This work reveals that Flot1 functions in Arabidopsis thaliana seedling development and is involved in a membrane microdomain-dependent, but clathrin-independent, endocytic pathway. Moreover, the dynamic behavior of Flot1-positive puncta differs considerably from that of clathrin light chain–positive puncta.
Abstract
Endocytosis is essential for the maintenance of protein and lipid compositions in the plasma membrane and for the acquisition of materials from the extracellular space. Clathrin-dependent and -independent endocytic processes are well established in yeast and animals; however, endocytic pathways involved in cargo internalization and intracellular trafficking remain to be fully elucidated for plants. Here, we used transgenic green fluorescent protein–flotillin1 (GFP-Flot1) Arabidopsis thaliana plants in combination with confocal microscopy analysis and transmission electron microscopy immunogold labeling to study the spatial and dynamic aspects of GFP-Flot1–positive vesicle formation. Vesicle size, as outlined by the gold particles, was ∼100 nm, which is larger than the 30-nm size of clathrin-coated vesicles. GFP-Flot1 also did not colocalize with clathrin light chain-mOrange. Variable-angle total internal reflection fluorescence microscopy also revealed that the dynamic behavior of GFP-Flot1–positive puncta was different from that of clathrin light chain-mOrange puncta. Furthermore, disruption of membrane microdomains caused a significant alteration in the dynamics of Flot1-positive puncta. Analysis of artificial microRNA Flot1 transgenic Arabidopsis lines established that a reduction in Flot1 transcript levels gave rise to a reduction in shoot and root meristem size plus retardation in seedling growth. Taken together, these findings support the hypothesis that, in plant cells, Flot1 is involved in a clathrin-independent endocytic pathway and functions in seedling development.
INTRODUCTION
Endocytosis occurs at the cell surface and involves the formation of membrane invaginations, followed by vesicle separation (pinching off) from the plasma membrane. This process plays a central role in the maintenance of the protein and lipid composition within the plasma membrane; endocytosis also mediates in the removal or recycling of materials from the apoplasm or extracellular space (Conner and Schmid, 2003; Dhonukshe et al., 2008; Tanaka et al., 2009; Chen et al., 2011).
During the past decade, cell biologists have focused a considerable amount of effort toward elucidating the molecular mechanisms underlying the process of endocytosis. Based on these studies, several distinct pathways have been characterized, including clathrin-mediated and clathrin-independent endocytosis (Pelkmans and Helenius, 2003; Benmerah and Lamaze, 2007; Dhonukshe et al., 2007). The latter pathway is a broad term used to describe a number of endocytic pathways that internalize external cargos. Here, membrane rafts, specific plasma membrane microdomains that facilitate protein recruitment in cellular trafficking events, have also been shown to participate in clathrin-independent endocytosis and signaling (Brown and London, 2000; Simons and Toomre, 2000; Simons and Gerl, 2010).
In mammalian cells, arguably the best characterized protein in the microdomains is the integral membrane protein caveolin; this protein is associated with specialized pit-forming regions termed caveolae (Kurzchalia and Parton, 1999). Caveolae are thought to play important roles in signaling, transendothelial transport, and lipid regulation (Drab et al., 2001; Razani et al., 2002a, 2002b). The reggie/flotillin family of proteins also represent common markers for microdomains in animal cells (Morrow et al., 2002). In recent studies, flotillins were shown to induce membrane curvature, an event necessary for the formation of plasma membrane invaginations (Glebov et al., 2006; Frick et al., 2007; Langhorst et al., 2008).
Detergent-resistant membranes (DRMs) are thought to be the biochemical counterpart of membrane rafts (Raffaele et al., 2009). Even though the membrane raft hypothesis is still a matter of debate, few researchers doubt the existence of large-scale lateral membrane compartmentalization (Keinath et al., 2010). In the past few years, numerous studies have demonstrated a close correlation between the association of membrane components with DRMs and their specific localization in membrane microdomains in vivo (Raffaele et al., 2009; Mongrand et al., 2010; Carmona-Salazar et al., 2011).
Although the concept of membrane microdomains and their potential roles in cellular processes have been widely studied in yeast and mammalian systems (Simons and Toomre, 2000; Garcia et al., 2003; Parton, 2003; Falk et al., 2004; Lafont et al., 2004; Salaün et al., 2004; Hansen et al., 2005 ; Lajoie and Nabi, 2010; Lingwood and Simons, 2010; Simons and Gerl, 2010), current knowledge on the composition and roles of such membrane microdomains remains sparse in plant cells (Bhat et al., 2005).
A number of studies have been performed on Arabidopsis thaliana (Borner et al., 2005), tobacco (Nicotiana tabacum; Mongrand et al., 2004; Morel et al., 2006), Sinapis alba (Shahollari et al., 2004), and Medicago truncatula (Lefebvre et al., 2007; Haney and Long, 2010). However, generally, these studies have focused on the analysis of protein and lipid components within membrane microdomains, using a variety of biochemical and cytological tools.
Recent studies reported that misexpression of the membrane raft protein remorin altered virus propagation (Borner et al., 2005; Raffaele et al., 2009; Titapiwatanakun et al., 2009), and plant flotillins appear to play a critical role in symbiotic bacterial infection in M. truncatula (Haney and Long, 2010; Haney et al., 2011). Arabidopsis flotillin1 (Flot1), a protein first identified in DRMs using subcellular fractionation, two-dimensional electrophoresis, and mass spectrometry, is considered to be one such membrane microdomain protein (Borner et al., 2005). Nevertheless, the underlying mechanism and functional significance of this plant protein, in the overall process of endocytosis, remains to be established.
In this study, we provide several lines of evidence that Arabidopsis Flot1 is localized to specific domains within both the plasma membrane and intracellular compartments. Subcellular localization experiments demonstrated that Flot1 and clathrin light chain (CLC) are present in discrete and separate regions of the cell. In addition, our analyses employing variable-angle total internal reflection fluorescence microscopy (VA-TIRFM) revealed that the dynamic behavior of green fluorescent protein (GFP)-Flot1 puncta was different from that of CLC-mOrange puncta. Furthermore, it was found that seedling development of Flot1 artificial microRNA (amiRNA) knockdown transgenic lines was retarded. These findings support the hypothesis that, in plants, Flot1 participates in a clathrin-independent endocytic pathway and functions in seedling development.
RESULTS
Characterization of Transgenic Arabidopsis GFP-Flot1 Plant Lines
To investigate the functional roles of Flot1 in plants, we first transformed wild-type Arabidopsis plants (ecotype Columbia-0) with a construct encoding full-length Flot1 fused with GFP under the control of its endogenous promoter. Since the fluorescence signals after expression using this promoter were too weak to be analyzed, we next employed the cauliflower mosaic virus 35S promoter to express a fluorescently tagged construct for our studies (see Supplemental Figure 1A online). Some 30 independent transgenic plant lines were generated, and plants from T2 seeds were used for experimental analysis. Protein gel blot assays were first performed on membrane fractions extracted from Arabidopsis seedlings to confirm GFP-Flot1 expression in these transgenic lines. As shown in Supplemental Figure 1B online, no signal was detected with preimmune serum for proteins extracted from either transgenic (lane 1) or wild-type (lane 2) seedlings. By contrast, anti-GFP antibodies detected a 78-kD band, the predicted molecular mass of the full-length GFP-Flot1 fusion protein, in the membrane fraction from transgenic plants (lane 3). No such band was detected with wild-type (control; lane 4) plants. Thus, the GFP-Flot1 fusion protein was successfully expressed in Arabidopsis seedlings with the correct molecular mass and membrane insertion.
An anti-Flot1 monoclonal antibody was next tested for specificity using protein gel blot assays; here, a 51-kD band, the expected size of Flot1, was detected in membrane fractions prepared from both transgenic (lane 5) and wild-type (lane 6) seedlings (see Supplemental Figure 1B online). Furthermore, a major band at 78 kD was also detected by the anti-Flot1 monoclonal antibody in transgenic Arabidopsis seedlings expressing GFP-Flot1 (lane 5).
Specificity of the anti-Flot1 monoclonal antibody was further tested by performing confocal immunofluorescence microscopy on chemically fixed wild-type Arabidopsis seedlings. No signal was detected when root cells were probed with preimmune serum (see Supplemental Figures 1C and 1D online). By contrast, the anti-Flot1 monoclonal antibody labeling showed that the protein was mostly localized at the plasma membrane of root cells (see Supplemental Figures 1E and 1F online). Equivalent experiments performed on root hair cells yielded similar results (see Supplemental Figures 1G and 1H online). In addition, when compared on an equivalent protein basis, Flot1 showed a significant enrichment in the DRMs fraction (see Supplemental Figures 2 and 3 online), indicating that Flot1 was associated to membrane microdomains. Taken together, these results indicated that the anti-Flot1 monoclonal antibody could be used to examine the distribution of endogenous Flot1.
Subcellular Localization of GFP-Flot1 Fusion Protein and Flot1 in Arabidopsis Root Cells
Subcellular localization of GFP-Flot1 was next examined in transgenic Arabidopsis root cells. As shown in Figure 1A, strong fluorescent signal localized to the cell periphery was readily detected in cells expressing GFP-Flot1 (Figure 1A, panels 1 and 2). However, no signal was detected in wild-type Arabidopsis root cells (data not shown). To determine whether these fluorescent signals were localized to the plasma membrane, cell wall, or periplasmic space, transgenic Arabidopsis root hair and root epidermal cells were plasmolyzed by treatment with 100 mM sorbitol. In such plasmolyzed cells, strong fluorescent signal appeared to be retained in the plasma membrane (Figure 1A, panels 3 and 4). These results suggested that GFP-Flot1 is not localized to the cell wall of root cells.
Figure 1.
Subcellular Localization of GFP-Flot1 in Roots of Transgenic Arabidopsis Seedlings.
(A) Confocal images of GFP signal in root tissues expressing GFP-Flot1. Control: root hair (panel 1) and epidermal (panel 2) cells. Plasmolysis experiment: roots treated with 100 mM sorbitol; root hair (panel 3) and epidermal (panel 4) cells. CW, cell wall; DIC, differential interference contrast microscopy; PM, plasma membrane.
(B) Seedlings expressing GFP-Flot1 (green) were incubated for 5 to 10 min with 5 µM FM4-64 (red). The merged image shows partial colocalization. Enlarged images of the boxed areas are presented in the right column. White arrows indicate examples of colocalizing punctate structures.
Bars = 20 μm in (A) and 10 μm in (B).
The similar patterns of fluorescence observed in root cells of wild-type (Figures 1E and 1G; anti-Flot1 monoclonal antibody-based detection) and transgenic GFP-Flot1 Arabidopsis plants suggested that GFP-Flot1 localization to the plasma membrane was unlikely to be a result of overexpression (see Supplemental Figure 4 online). Furthermore, our analysis of wild-type and GFP-Flot1–expressing roots indicated no differences in anatomical features (see Supplemental Figure 5 online).
To test whether the Flot1-containing dot structures could be an endosomal compartment, the localization of GFP-Flot1 was compared with that of the endosomal marker dye FM4-64. Cells were stained with 5 μM FM4-64 on ice and then washed with cold medium, after which they were returned to room temperature to resume intracellular trafficking. After a 5- to 10-min FM4-64 incubation, we observed that the GFP-Flot1–labeled compartments were partially colocalized with dotted structures having FM4-64 signal (Figure 1B). These results suggest that FM4-64 is rapidly internalized into plant cells, at room temperature, and is colocalized partially with Flot1-positive structures. We next pretreated GFP-Flot1–expressing root cells with brefeldin A to inhibit the secretory pathway. In response to this treatment, brefeldin A–induced aggregates derived from GFP-Flot1–labeled punctuate structures were found to be partially colocalized with internalized FM4-64 (see Supplemental Figure 6 online). Furthermore, we performed immunofluorescence studies using a trans-Golgi network marker (VHA-a1-GFP) (Dettmer et al., 2006). The results showed that the signal for the anti-Flot1 antibody was partially colocalized with the VHA-a1-GFP signal (see Supplemental Figure 7 online), revealing that a portion of the Flot1-labeled structures reflects an early endosomal compartment. These results suggest that internal GFP-Flot1–labeled structures likely represent an endosomal compartment in Arabidopsis root cells.
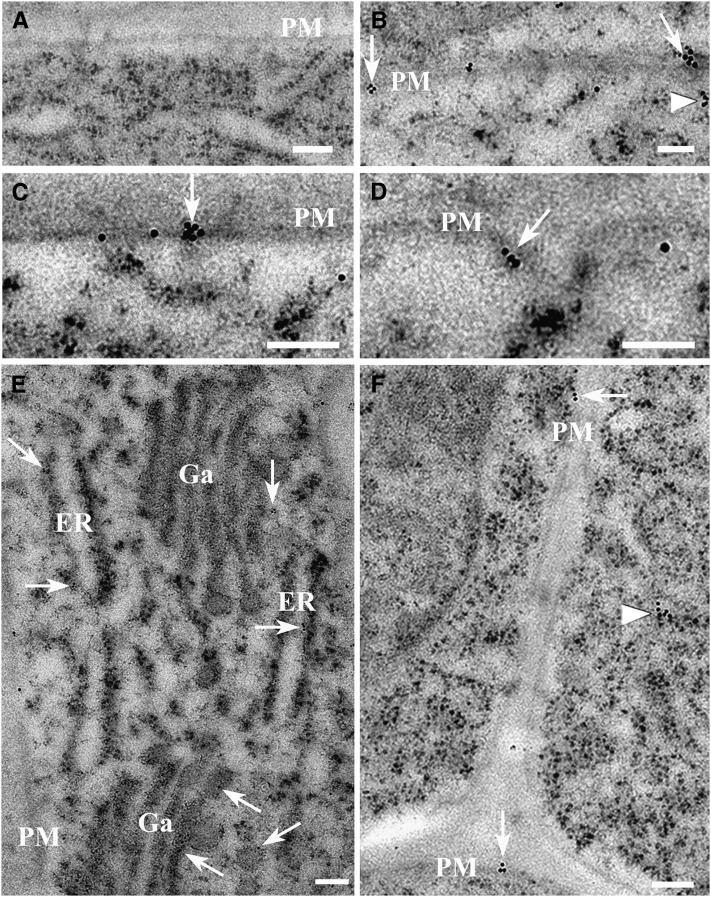
Localization of Flot1 in wild-type Arabidopsis root cells was further investigated by immunogold labeling experiments using electron microscopy. Preimmune controls showed no gold labeling (Figure 2A), whereas the anti-Flot1 monoclonal antibody clearly cross-reacted with protein localized to the plasma membrane (Figure 2B), either in flat regions (Figure 2C) or invaginations (Figure 2D). In addition, gold labeling was detected in the endoplasmic reticulum membranes (Figure 2E), in close proximity to the Golgi apparatus (Figure 2E), and cytoplasmic vesicles (Figures 2B and 2F). Based on these experiments, the size of the vesicles outlined by the gold particles was estimated to be ∼100 nm. Furthermore, we found that Flot1 is clustered in particular domains of the membrane (see Supplemental Figure 8A online). Statistical analysis showed that 75% of the gold particles appeared in a clustered pattern throughout the plasma membrane, whereas 25% showed a random distribution (see Supplemental Figure 8B online). Taken together, these studies demonstrated that Flot1 is localized to the plasma membrane and endosomal structures; presumably, Flot1 is involved in the formation of membrane microdomains, membrane invaginations, and vesicle budding.
Figure 2.
Immunolocalization Studies Performed on Wild-Type Arabidopsis Root Apices Establish That Flot1 Is Localized to the Plasma Membrane and Endosomal Structures.
(A) Preimmune controls showed no gold labeling in the cells.
(B) Flot1 signal appeared to be clustered on regions of the plasma membrane (PM; arrows) and to cytoplasmic structures (arrowhead).
(C) Flot1 localized to flat regions of the plasma membrane (arrow).
(D) Flot1 was detected in association with invaginations of the plasma membrane (arrow).
(E) Flot1 localized to the endoplasmic reticulum (ER) membranes and was in close proximity to the Golgi apparatus (Ga).
(F) Flot1 present in the plasma membrane (arrow) and cytoplasmic vesicle (arrowhead).
Bars = 100 nm.
Time-Course Experiments Reveal That Flot1 Is Associated with Endocytic Vesicle Formation
To provide further insight into the role of Flot1 in endocytosis, we next conducted time-course experiments in which we traced the path of GFP-Flot1–positive vesicles in root cells. These studies demonstrated that GFP dots move quickly from the plasma membrane into the cytoplasm (Figures 3A and 3B; see Supplemental Movie 1 online).
Figure 3.
Dynamics of GFP-Flot1–Labeled Compartments in Transgenic Arabidopsis Root Cells.
Confocal images were collected at the times indicated (seconds) from root cells expressing the GFP-Flot1 fusion protein.
(A) Low-magnification confocal image; inset shows the area imaged during the time course.
(B) Ten consecutive images taken at 12.8-s intervals. White arrows indicate an example of movement of a GFP-Flot1–labeled compartment from the PM to the cytoplasm (see Supplemental Movie 1 online).
Bars = 10 μm.
[See online article for color version of this figure.]
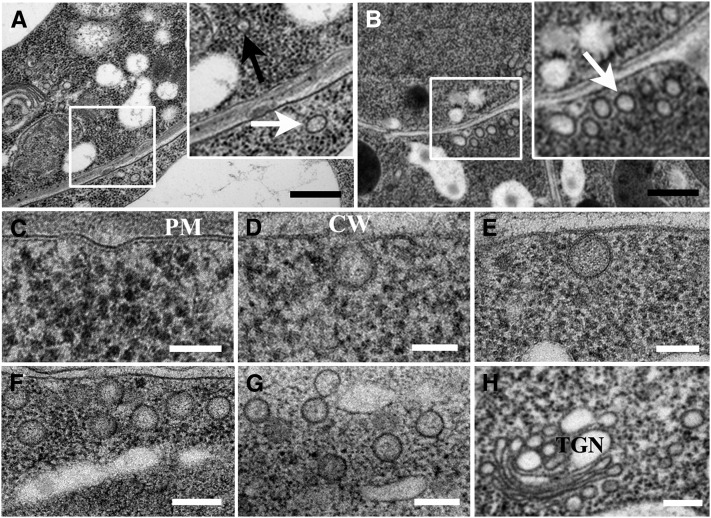
Morphological evidence of a role for Flot1 in endocytosis was also obtained from additional electron microscopy studies. The size distribution of vesicles was plotted in a histogram having spaced bins (see Supplemental Figure 9 online). The data were fitted using Gaussian methods, the position of the peak (noted as Ĝ) being considered the characteristic size of vesicles. As shown in Supplemental Figure 9A online, in wild-type plants, the size of vesicles was distributed in two subpopulations with Ĝ values of 43.6 ± 1.4 nm and 104.0 ± 3.0 nm (Figure 4A). By contrast, root cells overexpressing Flot1 had a dramatic increase in the number of larger vesicles with a Ĝ value of 98.5 ± 3.1 nm (Figure 4B; see Supplemental Figure 9B online). Here, the prominent features observed were plasma membrane invaginations (Figures 4C and 4D), accumulation of vesicles in close proximity to the plasma membrane (Figures 4E and 4F), and clusters of vesicles deeper within the cell interior (Figure 4G). Furthermore, vesicles were also in association with the trans-Golgi network (Figure 4H). In addition, the size of these vesicles was similar to the dimensions of the vesicle configuration outlined by the gold particles observed in our immunogold studies (Figure 2). Furthermore, a clear decrease in the number of larger vesicles was found in amiRNA root cells (see Supplemental Figure 9C online). Collectively, these results provide support for the hypothesis that Flot1 is associated with the formation of large endocytotic vesicles in cells of Arabidopsis roots.
Figure 4.
Flot1 Participates in the Formation of Endocytic Vesicles.
(A) Transmission electron micrographs showing that there are two types of vesicles in wild-type root cells, one being smaller (black arrows) and the other larger (white arrows) in size.
(B) Root cells overexpressing Flot1 have a significant increase in numbers of the large vesicles (white arrows).
(C) and (D) Plasma membrane (PM) invaginations observed in Arabidopsis root cells expressing GFP-Flot1. CW, cell wall.
(E) and (F) Sub–plasma membrane vesicles viewed in Arabidopsis root cells expressing GFP-Flot1.
(G) Clusters of putative endosomal vesicles that accumulated in Arabidopsis root cells expressing GFP-Flot1.
(H) Clusters of putative endosomal vesicles accumulated in the trans-Golgi network (TGN) of Arabidopsis root cells expressing GFP-Flot1.
Bars = 500 nm in (A) and (B), 100 nm in (C) to (E), and 200 nm in (F) to (H).
Flot1 Does Not Colocalize with CLC
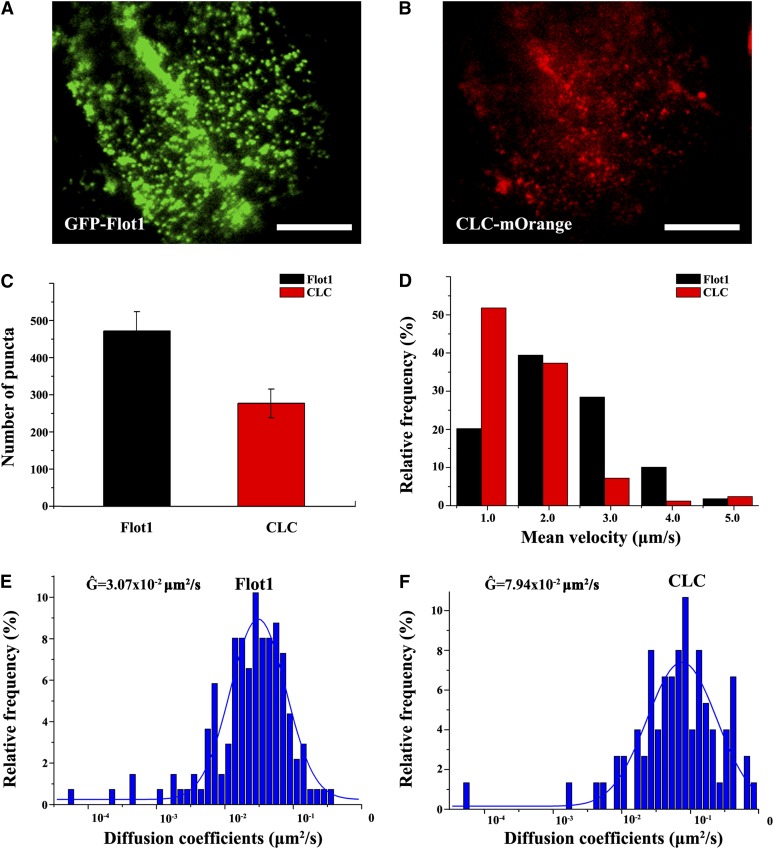
Transgenic Arabidopsis plants expressing both CLC-mOrange and GFP-Flot1 were next employed to test whether Flot1 functions in clathrin-mediated endocytosis. Confocal studies performed on root epidermal cells of these plants revealed that there was only a small degree of overlap between Flot1- and CLC-tagged signals (Figure 5A; see Supplemental Figure 10 online).
Figure 5.
In Vivo Confocal Fluorescence Analysis and Double Immunogold Electron Microscopy Studies Establish That Flot1 and CLC Are Not Colocalized.
(A) Confocal images of root epidermal cells within transgenic Arabidopsis plants expressing CLC-mOrange and GFP-Flot1. Boxed regions examined under higher magnification reveal only limited colocalization of the mOrange and GFP signals in the cortical cytoplasm (panel 1 to 3).
(B) to (E) Double immunogold labeling of Arabidopsis root epidermal cells performed using anti-Flot1 antibody (6-nm gold-conjugated secondary) and anti-CLC (15-nm gold-conjugated secondary) antibodies.
(B) and (C) Boundaries between three adjacent cells showing a lack of colocalization between Flot1 (white arrowheads) and CLC (black arrowheads). Inset in (C): small vesicular structure (inset: clathrin-coated vesicle) located within the cytoplasm. CW, cell wall.
(D) and (E) Regions covering two adjacent cells showing a lack of colocalization between Flot1 and CLC. Inset: Larger forming vesicular structure (Flot1-positive vesicle) located just beneath the plasma membrane.
Bars = 10 μm in (A), 2 μm in panels 1 to 3, and 200 nm in (B) to (E).
Double-labeling immunogold electron microscopy experiments performed on Arabidopsis root cells, using anti-Flot1 (6-nm gold-conjugated secondary) and anti-CLC (15-nm gold-conjugated secondary) monoclonal antibodies, provided further support that Flot1 and CLC are not colocalized either at the plasma membrane or in the cytoplasm (Figures 5B to 5E). A typical small cytoplasmic vesicular configuration (clathrin-coated vesicle, 15-nm gold) is illustrated in Figure 5C, whereas a larger Flot1-positive vesicle (6-nm gold) is shown in Figure 5E. These experiments demonstrate that Flot1 and CLC are rarely colocalized in Arabidopsis root cells.
Flot1- and Clathrin-Positive Puncta Display Different Dynamic Properties
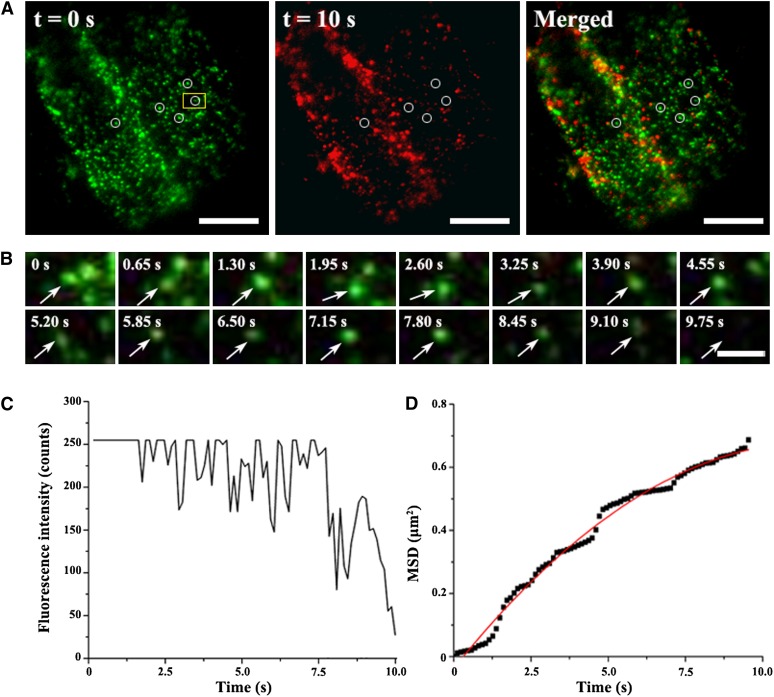
VA-TIRFM or variable-angle epifluorescence microscopy allows for imaging of the plant cell cortex with a high signal-to-noise ratio (Konopka et al., 2008; Konopka and Bednarek, 2008a, 2008b). VA-TIRFM was next employed to analyze the puncta dynamics associated with GFP-Flot1. These studies were performed on cotyledon leaf pavement cells, guard cells, hypocotyl and root epidermal cells, and root hairs. In all cell types, GFP-Flot1 was prominently organized into punctuate structures (Figure 6A; see Supplemental Figure 11 online). The Flot1-positive punctate structures that formed at the plasma membrane were highly dynamic (see Supplemental Movie 2 online); the fluorescence signal associated with these membrane structures underwent gradual loss as individual structures budded off from the plasma membrane and entered the cytoplasm (Figure 6B; see Supplemental Movie 3 online).
Figure 6.
Flot1-Positive Puncta Are Dynamic and Appear to Bud into Living Root Epidermal Cells.
(A) Transgenic Arabidopsis root epidermal cell expressing GFP-Flot1 imaged by VA-TIRFM. Shown are green and red images from a time-lapse sequence taken 10 s apart. The merged image was used to identify puncta that disappeared during this 10-s interval.
(B) Individual budding event of a GFP-Flot1–positive spot. Shown is a time series for the yellow boxed area marked in (A) (see Supplemental Movie 3 online).
(C) Fluorescence intensity profile for the tracked GFP-Flot1–positive spot documented in (B).
(D) MSD of the budding GFP-Flot1–positive spot plotted as a function of time. The resulting MSD-t curves (black squares) were fitted with restricted diffusion modes.
Bars = 10 μm in (A) and 2 μm in (B).
The budding event associated with individual Flot1-positive punctate structures was characterized by a sharp decrease in the fluorescence intensity profile (Figure 6C). This pattern differed greatly from those obtained on equivalent Flot1-positive punctate structures present in chemically fixed cells (see Supplemental Figure 12 online). Here, a slow and gradual decrease in fluorescence intensity was observed, reflecting photobleaching of the GFP signal. This difference in the time course of fluorescence intensity loss indicates that the change in signal illustrated in Figure 6C is not consistent with photobleaching.
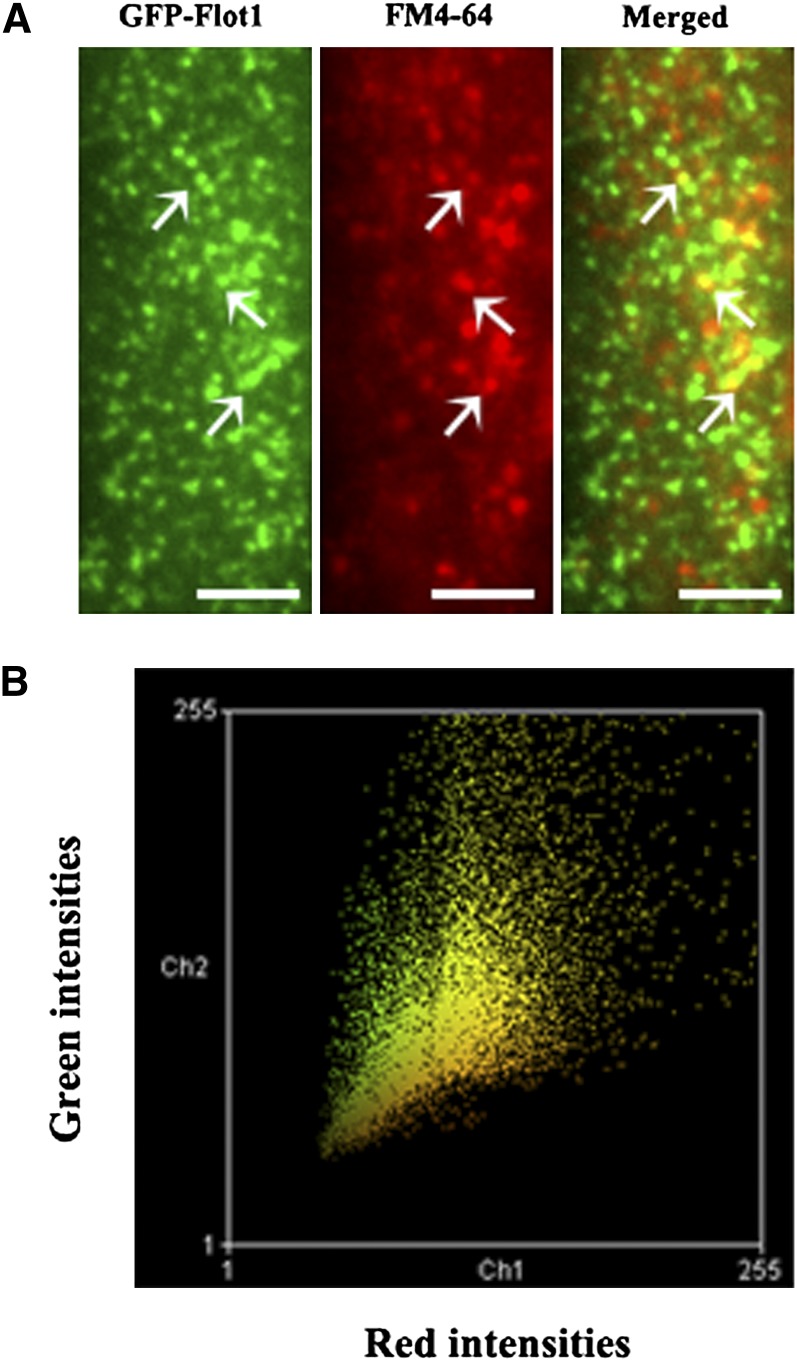
Next, we calculated the diffusion coefficient for GFP-Flot1 puncta using a linear fit to the mean squared displacement (MSD) versus time plots. The resulting MSD-t curve of the tracked budding GFP-Flot1–positive structures was best fitted using a restricted diffusion mode (Figure 6D), indicating that diffusion of such structures was being confined within a limited area. Finally, simultaneous monitoring of GFP-Flot1 and FM4-64 in root cells (Figures 7A and 7B; see Supplemental Movie 4 online) again revealed that, generally, these two signals were often colocalized. To rule out the possibility that the colocalization was due to random overlap of the highly dense foci in the root epidermal cells, the red channel image from five different cells was rotated 180° with respect to the green channel, a statistical analysis with a technique reported previously to show nonrandom colocalization (Konopka et al., 2008). The average peak distance for the original image (3.13 ± 0.89 pixels; n = 290 from five cells) was significantly different from that of the rotated images (7.09 ± 1.98 pixels; n = 295 from five cells; P < 0. 0001), indicating that the colocalization was statistically significant.
Figure 7.
GFP-Flot1 and FM4-64 Puncta Are Often Colocalized at the Plasma Membrane of the Root Epidermal Cells.
(A) VA-TIRFM image of a root epidermal cell expressing GFP-Flot1 treated with FM4-64. White arrows indicate examples of colocalized puncta.
(B) Colocalization histogram of a plot for each pixel according to its red (x axis) and green (y axis) intensity. These data were analyzed using Manders coefficients.
Bars = 5 μm.
Plants expressing both GFP-Flot1 and CLC-mOrange were also imaged using dual fluorescence color VA-TIRFM (Figures 8A and 8B; see Supplemental Movie 5 online). The behavior of Flot1-containing structures was compared with that of clathrin-coated pits. GFP-Flot1 puncta were more abundant than the CLC-mOrange–labeled pits (Figure 8C), and they moved laterally with higher velocities (Figure 8D). The motion characteristics of both kinds of endocytotic puncta were also compared through measurements of their diffusion coefficients. Interestingly, both Flot1- and CLC-labeled puncta had average diffusion coefficients that were on the same order of magnitude; Ĝ values for GFP-Flot1 and CLC-labeled puncta were 3.07 ± 0.18 × 10−2 μm2 s−1 and 7.94 ± 0.88 × 10−2 μm2 s−1, respectively (Figures 8E and 8F). Taken together, these data demonstrate that Flot1-positive and clathrin-coated puncta can be differentiated based on their dynamic characteristics.
Figure 8.
Flot1-Positive Puncta Derived from the Plasma Membrane Can Be Differentiated from CLC-Positive Puncta by their Dynamic Characteristics.
(A) and (B) VA-TIRFM image of a root cell expressing GFP-Flot1 and CLC-mOrange.
(C) Number of GFP-Flot1 and CLC-mOrange labeled puncta observed in VA-TIRFM movies of transgenic double-labeled Arabidopsis root epidermal cells.
(D) Histogram of mean velocity of tracked GFP-Flot1 and CLC-mOrange–labeled puncta in VA-TIRFM movies of transgenic double-labeled Arabidopsis root epidermal cells.
(E) and (F) Distribution of diffusion coefficients of tracked GFP-Flot1 and CLC-mOrange–labeled puncta in VA-TIRFM movies of transgenic double-labeled Arabidopsis root epidermal cells.
Bars = 10 μm.
Membrane Microdomains and Cytoskeleton Disruption Alter Flot1-Positive Puncta Dynamics
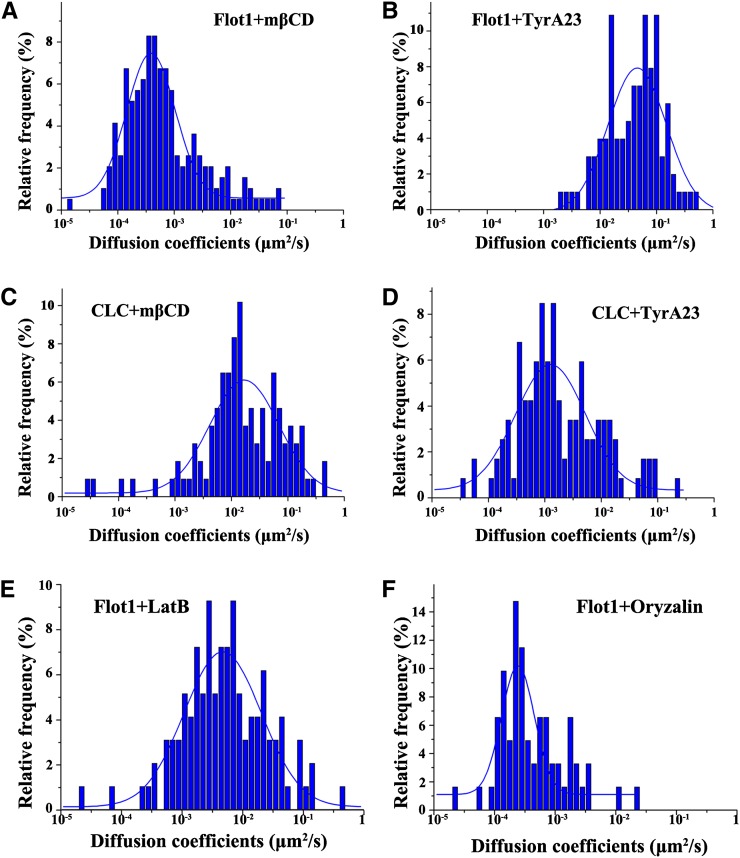
The sterol-depleting agent methyl-β-cyclodextrin (mβCD) was next employed to investigate the role of membrane microdomains on the dynamics of Flot1-positive puncta. A 30-min exposure to 10 mM mβCD was found to cause an overall decrease in the GFP-Flot1 diffusion coefficient, with the Ĝ value being greatly reduced from the control value of 3.07 ± 0.18 × 10−2 μm2 s−1 to 3.88 ± 0.26 × 10−4 μm2 s−1, which represents a 100-fold reduction (Figure 9A). Thus, disruption of sterol-rich membrane microdomains can cause a significant alteration in the dynamics of Flot1-positive structures.
Figure 9.
Histograms Showing the Different Dynamic Character of GFP-Flot1 and CLC-mOrange Puncta.
(A) Distribution of diffusion coefficients for GFP-Flot1 puncta after a 30-min treatment with 10 mM mβCD (n = 193 spots analyzed).
(B) Distribution of diffusion coefficients for GFP-Flot1 puncta after a 30-min treatment with 50 µM TyrA23 (n = 203 spots analyzed).
(C) Histogram showing the distribution for diffusion coefficients of CLC-mOrange puncta after a 30-min treatment with 10 mM mβCD (n = 208 spots analyzed).
(D) Histogram showing the distribution for diffusion coefficients of CLC-mOrange puncta after a 30-min treatment with 50 µM TyrA23 (n = 218 spots analyzed).
(E) Histogram showing the distribution of diffusion coefficients for GFP-Flot1 puncta after treatment with 1µM Lat-B (n = 163 spots analyzed).
(F) Histogram showing the distribution of diffusion coefficients for GFP-Flot1 puncta after treatment with10 µM Oryzalin (n = 122 spots analyzed).
[See online article for color version of this figure.]
We then investigated the effects of tyrphostin A23 (TyrA23), a well-characterized inhibitor of endocytic cargo recruitment into the clathrin-mediated pathway, both in vivo and in vitro (Banbury et al., 2003; Konopka et al., 2008) on GFP-Flot1 movement. A 30-min treatment of roots with 50 μM TyrA23 did not cause a significant change in the diffusion coefficient for GFP-Flot1 puncta (Ĝ = 2.44 ± 0.53 × 10−2 μm2 s−1) (Figure 9B). Taken together, these findings provide further support for the hypothesis that the Flot1-positive endocytic process functions independently of clathrin and requires the integrity of sterol-rich membrane microdomains within the plasma membrane.
The effects of mβCD and TyrA23 were also investigated in the same cells coexpressing CLC-mOrange. When the roots were pretreated with mβCD, the Ĝ value of CLC-mOrange underwent a small decrease to 1.69 ± 0.23 × 10−2 μm2 s−1 (Figure 9C). However, when the roots were pretreated with TyrA23, the CLC-mOrange Ĝ value decreased significantly to 1.26 ± 0.19 × 10−3 μm2 s−1 (Figure 9D). These results indicated that the dynamics of CLC-mOrange structures are less sensitive to disruption of sterol-rich membrane microdomains.
The involvement of the cytoskeleton in Flot1-positive endocytosis was also tested using two well-characterized agents. When treated with 1 μM latrunculin B (Lat-B), a specific inhibitor of actin polymerization, the trajectories of Flot1-positive puncta were found to be confined to smaller regions, in comparison to untreated control root cells. In addition, the Ĝ value of the GFP-Flot1 structures decreased by sixfold to 4.74 ± 0.48 × 10−3 μm2 s−1 (Figure 9E). Treatment with 10 μM oryzalin, an inhibitor of microtubule polymerization, led to an even greater decrease in the GFP-Flot1 diffusion coefficient, with the Ĝ value being reduced 120-fold to 2.43 ± 0.24 × 10−4 μm2 s−1 (Figure 9F).
Flot1 amiRNA Mutant Lines Display Retarded Seedling Development
To obtain insight into the physiological role of Flot1, we sought to obtain knockout T-DNA insertion lines through the SALK collection. Unfortunately, seeds for presumptive knockouts (e.g., SALK-034821) were found to be wild-type (see Supplemental Figure 13A online). We next obtained mutant line GK-357G06 from the GABI-Kat library; this mutant has a T-DNA insertion in the Flot1 promoter. After backcrossing, homozygous T-DNA lines were identified by PCR of genomic DNA; however, RT-PCR using Flot1-specific primers revealed that Flot1 transcript levels in these mutants were elevated above those of wild-type plants (see Supplemental Figure 13B online).
To resolve this situation, we developed amiRNA mutant lines expressing a 35S:amiRNA Flot1 construct. Both RT-PCR and real-time quantitative RT-PCR analysis of Flot1 showed that mRNA levels for Flot1a/b were decreased to varying levels in four amiRNA knockdown lines compared with wild-type plants (Figure 10A). Seedling growth, as reflected by root and hypocotyl lengths, was reduced in all four amiRNA knockdown lines compared with wild-type plants (Figures 10B and 10C). A phenotypic analysis performed on amiRNA15-5, the line displaying the most substantial effect on Flot1a/b mRNA levels, revealed a delay in development throughout vegetative growth (Figures 10D to 10F). Confirmation that these phenotypic changes were caused by a reduction in Flot1a/b mRNA levels was provided by complementation experiments in which the wild-type phenotype was restored in amiRNA15-5 plants expressing 35S:Flot1 (see Supplemental Figure 14 and Supplemental Table 1 online). These findings are consistent with the hypothesis that Flot1 is required for optimal growth of Arabidopsis.
Figure 10.
RNA Interference–Mediated Knockdown of Flot1 Caused Growth Defects in Arabidopsis Seedlings.
(A) Flot1a/b transcript levels measured from seedlings expressing an amiRNA construct and control (wild-type [WT]) plant lines. Quantitative RT-PCR assays were normalized to actin mRNA levels as an internal control; mean ± sd (n = ≥10 seedlings per plant line).
(B) Seedling (6 d old) root and hypocotyl length measured on wild-type and amiRNA plant lines. Mean ± sd (n = ≥30 seedlings per plant line).
(C) Comparative seedling growth characteristics between wild-type (left) and a homozygous amiRNA15-5 knockdown plant line (right); image taken 6 d after germination.
(D) and (E) Phenotypes of a wild-type seedling (D) and a homozygous amiRNA15-5 knockdown transgenic plant line (E); 23-d-old plants.
(F) Photograph of 7-week-old plants. Left, wild-type control plant; right, amiRNA15-5 transgenic plant.
Bars = 10 mm in (C) to (E) and 50 mm in (F).
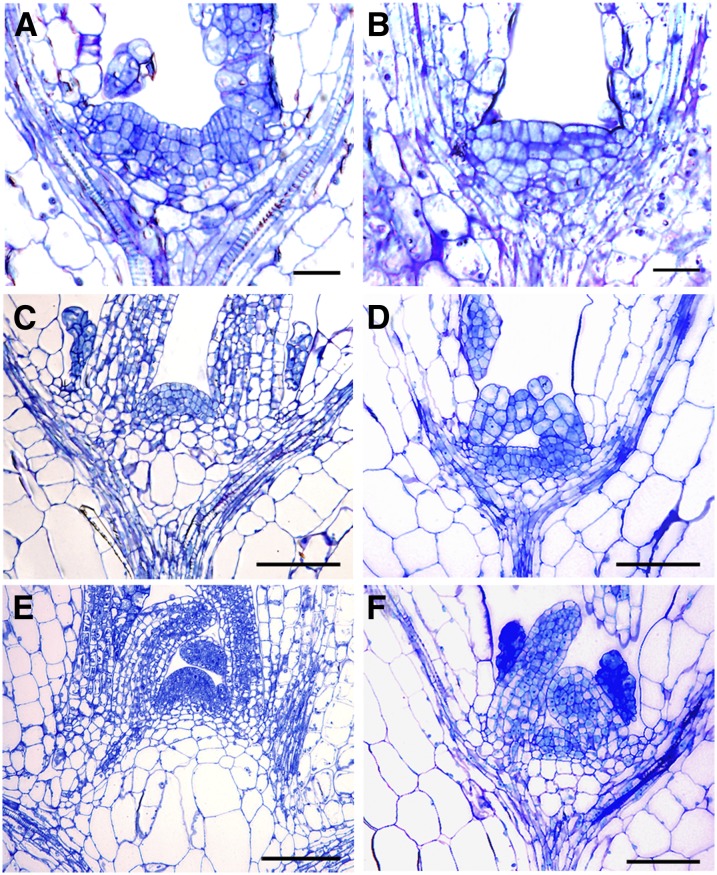
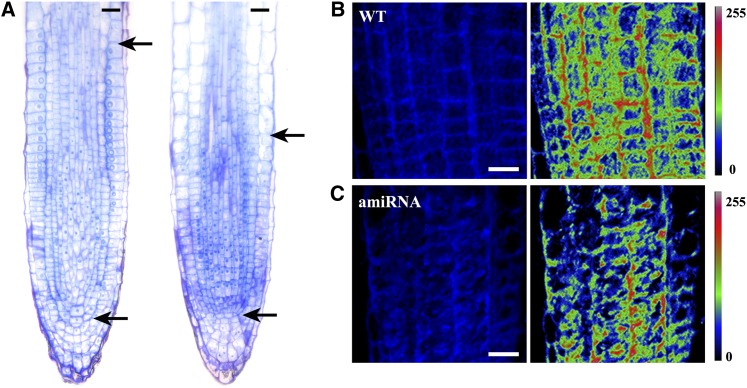
To investigate the basis for the observed stunted growth phenotype of our amiRNA lines, we next examined the effects of Flot1 mRNA reduction on cellular architecture within the shoot apical meristem (SAM) and root meristem (RM). Median longitudinal sections of the SAM from wild-type and amiRNA15-5 transgenic plants revealed that, compared with the wild-type SAM, this knockdown line had a smaller SAM during seedling development (Figures 11A to 11F). This difference was attributable to fewer meristematic cells within the SAM of the knockdown lines; in other words, the wild-type SAM contained 16.8 ± 0.7 cells across the L1 or epidermal layer, whereas this number was reduced to 11.2 ± 1.3 in amiRNA15-5 plants (mean and sd, n = 5 plants). However, the overall structural organization of the SAM was not altered in these Flot1 amiRNA knockdown plant lines. A similar situation was observed between the RM of wild-type and amiRNA15-5 plants; here, the number of meristematic cells in these knockdown miRNA RMs was ∼67% of that in wild-type RMs (Figure 12A).
Figure 11.
RNA Interference–Mediated Knockdown of Flot1 Retards SAM Development in Arabidopsis Seedlings.
Apices were fixed, embedded, and stained with toluidine blue. Longitudinal sections were examined under a bright-field light microscope.
(A), (C), and (E) Median longitudinal sections of SAMs of 3-, 8-, and 10-d-old wild-type Arabidopsis seedlings, respectively.
(B), (D), and (F) Median longitudinal sections of SAMs of 3-, 8-, and 10-d-old transgenic amiRNA15-5 Arabidopsis seedlings, respectively.
Bars = 20 μm.
[See online article for color version of this figure.]
Figure 12.
RM Size and Uptake of Filipin-Sterols Are Reduced in Transgenic amiRNA Arabidopsis Knockdown Plants.
(A) RMs of 3-d-old wild-type (left) and amiRNA15-5 (right) seedlings were fixed, embedded, and stained with toluidine blue. Longitudinal sections were examined under a bright-field light microscope. Top black arrow indicates the transition zone between meristem and elongation differentiation zone; bottom arrow marks the quiescent center of the RM.
(B) Filipin-sterol fluorescence detected in wild-type (WT) Arabidopsis root cells. Pseudocolor images show the fluorescence intensity of filipin (right). (Color code bar indicates the relative intensities.)
(C) Filipin-sterol fluorescence detected in amiRNA Arabidopsis knockdown root cells. Pseudocolor images show the fluorescence intensity of filipin (right). (Color code bar indicates the relative intensities.)
Bars = 20 μm.
To gain further insight into the basis for this retarded growth, we also compared the uptake of filipin-sterol in wild-type and amiRNA15-5 seedlings. As shown in Figures 12B and 12C, less filipin-sterol labeled intracellular puncta were observed in the amiRNA15-5 line compared with those in wild-type root cells, indicating that the internalization of sterols was affected in the Flot1 amiRNA knockdown plants. This finding is consistent with the hypothesis that At-Flot1 is required for efficient endocytosis. However, we cannot discount the possibility for general impairment in membrane turnover in these amiRNA15-5 seedlings.
DISCUSSION
Flot1 Is Associated with Membrane Microdomains
In recent years, the structure, composition, and possible functions of plant plasma membrane raft–like domains have been described (Raffaele et al., 2009; Haney and Long, 2010; Lefebvre et al., 2010). Some proteins involved in membrane trafficking and signal transduction were reported to be associated with membrane microdomains (Mongrand et al., 2010). A previous proteomic study revealed a tendency for Flot1 to partition into DRMs (Borner et al., 2005). However, whether this corresponds to an enrichment in bona fide plasma membrane microdomains remained to be determined. In our study, we found Flot1 was almost exclusively present in DRMs by immunoblotting, indicating that it was indeed a membrane microdomain–associated protein. Moreover, immunogold studies showed that Flot1 was clustered in particular domains of the membrane, further confirming the aforementioned conclusion. Based on these findings, we conclude that Flot1 is closely related to membrane microdomains.
Flot1 Participates in Non-Clathrin-Mediated Endocytosis
Plants appear to have at least four endocytic pathways. Apart from the well-studied clathrin-dependent endocytosis (Chen et al., 2011), other entry pathways, including membrane raft–mediated endocytosis, also operate in plant cells (Murphy et al., 2005; Moscatelli et al., 2007; Bandmann and Homann, 2011). Recent studies suggested that plant flotillins play a critical role in symbiotic bacterial infection of M. truncatula (Haney and Long, 2010; Haney et al., 2011), and the recycling of PIN proteins may depend on structural sterols and membrane rafts in Arabidopsis (Grebe et al., 2003; Willemsen et al., 2003; Men et al., 2008). Although these studies have begun to provide insights into the role of endocytosis in plants, the molecular mechanisms underlying these events remain to be elucidated. In this study, we show that the membrane microdomain–associated protein Flot1 was localized to the plasma membrane and intracellular compartments. In addition, GFP-Flot1 was also found to colocalize partially with FM4-64. Given that this FM4-64 dye is internalized, via endocytosis from the plasma membrane, and is considered to be a bona fide marker for the endocytic pathway in plant cells (Emans et al., 2002; Chintagari et al., 2006), our findings offer strong support for the hypothesis that Flot1 participates in an endocytic pathway.
Immunogold labeling with anti-Flot1 antibody provided strong confirmation of our confocal immunofluorescence findings, in that gold particles were clearly localized to plasma membrane invaginations and cytoplasmic vesicles in wild-type Arabidopsis root cells. Interestingly, we found the size of the vesicle configuration outlined by these gold particles to be on the order of 100 nm, which is different from the established 30-nm size of clathrin-coated vesicles (Dhonukshe et al., 2007). These findings suggest that the gold particle–labeled vesicles likely belong to a new type of endocytic vesicles. In agreement with the immunogold data, our transmission electron microscopy observations performed on high-pressure-frozen GFP-Flot1 root cells led to the detection of all the morphological intermediates for Flot1-positive vesicle formation at the plasma membrane, from invagination to internalization. These results offer strong support for the hypothesis that Flot1 is associated with the formation of these large endocytotic vesicles. The lack of spatial overlap between the fluorescent signals associated with GFP-Flot1 and CLC-mOrange structures within transgenic root cells provided further support for the notion that Flot1 is associated with a separate endocytotic pathway in plants. Similarly, raft-associated endocytosis, defined as the cholesterol-sensitive and clathrin-independent internalization of cargoes from the plasma membrane, is considered to be a new endocytic pathway in mammalian cells (Glebov et al., 2006; Lajoie and Nabi, 2007; Simons and Gerl, 2010).
TIRFM or evanescent wave microscopy is a powerful imaging technique to study biological processes occurring at the cell membrane. This technique is particularly suitable for the investigation of endocytosis and exocytosis, owing to its capacity to provide high signal-to-noise imaging of fluorescently labeled puncta located near the cover slip (Simons and Gerl, 2010). When the incident angle of the laser was adjusted, this technique can be successfully extended to the quantitative analysis of membrane-associated events within intact plant cells (Wang et al., 2006; Konopka et al., 2008; Konopka and Bednarek, 2008a, 2008b; Li et al., 2011; Wan et al., 2011). In our study, we used VA-TIRFM to clearly document the budding into the cell of GFP-Flot1 punctuate structures. Of equal importance, VA-TIRFM imaging revealed that the dynamic behavior of these Flot1-positive structures was different from that of CLC-positive puncta. Finally, we could demonstrate that interference with clathrin-dependent endocytosis by TyrA23 treatment did not change the dynamics of the GFP-Flot1 puncta, further confirming that Flot1 operates independently from clathrin-mediated endocytosis. Taken together, these findings offer strong support for the conclusion that Flot1 is involved in a different endocytic pathway in Arabidopsis.
Membrane Microdomains and the Cytoskeleton Are Required for Normal Flot1-Positive Puncta Dynamics
Membrane microdomains that are involved in many different cellular functions and processes, such as signaling, protein transport, cell adhesion, and movement, need to be connected with the cytoskeleton (Grzybek et al., 2005). Proteomic studies performed on membranes have shown that numerous cytoskeletal proteins coisolate with the DRM fraction (von Haller et al., 2001; Nebl et al., 2002; Bini et al., 2003; MacLellan et al., 2005; Yanagida et al., 2007; Yu et al., 2008). In contrast with the situation in mammalian and yeast cells (Langhorst et al., 2007), in which there is little evidence for microtubule involvement in flotillins, we found that microtubules were required for efficient GFP-Flot1 puncta dynamics. These findings are consistent with an earlier report that microtubules were required for efficient DRP1C and CLC dynamics (Konopka et al., 2008). Given the connection between formin and microtubule plus-ends and the link between formin and microfilaments (Cheung and Wu, 2004; Basu and Chang, 2007), we suggest that alterations in microtubule dynamics led directly to a redistribution of Flot1, which in turn would regulate the velocity and positioning of Flot1-positive puncta.
Role Played by Flot1 in Plant Growth
Endocytosis and vesicle recycling play crucial roles in plant developmental processes, including polar growth of root hair cells (Dhonukshe et al., 2006), cell-to-cell communication (Lewis and Lazarowitz, 2010), establishment of root cell polarity, and other auxin-dependent processes (Kleine-Vehn et al., 2008). As these studies have focused mainly on clathrin-dependent endocytosis, little information is currently available on the role of the non-clathrin-mediated pathway in seedling development. In this study, we demonstrated that Flot1 amiRNA knockdown lines exhibited retarded growth characteristics. This growth phenotype may reflect a limitation on recycling of plasma membrane proteins, which could then lead to a slowing of the cell cycle; this could explain the reduction in SAM and RM size. This model is consistent with an earlier report that root hair elongation depends on the highly regulated process of vesicle trafficking (Ovecka et al., 2010).
In summary, our study offers three novel findings: (1) Cellular and physiological data establish that Flot1 does not follow the prevailing endocytic pathway, as characterized by clathrin; (2) VA-TIRFM analysis revealed that the dynamic behavior of the Flot1-positive puncta differs from that of CLC-positive puncta; and (3) seedling development in Flot1 amiRNA knockdown transgenic lines is retarded. Taken together, our findings demonstrate that Flot1 is involved in a membrane microdomain–dependent, but clathrin-independent, endocytic pathway required for optimal seedling development.
METHODS
Plant Materials and Growth Conditions
All experiments were performed on Arabidopsis thaliana ecotype Columbia-0. Seeds were surface-sterilized and stratified at 4°C for 2 d in the dark, and seedlings were then grown on half-strength Murashige and Skoog (MS) medium supplemented with 1% Suc. Plants were grown under long-day (16 h light/8 h dark) conditions at a temperature of 20 to 22°C.
Plasmid Construction and Plant Transformation
The expression vector for GFP-Flot1 was constructed as follows: The coding sequence for Flot1 was PCR amplified (using primers 5′-CGCGGATCCATGTTCAAAGTTGCAAG-3′ and 5′-CCGGAATTCTTAGCTGCGAGTCACTT-3′) and subcloned as a BamHI-EcoRI fragment into a modified pCAMBIA1200 vector. Wild-type Arabidopsis was transformed with the GFP-Flot1 construct using the Agrobacterium tumefaciens–mediated floral dip method (Clough and Bent, 1998). Transgenic plants were selected on half-strength MS solid medium (1% agar) containing 50 µg/mL hygromycin.
Generation of Anti-Flot1 Antibodies
To generate polyclonal antibodies against Flot1, a full-length coding sequence was inserted into the pET-28a (+) expression vector (Novagen). After expression in Escherichia coli strain BL21, according to the manufacturer’s protocol (NEB), recombinant protein containing a 6xHis tag was purified using a nickel-nitrilotriacetic acid agarose column, according to the manufacturer’s instructions (Qiagen). Affinity-purified recombinant proteins were used to immunize rabbits. The polyclonal antiserum was affinity purified and tested for specificity using recombinant protein.
To generate monoclonal antibodies against Flot1, the recombinant proteins were used to immunize Balb/c mice (female, 4 to 6 weeks old) subcutaneously four times at 10- to 14-d intervals. Mice spleens were removed 3 d after the final immunization and cells isolated and fused with Sp2/0 myeloma cells at a ratio of 1:2 Sp2/0 cell/10 spleen cells using polyethylene glycol 1500. Fused cells were cultured in semisolid Iscove's Modified Dulbecco's Medium containing 25% newborn calf serum, 2% methyl cellulose, and 2% hypoxanthine-aminopterin-thymidine. Single hybridoma colonies were collected, individually, after 7 d of incubation. Appropriate amounts of supernatant generated from the hybridoma cell culture were used to screen for immune affinity by ELISA. Positive hybridomas were selected for further culture and titer determination.
Immunoblot Analysis
To determine protein levels in transgenic GFP-Flot1 plant lines, plasma membrane proteins were extracted from wild-type and transgenic GFP-Flot1 seedlings grown vertically on half-strength MS medium (1% agar). Twenty-day-old seedlings were homogenized in buffer (330 mM Suc, 50 mM MOPS-KOH, pH 7.5, 5 mM EDTA, 0.2% casein hydrolysate, 0.6% polyvinylpolypyrrolidone, 5 mM ascorbate, and 5 mM DTT). The homogenate was then filtered through a 200-μm nylon mesh and centrifuged at 10,000g for 15 min; the supernatant was saved and centrifuged at 50,000g for 60 min. The resulting microsomal pellet was resuspended in resuspension buffer (330 mM Suc, 5 mM K-phosphate, pH 7.8, 0.1 mM EDTA, and 1 mM DTT [freshly added]). The resuspended membranes were added to a phase mixture to produce an aqueous polymer two-phase system with a final composition of 6.1% (w/w) Dextran 500, 6.1% (w/w) polyethylene glycol 3350, 5 mM K-phosphate, pH 7.8, and 3 mM KCl. The final upper phases were diluted at least twofold with 330 mM Suc, 5 mM K-phosphate, pH 7.8, and 0.1 mM EDTA, and plasma membranes were pelleted by centrifugation at 100,000g for 1 h. PMs were further submitted to Triton X-100 treatment (final concentration 1% [v/v]) at 4°C for 30 min. Treated membranes were brought to a final concentration of 48% Suc (w/w), overlaid with successive 3-mL steps of 40, 35, 30, 10, and 5% Suc in TNE buffer (25 mM Tris-Cl, 150 mM NaCl, and 5 mM EDTA, pH 7.5), and then centrifuged for 18 h at 230,000g in a TST41 rotor (Sorvall). DRMs could be recovered below the 30 to 35% layers as an opaque band. This fraction was washed in TNE buffer to remove residual sucrose.
Isolated plasma membrane proteins were then separated by SDS-PAGE and analyzed by immunoblotting using our anti-Flot1 monoclonal antibody, biotin-conjugated anti-GFP antibody (Rockland Immunochemicals, Gilbertsville, PA) and anti-PM H+-ATPase antibody (Agrisera). Immunolabeling was performed according to (Konopka et al., 2008), with slight modifications. Horseradish peroxidase (HRP)-conjugated anti-mouse secondary antibodies (GE Healthcare) and HRP-conjugated streptavidin (Rockland Immunochemicals) were used to detect the primary antibodies, respectively.
Immunofluorescence and Confocal Microscopy
For indirect immunolocalization of Flot1 in wild-type or transgenic VHA-a1-GFP (Dettmer et al., 2006) plants, 4-d-old seedlings that were grown on half-strength MS medium (1% agar) were fixed in 4% (w/v) formaldehyde in PME (55 mM PIPES-KOH, 2.5 mM MgSO4, and 1 mM EGTA, pH 6.9) for 1 h under vacuum. Fixed seedlings were washed in PME and then in PBS, incubated in 1.5% (w/v) driselase for 40 min, and treated with permeabilization buffer (1% Nonidet P-40 and 10% DMSO in PBS) for 60 min. The roots were then washed in PBS. For immunolabeling, the roots were blocked with 2% BSA (w/v) in PBS for 1 h and probed with anti-Flot1 antibody in PBS + 2% BSA overnight at 4°C. Roots were then washed in PBS and incubated with tetramethylrhodamine-5-isothiocyanate–conjugated anti-mouse antibody (Sigma-Aldrich) in PBS + 2% BSA for 3 h. Afterward, roots were washed with PBS and observed by confocal microscopy (CLSM 510; Carl Zeiss).
High-Pressure Freezing, Freeze Substitution, Immunogold Labeling, and Electron Microscopy
Samples for transmission electron microscopy were prepared using high-pressure freezing. Three to four root apices (2 to 3 mm long) were placed in gold platelet carriers prefilled with 1-hexadecene and immediately frozen in a high-pressure freezing apparatus (HPM100; Bal-Tec). Freeze substitution in acetone, fixation in osmium, and embedding in Spurr’s resin were performed using a Leica EM AFS processor. Sections were poststained with uranyl acetate and lead citrate and then examined in a JEM-1230 (JEOL) operated at 80 keV.
Immunogold labeling was performed according to the procedure previously described (Müller et al., 2010), with slight modifications. In brief, Arabidopsis root tips were fixed using a high-pressure freeze fixation device (HPM010; Bal-Tec), cryo-substituted at −80°C, and embedded in Lowicryl HM20 (Polysciences). Ultrathin sections were blocked and subsequently incubated in a moist chamber with primary antibodies diluted 1:100 (anti-Flot1; polyclonal) or 1:150 (anti-CLC; monoclonal; Sigma-Aldrich) overnight at 4°C. This step was followed by rinsing and incubation with secondary antibodies conjugated to 15-nm (-labeling CLC) and 6-nm (-labeling Flot1) gold particles. Secondary antibodies were diluted 1:50 (Aurion) with PBS/BSA and incubated for 2 h at room temperature. Sections were extensively washed, stained with uranyl acetate, and examined with an LEO 912AB transmission electron microscope (Zeiss). Controls were performed using preimmune serum.
VA-TIRFM Imaging
The 4-d-old vertically grown GFP-Flot1 seedlings were stained with FM4-64 (Invitrogen, Molecular Probes; 2.5 μM) in half-strength MS medium supplemented with 1% Suc for 5 min on ice, washed twice with cold medium, and observed under VA-TIRFM. The data of GFP-Flot1 colocalization with FM4-64 were analyzed using Manders coefficients (Manders et al., 1993; Arimura et al., 2004).
Inhibitors, including mβCD, TyrA23, Lat-B, and oryzalin, were purchased from Sigma-Aldrich. Vertically grown 4-d-old Arabidopsis seedlings were incubated in half-strength liquid MS medium containing 50 μM TyrA23 or 10 mM MβCD for 30 min. For treatment with Lat-B or oryzalin, seedlings were incubated in half-strength liquid MS medium containing 1 μM Lat-B or 10 μM oryzalin for 30 min, then observed under VA-TIRFM.
An objective-type variable-angle total internal reflection fluorescence microscope, which was based on an inverted microscope (IX-71; Olympus) with a total internal reflective fluorescence illuminator and a ×100 oil-immersion objective (Olympus; numerical aperture = 1.45), was used for fluorescence imaging. The 473-nm/561-nm laser line from a diode laser (Changchun New Industries Optoelectronics Technology) was used to excite GFP and mOrange. Fluorescent signals were collected by the objective and passed through two filters, a BA 510IF long-pass filter (Chroma USA) and a HQ525/50 band-pass filter (Chroma), before being directed using a back-illuminated electron-multiplying charge-coupled device (EMCCD) camera (ANDOR iXon DV8897D-CS0-VP, Andor Technology) and high-quality filters (band-pass 525/45 and 609/54). We set the gain of our EMCCD camera at 300 throughout all single-molecule imaging experiments; the setting was in the linear dynamic range of the EMCCD camera. Images were acquired with 100-ms exposure time and analyzed with Image J software (NIH).
Cloning of 35S:amiRNA Flot1
The Flot1-directed amiRNA constructs were designed using a Web-based program (http://wmd2.weigelworld.org) (Schwab et al., 2006; Ossowski et al., 2008). The pRS300 plasmid was used as a template to create the amiRNA hairpin with an intron (Ossowski et al., 2008). Primer sequences are listed in Supplemental Table 2 online. The final amiRNA PCR product was digested at the KpnI and SpeI sites flanking the sequence encoding the amiRNA hairpin. The resultant product was ligated into the PTCK309 vector using the KpnI and SpeI sites. Transformation of Arabidopsis was conducted according to the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on half-strength MS medium containing 50 µg/mL hygromycin.
Real-Time Quantitative RT-PCR
Total RNA was extracted from seedlings using Trizol according to the manufacturer’s protocol (Invitrogen). Reverse transcription was performed using the First Strand cDNA Synthesis kit (Invitrogen) and an oligo(dT) primer. For Flot1, we designed specific primers (see Supplemental Table 3 online). PCR was performed using the SYBR Green Mix (Takara) in an optical 96-well plate with the ABI PRISM 7300 system (Applied Biosystems). In each reaction, 0.3 M primer and 10 ng of cDNA were used. PCR for each of three biological replicates was performed in triplicate. The initial denaturing time was 5 min, followed by 40 cycles of 95°C for 20 s, 54°C for 20 s, 72°C for 20 s, 84°C for 30 s, and 72°C for 10 min. A melting curve was run after the PCR cycles.
Characterization of Growth Phenotype
Seeds were treated at 4°C for 2 d and then grown for 6 d before measurements were taken of root and hypocotyl lengths. For adult plant phenotypes, wild-type and Flot1 amiRNA plants were grown side-by-side in a growth chamber. Mature plants (30 d old) were examined for the growth phenotype and photographed. To investigate the cellular architecture of the SAM and RM, shoot and root tips were fixed with 2.5% glutaraldehyde (w/v) in 0.1 M cacodylate buffer, pH 7.2, for 48 h at 4°C. Samples were then rinsed in 0.1 M cacodylate buffer and dehydrated in a graded ethanol series, infiltrated, and embedded using LR White resin (Electron Microscopy Sciences). Resin blocks were polymerized for 24 h at 60°C. Thick sections (1 μm) were cut using a Leica ultramicrotome. Sections were stained with 1% toluidine blue in 1% borax, and images were taken using a Zeiss Axioplan II compound microscope with an MRC digital camera and Axiovision software (Carl Zeiss).
Detection of Filipin-Sterol Fluorescence in Arabidopsis Roots
The filipin-sterol labeling was performed according to the procedure previously described (Boutté et al., 2011) with slight modifications. Seedlings were transferred to 4% paraformaldehyde, 150 μg/mL filipin III (Sigma-Aldrich) (from a 10 mg/mL stock in DMSO) in microtubule-stabilizing buffer (50 mM PIPES, 5 mM EGTA, and 5 mM MgSO4·7H2O, pH 7.0) for 1 h under vacuum. Specimens were washed three times in microtubule-stabilizing buffer and imaged with a multiphoton laser scanning microscope (FV 1000 MPE; Olympus). Filipin was excited with a 715-nm laser, and fluorescence was detected in an emission window at 460 to 500 nm. Fluorescence signals were collected by a ×20 water immersion objective (numerical aperture = 1.00). To compare sterol uptake levels in wild-type and amiRNA root cells, imaging was performed in parallel using exactly the same acquisition parameters for all samples. To create a pseudocolor image that visualizes the fluorescence intensity of filipin-stained structures, the original images were transferred to index color format using ImageJ software (NIH) with the Rainbow-LUT spectral setting.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Information Resource database under the following accession numbers: Flot1 (At5g25250), CLC (At2g40060), and VHA-a1 (At2g28520).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Characterization of Transgenic Arabidopsis Seedlings Expressing GFP-Flot1.
Supplemental Figure 2. Flot1 Was Detected in the Detergent-Resistant Membrane Fraction Isolated from a Plasma Membrane Preparation.
Supplemental Figure 3. Flot1 Is Enriched in the Detergent-Resistant Membrane Fraction.
Supplemental Figure 4. Localization of Flot1 Was Insensitive to Probe Level in a Range of Transgenic Lines.
Supplemental Figure 5. Anatomical Characterization of Overexpression and Wild-Type Roots.
Supplemental Figure 6. Four-Day-Old Arabidopsis Seedlings Expressing GFP-Flot1 or CLC-GFP Were Treated with 30 μM BFA.
Supplemental Figure 7. Immunofluorescence Labeling with Anti-Flot1 Antibody in a Root Hair Cell Containing a TGN Marker (VHA-a1-GFP).
Supplemental Figure 8. Flot1 Is Localized in Plasma Membrane Microdomains.
Supplemental Figure 9. Size Distribution Histogram of Vesicles in TEM Images of Wild-Type, Flot1 Overexpression, and amiRNA Root Cells.
Supplemental Figure 10. Quantitative Assessment of the Number of Colocalizing Flot1 and CLC Compartments.
Supplemental Figure 11. Flot1 Forms Punctuate Structures in Epidermal Cells from Various Tissues.
Supplemental Figure 12. Fluorescence Intensity Profile for Photobleaching of a GFP-Flot1 Positive Punctate Structure in a Chemically Fixed Root Cell.
Supplemental Figure 13. Identification of the Putative flot1 Mutant by PCR and RT-PCR.
Supplemental Figure 14. Phenotypes for the Wild Type, Transgenic amiRNA15-5, and Transgenic amiRNA15-5 Expressing 35S:GFP-rrFlot1 Plants.
Supplemental Table 1. PCR Primer Pairs Used for RNAi Resistant (rr)-Flot1 Cloning.
Supplemental Table 2. Primers Used for Cloning 35S:amiRNA Flot1.
Supplemental Table 3. Primers Employed for qRT-PCR and RT-PCR Analyses of Flot1 Expression.
Supplemental Movie 1. Time-Lapse Video of Living Root Cells Expressing GFP-Flot1 Imaged by Laser Scanning Confocal Microscopy.
Supplemental Movie 2. Time-Lapse Video of an Expanding Root Epidermal Cell Expressing GFP-Flot1 Imaged with VA-TIRFM.
Supplemental Movie 3. Apparent Budding of GFP-Flot1 Shown by VA-TIRFM Imaging.
Supplemental Movie 4. Colocalization of GFP-Flot1 and FM4-64 in Root Epidermal Cells Imaged Simultaneously with VA-TIRFM.
Supplemental Movie 5. Time-Lapse Images of an Expanding Root Epidermal Cell Expressing CLC-mOrange and GFP-Flot1 Imaged with Simultaneous Dual-Color VA-TIRFM.
Supplementary Material
Acknowledgments
We thank Sebastian Bednarek for kindly providing CLC-GFP/mOrange Arabidopsis seeds. This work was supported by the National Basic Research Program of China (973 Program 2011CB809103 and 2011CB944603), the Chinese Academy of Sciences/State Administration of Foreign Experts Affairs International Partnership Program for Creative Research Teams (20090491019), the National Natural Science Foundation of China (30730009, 30821007, 31000092, 31121065, and 30900072), the Knowledge Innovation Program of the Chinese Academy of Sciences (KJCX2-YW-L08 and KSCX2-EW-J-1), the Centre of the Region Haná for Biotechnological and Agricultural Research (ED0007/01/01), and the Czech Science Foundation Grant Agency of the Czech Republic (P501/11/1764).
AUTHOR CONTRIBUTIONS
R.L. and J.L. designed the research. R.L., P.L., and U.M. performed the experiments. R.L., Y.W., T.C., Q.W., and W.J.L analyzed data. X.F. contributed new analytic tools. R.L, F.B., J.S., W.J.L., and J.L. wrote the article. W.J.L. and J.L. contributed equally in supervising this project.
Glossary
- DRM
detergent-resistant membrane
- CLC
clathrin light chain
- VA-TIRFM
variable-angle total internal reflection fluorescence microscopy
- amiRNA
artificial microRNA
- GFP
green fluorescent protein
- MSD
mean squared displacement
- mβCD
methyl-β-cyclodextrin
- Lat-B
latrunculin B
- RM
root meristem
- SAM
shoot apical meristem
- MS
Murashige and Skoog
- TyrA23
tyrphostin A23
- EMCCD
electron-multiplying charge-coupled device
References
- Arimura S., Yamamoto J., Aida G.P., Nakazono M., Tsutsumi N. (2004). Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proc. Natl. Acad. Sci. USA 101: 7805–7808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banbury D.N., Oakley J.D., Sessions R.B., Banting G. (2003). Tyrphostin A23 inhibits internalization of the transferrin receptor by perturbing the interaction between tyrosine motifs and the medium chain subunit of the AP-2 adaptor complex. J. Biol. Chem. 278: 12022–12028 [DOI] [PubMed] [Google Scholar]
- Bandmann V., Homann U. (December 23, 2011). Clathrin-independent endocytosis contributes to uptake of glucose into BY-2 protoplasts. Plant J. http://dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- Basu R., Chang F. (2007). Shaping the actin cytoskeleton using microtubule tips. Curr. Opin. Cell Biol. 19: 88–94 [DOI] [PubMed] [Google Scholar]
- Benmerah A., Lamaze C. (2007). Clathrin-coated pits: Vive la différence? Traffic 8: 970–982 [DOI] [PubMed] [Google Scholar]
- Bhat R.A., Miklis M., Schmelzer E., Schulze-Lefert P., Panstruga R. (2005). Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc. Natl. Acad. Sci. USA 102: 3135–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bini L., Pacini S., Liberatori S., Valensin S., Pellegrini M., Raggiaschi R., Pallini V., Baldari C.T. (2003). Extensive temporally regulated reorganization of the lipid raft proteome following T-cell antigen receptor triggering. Biochem. J. 369: 301–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner G.H., Sherrier D.J., Weimar T., Michaelson L.V., Hawkins N.D., Macaskill A., Napier J.A., Beale M.H., Lilley K.S., Dupree P. (2005). Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 137: 104–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutté Y., Men S., Grebe M. (2011). Fluorescent in situ visualization of sterols in Arabidopsis roots. Nat. Protoc. 6: 446–456 [DOI] [PubMed] [Google Scholar]
- Brown D.A., London E. (2000). Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 275: 17221–17224 [DOI] [PubMed] [Google Scholar]
- Carmona-Salazar L., El Hafidi M., Enríquez-Arredondo C., Vázquez-Vázquez C., González de la Vara L.E., Gavilanes-Ruíz M. (2011). Isolation of detergent-resistant membranes from plant photosynthetic and non-photosynthetic tissues. Anal. Biochem. 417: 220–227 [DOI] [PubMed] [Google Scholar]
- Chen X., Irani N.G., Friml J. (2011). Clathrin-mediated endocytosis: The gateway into plant cells. Curr. Opin. Plant Biol. 14: 674–682 [DOI] [PubMed] [Google Scholar]
- Cheung A.Y., Wu H.M. (2004). Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell 16: 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintagari N.R., Jin N., Wang P., Narasaraju T.A., Chen J., Liu L. (2006). Effect of cholesterol depletion on exocytosis of alveolar type II cells. Am. J. Respir. Cell Mol. Biol. 34: 677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Conner S.D., Schmid S.L. (2003). Regulated portals of entry into the cell. Nature 422: 37–44 [DOI] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P., Aniento F., Hwang I., Robinson D.G., Mravec J., Stierhof Y.D., Friml J. (2007). Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr. Biol. 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., Baluska F., Schlicht M., Hlavacka A., Samaj J., Friml J., Gadella T.W., Jr (2006). Endocytosis of cell surface material mediates cell plate formation during plant cytokinesis. Dev. Cell 10: 137–150 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., et al. (2008). Generation of cell polarity in plants links endocytosis, auxin distribution and cell fate decisions. Nature 456: 962–966 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Drab M., et al. (2001). Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice. Science 293: 2449–2452 [DOI] [PubMed] [Google Scholar]
- Emans N., Zimmermann S., Fischer R. (2002). Uptake of a fluorescent marker in plant cells is sensitive to brefeldin A and wortmannin. Plant Cell 14: 71–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk J., Thoumine O., Dequidt C., Choquet D., Faivre-Sarrailh C. (2004). NrCAM coupling to the cytoskeleton depends on multiple protein domains and partitioning into lipid rafts. Mol. Biol. Cell 15: 4695–4709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick M., Bright N.A., Riento K., Bray A., Merrified C., Nichols B.J. (2007). Coassembly of flotillins induces formation of membrane microdomains, membrane curvature, and vesicle budding. Curr. Biol. 17: 1151–1156 [DOI] [PubMed] [Google Scholar]
- Garcia A., Cayla X., Fleischer A., Guergnon J., Alvarez-Franco Cañas F., Rebollo M.P., Roncal F., Rebollo A. (2003). Rafts: A simple way to control apoptosis by subcellular redistribution. Biochimie 85: 727–731 [DOI] [PubMed] [Google Scholar]
- Glebov O.O., Bright N.A., Nichols B.J. (2006). Flotillin-1 defines a clathrin-independent endocytic pathway in mammalian cells. Nat. Cell Biol. 8: 46–54 [DOI] [PubMed] [Google Scholar]
- Grebe M., Xu J., Möbius W., Ueda T., Nakano A., Geuze H.J., Rook M.B., Scheres B. (2003). Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr. Biol. 13: 1378–1387 [DOI] [PubMed] [Google Scholar]
- Grzybek M., Kozubek A., Dubielecka P., Sikorski A.F. (2005). Rafts—The current picture. Folia Histochem. Cytobiol. 43: 3–10 [PubMed] [Google Scholar]
- Haney C.H., Long S.R. (2010). Plant flotillins are required for infection by nitrogen-fixing bacteria. Proc. Natl. Acad. Sci. USA 107: 478–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney C.H., Riely B.K., Tricoli D.M., Cook D.R., Ehrhardt D.W., Long S.R. (2011). Symbiotic rhizobia bacteria trigger a change in localization and dynamics of the Medicago truncatula receptor kinase LYK3. Plant Cell 23: 2774–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen G.H., Dalskov S.M., Rasmussen C.R., Immerdal L., Niels-Christiansen L.L., Danielsen E.M. (2005). Cholera toxin entry into pig enterocytes occurs via a lipid raft- and clathrin-dependent mechanism. Biochemistry 44: 873–882 [DOI] [PubMed] [Google Scholar]
- Keinath N.F., Kierszniowska S., Lorek J., Bourdais G., Kessler S.A., Shimosato-Asano H., Grossniklaus U., Schulze W.X., Robatzek S., Panstruga R. (2010). PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J. Biol. Chem. 285: 39140–39149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Dhonukshe P., Sauer M., Brewer P.B., Wiśniewska J., Paciorek T., Benková E., Friml J. (2008). ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis. Curr. Biol. 18: 526–531 [DOI] [PubMed] [Google Scholar]
- Konopka C.A., Backues S.K., Bednarek S.Y. (2008). Dynamics of Arabidopsis dynamin-related protein 1C and a clathrin light chain at the plasma membrane. Plant Cell 20: 1363–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka C.A., Bednarek S.Y. (2008a). Comparison of the dynamics and functional redundancy of the Arabidopsis dynamin-related isoforms DRP1A and DRP1C during plant development. Plant Physiol. 147: 1590–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka C.A., Bednarek S.Y. (2008b). Variable-angle epifluorescence microscopy: A new way to look at protein dynamics in the plant cell cortex. Plant J. 53: 186–196 [DOI] [PubMed] [Google Scholar]
- Kurzchalia T.V., Parton R.G. (1999). Membrane microdomains and caveolae. Curr. Opin. Cell Biol. 11: 424–431 [DOI] [PubMed] [Google Scholar]
- Lafont F., Abrami L., van der Goot F.G. (2004). Bacterial subversion of lipid rafts. Curr. Opin. Microbiol. 7: 4–10 [DOI] [PubMed] [Google Scholar]
- Lajoie P., Nabi I.R. (2007). Regulation of raft-dependent endocytosis. J. Cell. Mol. Med. 11: 644–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie P., Nabi I.R. (2010). Lipid rafts, caveolae, and their endocytosis. Int. Rev. Cell Mol. Biol. 282: 135–163 [DOI] [PubMed] [Google Scholar]
- Langhorst M.F., Reuter A., Jaeger F.A., Wippich F.M., Luxenhofer G., Plattner H., Stuermer C.A. (2008). Trafficking of the microdomain scaffolding protein reggie-1/flotillin-2. Eur. J. Cell Biol. 87: 211–226 [DOI] [PubMed] [Google Scholar]
- Langhorst M.F., Solis G.P., Hannbeck S., Plattner H., Stuermer C.A. (2007). Linking membrane microdomains to the cytoskeleton: Regulation of the lateral mobility of reggie-1/flotillin-2 by interaction with actin. FEBS Lett. 581: 4697–4703 [DOI] [PubMed] [Google Scholar]
- Lefebvre B., Furt F., Hartmann M.A., Michaelson L.V., Carde J.P., Sargueil-Boiron F., Rossignol M., Napier J.A., Cullimore J., Bessoule J.J., Mongrand S. (2007). Characterization of lipid rafts from Medicago truncatula root plasma membranes: A proteomic study reveals the presence of a raft-associated redox system. Plant Physiol. 144: 402–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre B., et al. (2010). A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc. Natl. Acad. Sci. USA 107: 2343–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J.D., Lazarowitz S.G. (2010). Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc. Natl. Acad. Sci. USA 107: 2491–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wang X., Yang Y., Li R., He Q., Fang X., Luu D.T., Maurel C., Lin J. (2011). Single-molecule analysis of PIP2;1 dynamics and partitioning reveals multiple modes of Arabidopsis plasma membrane aquaporin regulation. Plant Cell 23: 3780–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D., Simons K. (2010). Lipid rafts as a membrane-organizing principle. Science 327: 46–50 [DOI] [PubMed] [Google Scholar]
- MacLellan D.L., Steen H., Adam R.M., Garlick M., Zurakowski D., Gygi S.P., Freeman M.R., Solomon K.R. (2005). A quantitative proteomic analysis of growth factor-induced compositional changes in lipid rafts of human smooth muscle cells. Proteomics 5: 4733–4742 [DOI] [PubMed] [Google Scholar]
- Manders E.M.M., Verbeek F.J., Aten J.A. (1993). Measurement of co-localization of objects in dual-color confocal images. J. Microsc. 169: 375–382 [DOI] [PubMed] [Google Scholar]
- Men S., Boutté Y., Ikeda Y., Li X., Palme K., Stierhof Y.D., Hartmann M.A., Moritz T., Grebe M. (2008). Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat. Cell Biol. 10: 237–244 [DOI] [PubMed] [Google Scholar]
- Mongrand S., Morel J., Laroche J., Claverol S., Carde J.P., Hartmann M.A., Bonneu M., Simon-Plas F., Lessire R., Bessoule J.J. (2004). Lipid rafts in higher plant cells: Purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem. 279: 36277–36286 [DOI] [PubMed] [Google Scholar]
- Mongrand S., Stanislas T., Bayer E.M., Lherminier J., Simon-Plas F. (2010). Membrane rafts in plant cells. Trends Plant Sci. 15: 656–663 [DOI] [PubMed] [Google Scholar]
- Morel J., Claverol S., Mongrand S., Furt F., Fromentin J., Bessoule J.J., Blein J.P., Simon-Plas F. (2006). Proteomics of plant detergent-resistant membranes. Mol. Cell. Proteomics 5: 1396–1411 [DOI] [PubMed] [Google Scholar]
- Morrow I.C., Rea S., Martin S., Prior I.A., Prohaska R., Hancock J.F., James D.E., Parton R.G. (2002). Flotillin-1/reggie-2 traffics to surface raft domains via a novel golgi-independent pathway. Identification of a novel membrane targeting domain and a role for palmitoylation. J. Biol. Chem. 277: 48834–48841 [DOI] [PubMed] [Google Scholar]
- Moscatelli A., Ciampolini F., Rodighiero S., Onelli E., Cresti M., Santo N., Idilli A. (2007). Distinct endocytic pathways identified in tobacco pollen tubes using charged nanogold. J. Cell Sci. 120: 3804–3819 [DOI] [PubMed] [Google Scholar]
- Müller J., Beck M., Mettbach U., Komis G., Hause G., Menzel D., Samaj J. (2010). Arabidopsis MPK6 is involved in cell division plane control during early root development, and localizes to the pre-prophase band, phragmoplast, trans-Golgi network and plasma membrane. Plant J. 61: 234–248 [DOI] [PubMed] [Google Scholar]
- Murphy A.S., Bandyopadhyay A., Holstein S.E., Peer W.A. (2005). Endocytotic cycling of PM proteins. Annu. Rev. Plant Biol. 56: 221–251 [DOI] [PubMed] [Google Scholar]
- Nebl T., Pestonjamasp K.N., Leszyk J.D., Crowley J.L., Oh S.W., Luna E.J. (2002). Proteomic analysis of a detergent-resistant membrane skeleton from neutrophil plasma membranes. J. Biol. Chem. 277: 43399–43409 [DOI] [PubMed] [Google Scholar]
- Ossowski S., Schwab R., Weigel D. (2008). Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Ovecka M., Berson T., Beck M., Derksen J., Samaj J., Baluska F., Lichtscheidl I.K. (2010). Structural sterols are involved in both the initiation and tip growth of root hairs in Arabidopsis thaliana. Plant Cell 22: 2999–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton R.G. (2003). Caveolae—From ultrastructure to molecular mechanisms. Nat. Rev. Mol. Cell Biol. 4: 162–167 [DOI] [PubMed] [Google Scholar]
- Pelkmans L., Helenius A. (2003). Insider information: What viruses tell us about endocytosis. Curr. Opin. Cell Biol. 15: 414–422 [DOI] [PubMed] [Google Scholar]
- Raffaele S., et al. (2009). Remorin, a Solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razani B., Combs T.P., Wang X.B., Frank P.G., Park D.S., Russell R.G., Li M., Tang B., Jelicks L.A., Scherer P.E., Lisanti M.P. (2002a). Caveolin-1-deficient mice are lean, resistant to diet-induced obesity, and show hypertriglyceridemia with adipocyte abnormalities. J. Biol. Chem. 277: 8635–8647 [DOI] [PubMed] [Google Scholar]
- Razani B., Wang X.B., Engelman J.A., Battista M., Lagaud G., Zhang X.L., Kneitz B., Hou H., Jr, Christ G.J., Edelmann W., Lisanti M.P. (2002b). Caveolin-2-deficient mice show evidence of severe pulmonary dysfunction without disruption of caveolae. Mol. Cell. Biol. 22: 2329–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salaün C., James D.J., Chamberlain L.H. (2004). Lipid rafts and the regulation of exocytosis. Traffic 5: 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahollari B., Peskan-Berghöfer T., Oelmüller R. (2004). Receptor kinases with leucine-rich repeats are enriched in Triton X-100 insoluble plasma membrane microdomains from plants. Physiol. Plant. 122: 397–403 [Google Scholar]
- Simons K., Gerl M.J. (2010). Revitalizing membrane rafts: New tools and insights. Nat. Rev. Mol. Cell Biol. 11: 688–699 [DOI] [PubMed] [Google Scholar]
- Simons K., Toomre D. (2000). Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1: 31–39 [DOI] [PubMed] [Google Scholar]
- Tanaka H., Kitakura S., De Rycke R., De Groodt R., Friml J. (2009). Fluorescence imaging-based screen identifies ARF GEF component of early endosomal trafficking. Curr. Biol. 19: 391–397 [DOI] [PubMed] [Google Scholar]
- Titapiwatanakun B., et al. (2009). ABCB19/PGP19 stabilises PIN1 in membrane microdomains in Arabidopsis. Plant J. 57: 27–44 [DOI] [PubMed] [Google Scholar]
- von Haller P.D., Donohoe S., Goodlett D.R., Aebersold R., Watts J.D. (2001). Mass spectrometric characterization of proteins extracted from Jurkat T cell detergent-resistant membrane domains. Proteomics 1: 1010–1021 [DOI] [PubMed] [Google Scholar]
- Wan Y., Ash W.M., III, Fan L., Hao H., Kim M.K., Lin J. (2011). Variable-angle total internal reflection fluorescence microscopy of intact cells of Arabidopsis thaliana. Plant Methods 7: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Teng Y., Wang Q., Li X., Sheng X., Zheng M., Samaj J., Baluska F., Lin J. (2006). Imaging of dynamic secretory vesicles in living pollen tubes of Picea meyeri using evanescent wave microscopy. Plant Physiol. 141: 1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen V., Friml J., Grebe M., van den Toorn A., Palme K., Scheres B. (2003). Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15: 612–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida M., Nakayama H., Yoshizaki F., Fujimura T., Takamori K., Ogawa H., Iwabuchi K. (2007). Proteomic analysis of plasma membrane lipid rafts of HL-60 cells. Proteomics 7: 2398–2409 [DOI] [PubMed] [Google Scholar]
- Yu X.M., Yu X.D., Qu Z.P., Huang X.J., Guo J., Han Q.M., Zhao J., Huang L.L., Kang Z.S. (2008). Cloning of a putative hypersensitive induced reaction gene from wheat infected by stripe rust fungus. Gene 407: 193–198 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.