This work describes biochemical, biophysical, structural, and genetic analyses of an Arabidopsis homolog of mammalian NEET proteins, which are involved in a wide range of cellular processes. It finds that At-NEET plays a key role in plant development, senescence, reactive oxygen species homeostasis, and iron metabolism.
Abstract
The NEET family is a newly discovered group of proteins involved in a diverse array of biological processes, including autophagy, apoptosis, aging, diabetes, and reactive oxygen homeostasis. They form a novel structure, the NEET fold, in which two protomers intertwine to form a two-domain motif, a cap, and a unique redox-active labile 2Fe-2S cluster binding domain. To accelerate the functional study of NEET proteins, as well as to examine whether they have an evolutionarily conserved role, we identified and characterized a plant NEET protein. Here, we show that the Arabidopsis thaliana At5g51720 protein (At-NEET) displays biochemical, structural, and biophysical characteristics of a NEET protein. Phenotypic characterization of At-NEET revealed a key role for this protein in plant development, senescence, reactive oxygen homeostasis, and Fe metabolism. A role in Fe metabolism was further supported by biochemical and cell biology studies of At-NEET in plant and mammalian cells, as well as mutational analysis of its cluster binding domain. Our findings support the hypothesis that NEET proteins have an ancient role in cells associated with Fe metabolism.
INTRODUCTION
The newly discovered NEET family class of proteins includes the human mitoNEET and Miner1 (Wiley et al., 2007a, 2007b). NEET proteins contain a unique homodimeric fold, the NEET fold, in which two protomers intertwine to form a two-domain structure, a β-cap, and a cluster binding domain (Hou et al., 2007; Lin et al., 2007; Paddock et al., 2007; Conlan et al., 2009a, 2009b, 2009c). Both parts of the homodimer harbor a redox-active 2Fe-2S cluster, each coordinated by 3 Cys and 1 His, defining a new class of 2Fe-2S proteins (Tirrell et al., 2009; Dicus et al., 2010; Conlan et al., 2011). This unique structure, which includes a single His-ligand at the 2Fe-2S cluster, is thought to make the cluster relatively labile and suggests that NEET proteins are involved in Fe-S cluster transfer, potentially facilitating cluster shuttling between proteins in intracellular organelles and the cytosol (Paddock et al., 2007; Conlan et al., 2009c).
MitoNEET and Miner1 were shown to be involved in human pathology and mammalian longevity (Colca et al., 2004; Chen et al., 2010). MitoNEET is thought to be involved in regulating redox reactions in the mitochondria, the accumulation of reactive oxygen species (ROS), and the control of apoptosis (Chang et al., 2010). MitoNEET was also found to bind the drug pioglitazone, a member of the thiazolidinedione class of insulin sensitizers, used in the treatment for type 2 diabetes (Colca et al., 2004; Zuris et al., 2011). Miner1, a protein essential for health and longevity, was recently identified as a mitochondrial integrity maintainer as well as a carrier of a mutation that causes Wolfram Syndrome 2, an autosomal recessive mitochondria-mediated disorder (Amr et al., 2007; Conlan et al., 2009b; Chen et al., 2010). Knockout of Miner1 in mice leads to premature aging, severe neurodegeneration, blindness, and muscle atrophy (Chen et al., 2009).
To facilitate the functional study of NEET proteins and to examine whether they have evolutionarily conserved functions, we identified and characterized a plant NEET protein. Here, we show that the Arabidopsis thaliana At5g51720 protein displays biochemical, biophysical, and structural characteristics common to NEET proteins, such as mitoNEET and Miner1. In contrast with human NEET proteins, At5g51720 does not appear to belong to a gene family and is localized to chloroplasts as well as mitochondria. At5g51720 was therefore named At-NEET. Phenotypic characterization using knockdown and RNA interference (RNAi) lines revealed a key role for At-NEET in plant development, senescence, ROS homeostasis, and Fe metabolism. Its proposed role in Fe metabolism is further supported by biochemical and cell biology studies of At-NEET in plant and mammalian cells as well as mutational analysis of the At-NEET cluster binding domain.
RESULTS
The Arabidopsis At5g51720 Protein Is a NEET Protein
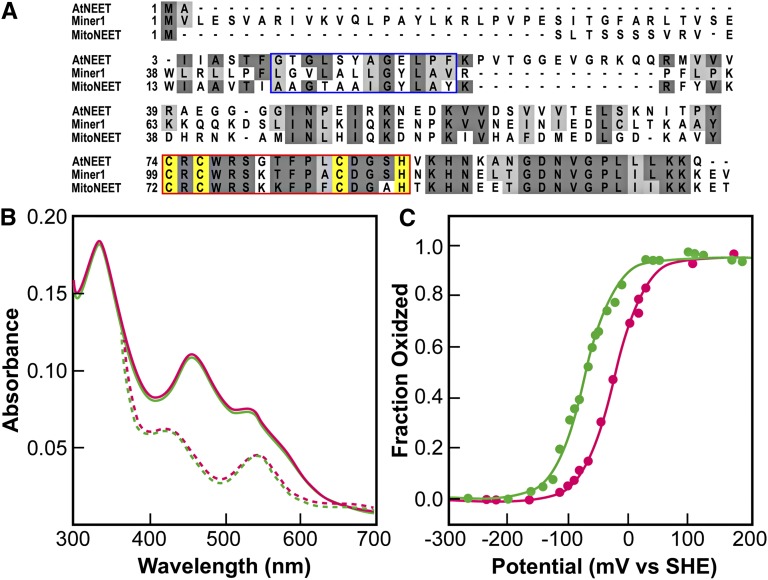
Arabidopsis contains a single copy gene (At5g51720) that shows 50 and 57% similarity to mitoNEET and Miner1, respectively (Figure 1A). A higher level of sequence identity to mitoNEET and Miner1 is found in the putative Fe-S binding domain of At5g51720 (75 and 88%, respectively; red box in Figure 1A). Similar to mitoNEET and Miner1, At5g51720 appears to contain a single transmembrane domain (blue box, Figure 1A).
Figure 1.
The At5g51720 Protein Exhibits Similar Properties to Human NEET Proteins.
(A) Alignment of the Arabidopsis At5g51720 protein with the human mitoNEET and Miner1 proteins. The blue box highlights the transmembrane region identified in the human proteins. The red box highlights the conserved 2Fe-2S cluster binding domain. The cluster binding side chains for the proteins are highlighted in yellow.
(B) UV-Vis optical spectra of At5g51720 (green) and human mitoNEET (magenta). The peaks at 458 and 530 nm (solid traces) are characteristic of ligand-to-charge transfer bands of the 2Fe-2S centers in human mitoNEET (Paddock et al., 2007; Wiley et al., 2007b). Upon addition of the reducing dithionite, the spectrum (reversibly) changed with peaks near 440 and 550 nm (dotted traces), nearly identical to that of the reduced human mitoNEET.
(C) Redox potential (EM) of the 2Fe-2S clusters of At5g51720 (green) and human mitoNEET (magenta) at pH 7.0. The EM of At5g51720 was measured using potentiometric redox titrations. The value for EM at pH 7.0 (EM, 7) was ?50 mV lower than that of human mitoNEET. At5g51720’s EM was found to be pH dependent (see Supplemental Figure 1 online), with a slope of −50 mV/pH.
To determine whether At5g51720 is a NEET protein, its biophysical properties were compared with those of mitoNEET and Miner1. For this purpose, the truncated soluble domain of At5g51720, from amino acid 39 to amino acid 108, was expressed in Escherichia coli and purified by nickel-nitrilotriacetic acid affinity chromatography. The purified protein was red in color with absorption peaks in the visible range at 458 and 530 nm (solid traces, Figure 1B), which are characteristic of the ligand-to-charge transfer bands of the 2Fe-2S centers in human mitoNEET (Paddock et al., 2007; Wiley et al., 2007b; Conlan et al., 2009b, 2009c). Upon addition of the reducing agent dithionite, the spectrum changed with peaks near 440 and 550 nm, nearly identical to that of the reduced mitoNEET (dotted traces, Figure 1B). These results show the presence of a redox active center and support the existence in At5g51720 of a 2Fe-2S center with a similar protein environment to that found in mitoNEET. The redox potential (EM) of At5g51720 was measured with potentiometric redox titrations (Zuris et al., 2010). The value for EM,7 at pH 7.0 was –50 mV lower than that of human mitoNEET (Figure 1C) but differs greatly from most ferredoxin (Fd) and Rieske 2Fe-2S centers (Meyer, 2008). At5g51720’s EM was found to be pH dependent, decreasing above pH 7.0 to 54 mV/pH, suggesting, as is the case for human mitoNEET, that its reduction is proton coupled (see Supplemental Figure 1 online).
Structural Characterization of At-NEET
To determine the structure of At-NEET, we crystallized the purified At5g51720 protein (amino acids 39 to 108) and solved its structure at 1.75 Å resolution. For the molecular replacement analysis, we used the structure of human mitoNEET (pdb ID code2QH7) (Paddock et al., 2007).
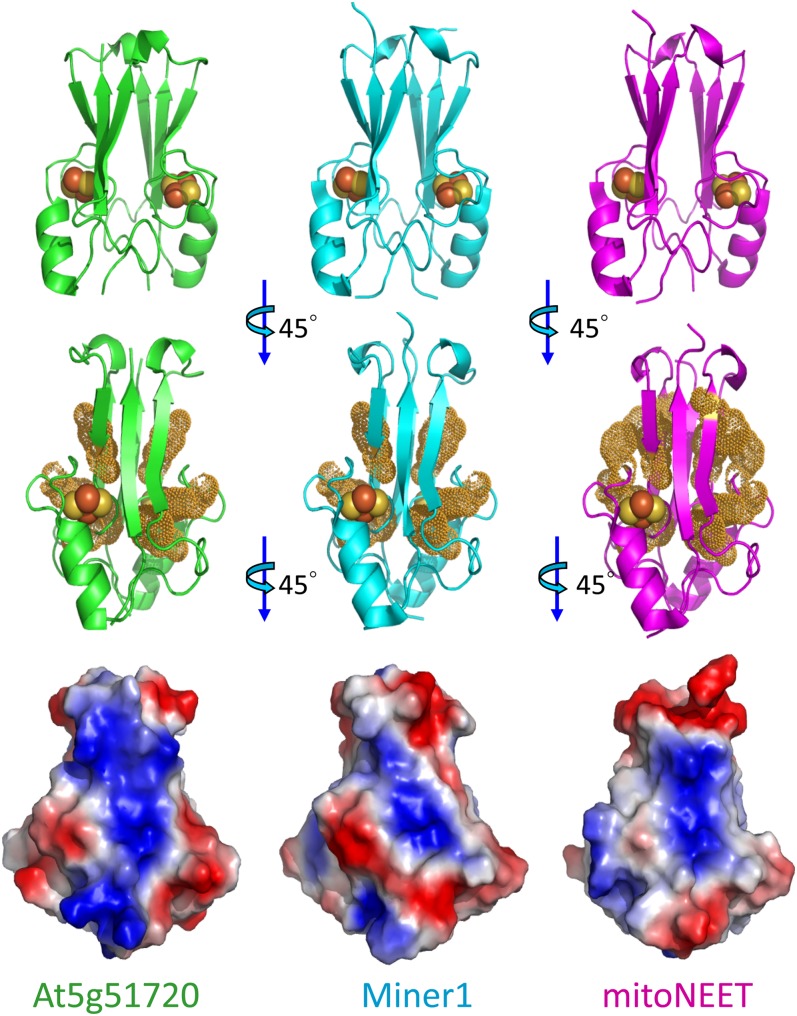
Figure 2 compares the structures of At5g51720 (pdb ID code3S2Q, left column, green), Miner1 (pdb ID code 3FNV, middle column, cyan), and mitoNEET (pdb ID code 2QH7, right column, magenta). As shown at the top panel of Figure 2, the At-NEET protein has a homodimeric structure similar to that of its human counterparts. Similar to Miner1 and mitoNEET, the plant protein contains a typical NEET fold having a strand-swapped β-cap domain and a cluster binding domain (Paddock et al., 2007). The At5g51720 protein can therefore be considered a plant NEET protein and will be referred to as At-NEET hereafter.
Figure 2.
Structural Comparison of At-NEET (At5g51720) with the Human Miner1 and mitoNEET 2Fe-2S Cluster–Containing NEET Proteins.
The top row shows the secondary structure of the different proteins with the 2Fe-2S centers shown as spheres (Fe is red and S is yellow). The middle row highlights the aromatic groups. Note that this view is rotated 45° from the top view to better illustrate the aromatic side chain distribution. The bottom row shows the overall surface charge based on vacuum electrostatics. All proteins are net positively charged at neutral pH and all are positively charged in the middle. By contrast, they all differ in the extent of continuous positive charge. Note that this view is rotated an additional 45° from the middle view to better illustrate the differences in the surface charges. At5g51720, green; human Miner1, cyan; mitoNEET, magenta.
The 2Fe-2S centers in each structure are shown as spheres (Fe is red and S is yellow). The membrane anchors, removed in the constructs used for the in vitro studies, would extend from the N termini located at the bottom of the structures. Although all three structures are relatively similar, both At-NEET and Hs-Miner1 are slightly wider at the top due to an extra amino acid located at the loop connecting the swapped strand to the remainder of the protomer. These structures also differ from that of Hs-mitoNEET in the arrangement of the aromatic groups, shown in the middle row of Figure 2 and highlighted by yellow dots. Both At-NEET and Hs-Miner1 have a similar aromatic distribution, whereas Hs-mitoNEET has several additional aromatic side chains. Although At-NEET exhibits a slightly higher similarity to Hs-Miner1, comparison of the overall surface charge of the molecules (Figure 2, bottom row), indicates that all three proteins (At-NEET, Hs-Miner1, and Hs-mitoNEET) differ in the extent of continuous positive/negative charges with At-NEET having the most continuity in positive charges. This finding suggests that despite the similar overall structural and biophysical characteristics, the three proteins may have unique molecular partners.
At-NEET Is Localized to Chloroplasts and Mitochondria
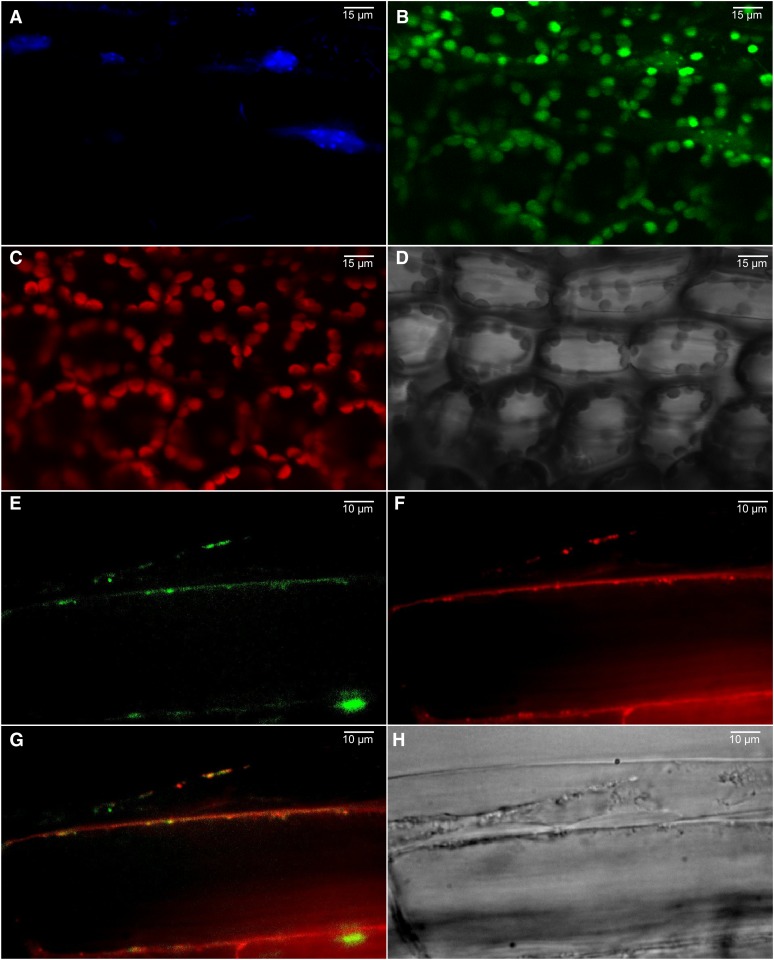
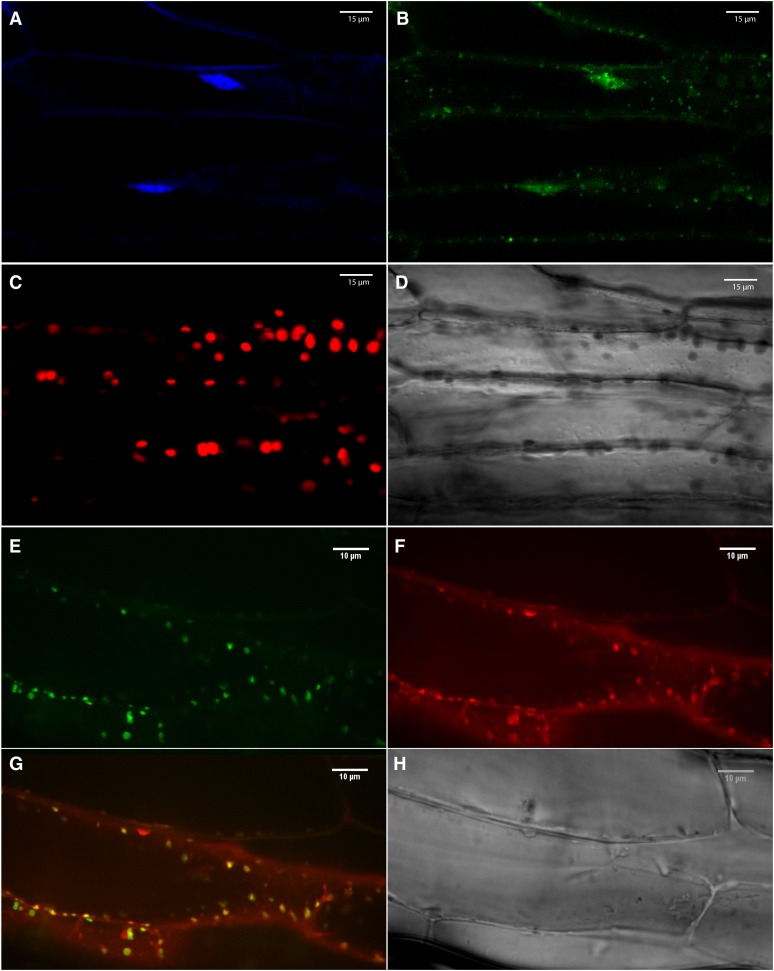
To determine the subcellular localization of the native At-NEET protein, we fused its C terminus in frame with green fluorescent protein (GFP) and expressed it as a stable transgene in transgenic Arabidopsis plants using the cauliflower mosaic virus (CaMV) 35S promoter (Luhua et al., 2008). N-terminal GFP fusions were not generated due to the existence of a transmembrane domain at the N-terminal region of At-NEET. Homozygous lines with low, moderate, or high expression levels were generated, and their leaf and root cells imaged using a conofocal microscope. As shown in Figure 3, the expressed At-NEET-GFP fusion protein was localized to chloroplasts, proplastids, and mitochondria. To examine whether human mitoNEET would have a similar localization when expressed in transgenic plants, we used the same cloning strategy to express the mitoNEET protein in transgenic Arabidopsis plants. As shown in Figure 4, the full-length human mitoNEET-GFP fusion protein was primarily localized to mitochondria with no detectable levels in chloroplasts.
Figure 3.
Localization of At-NEET in 3-Week-Old Transgenic Arabidopsis Plants with Low Expression Level of the At-NEET Protein Fused in Frame to GFP (C-Terminal Fusion).
(A) 4′,6-diamidino-2-phenylindole imaging showing the DNA of leaf cells.
(B) GFP imaging of leaf cells.
(C) Autofluorescence of chlorophyll (chloroplasts) of leaf cells.
(D) Bright field of leaf cells.
(E) GFP imaging of root cells.
(F) Mitotracker imaging showing the mitochondria of root cells.
(G) Merged image of (E) and (F).
(H) Bright field of root cells.
Images were taken using a confocal microscope and are representative of images obtained from six independent transgenic plants with low expression level of the fusion protein. All plants showed localization of the fusion protein to chloroplasts, proplastids, and mitochondria.
Figure 4.
Localization of Human mitoNEET in 3-Week-Old Transgenic Arabidopsis Plants with Low Expression Level of the Human mitoNEET Protein Fused in Frame to GFP (C-Terminal Fusion).
(A) 4′,6-diamidino-2-phenylindole imaging (DNA) of leaf cells.
(B) GFP imaging of leaf cells.
(C) Autofluorescence of chlorophyll (chloroplasts) of leaf cells.
(D) Bright field of leaf cells.
(E) GFP imaging of root cells.
(F) Mitotracker imaging (mitochondria) of root cells.
(G) Merged image of (E) and (F).
(H) Bright field of root cells.
Images were taken using a confocal microscope and are representative of images obtained from six independent transgenic plants with low expression level of the fusion protein. All plants showed localization of the fusion protein to mitochondria.
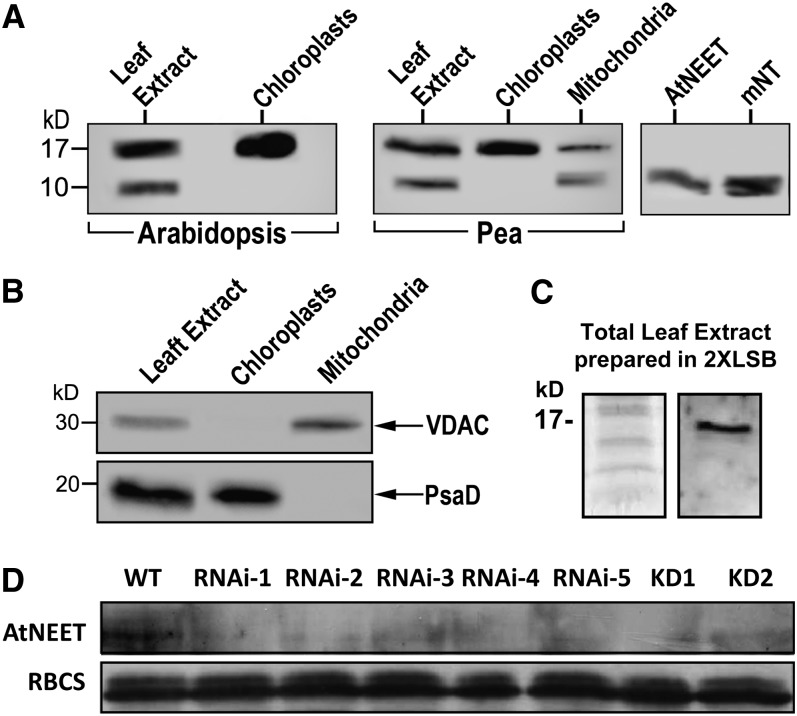
To examine the localization of At-NEET further, we raised polyclonal antibodies against its truncated form (amino acids 39 to 108) and used these antibodies to detect At-NEET in extracts from Arabidopsis and pea (Pisum sativum cv Dan) leaves. As shown in Figure 5, a protein reacting with the At-NEET antibody was detected in whole-cell extracts of Arabidopsis and pea leaves as well as in protein extracts obtained from isolated intact chloroplasts of these plants. To test whether a plant NEET protein could also be detected in mitochondria, we isolated intact mitochondria from pea plants and subjected their extracts to protein blots. As shown in Figure 5, At-NEET protein was detected in mitochondria. Interestingly, the At-NEET antibody reacted with a protein corresponding in molecular mass to the full-length At-NEET protein (15 kD) as well as with a protein corresponding in molecular mass to the truncated form of the protein (possibly missing the membrane anchor motif; 10 kD), in total extracts and in mitochondria, but not in chloroplasts. This observation could suggest that At-NEET is subjected in cells to posttranslational processing or that At-NEET is subjected to some level of proteolytic processing/degradation following cell disruption during sample preparation. When protein extracts were directly prepared under the denaturing conditions of the Laemmli sample buffer (Laemmli, 1970), only the full-length protein was detected (Figure 5C), supporting the latter option.
Figure 5.
Detection of Proteins That Cross-React with an At-NEET Antibody in Intact Chloroplasts, Mitochondria, and Plants with Suppressed Expression of At-NEET.
(A) Protein blot analyses of total leaf extract and isolated intact chloroplasts from Arabidopsis (left); total leaf extract, isolated intact chloroplasts, and isolated intact mitochondria from pea (middle); or purified truncated At-NEET or human mitoNEET (right), using antibodies raised against At-NEET.
(B) Detection of organelle-specific proteins in isolated intact chloroplasts and mitochondria from pea performed with antibodies raised against the outer mitochondrial membrane protein VDAC (top panel) or the chloroplast protein PsaD (subunit II of photosystem I; bottom panel).
(C) Protein gel (left) and immunoblot (right) of total leaf extract from wild-type Arabidopsis probed with antibodies raised against At-NEET showing that extraction under denaturing conditions prevents the appearance of the 10-kD reactive polypeptide. LSB, Laemmli sample buffer.
(D) Protein blot showing the expression of At-NEET in the wild type (WT) and the different knockdown and RNAi lines used in this study. RBCS, ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit was visualized with Coomassie blue.
Knockdown and RNAi Mutants of At-NEET Show Late Bolting and Early Senescence Phenotypes
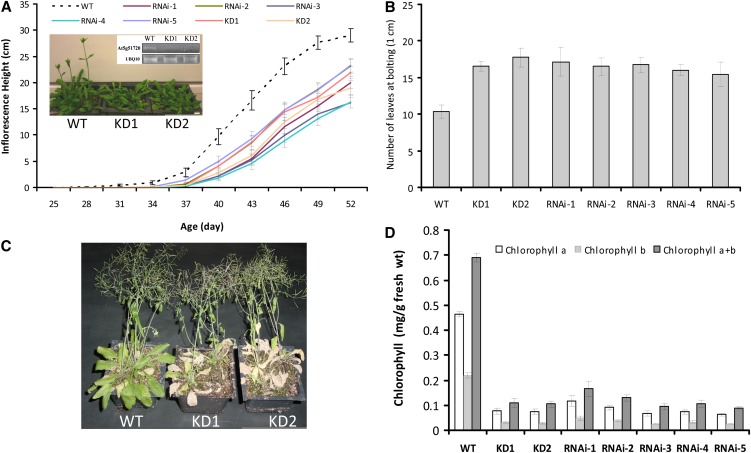
To study the function of At-NEET in Arabidopsis, we isolated two knockdown mutants (SALK_040013 and SALK_040019) and generated five homozygous RNAi lines with suppressed expression of At-NEET (Figure 5D). A complete knockout for At-NEET could not be found in any of the publicly available mutant collections, suggesting that it might be an essential protein. As shown in Figures 6A and 6B and Supplemental Figure 2 online, the knockdown and RNAi lines demonstrated a late bolting phenotype. When wild-type and At-NEET knockdown or RNAi lines were allowed to senesce under controlled growth conditions, it was found that the knockdown and RNAi lines senesced earlier (Figures 6C and 6D). Interestingly, mice lacking Miner1 demonstrated a significantly shorter life span (Chen et al., 2009). The results shown in Figure 6 suggest that suppression of NEET expression has a significant effect on plant development.
Figure 6.
Late Bolting and Early Senescence Phenotypes in Arabidopsis Plants with Suppressed Expression of NEET.
(A) Inflorescence height in the wild type (WT), knockdown plants, and RNAi lines for At-NEET. Plants were grown under controlled growth conditions in a mixed plot setting. A representative photograph of 38-d-old wild type and two independent knockdowns for At-NEET (KD1, SALK_040013; KD2, SALK_040019) is shown along with an RT-PCR gel for the expression of NEET in the plants photographed. UBIQUITIN was used as an internal reference
(B) Leaf number at the time inflorescence height reached 1 cm in wild-type knockdown plants and RNAi lines for NEET. Plants were grown under controlled growth conditions (long day) in a mixed plot setting.
(C) Photograph of 58-d-old wild type and two independent NEET knockdowns grown under controlled growth conditions.
(D) Chlorophyll content in wild-type knockdown plants and RNAi lines for NEET. Plants (58 d old) were gown under controlled growth conditions in a mixed plot setting.
All experiments were repeated at three different times with 60 plants per experiment. Error bars represent sd.
Because late bolting and early senescence are sometimes linked to an abiotic stress resistance phenotype (Mittler and Rizhsky, 2000; Luhua et al., 2008), we tested the tolerance of At-NEET knockdown and RNAi lines to different abiotic stresses, including heat, cold, osmotic, salinity, and oxidative stresses (Luhua et al., 2008). As shown in Supplemental Figure 3 online, the knockdown and RNAi lines were significantly more tolerant to osmotic and salinity stresses. A combination of early senescence, late bolting, and increased tolerance to abiotic stress is sometimes linked to impaired metabolic homeostasis in plants (Mittler and Rizhsky, 2000; Luhua et al., 2008). It is therefore possible that suppressed expression of At-NEET induces a metabolic imbalance in plants.
Cluster Transfer from At-NEET (Donor) to Apo-Ferredoxin (Acceptor)
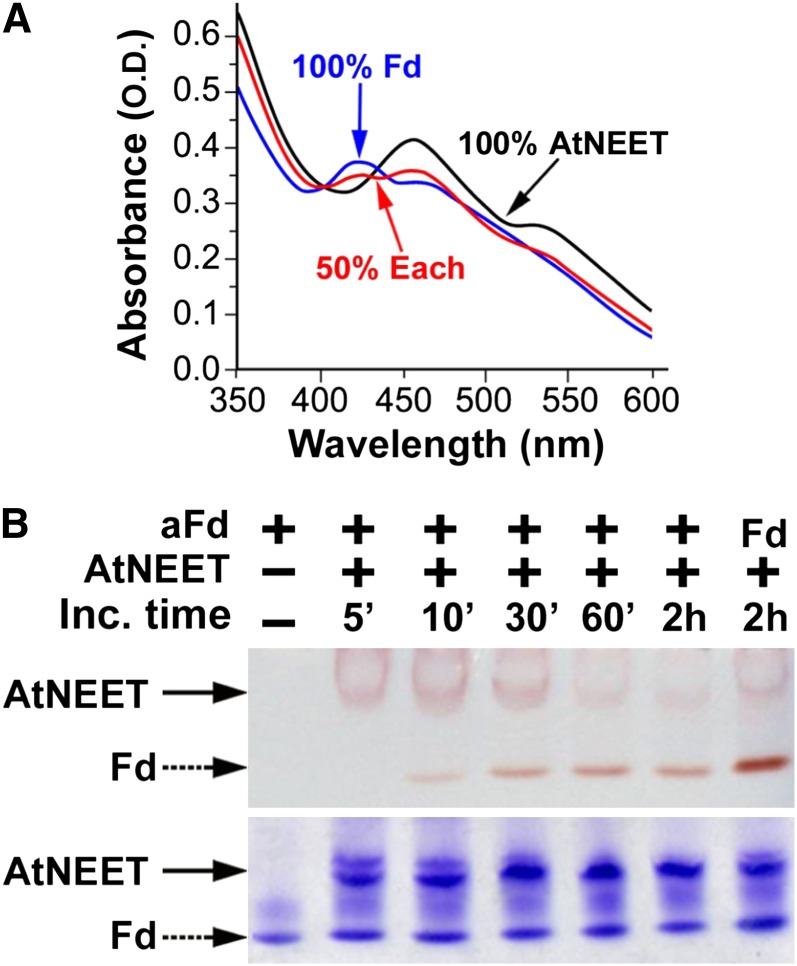
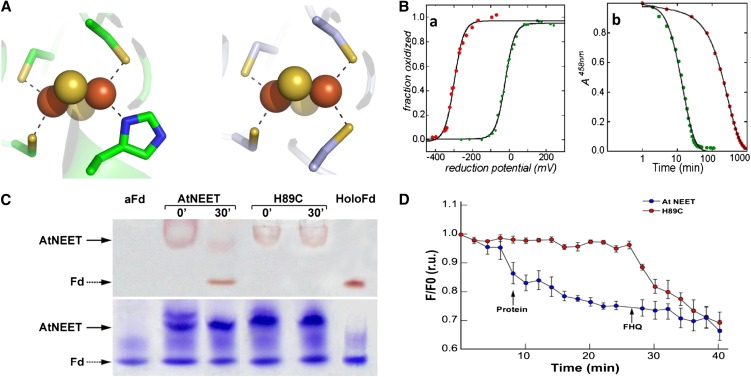
One of the suggested functions of NEET proteins in different biological systems involves [2Fe-2S] cluster transfer (Paddock et al., 2007; Zuris et al., 2011); we therefore assessed whether such transfer could occur between At-NEET and pea apo-ferredoxin (a chloroplastic protein). The apo form of ferredoxin (aFd), has classically served as a [2Fe-2S] cluster acceptor protein in different biochemical studies (Wu et al., 2002; Wu and Cowan, 2003; Chandramouli and Johnson, 2006). We therefore used At-NEET as a potential [2Fe-2S] cluster donor and aFd as a potential [2Fe-2S] cluster acceptor (Figure 7) to test the hypothesis that NEET could act as an Fe-S transfer protein. The distinct UV-Vis spectra and electrophoretic separation of NEET and Fd allow for the analysis of cluster transfer by monitoring the time-dependent change in UV-Vis absorption spectra (Figure 7A) or the visible chromophore change in proteins separated by native-PAGE (Figure 7B). A reduction in 458-nm absorption peak (characteristic of At-NEET) and a concomitant appearance of a 423-nm peak (characteristic of holo-Fd) was observed, indicating an interprotein transfer of the [2Fe-2S] cluster (Figure 7A). The cluster transfer was corroborated by native gel electrophoresis (Figure 7B, top panel), in which the holo-NEET’s red band intensity decreases with time of incubation with aFd, whereas the latter is concomitantly converted to the red-colored Fd band. These changes in red-colored band positions occur while the protein levels of NEET and aFd/Fd remained unchanged (Figure 7B, bottom panel).
Figure 7.
Cluster [2Fe-2S] Transfer from At-NEET to aFd.
(A) UV-VIS analysis. Absorption spectra of purified aFd (black), At-NEET after incubation with aFd that caused 50% of its cluster to be transferred to aFd (red), and At-NEET that transferred all of its cluster to (blue).
(B) Native-PAGE analysis showing cluster transfer from NEET to aFd. At-NEET and aFd were incubated for the indicated periods of time in an interaction buffer (5mM sodium dithionite, 2% β-mercapto ethanol, and 5 mM EDTA) and separated by native-PAGE. Native gels are shown before (top) and after (bottom) staining with Coomassie blue. The red colored bands of the 2Fe-2S cluster–containing proteins are indicated by arrows. Holo Fd is indicated as Fd.
At-NEET as an Fe-S/Fe Donor in Permeabilized Cells
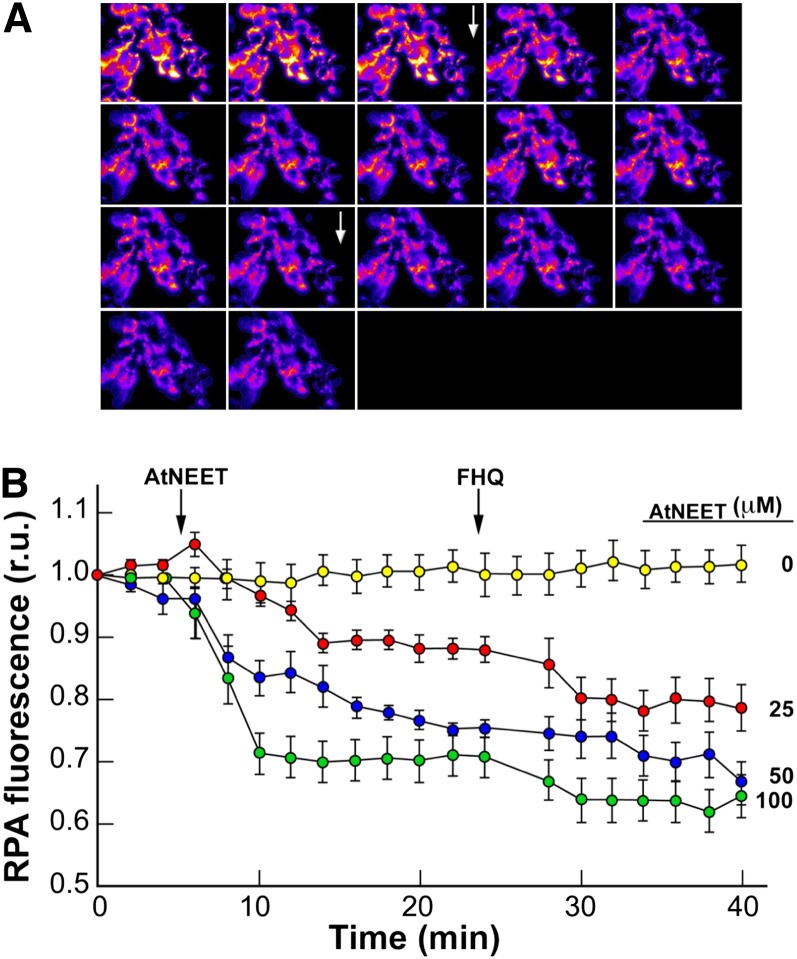
The demonstration that At-NEET is capable of transferring its [2Fe-2S] in vitro led us to test whether transfer of labile Fe-S/Fe from At-NEET could be demonstrated in situ in the cellular environment. Because At-NEET appears to be similar to human Miner1 and mitoNEET, and it localizes to plant mitochondria (Figures 3 and 5), we addressed this question in an established heterologous assay system (Petrat et al., 2002; Zuris et al., 2011), in which the Fe donor (At-NEET) was added to gently permeabilized HEK293 cells that were double labeled with the fluorescent Fe sensor calcein-green (CALG), as tracer for residual cytosolic Fe, and the mitochondrial Fe sensor rhodamine B-[(1,10- phenanthrolin-5-yl) aminocarbonyl] benzyl ester (RPA), as a measure of mitochondrial Fe levels. We then followed changes in relative fluorescence in response to the addition of At-NEET to cells. As shown in Figure 8, quenching of RPA by At-NEET, an indication of Fe-S/Fe transfer into mitochondria, rapidly occurred in the absence of externally added reductants (Figure 8A) and was directly proportional to the added At-NEET’s concentration (Figure 8B). Moreover, the addition of a reducing agent had no effect on the RPA fluorescence quenching rate (see Supplemental Figure 4 online). The findings shown in Figures 7 and 8 support a hypothesis that At-NEET is involved in Fe-S/Fe management in cells.
Figure 8.
Transfer of Labile Fe/FeS from At-NEET to Mitochondria in Permeabilized HEK293 Cells.
HEK293 cells labeled with RPA (for mitochondria) and CALG (for cytosol) were permeabilized and used for tracing changes in fluorescence following addition of At-NEET in a K-succinate medium without added reducing agents.
(A) A time-lapse series of fluorescence imaging recorded every 2 min. Zero time point at the top left corner with the highest fluorescence imaging (pseudocolor images). After 6 min, 25 µM At-NEET was added (top arrow), and 6 µM FHQ (bottom arrow) was added after 24 min.
(B) The fluorescence traces (relative units [r.u.]) represent a time series taken for cells exposed to different concentrations of At-NEET (no At-NEET added [yellow], 25 µM [red], 50 µM [blue], and 100 µM [green]). All data are represented as mean ± se (n = 12).
Involvement of NEET in Fe Metabolism and ROS Homeostasis in Plants
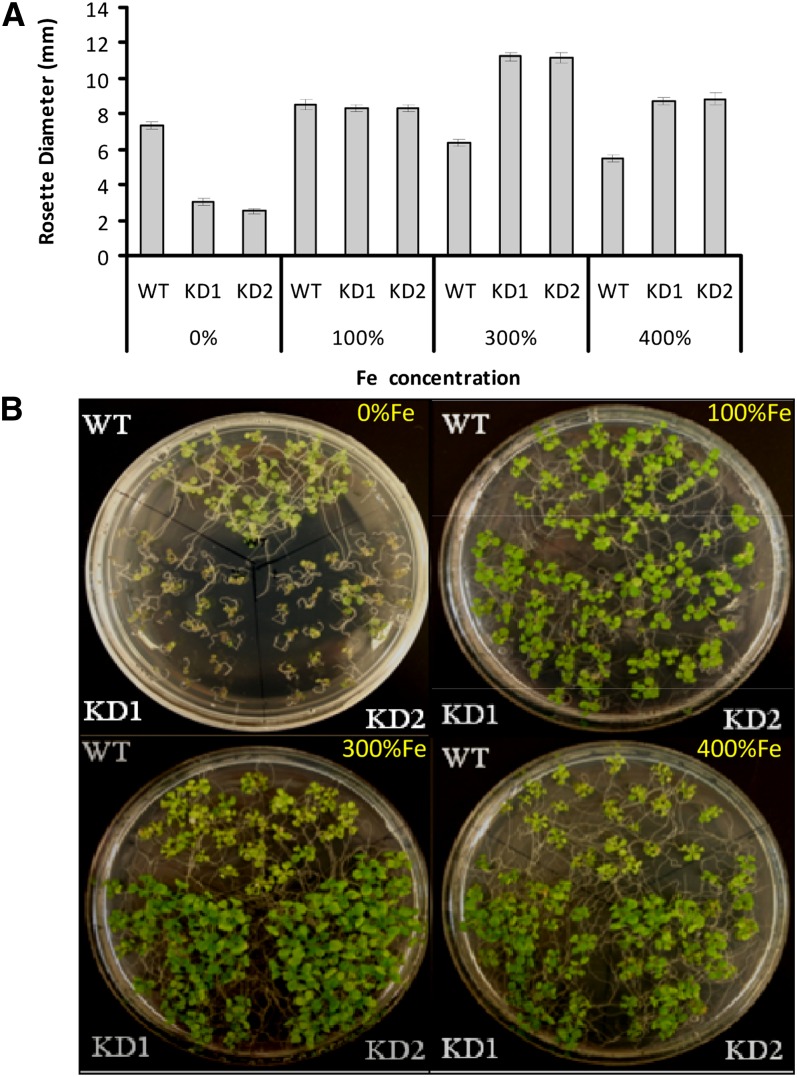
To test further whether At-NEET is involved in Fe transfer/management in cells, we grew wild-type and At-NEET knockdown seedlings on agar plates containing different concentrations of Fe. As shown in Figure 9, the growth of wild-type and NEET knockdown seedlings was similar on agar plates containing control levels of Fe (100%). By contrast, the growth of At-NEET knockdown seedlings was suppressed on plates with decreased Fe (0%). Moreover, while the growth of wild-type seedlings was suppressed on plates containing high concentrations of Fe (300 or 400%), the growth of NEET knockdown seedlings was not. The finding that growth of NEET knockdown seedlings is insensitive to high Fe levels, but sensitive to low levels of Fe, strongly suggests that NEET is involved in Fe transfer, distribution, and/or management in plant cells.
Figure 9.
Altered Sensitivity to Fe Levels in the Growth Media of Arabidopsis Seedlings with Suppressed Expression of NEET.
(A) Measurements of rosette diameter in wild-type (WT) and NEET knockdown plants. Seedlings were grown on agar plates with different concentrations of Fe under controlled growth conditions.
(B) Photograph of 12-d-old seedlings of the wild type and two independent NEET knockdowns. Seedlings were grown on agar plates with different concentrations of Fe under controlled growth conditions.
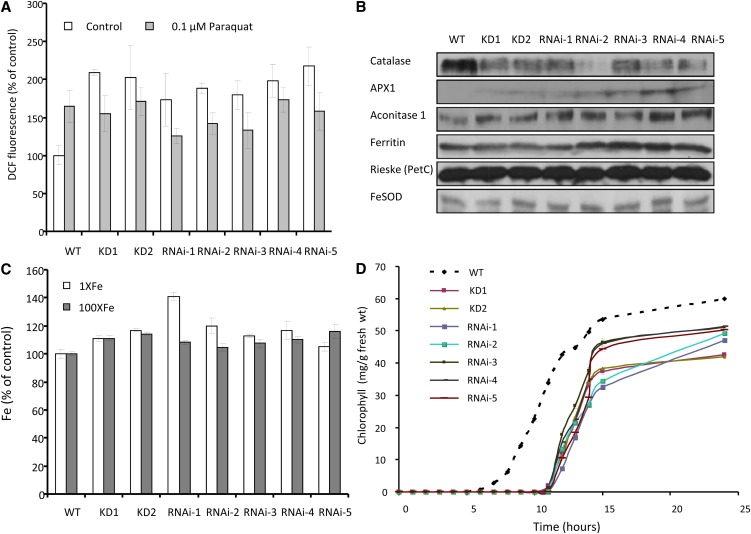
Because Fe metabolism is intimately linked with ROS homeostasis and photosynthesis in cells (Asada, 2006; Halliwell and Gutteridge, 2007), we measured the level of ROS using dichlorodihydrofluorescein acetate (Luhua et al., 2008) in wild-type and At-NEET knockdown or RNAi seedlings. As shown in Figure 10A, plants with reduced levels of NEET accumulated higher levels of ROS compared with the wild type when grown under controlled growth conditions. Whereas the addition of paraquat, a superoxide-generating agent, to wild-type seedlings resulted in enhanced accumulation of ROS, a similar increase was not observed in seedlings with reduced levels of At-NEET (Figure 10A), suggesting that the ROS levels measured under controlled growth conditions in seedlings with suppressed levels of NEET are already very high and could not be increased any further by the addition of an oxidizing agent.
Figure 10.
ROS and Fe Accumulation, Suppression of Catalase, and Late Greening Phenotypes in Arabidopsis Seedlings with Suppressed Expression of NEET.
(A) ROS accumulation in seedlings with wild-type (WT) or suppressed expression of NEET. ROS levels were measured in seedlings grown under controlled conditions or incubated with 0.1 µM of the ROS-generating compound paraquat.
(B) Protein blot analyses of different Fe- and ROS-associated proteins in seedlings with wild-type or suppressed expression of NEET.
(C) Fe content of 2-week-old plants with wild-type or suppressed expression of NEET. Plants grown under controlled conditions and fertilized with 1× or 100× Fe.
(D) Time course of greening in etiolated seedlings with wild-type or suppressed expression of NEET.
Alterations in Fe metabolism could cause changes in the steady state level of certain proteins that contain Fe in cells. To examine the relative level of certain Fe-containing proteins in wild-type and At-NEET mutants, we performed protein blots with antibodies to catalase, ascorbate peroxidase 1, aconitase 1, ferritin, Rieske, and Fe superoxide dismutase. As shown in Figure 10B, a dramatic decrease was found in the level of the heme protein catalase, a key ROS detoxifying enzyme in plant cells (Mittler et al., 2004), in plants with suppressed levels of NEET. This decrease could explain the accumulation of ROS in plants with reduced levels of NEET (Figure 10A). It is therefore possible that At-NEET is required for the supply of heme for catalase biosynthesis and that its deficiency causes the accumulation of ROS in cells.
To examine the total levels of Fe in the plants, we conducted elemental analysis of 2-week-old wild-type plants and plants with suppressed levels of At-NEET grown under controlled growth conditions, with or without the addition of 100-fold more Fe to the fertilizer solution. As shown in Figure 10C, compared with wild-type plants, plants with suppressed levels of NEET, fertilized with 1× or 100× Fe, accumulated higher levels of Fe. These findings support a role for NEET in Fe homeostasis or distribution in plant cells.
Alterations in Fe metabolism and ROS homeostasis are likely to affect different processes in plants, especially processes that require tight coordination between the biosynthesis of Fe-containing prosthetic groups and their corresponding polypeptides. One of the most tightly regulated processes in plants, which requires such coordination, is the greening of etiolated seedlings. As shown in Figure 10D, the greening of etiolated seedlings with reduced levels of At-NEET was significantly delayed compared with etiolated wild-type seedlings. This finding supports our observation that At-NEET is involved in Fe metabolism and ROS homeostasis (Figures 9 and 10A to 10C).
The Involvement of His89 in the Cluster Binding Site of At-NEET
Human mitoNEET and Miner1 contain an unusual 3Cys – 1 His [2Fe-2S] coordination site, which is one of the hallmark features of NEET proteins (Wiley et al., 2007a, 2007b). To test whether a similar His residue is involved in the coordination of the 2Fe-2S cluster in At-NEET, His-89 was replaced with Cys. The H89C protein was purified, crystallized, and subjected to x-ray diffraction analysis with a resulting 1.15-Å resolution structure (see Supplemental Table 1 and Supplemental Figure 5 online). Major changes between the wild type and the H89C protein were found only in the cluster binding region (Figure 11A; see Supplemental Figure 5 online). In addition to the local changes in structure, the replacement of His-89 by Cys had two major biophysical effects on the At-NEET cluster: (1) The redox potential of the cluster was shifted by nearly 300 mV and became more negative (red trace in left panel of Figure 11B); (2) the stability of the cluster was dramatically altered; as shown in Figure 11B (right panel), the mutant cluster (red dots) is nearly 10-fold more stable at pH 6.0 compared with the wild-type cluster (green trace).
Figure 11.
Characterization of the 2Fe-2S Cluster in the At-NEET H89C Mutant Protein.
(A) Comparison of the cluster binding structures of wild-type At-NEET (pdb ID code 3S2Q; left) and the H89C mutant (pdb ID code 3S2R; right). As the overall secondary structure of the mutant remains the same as that of the wild type (see Supplemental Figure 5 online), the major structural changes are observed in the immediate vicinity of the mutation.
(B) Biophysical characteristics of the At-NEETH89C 2Fe-2S cluster. (a) Decrease in redox potential. Measurement of the fraction oxidized versus reduction potential showing that the redox potentials (given by reduction potential at which the 2Fe-2S clusters are half reduced) differ by ∼300 mV in magnitude with the H89C mutant being lower (red trace versus green trace of the wild-type At-NEET). (b) Increase in cluster stability. A time course of cluster stability at pH 6.0 showing that the 2Fe-2S cluster of the H89C mutant is more stable than the wild type by approximately an order of magnitude (red trace versus green trace of the wild-type At-NEET).
(C) Cluster transfer analysis of wild-type At-NEET and the H89C mutant. At-NEET or the At-NEETH89C mutant were incubated with aFd for 30 min in a transfer assay buffer and analyzed by native-PAGE. Gels are shown before (top) and after (bottom) staining with Coomassie blue. The red-colored bands of the Fe-S cluster–containing proteins are indicated by arrows.
(D) Transfer of labile Fe/FeS from wild-type or mutated At-NEET to RPA-labeled permeabilized HEK293 cells. RPA fluorescence was measured 20 min after the addition of 10 µM wild-type (blue) or At-NEETH89C (red) proteins. Data are represented as mean ± se (n = 10). r.u., relative units.
To study the effect of the H89C mutation on the Fe-S/Fe transfer properties of At-NEET (Figures 7 and 8), native-PAGE (Figure 11C) and permeabilized HEK293 cells (Figure 11D) were used to compare the Fe-S/Fe transfer properties of wild-type At-NEET with that of the H89C mutant. Both assays indicated that compared with the wild-type At-NEET protein, the H89C mutant, with its more stable FeS cluster (Figure 11B), is impaired in transferring its Fe-S/Fe to an aFd acceptor protein (Figure 11C) or to mitochondria in permeabilized HEK293 cells (Figure 11D). His-89 could therefore play a key role in the Fe-S/Fe transfer process of At-NEET.
DISCUSSION
NEET proteins are involved in a large array of biological processes in mammalian cells (Colca et al., 2004; Amr et al., 2007; Wiley et al., 2007a, 2007b; Chen et al., 2009, 2010; Chang et al., 2010). Nevertheless, their mode of function is largely unknown. The unique 3Cys-1His cluster binding geometry of NEET proteins, which makes their 2Fe-2S cluster labile, could indicate a role for NEET proteins in Fe-S/Fe transfer within cells (Paddock et al., 2007; Zuris et al., 2011). Such a function could explain their wide-ranging activity because Fe is linked to many different biological processes (Lill, 2009). Alternatively, a role for NEET proteins in Fe metabolism/homeostasis could represent an ancient function of these proteins that is conserved between different organisms and is distinguished from their other functions. Our identification and function-structure characterization of a plant NEET protein (At-NEET, At5g51720) indicates that NEET proteins are conserved between plants and mammals. Our demonstration that the plant NEET protein is capable of transferring its Fe-S/Fe cluster to an acceptor protein (Figure 7), or to mitochondria in permeabilized cells (Figure 8), coupled with our genetic analysis showing altered sensitivity to Fe (Figure 9), enhanced accumulation of ROS (Figure 10A), altered accumulation of an Fe-containing enzyme (catalase; Figure 10B), enhanced accumulation of Fe (Figure 10C), and a late-greening phenotype (Figure 10D) in Arabidopsis NEET mutants strongly suggest a role for plant NEET proteins in Fe homeostasis/metabolism. These findings support a hypothesis that NEET proteins have an ancient role in cells associated with Fe homeostasis/metabolism.
Despite the overall low sequence similarity between At-NEET and the human mitoNEET or Miner1, our structural analysis of At-NEET clearly demonstrates that the three proteins have a common NEET structure (Figure 2). This finding is supported by our mutational analysis that demonstrates a functional role for At-NEET His-89 in the unique 3Cys-1His geometry of NEET proteins (Figure 11). Our structural analysis therefore confirms that At-NEET is a member of the NEET family along with the mammalian proteins. The higher structural similarity observed between At-NEET and Hs-Miner1 could suggest that, similar to Hs-Miner1, At-NEET is involved in processes such as cell death, ROS accumulation, and aging. Indeed, mutants deficient in At-NEET accumulate high levels of ROS and have a shorter life span compared with wild-type plants (Figures 6C, 6D, and 10A). In addition, At-NEET expression is enhanced in response to chloroplastic accumulation of singlet oxygen (op den Camp et al., 2003), and mutants deficient in At-NEET expression have higher tolerance of some abiotic stresses (see Supplemental Figure 3 online). One possible explanation for these combined phenotypes could be that NEET is involved in the Fe-dependent synthesis of heme required for catalase biosynthesis. In the absence of sufficient NEET protein, catalase levels would decrease and ROS accumulate (Figure 10). The resulting accumulation of ROS in mutants with suppressed NEET in turn could cause these plants to be more tolerant to abiotic stresses and to senesce earlier (Mittler et al., 2004; Luhua et al., 2008). Enhanced ROS accumulation in At-NEET mutants (Figure 10A) could also be a result of the enhanced accumulation of Fe in these plants (Figure 10C; Halliwell and Gutteridge, 2007). Although we could not obtain plants with complete suppression of At-NEET, our analysis of NEET knockdown and RNAi lines demonstrated a role for this protein in plant development, ROS and Fe homeostasis, abiotic stress tolerance, and senescence (Figures 6, 9, and 10; see Supplemental Figure 3 online). Such a broad phenotype could indicate involvement of At-NEET in key biological processes required for cellular homeostasis (Mittler and Rizhsky, 2000; Luhua et al., 2008), which could include involvement of NEET in the cellular regulation of Fe metabolism and/or distribution. Indeed, mutants with suppressed expression of At-NEET displayed reduced growth on low Fe as well as insensitivity of growth to the presence of high levels of Fe (Figure 9), suggesting that they are impaired in Fe uptake and/or distribution. The subcellular localization of NEET to chloroplasts and mitochondria (Figures 3 and 5) may be central to the proposed role of this protein in Fe or Fe-S/Fe transfer. Chloroplasts and mitochondria contain many Fe cluster proteins and require a supply of Fe from the cytosol for the biogenesis of different Fe cluster proteins. The finding that mutants deficient in NEET display late greening (Figure 10D) and early senescence that is accompanied by loss of chlorophyll (Figure 6) supports such a role. The finding that NEET mutants accumulated more Fe (Figure 10C) but could survive on higher levels of Fe in their growth medium (Figure 9) could further indicate a role for NEET in the distribution of Fe between the cytosol, vacuole, or apoplast and different organelles, such as mitochondria, chloroplast, or peroxisomes. Thus, although the total level of Fe in cells with suppressed At-NEET could be high (Figure 10C), the availability of this Fe for processes such as chloroplast biogenesis (Figure 10D) or catalase biosynthesis (Figure 10B) could be low.
The possible involvement of NEET in Fe metabolism in plants was further supported by biophysical and biochemical studies demonstrating the capability of the At-NEET protein to donate its Fe-S/Fe to acceptor proteins in vitro and in situ, a capability that was dependent upon the lability of its cluster (Figures 7, 8, and 11). Interestingly, At-NEET, which is localized to chloroplasts and mitochondria (Figure 5), was capable of donating its Fe-S/Fe to mammalian mitochondria, suggesting that this process is a conserved function of NEET proteins between plants and mammals (Figure 8; Zuris et al., 2011).
Assembly of Fe-S clusters in plants was recently divided into three distinct pathways: (1) the chloroplastic S mobilization pathway, (2) the mitochondrial Fe-S pathway, and (3) the cytosolic Fe-S cluster assembly pathway (Balk and Pilon, 2011). All pathways were divided into a first step in which S and Fe are combined on a scaffold protein and a second step in which the Fe-S cluster is transferred to a target protein pathway, possibly with the involvement of a carrier protein (Balk and Pilon, 2011). At-NEET, expressed in almost all different plant tissues and developmental stages (http://www.Arabidopsis.org/), might be a yet unidentified Fe-S carrier protein involved in these assembly pathways.
Our findings support an ancient role for NEET proteins in Fe metabolism in cells. Such a role could provide an insight into the function of NEET proteins in human diseases, such as Wolfram Syndrome 2 and diabetes (Colca et al., 2004; Amr et al., 2007; Conlan et al., 2009b; Chen et al., 2010). Due to the similarity in function and structure between the plant and mammalian NEET proteins, it would be interesting to find out whether the plant system could be used as an experimental tool to study different drugs or mutants associated with NEET proteins and human pathology.
METHODS
All materials used in this work were from best available commercial grade. The acetoxymethyl ester of CALG was obtained from Molecular Probes. The mitochondrial metal sensor RPA and its phenanthroline-lacking analog RPC were obtained as described elsewhere (Petrat et al., 2002; Rauen et al., 2003).
Expression and Purification of At-NEET Proteins
At-NEET cDNA was amplified by PCR and subcloned into a modified pET28-a(+) vector (Novagen) as previously described (Conlan et al., 2009c). The H89C mutant of At-NEET was generated by site-directed mutagenesis as previously described (Conlan et al., 2011). The recombinant wild-type and mutant proteins were expressed in Escherichia coli BL21-RIL grown in Luria-Bertani supplemented with 30 µg mL−1 kanamycin and 34 µg mL−1 chloramphenicol. At OD600 of 0.4, 0.75 mM FeCl3 was added and cell growth proceeded for an additional 18 to 24 h at 23°C. The truncated At-NEET or H89C mutant proteins were purified from lysed cells using Ni-agarose and size exclusion chromatography as previously described (Conlan et al., 2009c, 2011; Zuris et al., 2010). Human Miner1 and mitoNEET were expressed and purified as described by Paddock et al. (2007) and Conlan et al. (2009b).
Optical Spectroscopy and Stability Measurements
All UV-Vis absorption spectra were measured from the near UV to the near IR (250 to 700 nm) on a Cary50 spectrometer (Varian) equipped with a temperature-controlled cell (T = 35°C) (protein concentration 10 to 20 μM in 25 mM Tris-HCl, pH 8.0, and 100 mM NaCl). Stability of the 2Fe-2S cluster of At-NEET was determined by monitoring its characteristic absorbance at 460 nm as a function of time following dilution of the protein into the assay buffer (∼20 μM protein in 100 mM buffer at 35°C). The following buffers were used at the pH indicated: Tris-HCl at pH 7.1 and 6.8, and bis-Tris-HCl at pH 6.3, 5.8, and 5.3. Buffers and protein were filtered prior to the start of the measurement, and a layer of 100% paraffin oil was placed over the solution to minimize evaporation. The half-life, corresponding to decay by 50%, was determined at each pH.
Redox Measurements
The redox state of At-NEET was determined by monitoring the absorbance near 460 nm with 100 μM protein in 25 mM Tris-HCl, pH 7.5, and 100 mM NaCl at 25°C using a quartz cuvette containing a side arm under positive argon pressure (Airgas-West). An Ag/AgCl dual reference and working electrode (Microelectrodes) was used to measure the ambient redox potential (its calibration was checked using quinhydrone at pH 4.0 and 7.0 as recommended). pH-buffered dithionite (50 mM; Fisher Scientific) was added via a Hamilton syringe to adjust the redox potential. Ubiquinone (50 μM; Sigma-Aldrich), 50 μM duroquinone (Sigma-Aldrich), 100 μM menadione (Sigma-Aldrich), and 100 μM naphthaquinone (Sigma-Aldrich) were added to ensure efficient electron transfer between the solution components and the electrode. The midpoint/redox potential (Em) was determined from a fit of the fraction oxidized to the Nernst equation (A= Aox/{1 + 10 ^ [(Em – E) (n / 59.1 mV)]} + Ared) using the graphing program Origin 6.1 (OriginLab), where Aox is the absorbance of the fully oxidized sample, Em is the midpoint potential in mV, E is the measured cell potential in mV, n is the number of electrons being transferred in the redox reaction, and Ared is the lowest absorbance level corresponding to the fully reduced 2Fe-2S center in either native or mutant proteins. Potentials were adjusted to standard hydrogen electrode for presentation. Samples were tested for their ability to reoxidize upon exposure to ambient oxygen.
Cluster Transfer Optical Spectroscopy Assay
Absorption spectra were recorded at 400 to 650 nm (CARY, 300Bio; Varian) with a temperature control apparatus set to 37°C. Special attention was given to changes in 458-nm absorbance (At-NEET signature [2Fe-2S] absorbance peak) vis-à-vis the 423-nm peak characteristic of the [2Fe-2S] cluster in Fd. Kinetic data were obtained by taking the ratio of the 423 peak to the 458 peak over time. Data were then normalized and fit to a single exponential. Kinetic measurements were performed using equimolar concentrations of mNT (the wild type and mutants) and aFd in the presence of 2% β-mercaptoethanol, 5 mM EDTA, and 5 mM sodium dithionite. The aFd was preincubated for 30 min prior to the start of the experiment with the reducing agents. Decays were performed at 37°C and determined by monitoring loss of the 458-nm peak with time.
Crystallization, Data Collection, and Structure Solution of At-NEET and the H89C Mutant
Crystallization assays were initially conducted using the ORYX6 automatic crystallization system (Douglas Instruments) using the microbatch method. Commercial crystallization screens (index screen of Hampton Research) (Jancarik and Kim, 1991; D'Arcy et al., 1996) were employed and set at 4 and 20°C. Crystals appeared within 2 d under two crystallization conditions consisting either of 50 mM ZnAc2 and 20% polyethylene glycol (PEG) 3350 or 30% Jeffamine ED-2001 and 100 mM HEPES, pH 7.0, where the protein solution contained 33 mg/mL protein, 100 mM NaCl, 20 mM Tris, pH 8.0, and 0.02% NaN3 at both temperatures. Crystallization conditions were further refined using the sitting drop vapor diffusion method at 4°C. The 4-mL drop consisted of equal amounts of protein solution (33 mg/mL) and a reservoir solution containing 100 mM ZnAc2 and 20% PEG 3350. Crystals appeared within a day and reached the final size of 0.3 mm in the longest dimension within 2 d. Prior to data collection, crystals were cryoprotected in a solution containing 20% ethylene glycol and the reservoir solution and immediately flash cooled.
Crystallographic data were collected at the European Synchrotron Radiation Facility beamline ID29 (λ= 0.92 Å) on a PILATUS 6M charge-coupled device detector with an oscillation range of 1.0° at a temperature of 100K. Data were integrated and scaled using the HKL2000 suite (Otwinowski and Minor, 1997). The crystals belonged to the orthorhombic P212121 space group, with unit cell parameters a = 41.6 Å, b = 60.1 Å, and c = 66.4 Å with two molecules in the asymmetric unit (see Supplemental Table 1 online).
Crystals of the At-NEETH89C mutant were obtained using the microbatch method using the ORYX6 automatic crystallization system and commercial crystal screens (index screen, Crystal ScreenHT, SaltRx HT, and PEGRxHT of Hampton Research). Crystals appeared within 2 d under conditions containing equal amounts of protein solution (76 mg/mL, 100 mM NaCl, 20 mM Tris, pH 8.0, and 0.02% NaN3) and a solution containing 30% PEG 6000 and 0.1 M Bis Tris propane, pH 9.0, at 20°C. Prior to freezing, cryosolution containing 20% ethylene glycol and the crystallization solution was added and mounted directly from the 96-well plate and immediately flash frozen. Crystallographic data were collected at European Synchrotron Radiation Facility beamline ID14-4 (λ = 0.932 Å) on a Q315R ADSC charge-coupled device detector with an oscillation range of 1.0° at a temperature of 100K. Data were integrated and scaled using the HKL2000 suite (Otwinowski and Minor, 1997). The crystals belonged to the orthorhombic P212121 space group, with unit cell parameters a = 33.3 Å, b = 54.1 Å, and c = 72.7 Å with two molecules in the asymmetric unit.
The structure of wild-type At-NEET was solved via molecular replacement methods using MOLREP (Vagin and Teplyakov, 2010) implemented in CCP4i (Potterton et al., 2003) at a resolution range of 44.5 to 3.5 Å using the atomic coordinates of the human mitoNEET (PDB code 2QH7) (Paddock et al., 2007) as search model after removing all solvent molecules. The solution resulted in Rvalue of 49.9% and correlation coefficient of 45.2%. After 10 cycles of restrained refinement in REFMAC (Murshudov et al., 1997), the Rvalue converged to 35.9% (Rfree = 38.9%). The structure was fitted into electron density maps using the graphics program Coot (Emsley and Cowtan, 2004) and was initially mutated into the correct amino acids. In this regard, the sequence identity between the mitoNEET and At-NEET is 33.9% for the full-length proteins and 67% in the cluster binding domain, leaving 33 substitutions and a single insertion to be conducted. The structure was further refined using REFMAC5 restrained refinement with the maximum likelihood option, and solvent molecules were added using ARP/WARP (Morris et al., 2003).
The structure of the At-NEETH89C mutant was solved via molecular replacement methods using PHASER (Read and McCoy, 2011) at a resolution range of 44.5 to 3.5 Å using the coordinates of the refined wild-type At-NEET as the search model after removing solvent molecules. The search resulted in an RFZ (for rotation function Z) value of 8.7 (TFZ [for translation function Z] value = 13.8), indicating a valid solution, which was then shown to pack well without any clashes. The model was then refined using REFMAC restrained option at a resolution range of 43.4 to 1.15 Å for 10 cycles where the R value converged from 37.2 to 27.9% (Rfree from 36.0 to 30.7%). The structure was fitted into electron density maps using the graphics program Coot (Emsley and Cowtan, 2004) and was initially mutated into the correct amino acids. The structure was further refined using REFMAC5 restrained refinement with the maximum likelihood option, and solvent molecules were added using ARP/WARP (Morris et al., 2003).
[2Fe-2S] Cluster Transfer in Vitro Assay
At-NEET (the wild type or the H89C mutant) (1 mg/mL) was incubated (in a rolling shaker) with the apo-acceptor protein probe, aFd (1 mg/mL), obtained as described by Fish et al. (2005). The NEET and aFd were incubated in the presence of 2% mercaptoethanol, 5 mM Na-dithionite, and 5 mM EDTA for the specified time lengths (5 to 120 min). Transfer of the [2Fe-2S] cluster from NEET to aFd was analyzed by absorption spectroscopy and native-PAGE using 15% acrylamide precast gels according to Laemmli (1970).
Cell Culture
Human embryonic kidney cells (line HEK293) were grown at 37°C in 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; Biological Industries) supplemented with 1% antibiotics (penicillin, streptomycin, and amphotericin), 1% Gln, and 10% fetal calf serum. A day before experimentation, the cells were washed with PBS and then detached from plates with 1 mL trypsin-ethylene diamine tetraacetic acid. The cells were diluted to an optimal density of 1 to 2 million cells per plate (3-cm perforated plate with microscopic slides attached). The cells were further grown for additional 24 h and subjected to epifluorescence imaging on a Zeiss Axiovert 35 microscope(Carl Zeiss) attached to a Polychrome V image system (Till Photonics) (Glickstein et al., 2005).
Fluorescence Measurement
HEK293 cells were labeled for 15 min at 37°C with 1 µM RPA and/or 0.1 µM CALG-acetoxymethyl ester or 0.25 µM RPC in DMEM medium containing 10 mM Na-HEPES and supplemented with 10 µM desferrioxamine to prevent quenching of the probe by contaminant Fe from the medium. After washing with DMEM-HEPES medium and HBS, the cells were permeabilized in HBS buffer (37°C HEPES-buffered saline, pH 7.4) for 180 s with 25 µM digitonin (Nishikawa et al., 1984). The permeabilized cells were washed with permeabilization buffer (100 mM KCl, 5 mM Na2HPO4, Eagle's minimal essential medium–amino acids mix diluted 1:500, 10 mM HEPES, 1 µM CaCl2, and 1 mM MgSO4, pH 7.2) and used for fluorescence microscopy measurements in permeabilization buffer containing 1 mM succinate. For CALG (480-nm excitation; 520-nm emission) (Espósito et al., 2002; Glickstein et al., 2006) and RPA (560-nm excitation; 610-nm emission) (Petrat et al., 2002; Rauen et al., 2003), as described elsewhere (Espósito et al., 2002; Glickstein et al., 2005, 2006). Desferrioxamine (1 µM) was present in all solutions during permeabilization and fluorescence measurements to prevent RPA quenching by contaminant Fe. Following the recording of a 6-min baseline, 10 to 50 µM At-NEET (the wild type or the H89C mutant) was added, and changes in fluorescence were recorded for 20 min at 37°C followed by addition of labile Fe-FHQ (5 µM FHQ [FeCl3-8-hydroxyquinoline] 1:1 complex) so as to attain maximal quenching. The sequence of fluorescence images acquired by microscopy was analyzed by the Image J program (National Institutes of Health) (Abramoff et al., 2004). ROS imaging and quantification in Arabidopsis thaliana seedlings were performed as described (Luhua et al., 2008; Miller et al., 2009)
Plant Growth, Transformation, and Molecular Analysis
Arabidopsis cv Columbia plants were grown under controlled conditions at 21°C, 100 µmol m−2 s−1 (Luhua et al., 2008), and monitored for growth, flowering time, and senescence as described (Suzuki et al., 2005; Miller et al., 2007). Plants were watered daily with Spalding’s solution (Spalding et al., 1999) or with Spalding’s solution supplemented with Fe (FeCl2) to 100×. RNA and protein were isolated and analyzed by gel blot analysis and quantitative PCR as described (Miller et al., 2007, 2009). cDNAs for mitoNEET and At-NEET were obtained as described above and cloned in frame with GFP (C-terminal fusion) into a modified pGreen vector (Luhua et al., 2008). SpeI and XhoI were used to clone At-NEET and Hs-mitoNEET into GFP vector using the primers described in Supplemental Table 2 online. GFP, or GFP fused in frame to the C terminus of the NEET proteins, was expressed in plants under the control of the 35S CaMV promoter (Luhua et al., 2008). Transgenic plants were generated using the floral dip method, and homozygous lines were selected using hygromycin resistance and RNA and protein blots (Luhua et al., 2008). GFP was visualized in transgenic plants using a Nikon Eclipse E400 epifluorescence microscope or an Olympus IX 81 FV 1000 confocal microscope as described (Luhua et al., 2008). Mitotracker was used to image mitochondria in roots (see Supplemental Figure 5 online). Knockdown lines for At-NEET (SALK_040013 and SALK_040019) were obtained, isolated, and characterized using RNA and protein blots as described (Miller et al., 2007). RNAi vectors for At-NEET were generated by cloning the entire At-NEET open reading frame without the start and stop codons into a pHANNIBAL vector (CSIRO Plant Industry). The hairpin construct was then mobilized into a pGreen vector and expressed under the control of the 35S CaMV promoter. HindIII, XbaI, KpnI, and XhoI were used to build the At-NEET hairpin RNAi cassette in pKANNIBAL, and XhoI and SacI were used to clone the hairpin RNAi cassette into pGreen binary vector, using the primers described in Supplemental Table 2 online. Homozygous lines were then selected on hygromycin and tested by protein blots. Plant transformation was conducted as described (Luhua et al., 2008). Elemental analysis was performed with a Varian 820-ICP-MS as described by Baxter et al. (2008).
Stress Assays
For the analysis of stress tolerance, seeds of the wild type, two independent homozygous knockdown lines, and five independent RNAi lines for At-NEET were surface sterilized with bleach and placed in rows on 1% agar plates (0.5× Murashige and Skoog [MS] medium), containing different concentrations of paraquat, t-butyl hydroperoxide, NaCl, or sorbitol (Sigma-Aldrich) as described (Luhua et al., 2008). Each row of seeds (25 to 30 seedlings) placed on a plate was divided into three parts: control seeds and seeds of the two independent knockdown lines. Thus, the different seeds were placed side by side on the same plate. Plates were maintained vertically in a growth chamber (21 to 22°C, constant light, 60 µmol m−2 s−1) and percentage of germination and root length were scored 5 d after seed plating. Four- or five-day-old seedlings grown on 0.5× MS agar plates were also subjected to heat stress (38°C; 24 h) or cold stress (10°C; 48 h) and scored for percentage of germination and root length as described by Luhua et al. (2008). For tolerance to Fe, wild-type and knockdown lines were germinated on plates containing 0.5× MS without Fe or 0.5× MS containing 0.1 mM (100%), 0.3 mM (300%), or 0.4 mM (400%) FeSO27H2O. All experiments were repeated at least three different times, each with at least three different technical repeats. Results are shown as mean and se. Statistical analysis was performed as described (Suzuki et al., 2005).
Greening Assays
Seeds of the wild type, two independent homozygous knockdown lines, and five independent RNAi lines for At-NEET were surface sterilized with bleach and placed in rows on 1% agar plates (0.5× MS medium) containing 1% Suc. Following 48 h chilling at 4°C, plates were placed at 22°C in complete darkness for 1 week to obtain etiolated seedlings. To follow the greening process, etiolated seedlings were exposed to continuous light (150 µmol m−2 s−1) at 22°C for 24 h. At specified times, total chlorophyll concentration was determined following an 80% acetone extractions (Yalovsky et al., 1992).
Chloroplast and Mitochondrion Isolation
About 120 g of seedlings were ground in a Waring blender (five pulses of 15 s each) in grinding buffer (50 mM Tris-HCl, pH 8.0, 0.33 M d-mannitol, 2 mM MgCl2, 2.5 mM EDTA, and 2.5 mM Na-ascorbate; Schwitzguebel and Siegenthaler, 1984). The ground material was filtered through five layers of cloth and centrifuged for 3 min at 1500g. The obtained pellet, which contained the chloroplasts, was kept on ice, and the supernatant that contained the mitochondria was further centrifuged at 20,000g for 8 min. The chloroplast and mitochondrial pellets were gently suspended in 30 mL of washing buffer (10 mM Tris-HCl, pH 7.2, 0.25 M Suc, and 0.2% BSA). The solutions were centrifuged for 5 min at 1500g, and the resulting supernatants were further centrifuged at 20,000g. The resulting pellets (crude chloroplasts/crude mitochondria) were each suspended in 2 mL of washing buffer and loaded onto separate Percoll gradients (prepared in washing buffer in Corex 15-mL glass tubes). For isolation of intact mitochondria, the Percoll gradient contained three layers: 4 mL 60% Percoll, 6 mL 27% Percoll, and 4 mL 21% Percoll. The gradients for isolation of intact chloroplasts contained 4 mL 70% Percoll, 6 mL 50% Percoll, and 4 mL 30% Percoll. About 1 to 1.5 mL of the suspended pellets was loaded on each gradient, and they were centrifuged for 30 min at 9000g. The intact mitochondria were obtained in the interphase between the 60% and the 27% Percoll, and the intact chloroplasts were at the interphase between 70 and 50% Percoll. Each fraction was collected, diluted into 30 mL of washing buffer, and pelleted at 20,000g. The resulting pellets were suspended in minimal washing buffer.
Other Methods
Antibodies against the purified At-NEET were prepared as described by Miller et al. (2007) and used at a dilution of 1:250. Antibodies for VDAC, PsaD, and Hs-mitoNEET were purchased from Agrisera or obtained as described (Zuris et al., 2011). Protein concentration was determined by Bradford (1976) and by spectroscopic methods using the 458-nm absorption peak characteristic of At-NEET and its extinction coefficient of 5000 cm−1 M−1. Proteins were separated on SDS polyacrylamide gels using 15% acrylamide gels according to Laemmli (1970). Native gel electrophoresis was performed according to Wittig and Schägger (2005).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At-NEET, At5g51720, pdb ID code3S2Q; mitoNEET, CISD1, CDGSH Fe-S domain 1 (Homo sapiens), gene ID 55847; Miner1, CISD2, CDGSH Fe-S domain 2 (Homo sapiens), gene ID 493856.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Redox Potential (EM) of At5g51720 as a Function of pH.
Supplemental Figure 2. Measurements of Rosette Diameter in Wild-Type Knockdown Plants and RNAi Lines for At-NEET.
Supplemental Figure 3. Tolerance of Knockdown and RNAi Mutants for At-NEET to Abiotic Stress.
Supplemental Figure 4. Sodium Dithionite Does Not Affect RPA Fluorescence.
Supplemental Figure 5. Structural Comparison of the At5g51720 Wild-Type Protein (pdb ID Code 3S2Q) with the At5g51720 H89C Mutant (pdb ID Code 3S2R).
Supplemental Table 1. Crystallographic Data.
Supplemental Table 2. Primers Used for Cloning.
Supplementary Material
Acknowledgments
This work was supported by the Israel Science Foundation (ISF 863/09 awarded to R.N.); by the National Science Foundation (IBN-0420033, NSF-0431327, IOS-0639964, and IOS-0743954), Israel Science Foundation Grant 214/08, and European Union Grant FP7–Marie Curie 447 (to R.M.); by CMG Grant 2T32GM007240-29 (to A.R.C.); by National Institutes of Health Grants GM41637 (to M.L.P.), GM54038, and DK54441 (to P.A.J.); and by Heme and Blood Proteins Training Grant 5T32DK007233-34 (to J.A.Z.). We thank Guido F. Verbeck, the analytical support of the University of North Texas, Laboratory of Imaging Mass Spectrometry, and the FA9550-09-1-0496 Defense University Research Instrumentation Program Fund for inductively coupled plasma analysis.
AUTHOR CONTRIBUTIONS
R.N. designed experiments, performed research, analyzed data, and wrote the article. A.R.C. and L.S. performed research and analyzed data. Y.H. and J.A.Z. performed research, analyzed data, and wrote the article. O.Y. and D.M. performed research. Y.E.-D. performed research and analyzed data. O.L. designed experiments and wrote the article. R.R. performed research and wrote the article. V.M. and Y.L. performed research and analyzed data. M.L.P., P.A.J., and R.M. analyzed data, designed experiments, and wrote the article. Z.I.C. designed experiments.
Glossary
- ROS
reactive oxygen species
- RNAi
RNA interference
- GFP
green fluorescent protein
- CaMV
cauliflower mosaic virus
- aFd
apo form of ferredoxin
- Fd
ferredoxin
- CALG
calcein-green
- RPA
rhodamine B-[(1,10- phenanthrolin-5-yl) aminocarbonyl] benzyl ester
- PEG
polyethylene glycol
- DMEM
Dulbecco’s modified Eagle’s medium
- MS
Murashige and Skoog
References
- Abramoff M.D., Magelhaes P.J., Ram S.J. (2004). Image processing with ImageJ. Biophotonics Int. 11: 36–42 [Google Scholar]
- Amr S., Heisey C., Zhang M., Xia X.J., Shows K.H., Ajlouni K., Pandya A., Satin L.S., El-Shanti H., Shiang R. (2007). A homozygous mutation in a novel zinc-finger protein, ERIS, is responsible for Wolfram syndrome 2. Am. J. Hum. Genet. 81: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K. (2006). Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 141: 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J., Pilon M. (2011). Ancient and essential: The assembly of iron-sulfur clusters in plants. Trends Plant Sci. 16: 218–226 [DOI] [PubMed] [Google Scholar]
- Baxter I.R., Vitek O., Lahner B., Muthukumar B., Borghi M., Morrissey J., Guerinot M.L., Salt D.E. (2008). The leaf ionome as a multivariable system to detect a plant’s physiological status. Proc. Natl. Acad. Sci. USA 105: 12081–12086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Chandramouli K., Johnson M.K. (2006). HscA and HscB stimulate [2Fe-2S] cluster transfer from IscU to apoferredoxin in an ATP-dependent reaction. Biochemistry 45: 11087–11095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N.C., Nguyen M., Germain M., Shore G.C. (2010). Antagonism of Beclin 1-dependent autophagy by BCL-2 at the endoplasmic reticulum requires NAF-1. EMBO J. 29: 606–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.F., Kao C.H., Chen Y.T., Wang C.H., Wu C.Y., Tsai C.Y., Liu F.C., Yang C.W., Wei Y.H., Hsu M.T., Tsai S.F., Tsai T.F. (2009). Cisd2 deficiency drives premature aging and causes mitochondria-mediated defects in mice. Genes Dev. 23: 1183–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.F., Wu C.Y., Kirby R., Kao C.H., Tsai T.F. (2010). A role for the CISD2 gene in lifespan control and human disease. Ann. N. Y. Acad. Sci. 1201: 58–64 [DOI] [PubMed] [Google Scholar]
- Colca J.R., McDonald W.G., Waldon D.J., Leone J.W., Lull J.M., Bannow C.A., Lund E.T., Mathews W.R. (2004). Identification of a novel mitochondrial protein (“mitoNEET”) cross-linked specifically by a thiazolidinedione photoprobe. Am. J. Physiol. Endocrinol. Metab. 286: E252–E260 [DOI] [PubMed] [Google Scholar]
- Conlan A.R., Axelrod H.L., Cohen A.E., Abresch E.C., Nechushtai R., Paddock M.L., Jennings P.A. (2009a). Structural comparison of a diabetes drug target, Mitoneet, a 2Fe-2S cluster protein to its more stable mutant, H87C. Biophys. J. 96: 67a18849413 [Google Scholar]
- Conlan A.R., Axelrod H.L., Cohen A.E., Abresch E.C., Zuris J., Yee D., Nechushtai R., Jennings P.A., Paddock M.L. (2009b). Crystal structure of Miner1: The redox-active 2Fe-2S protein causative in Wolfram Syndrome 2. J. Mol. Biol. 392: 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan A.R., Paddock M.L., Axelrod H.L., Cohen A.E., Abresch E.C., Wiley S., Roy M., Nechushtai R., Jennings P.A. (2009c). The novel 2Fe-2S outer mitochondrial protein mitoNEET displays conformational flexibility in its N-terminal cytoplasmic tethering domain. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65: 654–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan A.R., Paddock M.L., Homer C., Axelrod H.L., Cohen A.E., Abresch E.C., Zuris J.A., Nechushtai R., Jennings P.A. (2011). Mutation of the His ligand in mitoNEET stabilizes the 2Fe-2S cluster despite conformational heterogeneity in the ligand environment. Acta Crystallogr. D Biol. Crystallogr. 67: 516–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Arcy A., Elmore C., Stihle M., Johnston J.E. (1996). A novel approach to crystallising proteins under oil. J. Cryst. Growth 168: 175–180 [Google Scholar]
- Dicus M.M., Conlan A., Nechushtai R., Jennings P.A., Paddock M.L., Britt R.D., Stoll S. (2010). Binding of histidine in the (Cys)3(His)1-coordinated [2Fe-2S] cluster of human mitoNEET. J. Am. Chem. Soc. 132: 2037–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. (2004). Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Espósito B.P., Epsztejn S., Breuer W., Cabantchik Z.I. (2002). A review of fluorescence methods for assessing labile iron in cells and biological fluids. Anal. Biochem. 304: 1–18 [DOI] [PubMed] [Google Scholar]
- Fish A., Danieli T., Ohad I., Nechushtai R., Livnah O. (2005). Structural basis for the thermostability of ferredoxin from the cyanobacterium Mastigocladus laminosus. J. Mol. Biol. 350: 599–608 [DOI] [PubMed] [Google Scholar]
- Glickstein H., El R.B., Link G., Breuer W., Konijn A.M., Hershko C., Nick H., Cabantchik Z.I. (2006). Action of chelators in iron-loaded cardiac cells: Accessibility to intracellular labile iron and functional consequences. Blood 108: 3195–3203 [DOI] [PubMed] [Google Scholar]
- Glickstein H., El R.B., Shvartsman M., Cabantchik Z.I. (2005). Intracellular labile iron pools as direct targets of iron chelators: A fluorescence study of chelator action in living cells. Blood 106: 3242–3250 [DOI] [PubMed] [Google Scholar]
- Halliwell B., Gutteridge J.M. C. (2007). Free Radicals in Biology and Medicine, 5th ed (Oxford, UK: Clarendon) [Google Scholar]
- Hou X., Liu R., Ross S., Smart E.J., Zhu H., Gong W. (2007). Crystallographic studies of human MitoNEET. J. Biol. Chem. 282: 33242–33246 [DOI] [PubMed] [Google Scholar]
- Jancarik J., Kim S.H. (1991). Sparse matrix sampling: A screening method for crystallization of proteins. J. Appl. Cryst. 24: 409–411 [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lill R. (2009). Function and biogenesis of iron-sulphur proteins. Nature 460: 831–838 [DOI] [PubMed] [Google Scholar]
- Lin J., Zhou T., Ye K., Wang J. (2007). Crystal structure of human mitoNEET reveals distinct groups of iron sulfur proteins. Proc. Natl. Acad. Sci. USA 104: 14640–14645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhua S., Ciftci-Yilmaz S., Harper J., Cushman J., Mittler R. (2008). Enhanced tolerance to oxidative stress in transgenic Arabidopsis plants expressing proteins of unknown function. Plant Physiol. 148: 280–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J. (2008). Iron-sulfur protein folds, iron-sulfur chemistry, and evolution. J. Biol. Inorg. Chem. 13: 157–170 [DOI] [PubMed] [Google Scholar]
- Miller G., Schlauch K., Tam R., Cortes D., Torres M.A., Shulaev V., Dangl J.L., Mittler R. (2009). The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2: ra45. [DOI] [PubMed] [Google Scholar]
- Miller G., Suzuki N., Rizhsky L., Hegie A., Koussevitzky S., Mittler R. (2007). Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development, and response to abiotic stresses. Plant Physiol. 144: 1777–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R., Rizhsky L. (2000). Transgene-induced lesion mimic. Plant Mol. Biol. 44: 335–344 [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Morris R.J., Perrakis A., Lamzin V.S. (2003). ARP[+45 degree rule]wARP and automatic interpretation of protein electron density maps. In Methods in Enzymology, C.W. Carter, Jr. and R.M. Sweet, eds (Amsterdam: Academic Press/Elsevier), pp. 229–244 [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. (1997). Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nishikawa M., Nojima S., Akiyama T., Sankawa U., Inoue K. (1984). Interaction of digitonin and its analogs with membrane cholesterol. J. Biochem. 96: 1231–1239 [DOI] [PubMed] [Google Scholar]
- op den Camp R.G.L., Przybyla D., Ochsenbein C., Laloi C., Kim C., Danon A., Wagner D., Hideg E., Göbel C., Feussner I., Nater M., Apel K. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15: 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. (1997). Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Paddock M.L., Wiley S.E., Axelrod H.L., Cohen A.E., Roy M., Abresch E.C., Capraro D., Murphy A.N., Nechushtai R., Dixon J.E., Jennings P.A. (2007). MitoNEET is a uniquely folded 2Fe 2S outer mitochondrial membrane protein stabilized by pioglitazone. Proc. Natl. Acad. Sci. USA 104: 14342–14347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrat F., Weisheit D., Lensen M., de Groot H., Sustmann R., Rauen U. (2002). Selective determination of mitochondrial chelatable iron in viable cells with a new fluorescent sensor. Biochem. J. 362: 137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton E., Briggs P., Turkenburg M., Dodson E. (2003). A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59: 1131–1137 [DOI] [PubMed] [Google Scholar]
- Rauen U., Kerkweg U., Weisheit D., Petrat F., Sustmann R., de Groot H. (2003). Cold-induced apoptosis of hepatocytes: Mitochondrial permeability transition triggered by nonmitochondrial chelatable iron. Free Radic. Biol. Med. 35: 1664–1678 [DOI] [PubMed] [Google Scholar]
- Read R.J., McCoy A.J. (2011). Using SAD data in Phaser. Acta Crystallogr. D Biol. Crystallogr. 67: 338–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitzguebel J.-P., Siegenthaler P.-A. (1984). Purification of peroxisomes and mitochondria from spinach leaf by percoll gradient centrifugation. Plant Physiol. 75: 670–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding E.P., Hirsch R.E., Lewis D.R., Qi Z., Sussman M.R., Lewis B.D. (1999). Potassium uptake supporting plant growth in the absence of AKT1 channel activity: Inhibition by ammonium and stimulation by sodium. J. Gen. Physiol. 113: 909–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Rizhsky L., Liang H., Shuman J., Shulaev V., Mittler R. (2005). Enhanced tolerance to environmental stress in transgenic plants expressing the transcriptional coactivator multiprotein bridging factor 1c. Plant Physiol. 139: 1313–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirrell T.F., Paddock M.L., Conlan A.R., Smoll E.J., Jr, Nechushtai R., Jennings P.A., Kim J.E. (2009). Resonance Raman studies of the (His)(Cys)3 2Fe-2S cluster of MitoNEET: Comparison to the (Cys)4 mutant and implications of the effects of pH on the labile metal center. Biochemistry 48: 4747–4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A., Teplyakov A. (2010). Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66: 22–25 [DOI] [PubMed] [Google Scholar]
- Wiley S.E., Murphy A.N., Ross S.A., van der Geer P., Dixon J.E. (2007a). MitoNEET is an iron-containing outer mitochondrial membrane protein that regulates oxidative capacity. Proc. Natl. Acad. Sci. USA 104: 5318–5323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley S.E., Paddock M.L., Abresch E.C., Gross L., van der Geer P., Nechushtai R., Murphy A.N., Jennings P.A., Dixon J.E. (2007b). The outer mitochondrial membrane protein mitoNEET contains a novel redox-active 2Fe-2S cluster. J. Biol. Chem. 282: 23745–23749 [DOI] [PubMed] [Google Scholar]
- Wittig I., Schägger H. (2005). Advantages and limitations of clear-native PAGE. Proteomics 5: 4338–4346 [DOI] [PubMed] [Google Scholar]
- Wu G., Mansy S.S., Wu Sp S.P., Surerus K.K., Foster M.W., Cowan J.A. (2002). Characterization of an iron-sulfur cluster assembly protein (ISU1) from Schizosaccharomyces pombe. Biochemistry 41: 5024–5032 [DOI] [PubMed] [Google Scholar]
- Wu S.P., Cowan J.A. (2003). Iron-sulfur cluster biosynthesis. A comparative kinetic analysis of native and Cys-substituted ISA-mediated [2Fe-2S]2+ cluster transfer to an apoferredoxin target. Biochemistry 42: 5784–5791 [DOI] [PubMed] [Google Scholar]
- Yalovsky S., Ne’eman E., Schuster G., Paulsen H., Harel E., Nechushtai R. (1992). Accumulation of a light-harvesting chlorophyll a/b protein in the chloroplast grana lamellae. The lateral migration of the membrane protein precursor is independent of its processing. J. Biol. Chem. 267: 20689–20693 [PubMed] [Google Scholar]
- Zuris J.A., Halim D.A., Conlan A.R., Abresch E.C., Nechushtai R., Paddock M.L., Jennings P.A. (2010). Engineering the redox potential over a wide range within a new class of FeS proteins. J. Am. Chem. Soc. 132: 13120–13122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuris J.A., Harir Y., Conlan A.R., Shvartsman M., Tamir S., Paddock M.L., Mittler R., Cabantchik Z.I., Jennings P.A., Nechushtai R. (2011). Facile transfer of [2Fe-2S] clusters from the diabetes drug target mitoNEET to an apo-acceptor protein. Proc. Natl. Acad. Sci. USA 108: 13047–13052 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.