the activities of the MRX/N complex and Exo1 (Cromie and Smith, 2008; Mimitou and Symington, 2008; Farah et al., 2009; Manfrini et al., 2010; Zakharyevich et al., 2010; Garcia et al., 2011). The extensive single-stranded DNA (ssDNA) overhangs are bound by the heterotrimeric replication protein A (RPA) complex with high affinity, a prerequisite for the loading of the strand-exchange proteins Rad51 and Dmc1 (reviewed in Fanning et al., 2006; Broderick et al., 2010). Shown for various organisms, although not formally for plants, RPA-coated ssDNA filaments are potent activators of the Ataxia telangiectasia-mutated and Rad3-related (ATR) damage signaling kinase, which interacts with RPA via its associated partner, ATR-interactin protein (ATRIP) (Cortez et al., 2001; Zou and Elledge, 2003; Cimprich and Cortez, 2008; Sweeney et al., 2009). ATR belongs to the family of phosphatidylinositol 3-kinase–related kinases (PIKK) (Cimprich and Cortez, 2008), and its closest relative is the Ataxia telangiectasia mutated (ATM) kinase. An essential role for ATM and/or ATR in meiosis has been reported for a variety of organisms (reviewed in Carballo and Cha, 2007). ATM is indispensable for mouse meiosis, whereas Atr−/− mice are inviable; therefore, the effect of ATR on meiosis in mice is still unknown (Xu et al., 1996; Brown and Baltimore, 2000; O’Driscoll, 2009). The related yeast proteins Tel1 (ATM) and Mec1 (ATR) phosphorylate a variety of proteins involved in meiotic recombination, among them the axis protein Hop1 (Hollingsworth and Byers, 1989; Schwacha and Kleckner, 1994; Hollingsworth and Ponte, 1997; Niu et al., 2005). Only mec1Δ mutants show meiotic defects, whereas tel1Δ mutants divide normally in meiosis (Kato and Ogawa, 1994; Usui et al., 2001; Cha and Kleckner, 2002; Nakada et al., 2003; Carballo et al., 2008). Arabidopsis thaliana atm mutants display chromosomal fragmentation in most meiocytes, leading to reduced fertility (Garcia et al., 2003). atr mutant plants, by contrast, are fully fertile, but atm atr double mutants are completely sterile, because of severe chromatin fragmentation (Culligan et al., 2004; Culligan et al., 2006; Culligan and Britt, 2008).
In most organisms, many meiotic breaks are generated in a single meiocyte (Buhler et al., 2007; Sanchez-Moran et al., 2007; Vignard et al., 2007; Pan et al., 2011). Only a fraction of those is repaired in such a way that maternal and paternal chromosome arms get connected to form a crossover (Higgins et al., 2004; Mercier et al., 2005; Wijeratne et al., 2006; Chen et al., 2008; Mancera et al., 2008). The other breaks are repaired to yield noncrossover products, possibly utilizing a synthesis-dependent strand annealing pathway (Allers and Lichten, 2001; McMahill et al., 2007), or are repaired via intersister recombination. It has long been speculated that the two ends of a meiotic DSB have distinct fates during meiotic DNA repair (Blat et al., 2002; Goldfarb and Lichten, 2010; Storlazzi et al., 2010), mediated by protein complexes of different composition. Although the nucleoprotein filament formed at one side of a meiotic DSB may probe for a repair template (like a “search tentacle”) (Hunter and Kleckner, 2001; Blat et al., 2002; Kim et al., 2010), the nucleoprotein filament formed at the other side has to be temporally retained to avoid a deleterious second invasion into another template DNA. It remained unclear how the two ends of a meiotic DSB are differentially coordinated; however, RAD51 and DMC1 seemed instrumental (Bishop, 1994; Shinohara et al., 1997). Earlier work in yeast demonstrated that Rad51 and Dmc1 function in independent meiotic repair pathways and that their loading onto RPA-coated ssDNA is supported by distinct sets of proteins (Bishop, 1994; Dresser et al., 1997; Gasior et al., 2001; Hayase et al., 2004; Lao et al., 2008). Regulation of recombinase loading is different in higher eukaryotes (including plants), because, for instance, BRCA2 (a protein not found in yeast) is essential for RAD51 loading (Siaud et al., 2004) (reviewed in San Filippo et al., 2008). Other proteins instrumental for loading of Rad51 or Dmc1 in yeast, like the Mei5/Sae3 complex, have not been found in Arabidopsis (Ray and Langer, 2002; Liu and Heyer, 2011), and the molecular function of the recently discovered RAD52-like proteins has not yet been defined (Samach et al., 2011). In yeast, Rad51 and Dmc1 proteins have been found to localize side-by-side in some well-spread meiotic nuclei (Shinohara et al., 1997; Shinohara et al., 2000). However, a side-by-side localization of RAD51 and DMC1 has not been revealed in previous studies performed in higher eukaryotes (Terasawa et al., 1995; Tarsounas et al., 1999). The molecular activities of RAD51 and DMC1 proteins have been studied in depth in vitro. Both proteins form helical structures on ssDNA and mediate the invasion of those so-called presynaptic filaments into homologous duplex DNA to form displacement loops (Shinohara et al., 1992; Ogawa et al., 1993; Benson et al., 1994; Sung, 1994; Baumann et al., 1996; Li et al., 1997; Masson et al., 1999; Hong et al., 2001; Sehorn et al., 2004; Yu and Egelman, 2010). Although similar in structure, some differences between the two recombinases may exist. Recently, it has been shown that DMC1-mediated displacement loop structures are more resistant to helicase-mediated dissociation in vitro (Bugreev et al., 2010). Importantly, the activities of both proteins are modulated by a range of accessory proteins, accounting for the differences between RAD51 and DMC1 function in vivo (Sheridan and Bishop, 2006; Sheridan et al., 2008; Kagawa and Kurumizaka, 2010; Okorokov et al., 2010; Dray et al., 2011).
Below, we present data from extensive genetic and cytological analyses, demonstrating that RAD51 and DMC1 predominantly localize to different sides of a meiotic DSB, that ATR is involved in regulating DMC1 deposition, and that DMC1’s proficiency to promote meiotic DNA repair using the homologous chromosome is governed by the axial element protein ASY1 (Hop1).
RESULTS
A Mutation in ATR Suppresses the Severe Meiotic Defects of rad51 Mutants
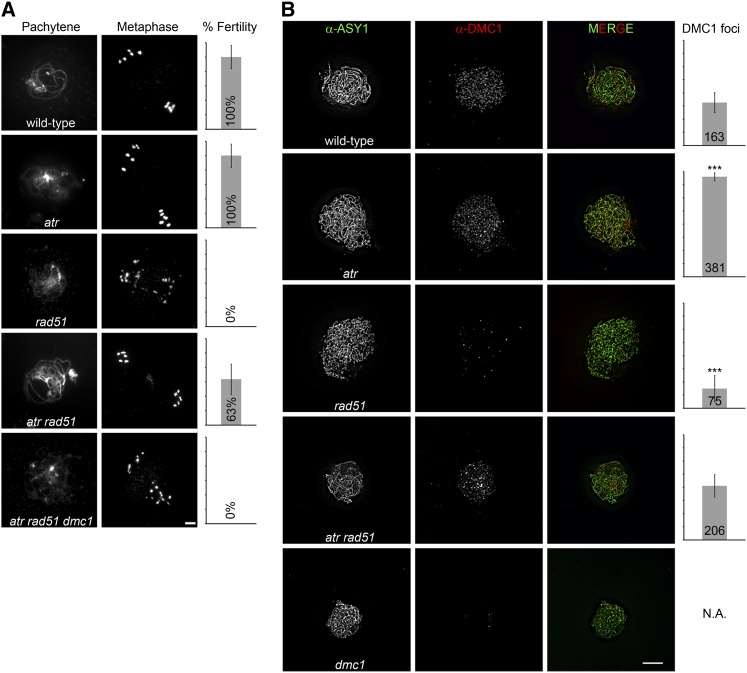
We have shown previously (Vignard et al., 2007) that the number of DMC1 foci, believed to represent the number of DMC1 nucleoprotein filaments, is sharply decreased in Arabidopsis rad51 null mutants (rad51-1 [Li et al., 2004]) (see Supplemental Figure 1 and Supplemental Methods 1 online) (75 ± 50, n = 11, P = 3.08 × 10−5) compared with the number of foci seen in wild-type plants (163 ± 40, n = 12) (Figure 1B). rad51 mutant plants are completely sterile and show severe DNA fragmentation (Li et al., 2004) (Figure 1A). We reasoned that plants lacking the RAD51 recombinase are deficient in forming nucleoprotein filaments containing DMC1, possibly because of persisting RPA-coated ssDNA (Fanning et al., 2006). As outlined above, RPA activates ATR, which may impede DMC1 deposition and may delay meiotic progression. To test this hypothesis, we crossed the rad51 mutant to a previously characterized atr null mutant (atr-2 [Culligan et al., 2004]). Indeed, in atr rad51 double mutants, DMC1 foci numbers were restored to wild-type levels (206 ± 43, n = 9; P = 0.197; number not significantly different from the wild type). Interestingly, significantly more DMC1 foci (381 ± 16, n = 6, P = 2.67 × 10−4) were observed in atr single mutants (Figure 1B). We further analyzed RAD51 foci formation in the atr mutant background and observed a minor but significant reduction (146 ± 27, n = 6, P = 0.01) compared with the wild type (186 ± 40, n = 12). RAD51 foci numbers were not altered in dmc1 mutants (dmc1 [Couteau et al., 1999]) (207 ± 40, n = 5, P = 0.9188) (see Supplemental Figure 2B online). Strikingly, the severe rad51 fertility defect (lack of seed formation; n = 258 siliques) was suppressed by atr (29 ± 10 seeds per silique; n = 715 siliques). Both wild-type and atr mutant plants produce ∼46 seeds per silique (plus or minus seven seeds, n = 483 siliques) (Figure 1A). Furthermore, the pervasive meiotic DNA fragmentation observed in all rad51 meiocytes was alleviated in atr rad51 double mutants (Figure 1A; see Supplemental Figure 2A online), with 36% (n = 117) of all observed meiocytes showing intact chromosomes. The reestablished meiotic DNA repair in atr rad51 mutant plants depended on the presence of functional DMC1 (Figure 1A). We reasoned that eliminating a cofactor of ATR—namely ATRIP (Cortez et al., 2001; Sweeney et al., 2009)—instead of ATR may recapitulate the rescue of the meiotic defects observed in atr rad51 double mutants. In line with our idea, atrip rad51 double mutants generate approximately two seeds per silique (n = 48 siliques), as opposed to the complete sterility seen in rad51 mutant plants. The lower efficiency in rescuing the rad51 deficiency seen in atrip rad51 double mutants compared with atr rad51 double mutants may be explained by residual activity of ATR in an atrip mutant background (Unsal-Kaçmaz and Sancar, 2004). Taking these results together, we conclude that successful RAD51 nucleoprotein filament formation during meiosis attenuates ATR signaling, which otherwise negatively regulates DMC1 nucleoprotein filament formation. The results furthermore demonstrate that Arabidopsis DMC1 can act independently of RAD51 as a bona fide recombinase in vivo, because it promotes DNA repair, chromosome pairing (inferred from observed pachytene stages) (Figure 1A), and synapsis (see Supplemental Figure 3 online) in atr rad51 double mutants. Our observations are consistent with the findings of in vitro recombinase activity of heterologously expressed and purified DMC1 proteins from various organisms and with Dmc1-mediated strand exchange activities observed in yeast rad51 mutants (Schwacha and Kleckner, 1997; Shinohara et al., 1997; Hong et al., 2001; Sheridan et al., 2008; Bugreev et al., 2010).
Figure 1.
A Mutation in ATR Suppresses the Severe Meiotic Defects of rad51 Mutants.
(A) Meiotic spreads of PMCs stained with DAPI and seed formation in percentage of the wild type. atr mutants are as fertile as wild-type plants and show normal meiotic progression. In rad51 mutant plants, homologous chromosomes do not pair, and severe chromatin fragmentation can be observed, leading to complete sterility. This phenotype is largely alleviated in atr rad51 double mutants. DNA repair in atr rad51 double mutants crucially depends on DMC1, because complete sterility and severe DNA fragmentation are observed in atr rad51 dmc1 triple mutants.
(B) Images of nuclear spreads of PMCs at zygotene stage with DMC1 (red) and ASY1 (green) detected by immunofluorescence. Bars represent DMC1 foci numbers per meiotic nucleus. DMC1 foci numbers significantly increase in the absence of ATR and are significantly reduced in the absence of RAD51. In atr rad51 double mutants, DMC1 foci numbers are similar to the wild type. Error bars represent sd; * P < 0.05, ** P < 0.01, *** P < 0.001; Wilcoxon-Mann-Whitney test.
Bars = 10 µm.
RAD51 and DMC1 Do Not Colocalize and Form Homotypic Foci during Meiosis
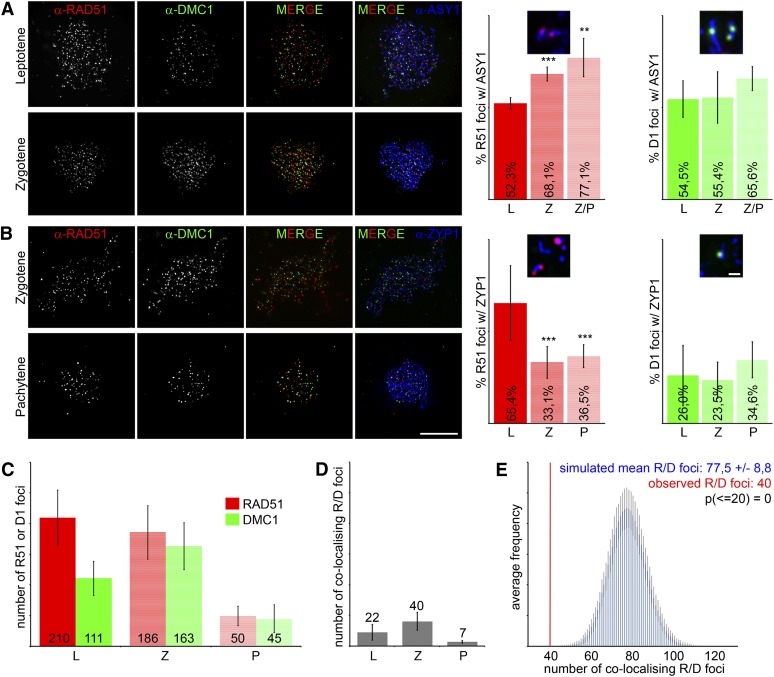
We consequently speculated that during wild-type meiosis, DMC1 and RAD51, respectively, form homogenous nucleoprotein filaments on ssDNA generated at DSB sites. To test this hypothesis, we simultaneously stained Arabidopsis RAD51 and DMC1 proteins on meiotic chromatin spreads (Figure 2). Before this experiment, we confirmed the specificity of the available antibodies against Arabidopsis RAD51 (Mercier et al., 2003; Kerzendorfer et al., 2006) and DMC1 (Chelysheva et al., 2007) (see Supplemental Figure 4A online). To distinguish early and late stages of meiotic prophase, we monitored axis formation by staining the axial element protein ASY1 (Armstrong et al., 2002; Higgins et al., 2004) (Figure 2A) or synaptonemal complex (SC) extension by staining the SC component ZYP1 (Higgins et al., 2005) (Figure 2B). We counted 210 ± 45 (n = 9) RAD51 foci in leptotene, with numbers decreasing to 186 ± 44 (n = 12) in zygotene and further to 50 ± 16 (n = 8) in pachytene. By contrast, DMC1 foci numbers were 111 ± 28 (n = 9) in leptotene, increasing to 163 ± 38 (n = 12) in zygotene and decreasing to 45 ± 23 (n = 8) in pachytene (Figure 2C). Even though the total number of RAD51 foci decreased from leptotene to pachytene, significantly more foci were found associated with ASY1 at the later zygotene/pachytene stage (77.1%, P = 5.9 × 10−3) than during earlier leptotene (52.3%) (Figure 2A). Notably, the relative number of RAD51 foci associated with ZYP1 decreased over time from 65.4% in leptotene to 36.5% in pachytene (P = 5.6 × 10−10) (Figure 2B). This suggests that RAD51 foci represent nucleoprotein filaments that stay predominantly axis-associated and furthermore that ZYP1 is loaded next to RAD51 at DSB sites, as suggested previously (Agarwal and Roeder, 2000). Decreasing total and ZYP1-associated RAD51 foci numbers during meiotic progression could be explained by depletion of RAD51 at sites of successful meiotic DNA repair (Franklin et al., 1999; Allers and Lichten, 2001; Bugreev et al., 2010). In accordance with our initial assumption that DMC1 foci formation depends on RAD51, the maximum numbers of DMC1 foci over time follow those of RAD51 with some delay (Figure 2C). The relative amount of axial element–associated and SC-associated DMC1 foci, respectively, remained constant during meiotic prophase (axial element–associated: 54.5% in leptotene and 65.7% in late zygotene; numbers not significantly different, P = 0.1343; SC-associated: 26% in leptotene and 34.6% in pachytene; numbers not significantly different, P = 0.8798) (Figures 2A and 2B). Many DMC1 foci are not axis-associated at all times and may therefore represent dynamic nucleoprotein filaments in search of homologous repair templates.
Figure 2.
RAD51 and DMC1 Display Different Dynamics of Appearance and Do Not Colocalize.
(A) At left, images of nuclear spreads of wild-type leptotene and zygotene PMCs with RAD51 (red), DMC1 (green), and ASY1 (blue) detected by immunofluorescence. At right, quantification of the observed association of either recombinase with the axial element protein ASY1. Relative numbers of ASY1-associated RAD51 foci were significantly enriched in zygotene and late zygotene stages compared with leptotene.
(B) At left, images of nuclear spreads of wild-type zygotene and pachytene PMCs with RAD51 (red), DMC1 (green), and ZYP1 (blue) detected by immunofluorescence. At right, quantification of the observed association of either recombinase with the synaptonemal complex protein ZYP1. Relative numbers of ZYP1-associated RAD51 foci were significantly reduced in zygotene and pachytene stages compared with leptotene.
(C) Mean numbers of RAD51 or DMC1 foci on chromatin spreads during leptotene, zygotene, and pachytene stages.
(D) Mean numbers of colocalizing RAD51 and DMC1 foci during leptotene, zygotene, and pachytene stages, related to the mean total numbers shown in (C).
(E) Computational Monte Carlo simulation for an exemplary, individually scored cell to determine the expected random frequency of colocalizing RAD51 and DMC1 foci. Error bars represent sd; * P < 0.05, ** P < 0.01, *** P <0.001; Fisher's exact test. D1, DMC1; L, leptotene; P, pachytene; R51, RAD51; R/D foci, colocalizing RAD51 and DMC1 foci; Z, zygotene; Z/P, late zygotene.
Bars = 10 µm.
In our experiments, RAD51 and DMC1 were not found to colocalize extensively during any meiotic stage (Figures 2A to 2D). Only a small number of all RAD51 and DMC1 foci colocalized in leptotene (22 ± 13, n = 7), zygotene (40 ± 14, n = 8), or pachytene (7 ± 3, n = 8) stages (Figure 2D), and a complete overlap was never observed. Two foci were counted as colocalizing foci when their centers were no further than 330 nm apart. This distance was chosen based on the mean recombinase focus diameter of 300 nm (n = 220 foci) and therefore includes all foci that would overlap or just touch each other. We added a tolerance of 10% to account for errors in measurement. We performed computational Monte Carlo simulations with parameters (foci numbers and diameters, nuclear areas) taken from individually scored cells to predict the expected random frequency of colocalization of RAD51 and DMC1 foci (Figure 2E; see Supplemental Figure 4B online). In the simulation, the two populations of foci were randomly distributed over the given nuclear area 200,000 times, revealing that random overlaps were significantly more frequent (77.5 ± 8.8 colocalizing signals) than observed ones (40 colocalizing signals; cumulative probability = 0; P = 2.5 × 10−2, n = 3). We therefore inferred that a specific mechanism keeps the two different classes of foci apart and that RAD51 and DMC1 bind to different microcompartments that tend not to overlap. We concluded that mixed nucleoprotein filaments, containing both RAD51 and DMC1 proteins, are highly unlikely to exist. Our findings instead suggest that either (1) individual meiotic DSBs are processed by either RAD51 or DMC1 or that (2) RAD51 and DMC1 occupy opposite DNA ends at a meiotic DSB. The second probability predicts the existence of a significantly higher proportion of side-by-side localizations (doublets) of RAD51 and DMC1, compared with RAD51–RAD51 or DMC1–DMC1 doublets.
Cytological Evidence That RAD51 and DMC1 Assemble Predominantly onto Opposite Sides of a Meiotic DSB
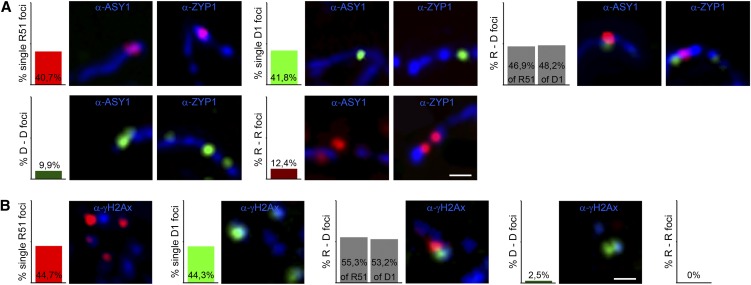
We analyzed our images for the occurrence of side-by-side localizations and counted foci signals as doublets when their centers were no further than 540 nm apart. This distance was based on the largest found recombinase foci being 490 nm in size (very rare; n = 220 foci). If, theoretically, two of them would be found to at least touch each other, their foci centers would be 490 nm apart. We added a tolerance of 10% to account for errors in measurement. We reasoned that meiotic DSBs are rare events on a genome-wide scale and that well-separated, single chromatin threads emanating from the mass of chromatin would allow us to characterize the configuration of recombinases at individual DSB sites. According to the definition above, 40.7% of the observed RAD51 signals (n = 145 foci) and 41.8% of the observed DMC1 signals (n = 141 foci) were single signals, whereas 46.9% of all RAD51 and 48.2% of all DMC1 foci were found in RAD51–DMC1 doublets. By contrast, only 10% of all DMC1 foci and 12.4% of all RAD51 foci, respectively, were arranged as homomeric doublets with only one type of recombinase present (RAD51–RAD51, DMC1–DMC1) (n = 79 singularized DNA threads from eight nuclei) (Figure 3A). Interestingly, the proportional numbers of observed DMC1–DMC1 doublets on single chromatin threads were 5.5-fold increased in atr (P = 3.9 × 10−5; n = 42 foci) and 4.3-fold increased in atr rad51 mutants (P = 0.01101; n = 60 foci) when compared with the wild type. The relative numbers of RAD51–RAD51 doublets were unchanged in atr mutants (data not shown).
Figure 3.
RAD51 and DMC1 Predominantly Occupy Opposite Sides of a Meiotic DSB.
(A) Picture details from nuclear spreads of wild-type PMCs with RAD51 (red), DMC1 (green), and ASY1 or ZYP1 (blue) detected by immunofluorescence. Each image shows an example of a DNA thread emanating from the mass of chromatin. The occurrences of single or doublet RAD51 or DMC1 foci in relation to all observed RAD51 or DMC1 foci within the chosen sections were quantified.
(B) Picture details of nuclear spreads of wild-type leptotene PMCs with RAD51 (red), DMC1 (green), and γH2Ax (blue) detected by immunofluorescence. The occurrences of single or doublet RAD51 or DMC1 foci associated with γH2Ax signals were quantified. In both analyses, RAD51–DMC1 doublets were significantly overrepresented compared with DMC1–DMC1 and RAD51–RAD51 doublets, respectively. D1, DMC1; D–D, DMC1 doublet; R51, RAD51; R–D, RAD51 and DMC1 doublet; R–R, RAD51 doublet.
Bars = 0.5 µm.
Next, we performed immunostaining of meiotic spreads using antibodies specific for RAD51, DMC1, and γH2AX. γH2AX is the phosphorylated form of the histone variant H2AX, transiently generated at and locally restricted to sites of DNA damage (Rogakou et al., 1998; Mahadevaiah et al., 2001; Amiard et al., 2010). We used γH2AX staining as a marker for individual DSB sites and quantified associated RAD51 and DMC1 foci. An association was positively scored when the focus center of at least one recombinase was no further than 330 nm apart from the γH2AX focus center. We found that of the γH2AX-associated signals, 44.7% of the RAD51 and 44.3% of the DMC1 foci were single signals, whereas 55.3% of all RAD51 and 53.2% of all DMC1 foci were found in R–D doublets. Only 2.6% of all DMC1 foci and none of the RAD51 foci were arranged as homomeric doublets (Figure 3B; see Supplemental Figure 5 online) (n = 112 γH2AX foci from six nuclei). γH2AX and both recombinases showed a high degree of colocalization, because 96.5% of all R–D foci, 75.1% of all single RAD51 foci, and 90.0% of all single DMC1 foci were associated with γH2AX staining (n = six nuclei). The interfoci distances between RAD51–RAD51, DMC1–DMC1, and RAD51–DMC1 doublets, respectively, did not significantly differ for any given foci distance between 150 and 450 nm (see Supplemental Table 1 online). The highly significant overrepresentation of RAD51–DMC1 doublets over homomeric doublets (P < 2.79 × 10−9 for all cases) strongly argues that most DSBs are flanked by two different filaments, comprised of either RAD51 or DMC1. Single RAD51 and DMC1 signals may represent intermediate steps during the repair process.
ASY1 Inhibits DMC1-Mediated Intersister DNA Repair
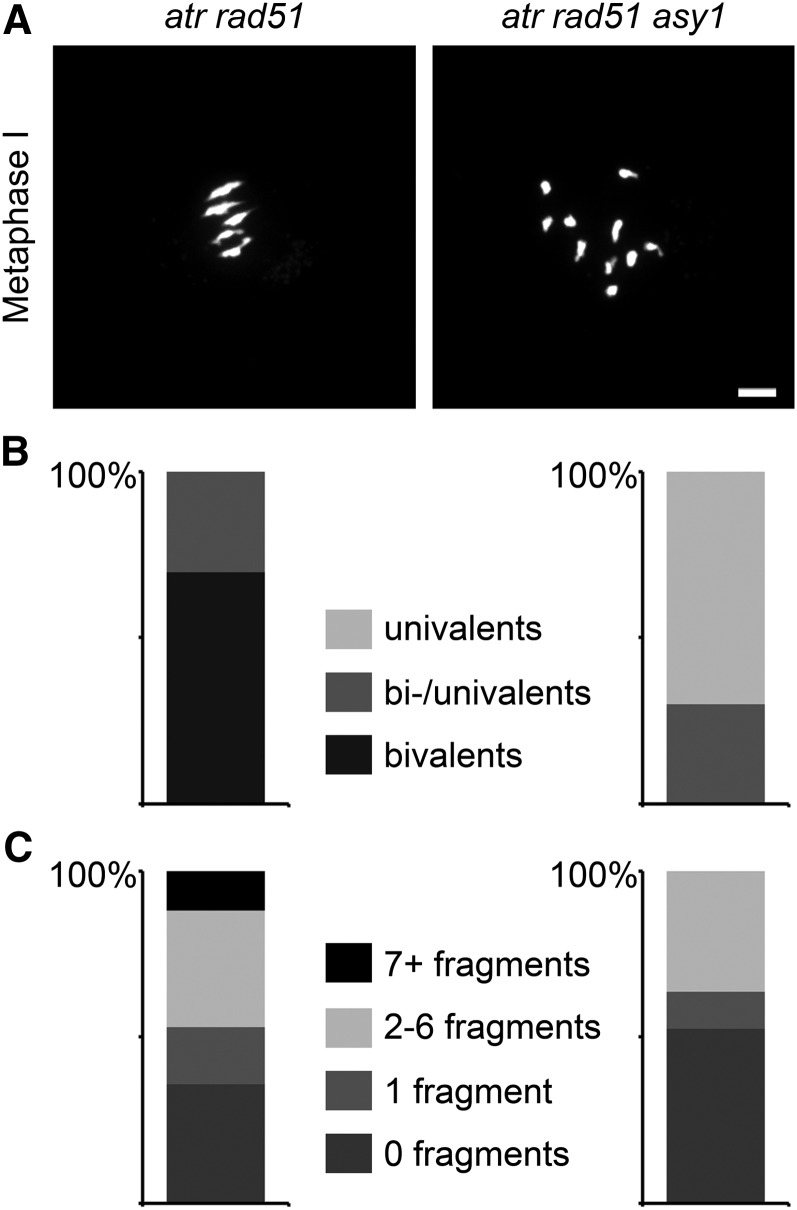
DMC1 nucleoprotein filaments may furthermore be highly dynamic, as suggested by the localization of DMC1 foci relative to ASY1 and ZYP1 signals (Figure 2). Our mutant combinations allow a detailed analysis of DMC1’s properties during DNA repair. In atr rad51 double mutants, DMC1 becomes essential for DNA repair (Figure 1A). Meiotic DSBs in atr rad51 were repaired to a certain extent via the homologous chromosome, resulting in bivalent formation in 69.7% of all meiocytes. The residual 30.3% of cells showed a mixture of univalents and bivalents (n = 33). Of all observed atr rad51 double mutant meiocytes, 36% displayed no DNA fragmentation (Figure 4). In Arabidopsis dmc1 mutants, meiotic DSBs are reliably repaired in a RAD51-dependent manner, most likely using the sister chromatid as a repair template, leading to 100% univalents (Couteau et al., 1999; Sanchez-Moran et al., 2007). Arabidopsis atr dmc1 double mutants were cytologically indistinguishable from dmc1 single mutants (see Supplemental Figure 2A online). Our observations are in accordance with previous findings (reviewed in Kagawa and Kurumizaka, 2010) establishing that the ability to repair meiotic DSBs using homologous chromosomes as repair templates is predominantly associated with the DMC1 recombinase. To test whether this competence is an intrinsic property of the DMC1 protein itself or determined by accessory factors, as suggested earlier (Schwacha and Kleckner, 1997; Sheridan et al., 2008), we generated the atr rad51 asy1 triple mutant. The HORMA-domain protein ASY1 is part of the meiosis-specific chromosome axes and displays highest similarity to the yeast Hop1 protein (Sanchez-Moran et al., 2008). Hop1 is instrumental for establishing the interhomolog bias during meiotic DNA repair. It has been shown that upon DSB formation, Tel1 (ATM) and/or Mec1 (ATR) phosphorylate Hop1, subsequently leading to activation of the Mek1 kinase. Mek1 suppresses Rad51-mediated strand invasion of sister chromatids by (transient) inhibition of Rad51/Rad54 complex formation and via a Rad54-independent mechanism (Niu et al., 2005; Carballo et al., 2008; Niu et al., 2009). The plant asy1 mutants show a high frequency of univalent formation (Caryl et al., 2000), and asy1 dmc1 double mutants exclusively form univalents and display efficient repair of meiotic DSBs. Different from yeast hop1 mutants, efficiency of DSB formation has not been found to be affected in Arabidopsis asy1 mutants (Sanchez-Moran et al., 2007). In Arabidopsis atr rad51 asy1 triple mutants, we observed univalent formation in 70% and a mixture of bivalent and univalent formation in 30% of all cells (n = 40) (Figures 4A and 4B). DNA repair was even more efficient in triple mutants than in atr rad51 double mutants (Figure 4C), with 36% (n = 117) of all meiocytes observed in atr rad51 double mutants and 53% (n = 80) of all cells in atr rad51 asy1 triple mutants displaying no DNA fragmentation. These results demonstrate that DMC1 may use both sister chromatids and homologous chromosomes as repair templates, as shown in earlier experiments performed in budding yeast (Shinohara et al., 1992; Schwacha and Kleckner, 1997). We therefore conclude that during wild-type meiosis, the bias for interhomolog DNA repair is supported by locally activated ASY1 inhibiting DMC1-coated nucleoprotein filaments from using sister chromatids as repair templates.
Figure 4.
ASY1 Inhibits DMC1-Mediated Intersister DNA Repair.
(A) Nuclear spreads of PMCs during metaphase I displaying five bivalents in atr rad51 and 10 univalents in atr rad51 asy1 triple mutants.
(B) Quantification of meiocytes with univalents, bivalents, or both (irrespective of DNA fragmentation). In atr rad51, most cells show bivalent formation, and only about one-third display bivalents and univalents (n = 33). In atr rad51 asy1, most cells show only univalents, one-third show bivalents and univalents, and cells containing only bivalents were never observed (n = 40).
(C) Quantification of chromosome fragmentation in the double and triple mutants. Of all observed atr rad51 double mutant meiocytes, 36% show no DNA fragmentation, 17% show only one additional DAPI-stained fragment, 35% show two to six additional DAPI-stained bodies, and 12% show severe DNA fragmentation with more than seven fragments (n = 117). DNA fragmentation is reduced in the triple mutants: 53% of cells show successful DNA repair, 11% display only one fragment, 35% have one to six fragments, and severe fragmentation with more than seven chromatin fragments was never observed in atr rad51 asy1 meiocytes (n = 80).
Bar = 10 µm.
DISCUSSION
Our data allow the extension of the current model of meiotic DSB repair (Figure 5). We established that Arabidopsis DMC1 is sufficient to promote interhomolog DNA repair of meiotic DSBs even in the absence of RAD51. We demonstrated that RAD51 and DMC1 foci do not colocalize, consistent with the formation of homogenous nucleoprotein filaments with RAD51 and DMC1 occupying opposite DNA ends at meiotic DSBs, as previously suggested (Shinohara et al., 1997; Shinohara et al., 2000). We furthermore revealed that DMC1-coated nucleoprotein filaments are in principle competent to repair from both sister chromatids and homologous chromosomes. In the wild type, they are impeded from accessing the sister chromatid, presumably by locally activated ASY1 (Hop1), consistent with earlier reports from yeasts (Niu et al., 2005; Carballo et al., 2008; Goldfarb and Lichten, 2010; Latypov et al., 2010). Building on these earlier reports, we infer that this local, DSB-dependent activation is mediated by ATM, because the interhomolog bias of meiotic DNA repair is still intact in atr rad51 mutants. Our data indicate that DMC1 abundance at DSB sites is negatively regulated by the ATR kinase and that the strand-exchange activity of RAD51 may be dispensable for meiosis. Nevertheless, in all organisms analyzed, RAD51 is an essential meiotic repair protein (Shinohara et al., 1992; Rinaldo et al., 2002; Li et al., 2004; Howard-Till et al., 2011).
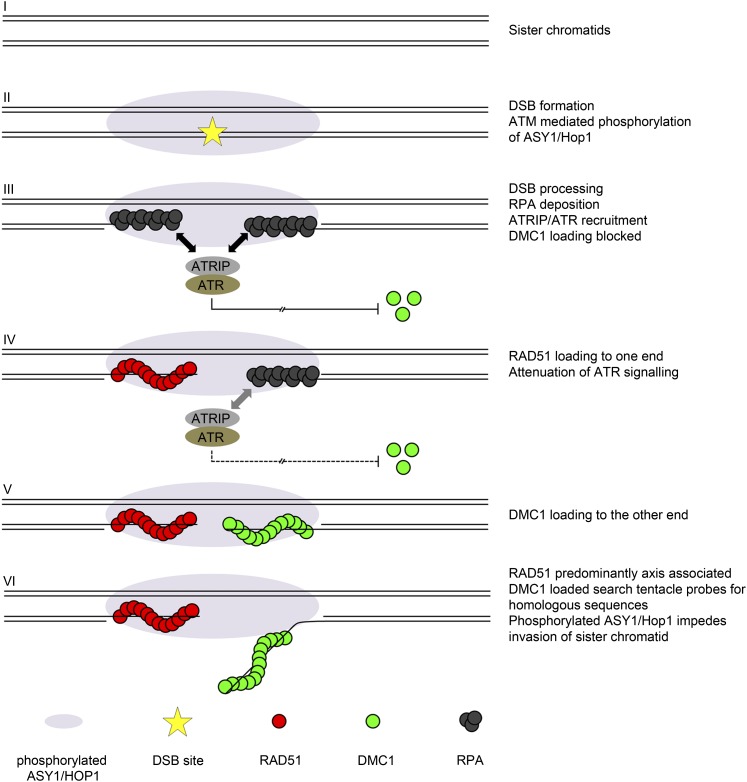
Figure 5.
Model for Early Events during Meiotic Recombination.
One of the sister chromatids (I) receives a programmed, SPO11-mediated DNA DSB early in meiotic prophase. The DNA damage leads to activation of the ATM kinase that phosphorylates the axial element protein ASY1 (Hop1), thereby generating a locally restricted signal in the vicinity of the DNA DSB site (II). DSB processing generates 3′ ssDNA overhangs that are rapidly bound by the trimeric RPA complex (III). RPA deposition recruits and activates ATR via its interaction partner ATRIP. ATR signaling blocks loading of the DMC1 recombinase but not of RAD51. RAD51 is loaded and replaces RPA at one end of the DSB (IV). Attenuated ATR-signaling allows subsequent loading of DMC1 to the other end of the DSB (V). The RAD51-coated end is retained, and the DMC1 nucleoprotein filament is released to search for homologous sequences. Locally activated ASY1 impedes access to the sister chromatid, thereby promoting interhomolog DNA repair and recombination (VI).
RAD51 may not only be instrumental in the early stage of filament assembly, but later as well, if a DMC1-coated filament fails to successfully invade a homologous sequence. In such cases, RAD51 may be needed for repairing the DSB via the sister chromatid (Goldfarb and Lichten, 2010). Total numbers of DMC1 foci were elevated in atr mutants, and thorough investigation of DMC1 foci in atr or atr rad51 mutants revealed that the proportional numbers of observed DMC1–DMC1 doublets on single chromatin threads were strongly increased when compared with the wild type. Total RAD51 foci numbers were reduced in atr, and relative numbers of RAD51–RAD51 doublets were unchanged. The total number of all recombinase foci (RAD51 and DMC1 together) in atr mutants (553 ± 80) is significantly higher than the total number of all recombinase foci in the wild type (350 ± 41). This increase in numbers cannot sufficiently be explained by the observed increase of DMC1–DMC1 doublets and the mild decrease of RAD51 foci but suggests that the number of meiotic DSBs may be increased in atr mutants. These observations are in accordance with a recent study demonstrating that yeast strains lacking the homolog of atr (mec1Δ) or atm (tel1Δ), respectively, form more meiotic breaks by losing the constraint of limiting DSB formation at a given locus to only one of the four sister chromatids (Zhang et al., 2011). We therefore postulate that ATR negatively regulates DMC1 abundance by limiting its deposition to only one end of a processed meiotic DSB site and by preventing the formation of supernumerary meiotic DSBs. Two recent publications demonstrate that ATM also acts as a negative regulator of meiotic DSB formation in mouse and fly (Joyce et al., 2011; Lange et al., 2011). Further efforts are needed to clarify to what extent ATM and ATR negatively control meiotic DSB formation in Arabidopsis. atr mutant plants are as fertile as wild-type plants, and the elevated levels of DMC1 filaments may only be detrimental in cases where DMC1 fails to find a homologous nonsister sequence for DNA repair. Meiosis in isogenic laboratory lines is not challenged by deletions and insertions, but genomes in nature may vary, and DSBs may form in regions without an appropriate homologous sequence present (Goldfarb and Lichten, 2010).
Our observations emphasize the notion that ATR is an integral part of the regular meiotic program (Cha and Kleckner, 2002; Di Giacomo et al., 2005; Carballo and Cha, 2007; MacQueen and Hochwagen, 2011). Future studies will focus on the mechanisms that promote differential RAD51 and DMC1 loading. In Figure 5, we propose a model in which RAD51 loading may attenuate ATR signaling and thereby allow DMC1 loading. However, it is still not known whether ATR regulates DMC1 directly by limiting its ATPase and ssDNA binding activity by phosphorylation, as recently proposed for yeast Rad51 by Mec1 (Flott et al., 2011). The highly conserved motif for ATM/ATR-dependent phosphorylation is also present in Arabidopsis RAD51 and DMC1. Several additional factors may influence RAD51 and DMC1 filament assembly. RPA, for instance, modulates meiotic recombination in yeast (Bartrand et al., 2006; Liaw et al., 2011), and several members for each of the three subunits, one of which is implicated in meiotic DSB repair, are encoded in the genome of Arabidopsis (Shultz et al., 2007; Osman et al., 2009; Sakaguchi et al., 2009). The yeast Swi/Snf2-remodelling factors Rad54 and Tid1/Rdh54 specifically promote strand exchange reactions of Rad51 and Dmc1, respectively (Shinohara et al., 2000; Niu et al., 2009; Mazin et al., 2010; Ceballos and Heyer, 2011). Several members of this family can be found in the genome of Arabidopsis, but so far none of them has been found to be required for meiosis (P. Schlögelhofer, unpublished data). In yeast, Rad51 nucleoprotein filament assembly requires the Rad51 paralogs Rad55 and Rad57, and Dmc1 nucleoprotein filament assembly depends on Mei5 and Sae3. None of the respective homologs have yet been identified in Arabidopsis. Five Rad51 paralogs (RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3) have been identified in higher eukaryotes, including plants. In Arabidopsis, only RAD51C and XRCC3 are involved in meiosis but most likely play a role in recombination after DMC1 and RAD51 loading (Bleuyard et al., 2005; Vignard et al., 2007). Consistently, RAD51C (Abe et al., 2005) is an essential factor for DMC1-mediated meiotic repair (data not shown). Finally, BRCA2, only found in higher eukaryotes, has been found to substitute to some extent for Rad52 function, stimulating loading of recombinases (reviewed in San Filippo et al., 2008; Liu and Heyer, 2011). Arabidopsis encodes two BRCA2 genes, and RNA interference lines that silence both copies show severe DNA fragmentation during meiosis (Siaud et al., 2004).
Alternatively, or additionally, ATR may indirectly influence the timing of the differential loading of recombinases by regulating meiotic progression. As shown, RAD51 loading is needed to overcome a meiotic checkpoint that depends on ATR. The replacement of RPA by RAD51 on one side of a DSB may attenuate ATR signaling and thereby allow DMC1 loading to the other side. In yeast, ATR (Mec1) ensures that meiotic DNA repair is completed before the first division can take place (reviewed in Carballo and Cha, 2007). In mammalian somatic cells, ATR triggers a signaling cascade that indirectly regulates the level of cyclin-dependent kinases in S-phase (Syljuåsen et al., 2005; Hurley and Bunz, 2007). Even though homologs of several key factors involved in these processes have not yet been identified in the genome of Arabidopsis, the conserved ATR kinase has been established as a central player during meiosis.
METHODS
Plant Growth Conditions
Plants were grown under long-day conditions (16 h light, 8 h dark, 21°C; 60 to 80% humidity, 5800 LUX, 3× Philips TLD 36W, and 2× Sylvana GroLUX 36W). All lines were Arabidopsis thaliana, ecotype Columbia, except when otherwise noted.
Mutant Plant Lines
The following mutant plant lines were used: atr-2 (SALK_032841 [Culligan et al., 2004]), dmc1 (Feldmann line 3668 [Couteau et al., 1999], ecotype Wassilewski), rad51-1 (GABI_134A01 [Li et al., 2004]), and asy1 (SALK_144182 [Sanchez-Moran et al., 2007]). Please refer to Supplemental Table 2 online for primer and genotyping information.
Cytology
Spreads of pollen mother cells (PMCs) for cytological detection of proteins were performed as described previously (Armstrong et al., 2002), with some modifications. In brief, anthers were dissected from several inflorescences and kept in artificial pond water (Miller and Gow, 1989) until further use. PMCs were released from anthers and collected in a small volume of artificial pond water (Chen et al., 2010). The liquid was exchanged for 15 μL of enzyme mixture (1% [w/v] cytohelicase [Pall], 1.5% [w/v] Suc, and 1% [w/v] polyvinylpyrrolidone, average molecular weight 40,000 [all Sigma-Aldrich]) (Albini et al., 1984), and meiocytes were incubated for 5 min at room temperature. A total of 8 μL of the enzyme mixture, containing extracted meiocytes, was transferred to a clean glass slide, and spreading was achieved by addition of 20 μL of 1% Lipsol (pH 9.0, borate buffer). After 4 min, 24 μL of 4% paraformaldehyde (pH 8.0) was added. Slides were air-dried and kept at room temperature until further use. Immunolocalization was performed as described before with some modifications (Armstrong et al., 2002). In brief, slides were rehydrated in 1× PBS before staining. Primary antibodies were applied, and slides were incubated at 4°C overnight in a humid atmosphere. After washing twice in 1× PBS, slides were sequentially incubated with appropriate secondary antibodies, each for 1 h at 37°C in a humid atmosphere. After washing again, slides were mounted with Vectashield (Vector Labs) supplied with 2 µg/mL 4′,6-diamidino-2-phenylindole (DAPI). In the case of the RAD51/DMC1/ASY1, RAD51/DMC1/ZYP1, and RAD51/DMC1/γH2AX sequential triple stainings, the first two primary antibodies (αDMC1 and αRAD51) were applied and incubated at 4°C overnight in a humid atmosphere. After washing twice in 1× PBS, slides were first incubated with the first two appropriate secondary antibodies and incubated for 1 h at 37°C. After washing, the third primary antibody (αASY1, αZYP1, or αγH2Ax) was added, and after a 3-h incubation at 37°C, slides were washed again, and the appropriate secondary antibody was added. Washing and mounting were then performed as described above. Analysis was performed on a Zeiss Axioplan microscope, where Z-stacks with 100-nm intervals were acquired with MetaMorph software. Z-stacks were deconvolved using AutoQuant software and are presented as projections done with HeliconFocus software. The RAD51/DMC1/ASY1, RAD51/DMC1/ZYP1, and RAD51/DMC1/γH2AX triple stainings were analyzed and processed on a Personal Deltavision system (Applied Precision). Recombinase foci were counted manually with the help of the Count tool in Adobe Photoshop CS4. The following primary antibodies were used as described previously: αASY1 raised in rabbit (Armstrong et al., 2002), αASY1 raised in rat (Higgins et al., 2004), αRAD51 raised in rat (Mercier et al., 2003; Kerzendorfer et al., 2006), αDMC1 raised in rabbit (Chelysheva et al., 2007), αZYP1 raised in rat (Higgins et al., 2005), and αγH2Ax raised in rabbit (Amiard et al., 2010). The secondary antibodies used were as follows: Goat-anti-rabbit conjugated to FITC (1:300; Sigma-Aldrich), goat-anti-rat conjugated to Cy3 (1:300; Chemicon), goat-anti-rabbit conjugated to Alexa Fluor 568 (1:500; Invitrogen), donkey-anti-rat conjugated to Alexa Fluor 488 (1:500; Invitrogen), and donkey-anti-rabbit conjugated to Cy5 (1:100; Jackson Immunoresearch).
Statistical Analysis
Monte Carlo Simulation of Foci Colocalization
To determine the expected number of RAD51 and DMC1 foci colocalizations, random distributions of two foci populations were simulated in silico (custom-made Java program). The same foci sizes, counts, and nucleus sizes (as measured in pixels) were applied, and 200 × 1000 simulations were performed. Foci were assumed to be circles, spreading areas (nuclei) as elliptic areas. Colocalization of a foci pair was defined by fulfillment of the condition (r1 + r2)2 ≥ (x2 − x1)2 + (y2 − y1)2, where r1 and r2 are the radii of focus 1 and focus 2, x1 and x2 are the respective x coordinates within the nucleus, and y1 and y2 are the respective y coordinates. Thus, two foci were said to colocalize when the distance between the two foci centers was smaller than or equal to the sum of their radii. To avoid distance measure artifacts, the numbers of observed colocalizations were calculated based on the observed radii, distances between centers, and percent overlapping areas of closely lying foci pairs. The average simulated overlap counts were subsequently compared with the observed counts. Three nuclei were treated as separate cases, because the spread areas (and thus foci sizes and counts) showed high variability. Each case resulted in a significantly lower number of observed colocalizations compared with the randomly distributed foci, with z values ranging from −4.237 to −2.678. For calculation of the overall P value, an unpaired, one-sided t test with confidence level = 0.95 was performed (R version 2.12.0; t.test).
Foci Counts
To test whether two foci size distributions are significantly different, a two-sided, unpaired Wilcoxon-Mann-Whitney test was performed (confidence level = 0.95; R 2.12.0, wilcox.test with exact P value and continuity correction). For testing the significance of RAD51 or DMC1 counts (as proportions of total RAD51 or DMC1 counts) associated with the axis or the synaptonemal complex during different meiotic phases, the means of the respective counts were subjected to the Fisher's exact test (R 2.12.0, fisher.test). The same test was applied for determination of the significant overrepresentation of RAD51–DMC1 doublets versus RAD51–RAD51 and DMC1–DMC1 doublets, respectively, and for determination of differences of interfoci distances comparing RAD51–RAD51, DMC1–DMC1, and RAD51–DMC1 doublets, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Molecular Characterization of the Arabidopsis rad51 Null Allele.
Supplemental Figure 2. A Mutation in ATR Suppresses the Severe Meiotic Defects of rad51 Mutants.
Supplemental Figure 3. Synaptonemal Complex Formation in atr rad51.
Supplemental Figure 4. Antibodies and Statistics.
Supplemental Figure 5. RAD51, DMC1, and γH2Ax Localization in Meiocytes.
Supplemental Table 1. Interfoci Distances.
Supplemental Table 2. Primer Information.
Supplemental Methods 1. Supplemental Methods for the Supplemental Data.
Supplementary Material
Acknowledgments
We thank Chris Franklin for providing antibodies against Arabidopsis RAD51, ASY1, and ZYP1, Raphael Mercier for providing an antibody against Arabidopsis DMC1, and Charles White for providing an antibody against Arabidopsis γH2AX. We thank Bernd Reiss for sending seeds of the rad51-1 mutant line, Anne Britt for sending seeds of the atr-2 mutant line, and Marie-Pascale Doutriaux for sending seeds of the dmc1 mutant line. We thank Juraj Gregan, Verena Jantsch, Franz Klein, Josef Loidl, Silvia Panizza, Martin Xaver, and the entire Schlögelhofer lab for discussions and/or for critically reading the article. This project was funded in part by the Austrian Academy of Sciences (Austrian Programme for Advanced Research and Technology 11230), the Austrian Science Fund (Special Research Program 3408), and the University of Vienna (I031-B).
AUTHOR CONTRIBUTIONS
M.-T.K. performed the genetic and cytological experiments, analyzed the data, and wrote parts of the article, C.U. generated and analyzed RAD51 and DMC1 antibodies, D.C. performed the statistical significance tests and the Monte Carlo simulations, and P.S. conceived the experiments, analyzed the data, and wrote the article.
Glossary
- DSB
double strand break
- ssDNA
single-stranded DNA
- SC
synaptonemal complex
- PMCs
pollen mother cells
- DAPI
4′,6-diamidino-2-phenylindole
References
- Abe K., Osakabe K., Nakayama S., Endo M., Tagiri A., Todoriki S., Ichikawa H., Toki S. (2005). Arabidopsis RAD51C gene is important for homologous recombination in meiosis and mitosis. Plant Physiol. 139: 896–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S., Roeder G.S. (2000). Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 102: 245–255 [DOI] [PubMed] [Google Scholar]
- Albini S.M., Jones G.H., Wallace B.M. (1984). A method for preparing two-dimensional surface-spreads of synaptonemal complexes from plant meiocytes for light and electron microscopy. Exp. Cell Res. 152: 280–285 [DOI] [PubMed] [Google Scholar]
- Allers T., Lichten M. (2001). Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57 [DOI] [PubMed] [Google Scholar]
- Amiard S., Charbonnel C., Allain E., Depeiges A., White C.I., Gallego M.E. (2010). Distinct roles of the ATR kinase and the Mre11-Rad50-Nbs1 complex in the maintenance of chromosomal stability in Arabidopsis. Plant Cell 22: 3020–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong S.J., Caryl A.P., Jones G.H., Franklin F.C. (2002). Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 115: 3645–3655 [DOI] [PubMed] [Google Scholar]
- Bartrand A.J., Iyasu D., Marinco S.M., Brush G.S. (2006). Evidence of meiotic crossover control in Saccharomyces cerevisiae through Mec1-mediated phosphorylation of replication protein A. Genetics 172: 27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P., Benson F.E., West S.C. (1996). Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell 87: 757–766 [DOI] [PubMed] [Google Scholar]
- Benson F.E., Stasiak A., West S.C. (1994). Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 13: 5764–5771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D.K. (1994). RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 79: 1081–1092 [DOI] [PubMed] [Google Scholar]
- Blat Y., Protacio R.U., Hunter N., Kleckner N. (2002). Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111: 791–802 [DOI] [PubMed] [Google Scholar]
- Bleuyard J.Y., Gallego M.E., Savigny F., White C.I. (2005). Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. Plant J. 41: 533–545 [DOI] [PubMed] [Google Scholar]
- Broderick S., Rehmet K., Concannon C., Nasheuer H.P. (2010). Eukaryotic single-stranded DNA binding proteins: Central factors in genome stability. Subcell. Biochem. 50: 143–163 [DOI] [PubMed] [Google Scholar]
- Brown E.J., Baltimore D. (2000). ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 14: 397–402 [PMC free article] [PubMed] [Google Scholar]
- Bugreev D.V., Pezza R.J., Mazina O.M., Voloshin O.N., Camerini-Otero R.D., Mazin A.V. (2010). The resistance of DMC1 D-loops to dissociation may account for the DMC1 requirement in meiosis. Nat. Struct. Mol. Biol. 18: 56–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler C., Borde V., Lichten M. (2007). Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 5: e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo J.A., Cha R.S. (2007). Meiotic roles of Mec1, a budding yeast homolog of mammalian ATR/ATM. Chromosome Res. 15: 539–550 [DOI] [PubMed] [Google Scholar]
- Carballo J.A., Johnson A.L., Sedgwick S.G., Cha R.S. (2008). Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell 132: 758–770 [DOI] [PubMed] [Google Scholar]
- Caryl A.P., Armstrong S.J., Jones G.H., Franklin F.C. (2000). A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma 109: 62–71 [DOI] [PubMed] [Google Scholar]
- Ceballos S.J., Heyer W.D. (2011). Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. Biochim. Biophys. Acta 1809: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha R.S., Kleckner N. (2002). ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297: 602–606 [DOI] [PubMed] [Google Scholar]
- Chelysheva L., Gendrot G., Vezon D., Doutriaux M.P., Mercier R., Grelon M. (2007). Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet. 3: e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Farmer A.D., Langley R.J., Mudge J., Crow J.A., May G.D., Huntley J., Smith A.G., Retzel E.F. (2010). Meiosis-specific gene discovery in plants: RNA-Seq applied to isolated Arabidopsis male meiocytes. BMC Plant Biol. 10: 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.Y., Tsubouchi T., Rockmill B., Sandler J.S., Richards D.R., Vader G., Hochwagen A., Roeder G.S., Fung J.C. (2008). Global analysis of the meiotic crossover landscape. Dev. Cell 15: 401–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich K.A., Cortez D. (2008). ATR: An essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D., Guntuku S., Qin J., Elledge S.J. (2001). ATR and ATRIP: Partners in checkpoint signaling. Science 294: 1713–1716 [DOI] [PubMed] [Google Scholar]
- Couteau F., Belzile F., Horlow C., Grandjean O., Vezon D., Doutriaux M.P. (1999). Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell 11: 1623–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie G., Smith G.R. (2008). Meiotic recombination in Schizosaccharomyces pombe: A paradigm for genetic and molecular analysis. Genome Dyn Stab 3: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan K., Tissier A., Britt A. (2004). ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16: 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan K.M., Britt A.B. (2008). Both ATM and ATR promote the efficient and accurate processing of programmed meiotic double-strand breaks. Plant J. 55: 629–638 [DOI] [PubMed] [Google Scholar]
- Culligan K.M., Robertson C.E., Foreman J., Doerner P., Britt A.B. (2006). ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 48: 947–961 [DOI] [PubMed] [Google Scholar]
- Di Giacomo M., Barchi M., Baudat F., Edelmann W., Keeney S., Jasin M. (2005). Distinct DNA-damage-dependent and -independent responses drive the loss of oocytes in recombination-defective mouse mutants. Proc. Natl. Acad. Sci. USA 102: 737–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray E., Dunlop M.H., Kauppi L., San Filippo J., Wiese C., Tsai M.S., Begovic S., Schild D., Jasin M., Keeney S., Sung P. (2011). Molecular basis for enhancement of the meiotic DMC1 recombinase by RAD51 associated protein 1 (RAD51AP1). Proc. Natl. Acad. Sci. USA 108: 3560–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresser M.E., Ewing D.J., Conrad M.N., Dominguez A.M., Barstead R., Jiang H., Kodadek T. (1997). DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics 147: 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlinger B., Schlögelhofer P. (2011). Have a break: Determinants of meiotic DNA double strand break (DSB) formation and processing in plants. J. Exp. Bot. 62: 1545–1563 [DOI] [PubMed] [Google Scholar]
- Fanning E., Klimovich V., Nager A.R. (2006). A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 34: 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah J.A., Cromie G.A., Smith G.R. (2009). Ctp1 and Exonuclease 1, alternative nucleases regulated by the MRN complex, are required for efficient meiotic recombination. Proc. Natl. Acad. Sci. USA 106: 9356–9361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flott S., Kwon Y., Pigli Y.Z., Rice P.A., Sung P., Jackson S.P. (2011). Regulation of Rad51 function by phosphorylation. EMBO Rep. 12: 833–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin A.E., McElver J., Sunjevaric I., Rothstein R., Bowen B., Cande W.Z. (1999). Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell 11: 809–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V., Bruchet H., Camescasse D., Granier F., Bouchez D., Tissier A. (2003). AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15: 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V., Phelps S.E., Gray S., Neale M.J. (2011). Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 479: 241–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior S.L., Olivares H., Ear U., Hari D.M., Weichselbaum R.R., Bishop D.K. (2001). Assembly of RecA-like recombinases: Distinct roles for mediator proteins in mitosis and meiosis. Proc. Natl. Acad. Sci. USA 98: 8411–8418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb T., Lichten M. (2010). Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol. 8: e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Wurz-Wildersinn R., Fuchs J., Schubert I., Suer S., Puchta H. (2007). The catalytically active tyrosine residues of both SPO11-1 and SPO11-2 are required for meiotic double-strand break induction in Arabidopsis. Plant Cell 19: 3090–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayase A., Takagi M., Miyazaki T., Oshiumi H., Shinohara M., Shinohara A. (2004). A protein complex containing Mei5 and Sae3 promotes the assembly of the meiosis-specific RecA homolog Dmc1. Cell 119: 927–940 [DOI] [PubMed] [Google Scholar]
- Higgins J.D., Armstrong S.J., Franklin F.C., Jones G.H. (2004). The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: Evidence for two classes of recombination in Arabidopsis. Genes Dev. 18: 2557–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.D., Sanchez-Moran E., Armstrong S.J., Jones G.H., Franklin F.C. (2005). The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 19: 2488–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N.M., Byers B. (1989). HOP1: A yeast meiotic pairing gene. Genetics 121: 445–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N.M., Ponte L. (1997). Genetic interactions between HOP1, RED1 and MEK1 suggest that MEK1 regulates assembly of axial element components during meiosis in the yeast Saccharomyces cerevisiae. Genetics 147: 33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E.L., Shinohara A., Bishop D.K. (2001). Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J. Biol. Chem. 276: 41906–41912 [DOI] [PubMed] [Google Scholar]
- Howard-Till R.A., Lukaszewicz A., Loidl J. (2011). The recombinases Rad51 and Dmc1 play distinct roles in DNA break repair and recombination partner choice in the meiosis of Tetrahymena. PLoS Genet. 7: e1001359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N., Kleckner N. (2001). The single-end invasion: An asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 106: 59–70 [DOI] [PubMed] [Google Scholar]
- Hurley P.J., Bunz F. (2007). ATM and ATR: Components of an integrated circuit. Cell Cycle 6: 414–417 [DOI] [PubMed] [Google Scholar]
- Joyce E.F., Pedersen M., Tiong S., White-Brown S.K., Paul A., Campbell S.D., McKim K.S. (2011). Drosophila ATM and ATR have distinct activities in the regulation of meiotic DNA damage and repair. J. Cell Biol. 195: 359–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa W., Kurumizaka H. (2010). From meiosis to postmeiotic events: Uncovering the molecular roles of the meiosis-specific recombinase Dmc1. FEBS J. 277: 590–598 [DOI] [PubMed] [Google Scholar]
- Kato R., Ogawa H. (1994). An essential gene, ESR1, is required for mitotic cell growth, DNA repair and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 22: 3104–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S. (2001). Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52: 1–53 [DOI] [PubMed] [Google Scholar]
- Keeney S., Giroux C.N., Kleckner N. (1997). Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384 [DOI] [PubMed] [Google Scholar]
- Kerzendorfer C., Vignard J., Pedrosa-Harand A., Siwiec T., Akimcheva S., Jolivet S., Sablowski R., Armstrong S., Schweizer D., Mercier R., Schlögelhofer P. (2006). The Arabidopsis thaliana MND1 homologue plays a key role in meiotic homologous pairing, synapsis and recombination. J. Cell Sci. 119: 2486–2496 [DOI] [PubMed] [Google Scholar]
- Kim K.P., Weiner B.M., Zhang L., Jordan A., Dekker J., Kleckner N. (2010). Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell 143: 924–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange J., Pan J., Cole F., Thelen M.P., Jasin M., Keeney S. (2011). ATM controls meiotic double-strand-break formation. Nature 479: 237–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao J.P., Oh S.D., Shinohara M., Shinohara A., Hunter N. (2008). Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Mol. Cell 29: 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latypov V., Rothenberg M., Lorenz A., Octobre G., Csutak O., Lehmann E., Loidl J., Kohli J. (2010). Roles of Hop1 and Mek1 in meiotic chromosome pairing and recombination partner choice in Schizosaccharomyces pombe. Mol. Cell. Biol. 30: 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Chen C., Markmann-Mulisch U., Timofejeva L., Schmelzer E., Ma H., Reiss B. (2004). The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl. Acad. Sci. USA 101: 10596–10601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Golub E.I., Gupta R., Radding C.M. (1997). Recombination activities of HsDmc1 protein, the meiotic human homolog of RecA protein. Proc. Natl. Acad. Sci. USA 94: 11221–11226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw H., Lee D., Myung K. (2011). DNA-PK-dependent RPA2 hyperphosphorylation facilitates DNA repair and suppresses sister chromatid exchange. PLoS ONE 6: e21424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Heyer W.D. (2011). Who’s who in human recombination: BRCA2 and RAD52. Proc. Natl. Acad. Sci. USA 108: 441–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A.J., Hochwagen A. (2011). Checkpoint mechanisms: The puppet masters of meiotic prophase. Trends Cell Biol. 21: 393–400 [DOI] [PubMed] [Google Scholar]
- Mahadevaiah S.K., Turner J.M., Baudat F., Rogakou E.P., de Boer P., Blanco-Rodríguez J., Jasin M., Keeney S., Bonner W.M., Burgoyne P.S. (2001). Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27: 271–276 [DOI] [PubMed] [Google Scholar]
- Mancera E., Bourgon R., Brozzi A., Huber W., Steinmetz L.M. (2008). High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfrini N., Guerini I., Citterio A., Lucchini G., Longhese M.P. (2010). Processing of meiotic DNA double strand breaks requires cyclin-dependent kinase and multiple nucleases. J. Biol. Chem. 285: 11628–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson J.Y., Davies A.A., Hajibagheri N., Van Dyck E., Benson F.E., Stasiak A.Z., Stasiak A., West S.C. (1999). The meiosis-specific recombinase hDmc1 forms ring structures and interacts with hRad51. EMBO J. 18: 6552–6560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin A.V., Mazina O.M., Bugreev D.V., Rossi M.J. (2010). Rad54, the motor of homologous recombination. DNA Repair (Amst.) 9: 286–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahill M.S., Sham C.W., Bishop D.K. (2007). Synthesis-dependent strand annealing in meiosis. PLoS Biol. 5: e299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R., Armstrong S.J., Horlow C., Jackson N.P., Makaroff C.A., Vezon D., Pelletier G., Jones G.H., Franklin F.C. (2003). The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis. Development 130: 3309–3318 [DOI] [PubMed] [Google Scholar]
- Mercier R., Jolivet S., Vezon D., Huppe E., Chelysheva L., Giovanni M., Nogué F., Doutriaux M.P., Horlow C., Grelon M., Mézard C. (2005). Two meiotic crossover classes cohabit in Arabidopsis: One is dependent on MER3, whereas the other one is not. Curr. Biol. 15: 692–701 [DOI] [PubMed] [Google Scholar]
- Miller A.L., Gow N.A. (1989). Correlation between root-generated ionic currents, pH, fusicoccin, indoleacetic acid, and growth of the primary root of Zea mays. Plant Physiol. 89: 1198–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milman N., Higuchi E., Smith G.R. (2009). Meiotic DNA double-strand break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes. Mol. Cell. Biol. 29: 5998–6005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E.P., Symington L.S. (2008). Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D., Shimomura T., Matsumoto K., Sugimoto K. (2003). The ATM-related Tel1 protein of Saccharomyces cerevisiae controls a checkpoint response following phleomycin treatment. Nucleic Acids Res. 31: 1715–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale M.J., Pan J., Keeney S. (2005). Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436: 1053–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., Wan L., Baumgartner B., Schaefer D., Loidl J., Hollingsworth N.M. (2005). Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol. Biol. Cell 16: 5804–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., et al. (2009). Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol. Cell 36: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll M. (2009). Mouse models for ATR deficiency. DNA Repair (Amst.) 8: 1333–1337 [DOI] [PubMed] [Google Scholar]
- Ogawa T., Yu X., Shinohara A., Egelman E.H. (1993). Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science 259: 1896–1899 [DOI] [PubMed] [Google Scholar]
- Okorokov A.L., Chaban Y.L., Bugreev D.V., Hodgkinson J., Mazin A.V., Orlova E.V. (2010). Structure of the hDmc1-ssDNA filament reveals the principles of its architecture. PLoS ONE 5: e8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman K., Sanchez-Moran E., Mann S.C., Jones G.H., Franklin F.C. (2009). Replication protein A (AtRPA1a) is required for class I crossover formation but is dispensable for meiotic DNA break repair. EMBO J. 28: 394–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Sasaki M., Kniewel R., Murakami H., Blitzblau H.G., Tischfield S.E., Zhu X., Neale M.J., Jasin M., Socci N.D., Hochwagen A., Keeney S. (2011). A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144: 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A., Langer M. (2002). Homologous recombination: Ends as the means. Trends Plant Sci. 7: 435–440 [DOI] [PubMed] [Google Scholar]
- Rinaldo C., Bazzicalupo P., Ederle S., Hilliard M., La Volpe A. (2002). Roles for Caenorhabditis elegans rad-51 in meiosis and in resistance to ionizing radiation during development. Genetics 160: 471–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. (1998). DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273: 5858–5868 [DOI] [PubMed] [Google Scholar]
- Sakaguchi K., Ishibashi T., Uchiyama Y., Iwabata K. (2009). The multi-replication protein A (RPA) system—a new perspective. FEBS J. 276: 943–963 [DOI] [PubMed] [Google Scholar]
- Samach A., Melamed-Bessudo C., Avivi-Ragolski N., Pietrokovski S., Levy A.A. (2011). Identification of plant RAD52 homologs and characterization of the Arabidopsis thaliana RAD52-like genes. Plant Cell 23: 4266–4279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J., Sung P., Klein H. (2008). Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77: 229–257 [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran E., Osman K., Higgins J.D., Pradillo M., Cuñado N., Jones G.H., Franklin F.C. (2008). ASY1 coordinates early events in the plant meiotic recombination pathway. Cytogenet. Genome Res. 120: 302–312 [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran E., Santos J.L., Jones G.H., Franklin F.C. (2007). ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 21: 2220–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N. (1994). Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell 76: 51–63 [DOI] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N. (1997). Interhomolog bias during meiotic recombination: Meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90: 1123–1135 [DOI] [PubMed] [Google Scholar]
- Sehorn M.G., Sigurdsson S., Bussen W., Unger V.M., Sung P. (2004). Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature 429: 433–437 [DOI] [PubMed] [Google Scholar]
- Sheridan S., Bishop D.K. (2006). Red-Hed regulation: Recombinase Rad51, though capable of playing the leading role, may be relegated to supporting Dmc1 in budding yeast meiosis. Genes Dev. 20: 1685–1691 [DOI] [PubMed] [Google Scholar]
- Sheridan S.D., Yu X., Roth R., Heuser J.E., Sehorn M.G., Sung P., Egelman E.H., Bishop D.K. (2008). A comparative analysis of Dmc1 and Rad51 nucleoprotein filaments. Nucleic Acids Res. 36: 4057–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A., Ogawa H., Ogawa T. (1992). Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69: 457–470 [DOI] [PubMed] [Google Scholar]
- Shinohara A., Gasior S., Ogawa T., Kleckner N., Bishop D.K. (1997). Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells 2: 615–629 [DOI] [PubMed] [Google Scholar]
- Shinohara M., Gasior S.L., Bishop D.K., Shinohara A. (2000). Tid1/Rdh54 promotes colocalization of rad51 and dmc1 during meiotic recombination. Proc. Natl. Acad. Sci. USA 97: 10814–10819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz R.W., Tatineni V.M., Hanley-Bowdoin L., Thompson W.F. (2007). Genome-wide analysis of the core DNA replication machinery in the higher plants Arabidopsis and rice. Plant Physiol. 144: 1697–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaud N., Dray E., Gy I., Gérard E., Takvorian N., Doutriaux M.P. (2004). Brca2 is involved in meiosis in Arabidopsis thaliana as suggested by its interaction with Dmc1. EMBO J. 23: 1392–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A., Gargano S., Ruprich-Robert G., Falque M., David M., Kleckner N., Zickler D. (2010). Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell 141: 94–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P. (1994). Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265: 1241–1243 [DOI] [PubMed] [Google Scholar]
- Sweeney P.R., Britt A.B., Culligan K.M. (2009). The Arabidopsis ATRIP ortholog is required for a programmed response to replication inhibitors. Plant J. 60: 518–526 [DOI] [PubMed] [Google Scholar]
- Syljuåsen R.G., Sørensen C.S., Hansen L.T., Fugger K., Lundin C., Johansson F., Helleday T., Sehested M., Lukas J., Bartek J. (2005). Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol. Cell. Biol. 25: 3553–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarsounas M., Morita T., Pearlman R.E., Moens P.B. (1999). RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J. Cell Biol. 147: 207–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasawa M., Shinohara A., Hotta Y., Ogawa H., Ogawa T. (1995). Localization of RecA-like recombination proteins on chromosomes of the lily at various meiotic stages. Genes Dev. 9: 925–934 [DOI] [PubMed] [Google Scholar]
- Uanschou C., Siwiec T., Pedrosa-Harand A., Kerzendorfer C., Sanchez-Moran E., Novatchkova M., Akimcheva S., Woglar A., Klein F., Schlögelhofer P. (2007). A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J. 26: 5061–5070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsal-Kaçmaz K., Sancar A. (2004). Quaternary structure of ATR and effects of ATRIP and replication protein A on its DNA binding and kinase activities. Mol. Cell. Biol. 24: 1292–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T., Ogawa H., Petrini J.H. (2001). A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7: 1255–1266 [DOI] [PubMed] [Google Scholar]
- Vignard J., Siwiec T., Chelysheva L., Vrielynck N., Gonord F., Armstrong S.J., Schlögelhofer P., Mercier R. (2007). The interplay of RecA-related proteins and the MND1-HOP2 complex during meiosis in Arabidopsis thaliana. PLoS Genet. 3: 1894–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijeratne A.J., Chen C., Zhang W., Timofejeva L., Ma H. (2006). The Arabidopsis thaliana PARTING DANCERS gene encoding a novel protein is required for normal meiotic homologous recombination. Mol. Biol. Cell 17: 1331–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Ashley T., Brainerd E.E., Bronson R.T., Meyn M.S., Baltimore D. (1996). Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 10: 2411–2422 [DOI] [PubMed] [Google Scholar]
- Yu X., Egelman E.H. (2010). Helical filaments of human Dmc1 protein on single-stranded DNA: A cautionary tale. J. Mol. Biol. 401: 544–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K., Ma Y., Tang S., Hwang P.Y., Boiteux S., Hunter N. (2010). Temporally and biochemically distinct activities of Exo1 during meiosis: Double-strand break resection and resolution of double Holliday junctions. Mol. Cell 40: 1001–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Kim K.P., Kleckner N.E., Storlazzi A. (2011). Meiotic double-strand breaks occur once per pair of (sister) chromatids and, via Mec1/ATR and Tel1/ATM, once per quartet of chromatids. Proc. Natl. Acad. Sci. USA 108: 20036–20041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Elledge S.J. (2003). Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.