PHO2, a ubiquitin-conjugating E2 enzyme, maintains phosphate homeostasis by regulating phosphate uptake and translocation. This work identifies PHO1, a membrane-spanning protein implicated in xylem loading of phosphate, as a PHO2-interacting protein in the endomembrane system and shows that PHO1 degradation is PHO2 dependent and involves multivesicular body–mediated vacuolar proteolysis.
Abstract
The Arabidopsis thaliana pho2 mutant, which is defective in a ubiquitin-conjugating E2 enzyme, displays inorganic phosphate (Pi) toxicity as a result of enhanced uptake and root-to-shoot translocation of Pi. To elucidate downstream components of the PHO2-dependent regulatory pathway, we identified two pho2 suppressors as carrying missense mutations in PHO1, which has been implicated in Pi loading to the xylem. In support of the genetic interaction between PHO1 and PHO2, we found that the protein level of PHO1 is increased in pho2, whereas such accumulation is ameliorated in both pho2 suppressors. Results from cycloheximide and endosomal Cys protease inhibitor E-64d treatments further suggest that PHO1 degradation is PHO2 dependent and involves multivesicular body–mediated vacuolar proteolysis. Using the transient expression system of tobacco (Nicotiana tabacum) leaves, we demonstrated that PHO1 and PHO2 are partially colocalized and physically interact in the endomembranes, where the ubiquitin conjugase activity of PHO2 is required for PHO1 degradation. In addition, reduced PHO1 expression caused by PHO1 mutations impede Pi uptake, indicating a functional association between xylem loading and acquisition of Pi. Together, our findings uncover a pivotal molecular mechanism by which PHO2 modulates the degradation of PHO1 in the endomembranes to maintain Pi homeostasis in plants.
INTRODUCTION
Like other nutrients in the soil, inorganic phosphate (H2PO4–; Pi) has to be transported across the plasma membrane (PM) of the outermost layers of root cells before entering the plant body. As the concentration of available Pi in most soils is 1000- to 10,000-fold lower than the intracellular concentration (∼10 mM), uptake of the soil Pi against the steep Pi concentration, as well as electrical gradients between the soil solution and the cytosol, is an energy-requiring process that involves the H+-coupled Pi transport system (Rausch and Bucher, 2002).
Once taken up into the root symplast, Pi can reach the metabolic pools and be used for processes such as biosynthesis of nucleic acids and phospholipids, energy transfer, or protein activation or can be sequestered into the vacuole for storage. Alternatively, Pi can move toward the stele, either intracellularly via plasmodesmata in the symplastic pathway or through the extracellular space in the apoplastic pathway (Poirier and Bucher, 2002). Given that the Casparian strip, a zone in which the cell wall is impregnated with lignin and suberin, impedes the apoplastic passage of Pi beyond the endodermis, radial transfer of Pi into the vascular tissue must proceed via the symplastic pathway. Subsequently, Pi efflux out of the stellar cells is a prerequisite to xylem loading for the long-distance transport of Pi to the aboveground tissues (Poirier and Bucher, 2002; Rausch and Bucher, 2002).
Over the past few years, the intricate mechanisms involved in maintaining whole-plant Pi homeostasis have emerged and revealed the complexity of the regulation of Pi uptake and Pi translocation (Chiou and Lin, 2011). In Arabidopsis thaliana, at least nine PHOSPHATE TRANSPORTER1 (PHT1) genes encoding high-affinity H+/Pi cotransporters of PMs (PHT1;1 to PHT1;9) have been identified (Poirier and Bucher, 2002). Most of the PHT1 genes are preferentially expressed in the root epidermal or cortical cells, implying their direct involvement in Pi acquisition (Muchhal et al., 1996; Mudge et al., 2002). In particular, the functional characterization of the Arabidopsis pht1;1 and pht1;4 single and double mutants suggested that PHT1;1 and PHT1;4 play significant roles in Pi uptake from both low and high Pi environments (Misson et al., 2004; Shin et al., 2004). Besides being transcriptionally induced by Pi starvation, the PHT1 genes are also regulated at the posttranscriptional level. PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 (PHF1) is a SEC12-related plant-specific protein that modulates the endoplasmic reticulum (ER) exit of PHT1 transporters, thus regulating the activity of PHT1 transporters along their intracellular trafficking en route to the PM (González et al., 2005; Bayle et al., 2011; Chen et al., 2011).
Compared with our understanding of Pi uptake in plants, the molecular mechanism underlying the regulation of Pi allocation among different organs during plant development and in response to stress is relatively unexplored. The Arabidopsis pho1 and pho2 mutants were originally identified in the genetic screens for mutants with abnormal distribution of Pi, namely, altered Pi levels in leaves (Poirier et al., 1991; Delhaize and Randall, 1995). The pho1 mutant has low leaf Pi levels and shows severe phenotypes of shoot Pi deficiency due to defective Pi loading into the xylem (Poirier et al., 1991). Mapping and sequence analysis of PHO1 suggested that it encodes a potential membrane-spanning transporter protein (Hamburger et al., 2002). However, PHO1 shows no homology to any previously characterized solute transporters and only bears low homology with the human retrovirus receptor Rmc1 and the yeast Syg1 (Hamburger et al., 2002). The 11 members of the Arabidopsis PHO1 gene family have the same topology and harbor a SPX (for SYG1/PHO81/XPR1) tripartite domain in the N-terminal hydrophilic region and an EXS (for ERD1/XPR1/SYG1) domain in the C-terminal hydrophobic tail (Wang et al., 2004). In yeast, the SPX domain of the low-affinity Pi transporters, Pho87 and Pho90, was shown to modulate Pi uptake activity and affect the regulation of the Pi signaling pathway (Hürlimann et al., 2009). Although PHO1 was proposed to function as a Pi transporter, neither Pi influx nor Pi efflux activity has been directly demonstrated (Hamburger et al., 2002), leaving its mode of action in root-to-shoot transfer of Pi as a puzzle yet to be solved.
The pho2 mutant exhibits high shoot Pi levels and displays symptoms of Pi toxicity when Pi supply is replete (Delhaize and Randall, 1995; Dong et al., 1998). The mutation of pho2 is caused by a single nucleotide change, resulting in early termination within UBC24, which encodes a putative ubiquitin-conjugating (UBC) E2 enzyme (Aung et al., 2006; Bari et al., 2006). Previously, we and other groups revealed a mechanism by which plants regulate Pi homeostasis to adapt to fluctuations in Pi availability. The mechanism involves the suppression of PHO2 by a specific microRNA, miR399. Upon Pi starvation, miR399 is upregulated by the transcription factor PHR1 and directed to cleave the transcripts of its target gene, PHO2 (Aung et al., 2006; Bari et al., 2006; Chiou et al., 2006). In accordance with the inverse correlation between miR399 and PHO2 mRNA levels, the transgenic plants overexpressing miR399 phenocopied pho2 mutants. Consistent with the expression patterns of MIR399 and PHO2 in vascular tissues (Aung et al., 2006), results from reciprocal grafting between wild-type and miR399-overexpressing plants suggested that the shoot-derived mature miR399s move across the grafting junction to repress the expression of PHO2 in the root (Lin et al., 2008b; Pant et al., 2008). Furthermore, induction of miR399 expression by Pi deficiency occurred earlier in the shoot than in the root, indicating that the shoot-to-root movement of miR399 may serve as a systemic signal of Pi deficiency (Lin et al., 2008b).
Although our understanding of Pi starvation signaling involving PHR1, miR399, and PHO2 is well established, there remain missing links in our knowledge as to how PHO2 as a putative UBC enzyme mediates the downstream activities/responses, including the enhanced Pi uptake and the facilitated translocation of Pi from roots to shoots. In this article, we identified two pho2 suppressors as carrying missense mutations in the PHO1 gene. In support of our genetic results, the immunoblot analysis of PHO1 revealed that the accumulation of PHO1 in pho2 is suppressed in both pho2 suppressors. Moreover, we showed that functional PHO2 with UBC activity is required for the degradation of PHO1 in the endomembranes (EMs). Our findings provide compelling evidence that PHO1 is a crucial downstream component of the PHO2 regulatory pathway.
RESULTS
Two pho2 Suppressors Carry Missense Mutations in PHO1
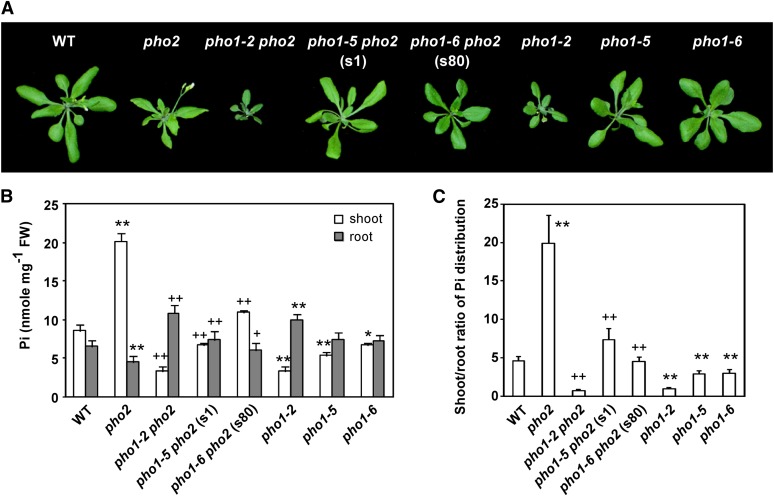
The genetic screening for the pho2 suppressor was performed using ethyl methanesulfonate–induced mutagenesis. From the M2 population of ∼13,000 plants grown in fertilized soils mixed with 1 mM KH2PO4, we identified two suppressors, s1 and s80, which did not display the Pi toxicity phenotype of pho2 but showed improved growth (Figure 1A). By measuring the shoot Pi concentration of s1 and s80 M3 plants and verifying that the pho2 point mutation remained in the mutants, we confirmed that both are indeed pho2 suppressors. Compared with pho2 grown under Pi-sufficient conditions (+Pi; 250 μM KH2PO4), s1 and s80 seedlings had lower shoot Pi concentrations yet higher root Pi concentrations (Figure 1B). In addition, while pho2 exhibited a higher shoot-to-root ratio of total Pi distribution than the wild-type controls under +Pi conditions, the tendency was ameliorated in s1 and s80 (Figure 1C). Notably, when s1 was grown hydroponically to an adult stage under +Pi conditions, the shoot Pi concentration of s1 was nearly as low as that of the wild type under –Pi conditions (see Supplemental Figure 1 online). However, s1 did not show the characteristic symptoms of shoot Pi deficiency, such as anthocyanin accumulation (Figure 1A). By contrast, the adult s80 grown hydroponically under +Pi conditions was slightly smaller than the wild type and had an intermediate shoot Pi concentration between that of pho2 and the wild type (Figure 1A; see Supplemental Figure 1 online).
Figure 1.
Phenotype and Pi Concentrations of the pho2 Suppressors.
(A) The phenotype of 20-d-old wild-type (WT), pho2, pho1-2 pho2, pho1-5 pho2 (s1), pho1-6 pho2 (s80), pho1-2, pho1-5, and pho1-6 plants grown in +Pi hydroponic medium.
(B) and (C) The Pi concentrations of the shoot and root (B) and the shoot-to-root ratio of total Pi distribution (C) of 13-d-old wild-type, pho2, pho1-2 pho2, pho1-5 pho2 (s1), pho1-6 pho2 (s80), pho1-2, pho1-5, and pho1-6 seedlings under +Pi conditions. The distribution ratio of total Pi between shoot and root was calculated from the shoot/root Pi concentrations multiplied by the total shoot/root masses. Error bars represent se (n = 6 in [B] and 9 in [C], pooled from three independent experiments). Data significantly different from the corresponding controls are indicated (mutant versus the wild type, *P < 0.05, **P < 0.01; mutant versus the pho2 mutant, +P < 0.05, ++P < 0.01; Student’s t test). FW, fresh weight.
[See online article for color version of this figure.]
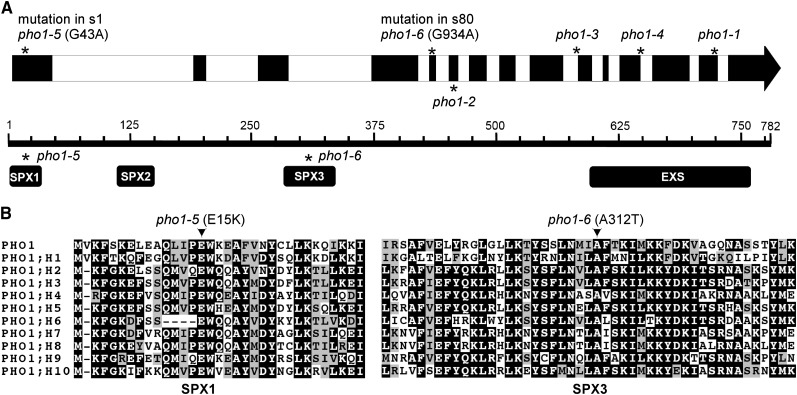
As the pho1-2 pho2 double mutant has been shown to display the pho1-2–like phenotype, pho1-2 was suggested to be epistatic to pho2 (Delhaize and Randall, 1995). It is tempting to suspect that disruption of PHO1 in the pho2 mutant may disrupt an inevitable step of Pi accumulation, the root-to-shoot translocation of Pi, thereby rendering the phenotype of pho1-2 pho2 similar to that of pho1-2 (Figures 1A and 1B). Considering that mutations in PHO1 may hamper Pi accumulation in the shoot of pho2 through a similar mechanism as assumed in pho1-2 pho2, we then sequenced the coding sequence of PHO1 in s1 and s80 before proceeding with the map-based cloning. Intriguingly, the point mutations G43A and G934A of PHO1 were identified in s1 and s80, respectively. Both mutations reside in the SPX domain of PHO1, causing substitution of two amino acid residues conserved among the Arabidopsis PHO1 family: Glu for Lys at position 15 (E15K) in s1 and Ala for Thr at position 312 (A312T) in s80 (Figures 2A and 2B).
Figure 2.
The pho2 Suppressors Carry Missense Mutations in PHO1.
(A) Genome structure of Arabidopsis PHO1 along with the point mutation positions of the PHO1 alleles. The bottom panel depicts the domain architecture of PHO1, containing SPX and EXS domains. Asterisks indicate the positions of the different point mutations.
(B) The alignment of the corresponding amino acid sequences of subdomains SPX1 and SPX3 among members of the Arabidopsis PHO1 gene family (Wang et al., 2004). Identical amino acids are shaded in black. Note that the amino acid residues Glu-15 and Ala-312 of PHO1, where pho1-5 and pho1-6 mutations reside, respectively, are conserved among the PHO1 gene members.
To determine the genetic inheritance of s1, we backcrossed s1 to pho2. By examining the distribution and range of shoot Pi concentrations among the F2 progeny, we ensured that the pho2 suppressor phenotype was segregated in the F2 population as a single locus (see Supplemental Figure 2A online). Interestingly, we found that one peak representing the s1-like population within the range of wild-type controls was followed by two close pho2-like peaks (see Supplemental Figure 2B online), indicating that s1 is not a simple recessive mutation. Next, we introduced the genomic fragment encompassing the PHO1 coding region, spanning from 3587 bp upstream of the start codon to 523 bp downstream of the stop codon, to complement s1 and s80. Because the exogenous expression of wild-type PHO1 restored the capacity of both pho2 suppressors to accumulate more Pi in the shoot (see Supplemental Figures 3A and 3B online), we inferred that the missense mutations of PHO1 in s1 and s80 would contribute to the suppression of Pi accumulation. In fact, these complemented lines accumulated more shoot Pi than pho2 due to the overexpression of PHO1 (see below). Four PHO1 alleles with point mutations have been previously isolated (pho1-1 to pho1-4), presumably leading to the synthesis of truncated PHO1 proteins (Hamburger et al., 2002). Here, with the identification of s1 and s80, we isolated two additional PHO1 alleles designated pho1-5 and pho1-6, respectively (Figure 2A), and therefore used pho1-5 pho2 and pho1-6 pho2 designations, respectively, for s1 and s80 in the following text.
PHO1 Alleles Exhibit Gene Dosage Effects on Shoot Pi Accumulation
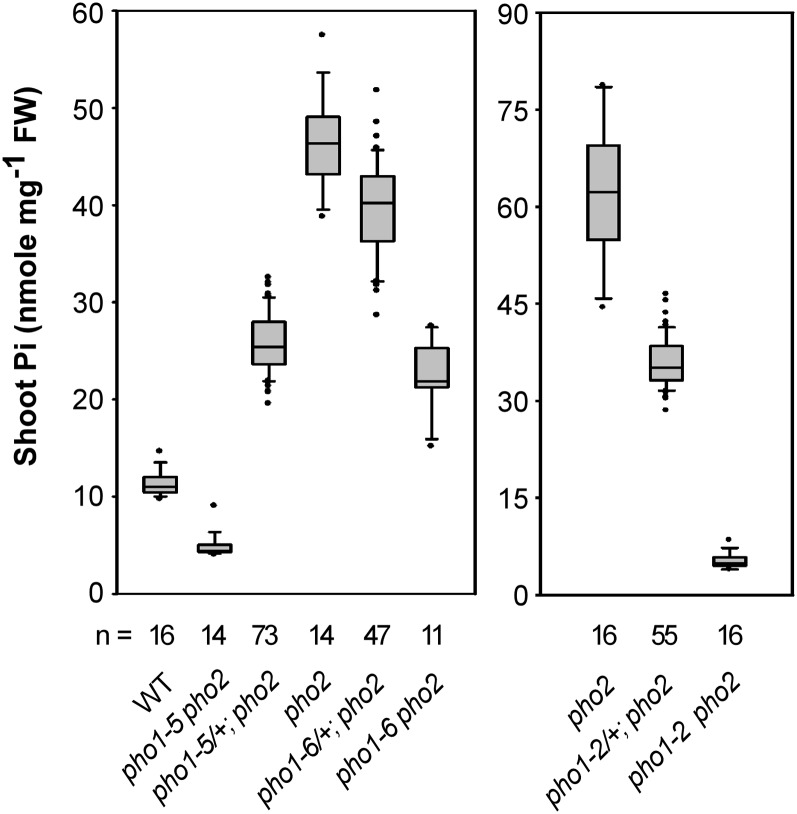
To further explore the genetic inheritance of pho1-5 and pho1-6 mutations, we grew the F1 progeny of pho1-5 pho2 × pho2 and pho1-6 pho2 × pho2 in hydroponic culture to an adult stage. Interestingly, the pho1-5 and pho1-6 heterozygotes in the pho2 background (pho1-5/+; pho2/pho2 and pho1-6/+; pho2/pho2) showed intermediate shoot Pi concentrations between their respective parents (Figure 3), suggesting that one copy of the pho1-5 or pho1-6 allele is sufficient to partially suppress the Pi toxicity of pho2. These results also confirmed our speculation that the two close pho2-like peaks in the F2 segregation analysis of s1 (pho1-5 pho2) × pho2 represented the pho1-5 heterozygotes and pho2, respectively (see Supplemental Figure 2B online).
Figure 3.
PHO1 Alleles Exert Gene Dosage Suppression of Pi Toxicity in pho2.
The shoot Pi concentrations of 23-d-old plants grown in hydroponic medium containing 1 mM KH2PO4 are presented as box plots. The boundaries of the boxes indicate the 25th and 75th percentiles. The median is marked by a black line within the box. Error bars above and below the box indicate the 90th and 10th percentiles. Each individual plant outside the 10th and 90th percentiles is displayed as a single dot. FW, fresh weight; n, the total number of plants; WT, the wild type.
In a similar fashion, the pho2 mutants harboring one copy of the pho1-2 allele (pho1-2/+; pho2/pho2) also displayed intermediate shoot Pi concentrations between pho2 and pho1-2 pho2 (Figure 3). Because these observations appeared contradictory to the earlier report showing that pho1-2 is a recessive allele (Poirier et al., 1991), we thus reexamined the shoot Pi concentration of pho1-2/+ in the wild-type background. The shoot Pi level of pho1-2/+ was only slightly but significantly lower than that of wild-type plants (see Supplemental Figure 4 online). Apparently, the effects of the pho1-2 alleles on the suppression of shoot Pi accumulation are more prominent in the pho2 background than in the wild-type background.
Next, we obtained the pho1-5 and pho1-6 homozygotes (pho1-5/pho1-5 and pho1-6/pho1-6) to explore how pho1-5 and pho1-6 would affect Pi homeostasis in the wild-type background. Under +Pi conditions, pho1-5 and pho1-6 exhibited reduced shoot Pi concentrations and shoot-to-root ratios of total Pi distribution compared with the wild-type plants (Figures 1B and 1C). These results suggest that both pho1-5 and pho1-6 are weak PHO1 alleles with putative roles in Pi translocation from root to shoot, resulting in minor growth retardation (Figure 1A).
PHO1 Mutations Impede Pi Uptake
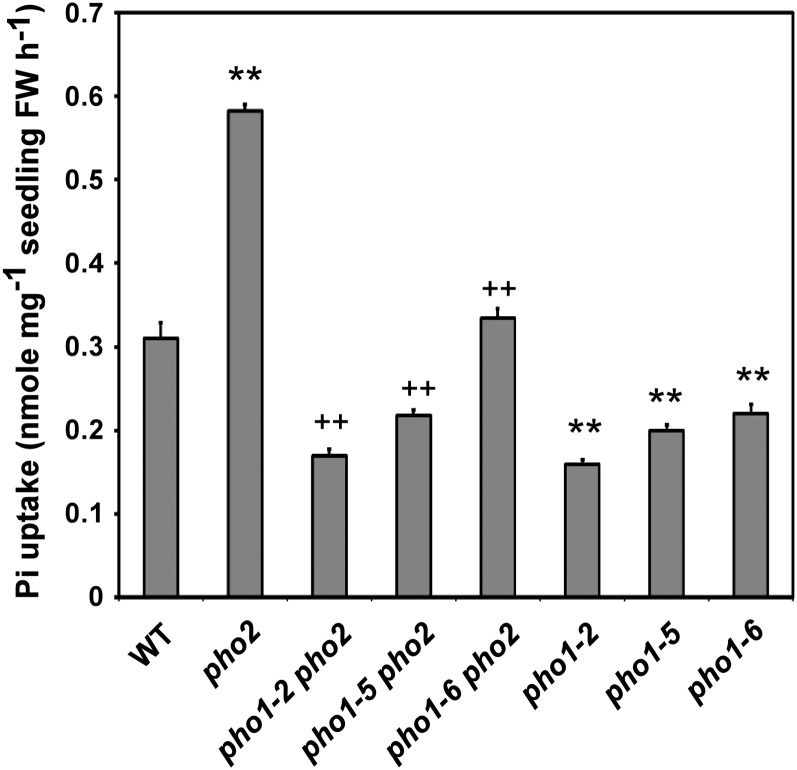
Since both enhanced Pi acquisition and root-to-shoot translocation of Pi account for Pi overaccumulation in pho2, we wondered whether the Pi uptake activity of these pho2 suppressors was also affected. Under +Pi conditions, both pho1-5 pho2 and pho1-6 pho2 exhibited reduced Pi uptake activity compared with pho2 (Figure 4). Furthermore, we found that the Pi uptake activity of pho1-2, pho1-5, and pho1-6 mutants was reduced compared with that of the wild type (Figure 4). Similar to the magnitude of shoot Pi concentrations in these mutants, the order of the mutants regarding their respective aberrant Pi uptake activity was as follows: pho2 > pho1-6 pho2 > pho1-5 pho2 > pho1-2 pho2 in the pho2 background; wild type > pho1-6 > pho1-5 > pho1-2 in the wild-type background (Figure 4). Overall, pho1-2, pho1-5, and pho1-6 alleles impair not only Pi translocation from roots to shoots, as expected on the basis of the role of PHO1 in xylem loading, but also the process of Pi acquisition.
Figure 4.
The Pi Uptake Activity of the pho2 Suppressors.
The [P33] Pi uptake rate of 13-d-old wild-type (WT), pho2, pho1-2 pho2, pho1-5 pho2, pho1-6 pho2, pho1-2, pho1-5, and pho1-6 seedlings under +Pi conditions. The Pi uptake rate was calculated per fresh weight of the whole seedling. Error bars represent se (n = 9, pooled from three independent experiments). Data significantly different from the corresponding controls are indicated (mutant versus the wild type, *P < 0.05, **P < 0.01; mutant versus the pho2 mutant, +P < 0.05, ++P < 0.01; Student’s t test). FW, fresh weight.
PHO2 Mediates PHO1 Degradation in Response to Pi Availability
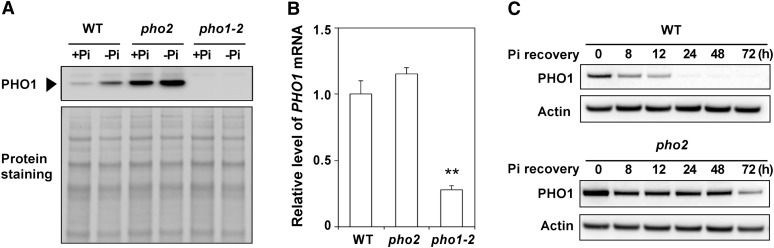
To identify the role of PHO1 downstream of the PHO2 regulatory pathway, we were prompted to check whether PHO1 is upregulated in pho2. A specific polyclonal antibody against PHO1 was raised and able to detect a band in the wild type at ∼68 kD that was absent in the pho1-2 mutant, likely corresponding to PHO1, despite its predicted molecular mass of 90 kD (Figure 5A). The relatively fast mobility of PHO1 in SDS-PAGE, a common observation for hydrophobic membrane proteins, probably resulted from incomplete unfolding or binding of more SDS per mass unit protein. Strikingly, we found that more PHO1 protein accumulated in the +Pi root of pho2 compared with the wild type (Figure 5A), which was apparently not attributed to a change in transcript abundance (Aung et al., 2006; Figure 5B). Next, we compared the protein level of PHO1 between the wild type and pho2 following Pi recovery. While Pi deficiency-induced accumulation of PHO1 gradually decreased in the wild type over the first 48 h of Pi resupply, the level of PHO1 in pho2 declined in the first 8 h and then remained unchanged until the 72-h time point (Figure 5C). These results suggest that PHO2 is required for downregulation of PHO1 in response to Pi availability.
Figure 5.
Upregulation of PHO1 at the Protein Level in pho2.
(A) Immunoblot analysis of PHO1 in the root of 12-d-old wild-type (WT), pho1-2, and pho2 seedlings under +Pi and −Pi (6 d of Pi deficiency) conditions. The bottom panel shows protein staining on the membrane.
(B) Quantitative RT-PCR analysis of PHO1 expression in the root of 12-d-old wild-type, pho1-2, and pho2 seedlings under +Pi conditions. The value is presented as the fold change relative to the expression of wild-type plants. Error bars represent se (n = 3). Data significantly different from the wild-type controls are indicated (**P < 0.01; Student’s t test).
(C) The expression level of PHO1 protein in the root of 13-d-old wild-type and pho2 seedlings over 72 h of Pi recovery (250 µM KH2PO4) following 8 d of Pi deficiency. Actin is used as a loading control.
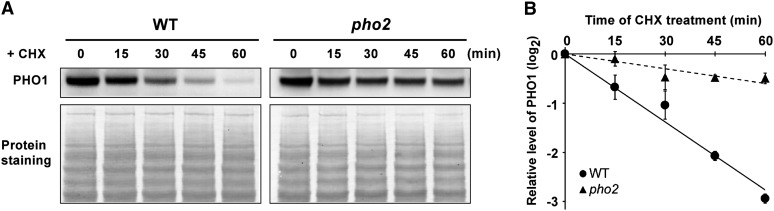
Since PHO2 is assumed to participate in protein modification due to its putative role as a UBC enzyme, we then evaluated whether downregulation of PHO1 by PHO2 is at the posttranslational level. Under +Pi conditions, PHO1 rapidly declined in the wild type in a time course–dependent manner in the presence of the protein synthesis inhibitor, cycloheximide (CHX), with a half-life calculated to be 21.7 min (Figures 6A and 6B). By contrast, the degradation rate of PHO1 in pho2 was slower than in the wild type (Figures 6A and 6B), suggesting that PHO2 mediates the downregulation of PHO1 at the posttranslational level.
Figure 6.
PHO2-Dependent Degradation of PHO1.
(A) The expression of PHO1 in the root of 12-d-old wild-type (WT) and pho2 seedlings over a 60-min period of CHX treatment (200 µM) under +Pi conditions.
(B) The relative remaining amount of PHO1 upon CHX treatment was calculated and plotted on a semilog graph. The expression level of PHO1 was normalized with the corresponding actin and DMSO controls. Error bars represent se (n = 4).
PHO1 Missense Mutations Cause Decreased Level of PHO1 in pho2 Suppressors
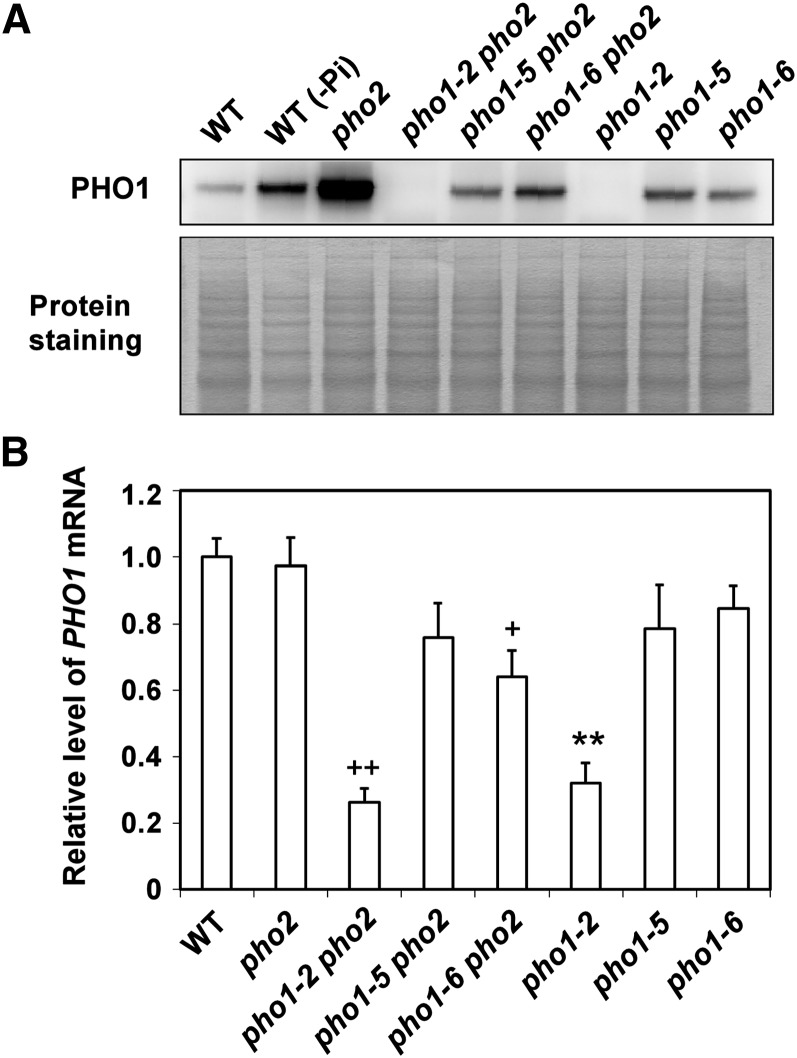
As one copy of the pho1-5 or pho1-6 allele is sufficient to reduce the shoot Pi accumulation of pho2 (Figure 3), we speculated that the reduced abundance of PHO1 variants in pho1-5 pho2 and pho1-6 pho2 would contribute to suppression of shoot Pi accumulation. Indeed, the level of PHO1 variants was remarkably decreased in both pho2 suppressors compared with the level of wild-type PHO1 in pho2 (Figure 7A). However, the transcript level of PHO1 was unchanged in pho1-5 pho2 but slightly decreased in pho1-6 pho2 (P < 0.05) compared with that of pho2 (Figure 7B), suggesting that the decreased protein level of PHO1A312T would have resulted, in part, from the reduced expression of transcript. To gain insight into the mechanism behind the reduced steady state level of PHO1 variants in the pho2 suppressors, we also examined the stability of PHO1E15K and PHO1A312T by application of CHX. We found that PHO1E15K is more susceptible to degradation than PHO1WT in the pho2 background, indicative of the PHO2-independent intrinsic instability of PHO1E15K (see Supplemental Figure 5 online). By contrast, the protein stability of PHO1A312T was not affected compared with that of PHO1WT in the pho2 background (see Supplemental Figure 5 online). To our surprise, the protein but not the transcript level of PHO1 variants was increased in the +Pi root of pho1-5 and pho1-6 compared with that of the wild type (Figures 7A and 7B), hinting at different factors involved in regulating the expression of PHO1E15K and PHO1A312T in the wild type and pho2.
Figure 7.
Suppression of PHO1 Expression in the pho2 Suppressors.
(A) Immunoblot analysis of PHO1 in the root of 14-d-old wild-type (WT), pho2, pho1-2 pho2, pho1-5 pho2, pho1-6 pho2, pho1-2, pho1-5, and pho1-6 seedlings under +Pi conditions. A root sample from the wild-type seedling subjected to 5 d of Pi deficiency (−Pi) was used as comparison.
(B) Quantitative RT-PCR analysis of PHO1 expression in the root under +Pi conditions. Error bars represent se (n = 4 to 6, pooled from three independent experiments). Data significantly different from the corresponding controls are indicated (mutant versus the wild type, **P < 0.01; mutant versus the pho2 mutant, +P < 0.05, ++P < 0.01; Student’s t test).
Ubiquitin Conjugase Activity of PHO2 Is Required for Degradation of PHO1
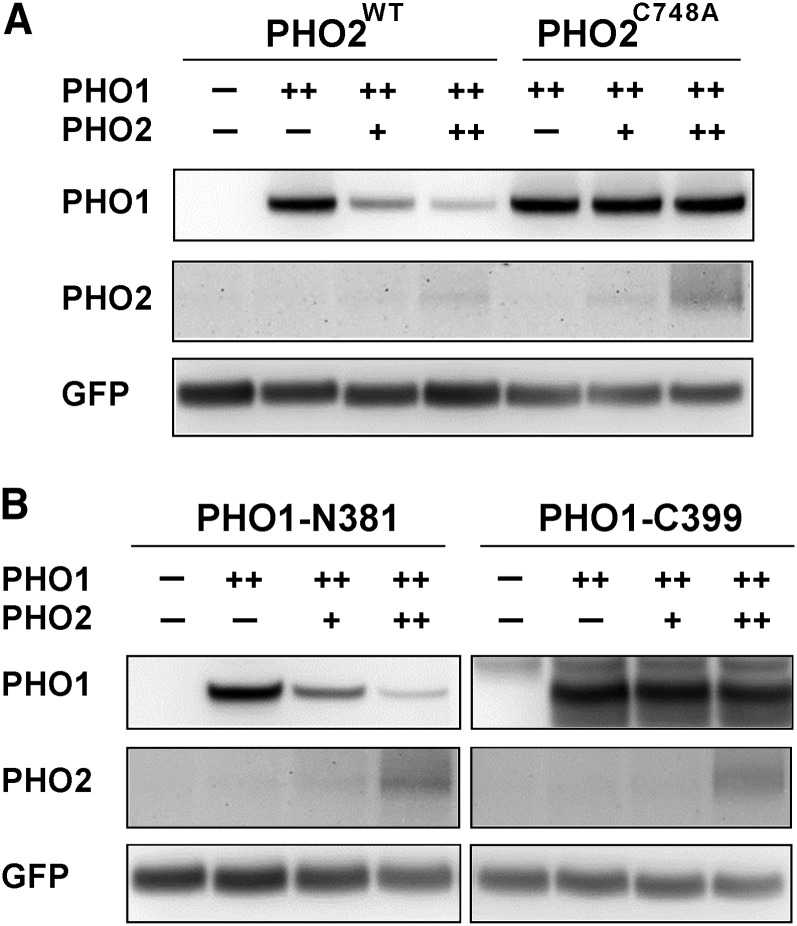
To determine whether PHO2 is an intrinsic UBC enzyme involved in PHO1 degradation, we generated a mutant construct in which the conserved catalytic active residue Cys-748 of PHO2 was replaced with Ala. As expected, the expression of wild-type PHO2, but not of the PHO2C748A mutant variant, driven by the 35S promoter, was able to rescue the phenotype of pho2 in terms of the reduction in shoot Pi concentrations (see Supplemental Figure 6A online). Accordingly, when PHO1 and wild-type PHO2 were transiently coexpressed in tobacco (Nicotiana tabacum) leaves, the expression of PHO1 was strikingly reduced in a PHO2 dosage-dependent manner (Figure 8A). By contrast, the level of PHO1 remained relatively constant when it was coexpressed with the PHO2C748A mutant variant (Figure 8A). The unstable nature of PHO1 when it was coexpressed with the catalytically active PHO2 suggests that PHO2 acts as a bona fide UBC enzyme sufficient for downregulation of PHO1 in planta.
Figure 8.
The N-Terminal Part of PHO1 Is Required for PHO2 Activity-Mediated Degradation of PHO1.
Immunoblot analyses of the expression of full-length (A) or truncated PHO1 variants (B) when coexpressed with the functional PHO2WT (35S:PHO2; in [A] and [B]) or catalytically defective PHO2C748A (35S:PHO2C748A; in [A]) in tobacco leaves. Coinfiltration of the 35S:GFP construct was used as an internal control. Note that ++ denotes a 10-fold increase in volume of infiltration relative to +. WT, the wild type.
To determine which part of PHO1 is subject to PHO2-dependent degradation, we further generated two truncated variants of PHO1 on the basis of the membrane topology predicted by the ConPred II algorithm (Arai et al., 2004). These variants consisted of the N-terminal cytosolic region of PHO1 (PHO1-N381) and the C-terminal transmembrane domain region of PHO1 (PHO1-C399). While there was a negative correlation between the expression level of PHO1-N381 and PHO2, the expression level of PHO1-C399 remained unchanged when it was coexpressed with increasing amounts of PHO2 (Figure 8B). These results demonstrated that the N-terminal part of PHO1 is required for the recognition and/or interaction by PHO2.
Subcellular Colocalization and Interaction of PHO1 and PHO2 in the EMs
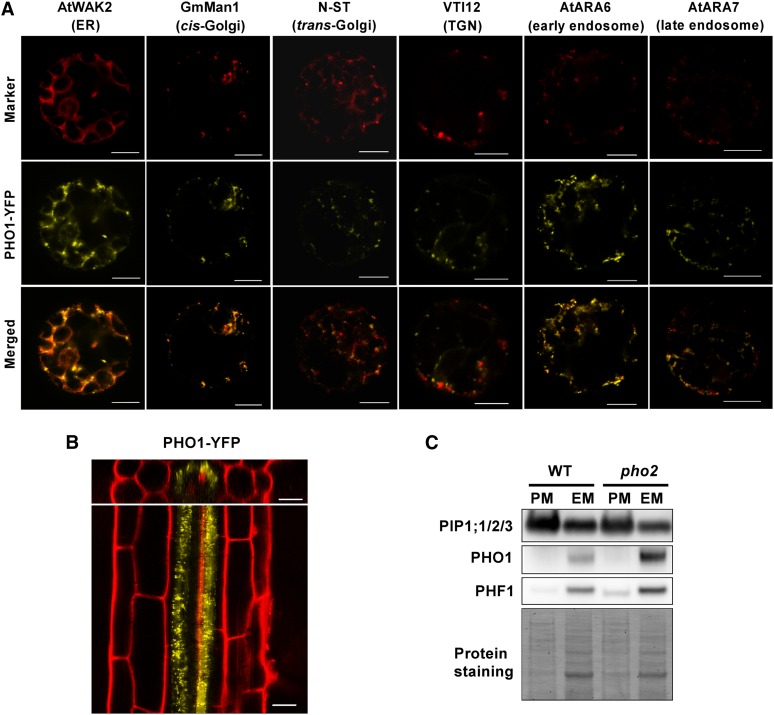
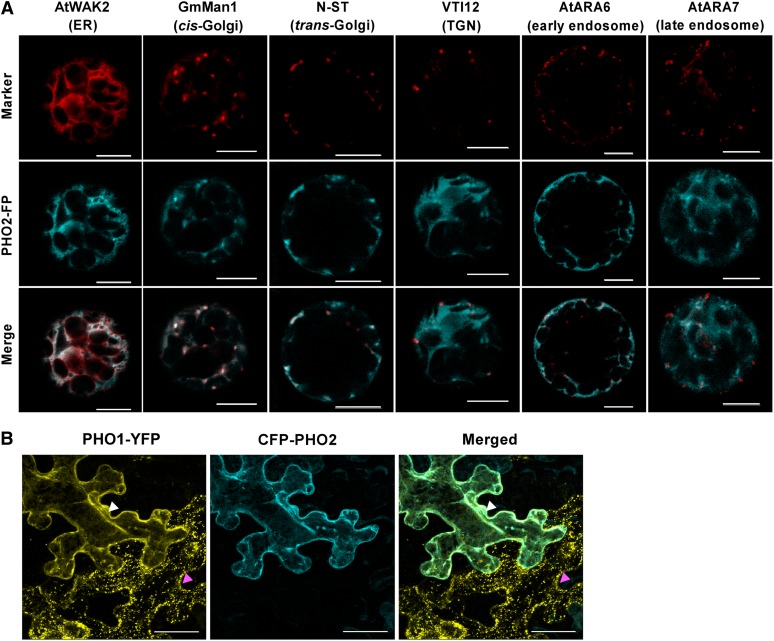
It has been shown that PHO1 and PHO2 are both expressed in the vascular tissue of the roots, hinting at their potential for direct interaction (Hamburger et al., 2002; Aung et al., 2006). We then examined the respective subcellular localizations of fluorescent protein–tagged PHO1 and PHO2. When transiently expressed in Arabidopsis protoplasts, PHO1-YFP (for yellow fluorescent protein) was detected in both reticular and punctate patterns (Figure 9A). The fluorescence signals were mainly colocalized with the ER and Golgi but were occasionally colocalized with the endosomal markers, suggesting that PHO1-YFP was widely distributed in the EMs (Figure 9A). Consistent with these observations, the functional PHO1-YFP under its native promoter, which rescued the pho1-2 phenotype (see Supplemental Figure 6B online), primarily displayed a dotted pattern in the root pericycle cells of the Arabidopsis transgenic lines (Figure 9B). Moreover, results from the membrane fractionation of the native PHO1 in both the wild type and pho2 reinforced that PHO1 resided predominantly in the EM rather than the PM (Figure 9C). By comparison, the functional C-terminal fluorescent protein fusion of PHO2 (see Supplemental Figure 7 online) colocalized mainly with the ER and Golgi but rarely with the endosomal markers (Figure 10A). Of note, in contrast with the punctate pattern of PHO1-YFP when expressed alone in the adjacent cell (Figure 10B, magenta arrowhead), coexpression of PHO1-YFP and cyan fluorescent protein (CFP)-PHO2 in tobacco leaves gave rise to ER-like signals in the EM (Figure 10B, white arrowhead), indicating that the subcellular distribution of PHO1 was altered in the presence of PHO2.
Figure 9.
Subcellular Localization of PHO1.
(A) Analyses of subcellular localization of PHO1-YFP (35S:gPHO1insYFP) using the ER, Golgi, trans-Golgi network (TGN), and endosomal markers in Arabidopsis protoplasts. Bars = 10 μm.
(B) Cellular and subcellular localization of PHO1-YFP (PHO1pro:gPHO1insYFP) in the Arabidopsis roots of a 5-d-old wild-type transgenic seedling under +Pi conditions. Bars = 20 µm.
(C) The majority of PHO1 is detected in the EM-enriched fraction. PIP1;1/2/3 and PHF1 are used as PM- and EM-localized markers, respectively. The bottom panel shows the protein staining on the membrane. WT, the wild type.
Figure 10.
Subcellular Localization of PHO2 and Colocalization of PHO1 and PHO2.
(A) Analyses of subcellular localization of PHO2 fluorescence protein (35S:PHO2-FP) using the ER, Golgi, trans-Golgi network (TGN), and endosomal markers in Arabidopsis protoplasts. PHO2-CFP proteins were used to analyze along with all of the subcellular markers, except that PHO2-RFP was employed in conjunction with the TGN marker. Bars = 10 µm.
(B) Colocalization of PHO1-YFP (35S:gPHO1insYFP) and CFP-PHO2 (35S:CFP-PHO2) in tobacco leaves. Note the punctate pattern of PHO1-YFP when expressed alone as indicated by the magenta arrowhead; the subcellular localization of PHO1 displayed an ER-like pattern when coexpressed with PHO2 as indicated by the white arrowhead. The displayed image is a maximum projected z-stack of 24 confocal slices (total z depth = 24 µm). Bars = 10 µm.
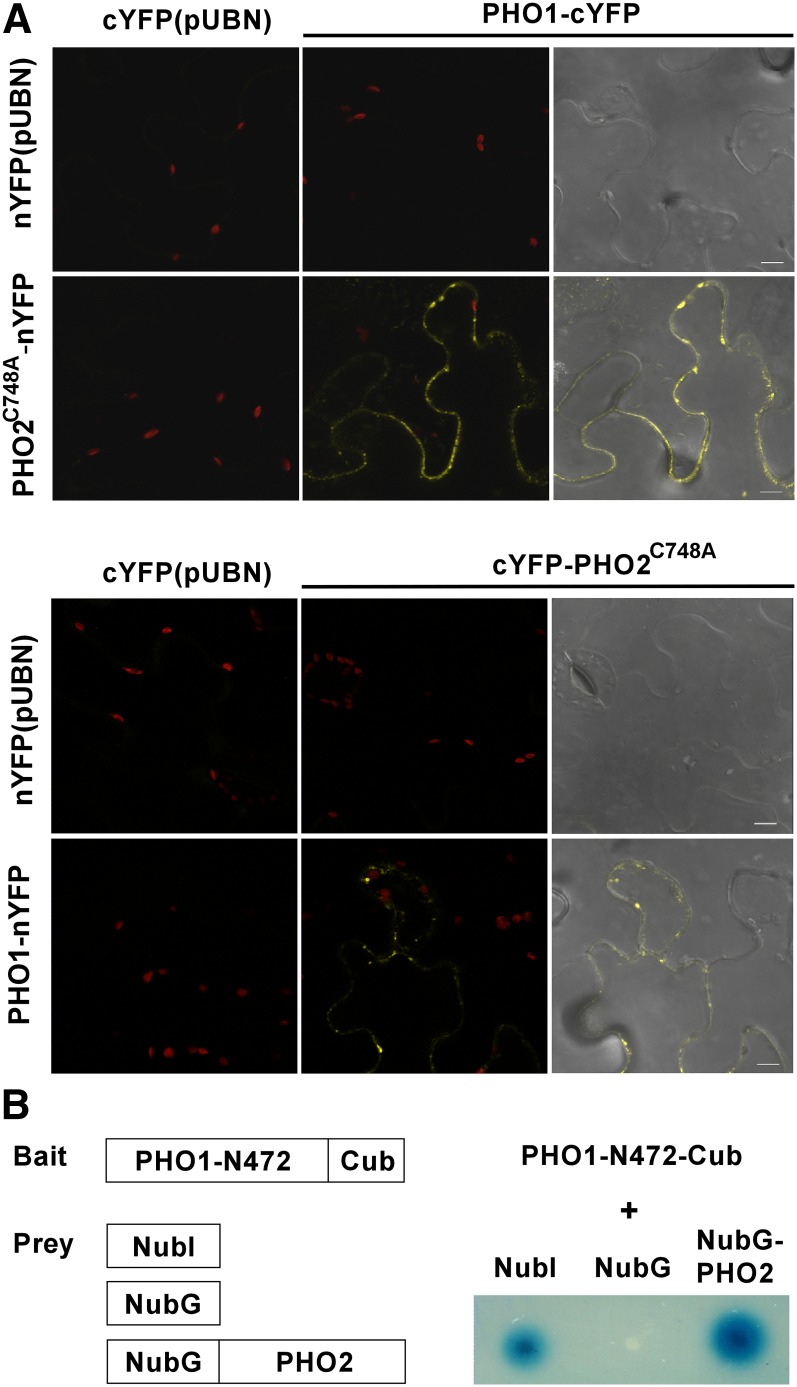
Next, we tested whether PHO2 directly interacts with PHO1 using bimolecular fluorescence complementation (BiFC) assays in tobacco leaves and split-ubiquitin yeast two-hybrid assays. In the BiFC assay, while the empty vector controls did not produce any detectable fluorescence signal, coexpression of PHO2C748A-nYFP and PHO1-cYFP or PHO1-nYFP and cYFP-PHO2C748A gave rise to intracellular signals (Figure 11A). Results from the yeast two-hybrid system further showed the interaction between the N-terminal portion of PHO1 and full-length PHO2 (Figure 11B), supporting our previous conclusion that the N-terminal region of PHO1 is required for PHO2-mediated degradation of PHO1 (Figure 8B). Taken together, these data suggest that PHO1 and PHO2 physically interact in the EM.
Figure 11.
Interaction of PHO1 and PHO2.
(A) Coexpression of PHO1-cYFP and PHO2C748A-nYFP or PHO1-nYFP and cYFP-PHO2C748A in tobacco leaves showed BiFC fluorescence in the endocompartment. Coexpression of nYFP or cYFP with the corresponding PHO1 or PHO2C748A constructs was used as negative controls. Bars = 10 μm.
(B) Interaction between the N-terminal part of PHO1 and full-length PHO2 in a split-ubiquitin yeast two-hybrid system. Coexpression of PHO1-N472 with NubI and NubG was used as positive and negative controls, respectively.
PHO2-Dependent Degradation of PHO1 Involves Multivesicular Body–Mediated Vacuolar Proteolysis
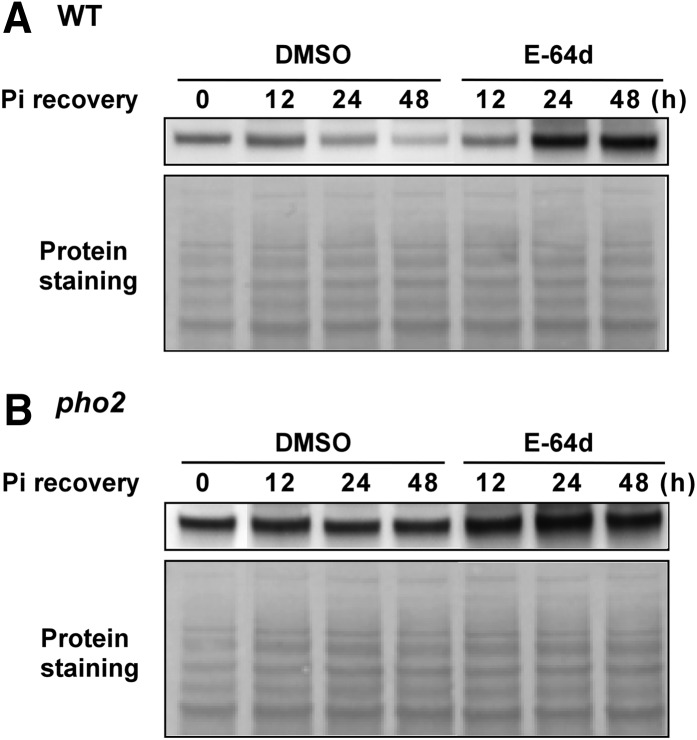
Turnover of membrane proteins relies on vacuolar proteolysis- or 26S proteasome–mediated degradation pathways. While mono- or diubiquitination has been reported to serve as signals for endocytosis of PM proteins and sorting of membrane proteins to the multivesicular bodies (MVBs), polyubiquitination has been mainly associated with the targeting of substrates to the 26S proteasome (Mukhopadhyay and Riezman, 2007). As PHO1 is a potential substrate for PHO2-dependent ubiquitin modification, we intended to determine which protein degradation pathway is involved in the PHO2-mediated downregulation of PHO1. It has been shown that the endosomal protease inhibitor E-64d can prevent degradation of the Arabidopsis root PM proteins by attenuating the final step of the endocytic pathway (i.e., the fusion of the late endosomes/MVB with vacuoles) (Yamada et al., 2005). Interestingly, the E-64d (50 µM) treatment of the wild type alleviated the downregulation of PHO1 upon Pi resupply and resulted in an increase in the expression of PHO1 after 24 h of Pi recovery, whereas the expression level of PHO1 in the roots of E-64d–treated pho2 was only slightly affected (Figures 12A and 12B). In accordance with these results, accumulation of PHO1 under +Pi conditions was increased in the 24-h E-64d–treated wild type in a dosage-dependent manner but not in pho2 (see Supplemental Figure 8A online). By contrast, the expression of PHO1 in wild-type seedlings treated with the proteasome inhibitor MG132 (50 µM) was gradually decreased in a Pi recovery time-dependent manner, as the DMSO controls (see Supplemental Figure 8B online), suggesting that downregulation of PHO1 was not blocked by inhibition of 26S proteasome activity. Collectively, our data support the hypothesis that PHO2 is required for MVB-mediated vacuolar degradation of PHO1 when Pi is adequate.
Figure 12.
Inhibition of PHO1 Degradation by Blocking MVB-to-Vacuole Trafficking.
The expression level of PHO1 in the root of 15-d-old wild-type (WT) (A) and pho2 (B) seedlings over 48 h of Pi recovery (250 µM KH2PO4) and E-64d treatment (50 µM) following 6 d of Pi deficiency. The bottom panel shows the protein staining on the membrane.
Overexpression of PHO1 Is Not Sufficient to Reproduce the Pi Toxicity Phenotype of pho2
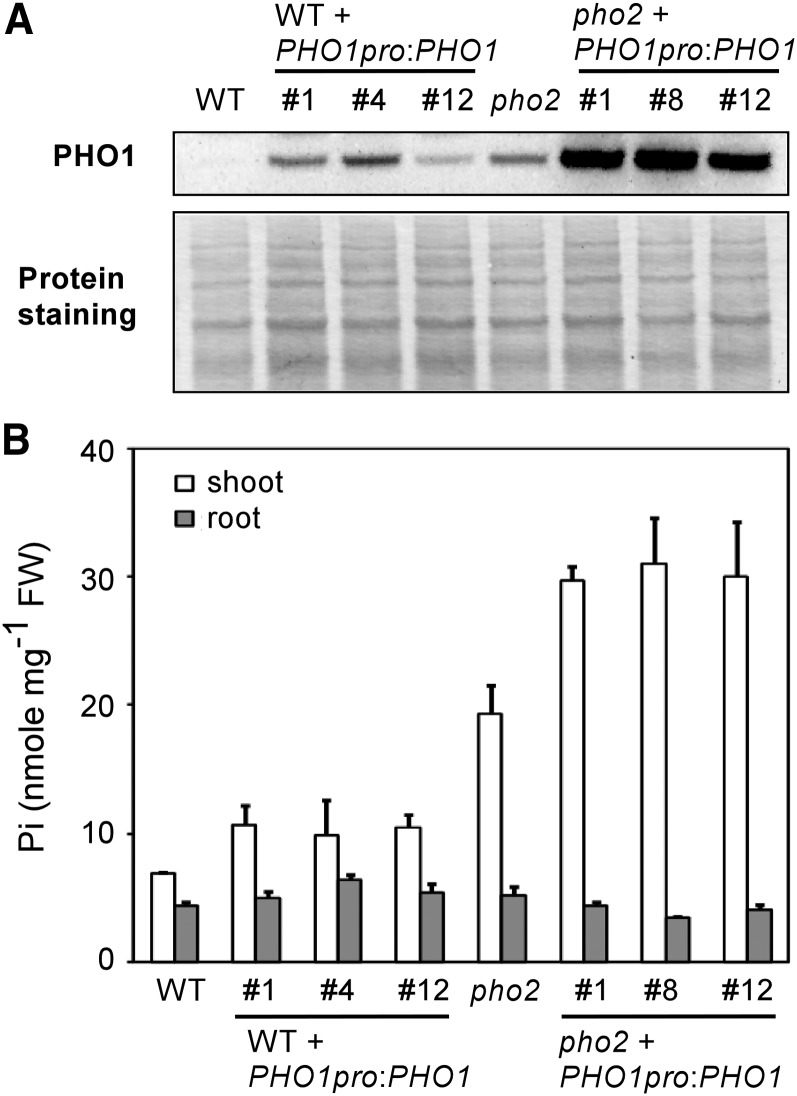
As the genomic PHO1 construct we used for complementation of pho1-5 pho2 and pho1-6 pho2 unexpectedly resulted in tissue-specific overexpression of PHO1 (see Supplemental Figure 3 online), we attempted to bring this construct into the wild type and pho2 background. Interestingly, although the abundance of PHO1 was elevated to the same extent in the PHO1-overexpressing lines as in pho2 (Figure 13A), the shoot Pi concentrations were increased by 40 to 55% at most compared with the wild-type control and did not reach the levels seen in pho2 (Figure 13B). By comparison, overexpression of PHO1 in the pho2 background aggravated Pi toxicity (Figures 13A and 13B), often resulting in the failure of seed development under +Pi conditions.
Figure 13.
Analysis of PHO1-Overexpressing Lines.
Immunoblot analysis of PHO1 in the root (A) and the shoot and root Pi concentrations (B) of 17-d-old wild-type (WT), pho2, and PHO1-overexpressing lines in the wild type or pho2 background under +Pi conditions. Note that the intensity of PHO1 signal in the wild type was minimized to avoid oversaturation of PHO1 detection in PHO1-overexpressing pho2 lines. Three independent transgenic lines in each genotype are shown. The bottom panel shows protein staining on the membrane. FW, fresh weight.
DISCUSSION
PHO2 Modulates PHO1 Expression at the Posttranslational Level
To cope with Pi limitation, plants have developed a series of adaptive mechanisms involving transcriptional regulation of Pi starvation-responsive genes. Among them, the expression of PHO1 is induced by Pi limitation, in part through the transcription factor WRKY6 (Chen et al., 2009). When Pi supply is adequate, the binding of WRKY6 to the promoter of PHO1 inhibits the transcription of PHO1; however, Pi limitation–induced downregulation of WRKY6, likely by 26S proteosome–mediated proteolysis, removes such transcriptional repression (Chen et al., 2009). In this study, we unveiled a layer of PHO1 regulation at the posttranslational level by PHO2. We found that PHO1 levels declined rapidly under +Pi conditions when the de novo protein synthesis was blocked by CHX and nearly disappeared within 1 h of treatment (Figures 6A and 6B). This suggested that PHO1 is subject to constitutive degradation when Pi is adequate. Remarkably, we showed that such degradation of PHO1 under +Pi conditions is circumvented in pho2, indicating that PHO2 mediates the posttranslational downregulation of PHO1. This is in agreement with the observation that downregulation of PHO1 is relieved under −Pi conditions (Figure 5A), during which PHO2 expression is suppressed by upregulation of miR399 (Aung et al., 2006; Bari et al., 2006; Chiou et al., 2006). In addition, it is noteworthy that the increased accumulation of PHO1 in pho2 was exacerbated by Pi deficiency (Figure 5A). We surmised that this was because the increased PHO1 protein resulting from the low Pi-induced transcriptional upregulation of PHO1 failed to undergo PHO2-mediated degradation.
In this study, using a genetic approach, we identified PHO1 as one of the downstream components of the PHO2-dependent Pi homeostasis regulatory pathway. While PHO1 accumulated in pho2, it was reduced in pho1-5 pho2 and pho1-6 pho2. Intriguingly, we found that both the missense mutations pho1-5 and pho1-6 are located within the N-terminal SPX domain of PHO1 (Figure 2B). Although the mechanism behind the reduced steady state level of these two PHO1 variants in pho2 remains to be studied, our current data revealed that PHO1-5 is regulated in a similar fashion as native PHO1 at the transcript level (Figure 7B). Furthermore, we showed that the PHO2-independent intrinsic stability of the PHO1 variants was reduced in pho1-5 pho2 but was relatively unaffected in pho1-6 pho2 compared with that of the native PHO1 in pho2 (see Supplemental Figure 5 online). Nevertheless, we cannot rule out that these PHO1 mutations may alter other inherent properties of PHO1 or unidentified aspects of PHO1 function, such as protein–protein interaction or its mode of action.
PHO2-Dependent Turnover of PHO1 in the EMs
By transient expression assays with Agrobacterium tumefaciens infiltration of tobacco leaves, we demonstrated that PHO1 degradation is genuinely mediated by PHO2 in a dosage-dependent manner and that both the full-length PHO1 and its N-terminal region are susceptible to degradation when coexpressed with catalytically active PHO2 (Figures 8A and 8B). It was previously reported that PHO2 has high sequence similarity with the human giant protein BRUCE/APOLLON, which harbors the ability to receive ubiquitin from ubiquitin-activating enzyme E1 enzyme as well as to bind a substrate directly (Bartke et al., 2004; Hao et al., 2004; Kraft et al., 2005). Since PHO2 directly interacts with PHO1 and the expression of functional PHO2 alone without additional supply of ubiquitin ligase E3 enzyme is sufficient for PHO1 degradation in tobacco leaves, we suspected that PHO2 could be a chimeric E2-E3 enzyme.
When the subcellular localization of PHO1 fusion protein was examined in the Arabidopsis protoplast, the fluorescence signals revealed prominent distributions in the EM (Figure 9A). As native PHO1 was also enriched in the EM fraction (Figure 9C), we inferred that the primary location of PHO1 is in the ER and Golgi. These observations were in agreement with the results from the recent publication by Arpat et al. (2012), showing that PHO1 is associated with punctate structures in the EM, except that the localization of PHO1 at the ER was only observed in our experiments. The discrepancy may have resulted from the different expression system used.
By comparison, despite lacking a membrane-spanning domain, PHO2 is membrane associated and mostly colocalized with the ER and Golgi markers (Figure 10A). Although whether PHO2 functions as a chimeric E2-E3 enzyme for PHO1 ubiquitination remains to be confirmed by an in vitro ubiquitination assay, our BiFC results suggested that direct interaction of PHO1 and PHO2 takes place in the EM (Figure 11A). This prompted the question of where and how PHO1 is selectively targeted for ubiquitination and degradation. Ubiquitin modification of transmembrane proteins has been shown to regulate several distinct intracellular processes, including ER-associated degradation, sorting in the trans-Golgi, endocytosis from the PM, and entry into the MVB (Bonifacino and Weissman, 1998; Komander, 2009). The ER, where both PHO1 and PHO2 localize, has the capacity to monitor protein activity to prevent nonfunctional proteins from proceeding to the Golgi. Such surveillance mechanisms within the ER are linked to the 26S proteasome proteolytic pathway, by which not only aberrant but also unneeded normal proteins are degraded (Vembar and Brodsky, 2008). Despite PHO2 being a UBC enzyme associated with the ER, we thought that PHO1 is unlikely to be degraded via the ER-associated degradation pathway, given that MG132 treatment was not effective in preventing the gradual degradation of PHO1 following Pi recovery (see Supplemental Figure 8B online).
In plants, endocytosis of several PM proteins for recycling or vacuolar degradation is a critical regulatory mechanism modulating their numbers/activities at the cell surface (Takano et al., 2005; Geldner et al., 2007; Laxmi et al., 2008; Barberon et al., 2011). As our current data demonstrated that PHO1 is not mainly destined for PM localization and that PHO2-dependent PHO1 degradation requires the MVB-mediated vacuolar degradation pathway as revealed by the E-64d treatment (Figures 12A and 12B), we inferred that the population of PHO1 subject to vacuolar proteolysis is derived from the EM rather than the PM pools. Interestingly, it was shown that treatment of barley (Hordeum vulgare) and Arabidopsis root tips with E-64d led to the accumulation of undegraded cytoplasmic components in the lytic compartments for autophagy (Moriyasu et al., 2003; Inoue et al., 2006). Since autophagy plays a pivotal role in responses to nutrient deprivation, it is counterintuitive to consider its involvement in degradation of PHO1 when Pi is adequate. Nevertheless, it is possible that E-64d disrupts the function of common molecular machinery that regulates the vacuolar fusion events shared by the MVB and autophagy pathways (Fader and Colombo, 2009).
In yeast, selective rerouting of the secretory trafficking is a strategy for destruction of the newly synthesized but unneeded proteins along the secretory pathway. Ubiquitination of the newly synthesized amino acid permease Gap1 at the Golgi in response to surplus nitrogen is required for its sorting to the vacuole without passing through the cell surface (Risinger and Kaiser, 2008). Similarly, ubiquitination serves as an early determinant for the delivery of the manganese transporter Smf1p along the biosynthetic route to the MVB and the vacuolar lumen (Eguez et al., 2004). We therefore postulate that the newly synthesized and ubiquitinated PHO1 at the ER or Golgi may be redirected en route to the vacuolar lumen for degradation in response to sufficient Pi availability.
Mode of Action of PHO1 in Mediating Pi Transfer into the Root Xylem
In Arabidopsis, several genes primarily expressed in the root pericycle and/or xylem parenchyma cells have been implicated in mediating ion influx or efflux that directs nutrient transport into the xylem. For instance, the low-affinity sulfate transporters SULTR2;1 and SULTR3;5 have been shown to participate in sulfate influx (Takahashi et al., 2000; Kataoka et al., 2004), whereas the SKOR1 channel and BOR1 transporter are involved in potassium and boron efflux, respectively (Gaymard et al., 1998; Takano et al., 2002). In addition, the low-affinity nitrate transporter NRT1.5 involved in xylem loading was reported to be a bidirectional solute transporter (Lin et al., 2008a). It has long been suspected that PHO1 is involved either in Pi transport into the pericycle and/or xylem parenchyma cells or in the release of Pi into the apoplastic space of vascular cylinder, including xylem vessels (Hamburger et al., 2002). In support of these assumptions, we showed here that PHO1-YFP under its native promoter was specifically localized to the root pericycle cells, despite its primary subcellular localization in the endocompartment (Figure 9B). Of note, an early report inferred that the transport of ions, such as potassium and chloride, into the xylem is mediated by a protein that is turned over relatively rapidly (Pitman et al., 1977). This seemed to fit our finding that PHO1 is a short-lived protein that undergoes Pi-dependent degradation and is implicated in xylem loading.
It was previously demonstrated that overexpression of PHO1 in leaves using the native promoter led to an increase in export of Pi into the xylem vessel and consequently out of the leaves, revealing a role for PHO1 in mediating Pi efflux (Stefanovic et al., 2011). In contrast with the drastic effect of PHO1 accumulation in leaves, overexpression of PHO1 in roots was only associated with a minor change in the rate of root-to-shoot Pi translocation. In this regard, Stefanovic et al. (2011) surmised that other factors limiting Pi efflux activity, such as the rate of Pi transfer across the endodermis, or mechanisms of posttranslational control of PHO1 activity present in roots but absent in shoots may explain the different effects of PHO1 when expressed in these two different tissues. The latter speculation is in agreement with the observations of predominant expression of PHO2 in the root (Chiou et al., 2006) and PHO2-dependent degradation of PHO1 shown here.
More recently, Arpat et al. (2012) showed that Pi was released to the extracellular medium when PHO1 was ectopically and transiently expressed in leaves and roots. Based on the subcellular localization of PHO1, we thought that its mode of action in mediating Pi transport seems to be different from the aforementioned PM-localized nutrient efflux transporters. The Arabidopsis Golgi-localized putative manganese transporter MTP11 was proposed to confer manganese tolerance by sequestering the metal into secretory vesicles that subsequently underwent exocytosis (Peiter et al., 2007). As the majority of PHO1 is detected in the EM fractions and rarely localizes to the PM, it would be interesting to explore whether a similar strategy is used to unload Pi into the apoplastic space through the exocytic vesicles. Nevertheless, Arpat et al. (2012) showed that PHO1 was able to partially relocalize to the PM of tobacco leaves infiltrated with a high concentration of Pi and thus interpreted PHO1 itself as a Pi efflux transporter. Although our conclusion regarding PHO2-dependent degradation of PHO1 in response to high Pi seems to contradict this observation, we cannot exclude the possibility that PHO1, if it indeed reaches the PM, may rapidly recycle, similar to the kiss-and-run mechanism seen in mammalian cells in which the transient fusion of a vesicle releases its interior contents (Schneider, 2001).
Additional Factors Involved in PHO2-Dependent Pi Homeostasis
The phenotype of Pi toxicity displayed by pho2 can be attributed to the misregulation of Pi homeostasis at three different levels: (1) enhanced Pi uptake, (2) increased root-to-shoot translocation of Pi, and (3) retention of Pi in older leaves (Aung et al., 2006). Our discovery of PHO1 as the downstream target of the PHO2-dependent Pi homeostasis regulatory pathway affirmed this notion. An early report showed that the Pi uptake rate of pho1-2 was similar to that of the wild type over a wide range of external Pi concentrations (Poirier et al., 1991). However, under our growth conditions, we found that defective xylem loading of Pi could affect the step of Pi acquisition. Regardless of the genetic background (the wild type or pho2), pho1-2, pho1-5, and pho1-6 mutations all led to a reduction in Pi uptake (Figure 4). In fact, a few early studies pointed out that ion influx appeared to be regulated by ion efflux to the xylem, as stimulating the transport of labeled ions to the shoot was accompanied by a greater ion influx to the root cells (Drew and Saker, 1978, 1984).
Interestingly, in our genomic PHO1-overexpressing lines in the wild-type background, the level of PHO1 in roots was comparable to or slightly greater than that detected in pho2, but the shoot Pi concentrations were only moderately increased (Figures 13A and 13B). By comparison, when PHO1 was overexpressed in pho2, the extent of Pi accumulation in shoots was further increased (Figures 13A and 13B). In fact, most of the PHO1-overexpressing pho2 lines displayed severe phenotypes of Pi toxicity and failed to set seeds during the reproductive stage. This means that mere overexpression of PHO1 in the wild-type background was not sufficient to give rise to the phenotype of pho2. No matter whether PHO1 is the bona fide root-to-shoot Pi transporter or only acts a cofactor that activates or facilitates transport of Pi, we envisage that additional factors regulated by PHO2, that either modulate the activity of PHO1 or limit the rate of Pi transfer across the endodermis, must be involved to account for the Pi toxicity of pho2.
METHODS
Plant Material and Growth Conditions
Seeds of the Arabidopsis thaliana wild type (Columbia-0), pho1-2 (Poirier et al., 1991), and pho2 (Delhaize and Randall, 1995) mutants used in this study were obtained from the ABRC. The s1 and s80 mutants were backcrossed to pho2 one time to clean genetic backgrounds after mutagenesis. Seeds were surface sterilized and germinated on agar plates with one-half modified Hoagland nutrient solution containing 250 μM KH2PO4, 1% Suc, and 1% Bactoagar (Chiou et al., 2006). The Pi-sufficient (+Pi) and Pi-deficient (−Pi) media were supplemented with 250 μM and 10 µM KH2PO4, respectively, unless specified otherwise. For hydroponic growth, the medium was prepared without supplement of Suc.
Constructs for Expression in Plants and Plant Transformation
All the insert fragments of interest were amplified and cloned into pCR8/GW/TOPO (Invitrogen) for sequencing and then recombined into the desired Gateway destination vectors via LR Clonase enzyme mix (Invitrogen). Site-directed mutagenesis of the catalytic active residue of PHO2 was performed according to the instructions of the Phusion site-directed mutagenesis kit (Thermo). All primers used are listed in Supplemental Table 1 online.
For unknown reasons, cloning of the full-length cDNA of PHO1 in Escherichia coli was unsuccessful. Therefore, all the constructs for expression of the full-length PHO1 protein were obtained by cloning the genomic sequences. For Arabidopsis complementation and overexpression experiments, a 10-kb genomic fragment of DNA containing the coding region of PHO1 was cloned and recombined into the Gateway destination vector pMDC100 (Curtis and Grossniklaus, 2003), designated as PHO1pro:PHO1.
For Arabidopsis PHO1pro:gPHO1insYFP transgenic lines, an 8-kb genomic fragment encoding the PHO1 coding region, spanning from 2.1 kb upstream of the start codon to 0.5 kb downstream of the stop codon, was cloned. The C-terminal YFP fusion of PHO1 was constructed with YFP inserted at nine amino acids from the C terminus of PHO1. pKGW was used as the Gateway destination vector (Karimi et al., 2002).
For transient expression of PHO1-YFP (35S:gPHO1insYFP) in Arabidopsis protoplasts and Nicotiana benthamiana tobacco leaves, the genomic PHO1 sequence with the C-terminal YFP fusion was amplified using PHO1pro:gPHO1insYFP as template and introduced into pMDC32 Gateway destination vector (Curtis and Grossniklaus, 2003). For transient expression of PHO2-CFP (35S:PHO2-CFP) in Arabidopsis protoplasts, the expression constructs were generated using p2CWG7 as the Gateway destination vectors (Karimi et al., 2005).
For the PHO1 degradation assay in tobacco leaves, the PHO1 genomic construct containing the coding region of PHO1 (35S:gPHO1), the N- and C-terminal truncated PHO1 hemagglutinin-tagged constructs expressing the partial coding sequences of PHO1-N381 (35S:PHO1N318-HA) and PHO1-C399 (35S:PHO1C399-HA), and the full-length PHO2 coding sequence constructs expressing the wild-type PHO2 (35S:PHO2) and the mutant variant PHO2C748A (35S:PHO2C748A), were generated using pMDC32 as the Gateway destination vector (Curtis and Grossniklaus, 2003).
For colocalization of PHO1-YFP (35S:gPHO1insYFP) and CFP-PHO2 in tobacco leaves, the CFP-PHO2 constructs (35S:CFP-PHO2) were generated using pK7WGC2 as the Gateway destination vector (Karimi et al., 2005).
For BiFC analysis, the full-length PHO2 coding sequence encoding the mutant variant PHO2C748A was recombined into pUBN-cYFP (UBQ10:cYFP-PHO2C748A) or pUBC-nYFP (UBQ10:PHO2C748A-nYFP). The genomic PHO1 sequence was recombined into pUBC-nYFP (UBQ10:gPHO1-nYFP) or pUBC-cYFP (UBQ10:gPHO1-cYFP) (Grefen et al., 2010).
Yeast Two-Hybrid Assay and Constructs
Split-ubiquitin yeast two-hybrid assay was performed according to the instructions provided with the DUALmembrane kit (Dualsystems Biotech). The N-terminal 1416-bp fragment of the PHO1 coding sequence corresponding to the first N-terminal 472 amino acids encompassing one putative transmembrane domain was cloned into the XbaI and StuI sites of the bait vector pAMBV4. The full-length PHO2 coding sequence was cloned into the BamHI and SmaI sites of the prey vector pDL-Nx. Yeast strain DSY-1 cells were cotransformed with the resulting constructs and plated onto synthetic medium lacking Leu, Trp, and His. The specificity of protein–protein interactions was confirmed by chloroform overlay β-galactosidase plate assay as described by Duttweiler (1996).
Transient Expression in Mesophyll Protoplasts of Arabidopsis
Transformation of Arabidopsis mesophyll protoplasts for transient expression of fluorescence fusion proteins was performed as described previously (Wu et al., 2009). For subcellular colocalization experiments, the fluorescent markers N-ST-RFP (for red fluorescent protein) and RFP-ARA6 used have been previously reported (Grebe et al., 2003). ARA7-RFP and VTI12-TFP (for teal fluorescent protein) constructs were derived from WAVE lines (Geldner et al., 2009) obtained from the Nottingham Arabidopsis Stock Centre. AtWAK2-mCherry and GmMAN1-mCherry constructs were derived from the multicolored organelle marker set (Nelson et al., 2007) obtained from the ABRC.
Agrobacterium tumefaciens–Mediated Infiltration of Tobacco Leaves
The Agrobacterium-mediated transient expression in tobacco leaves was conducted as described at http://www.plantsci.cam.ac.uk/research/baulcombe/induction.html, with minor modifications. A preculture of the Agrobacterium EHA105 strain harboring the constructs of genes of interest or p19 (Voinnet et al., 2003) was prepared in Luria-Bertani medium with the proper antibiotics and incubated overnight with shaking at 28°C. A 1-mL aliquot of preculture was used to inoculate 50 mL of Luria-Bertani medium with the appropriate antibiotics, 10 mM MES, and 20 µM acetosyringone, and the bacteria were allowed to grow overnight. After centrifugation at 5000 rpm for 10 min at 4°C, the cell pellet was resuspended in the infiltration medium (10 mM MgCl2, 10 mM MES, and 100 µM acetosyringone) to an OD600 of 1.0. The cell suspension was then left standing at room temperature for 2 to 3 h before infiltration of N. benthamiana tobacco leaves. A mix of cells containing the genes of interest and p19 was then prepared to infiltrate the second or third true leaves of 3-week-old tobacco plants. Infiltrated tobacco plants were grown for another 3 d before sample collection.
Phosphate Concentration and Phosphate Uptake Analysis
Pi concentration and uptake activity were determined as described (Chiou et al., 2006). To assay Pi uptake, the seedlings grown under +Pi conditions (i.e., 250 μM KH2PO4) were transferred to medium containing 250 μM [33P] KH2PO4 for the measurement of Pi uptake.
RNA Isolation and Quantitative RT-PCR
Total RNA from samples was isolated using TRIzol reagent (Invitrogen) followed by treatment with DNase I (Ambion) before quantitative RT-PCR to eliminate genomic DNA contamination. cDNA was synthesized from 0.5 to 1 μg total RNA using Moloney murine leukemia virus reverse transcriptase (Invitrogen) with oligo(dT) primer. Quantitative RT-PCR was performed using the Power SYBR Green PCR Master Mix kit (Applied Biosystems) on a 7300 Real-Time PCR system (Applied Biosystems) according to the manufacturer’s instructions. Relative expression levels were normalized to that of an internal control, UBQ10.
Immunoblot Analysis
For extraction of total root protein, the roots of wild-type and mutant seedlings were ground in liquid nitrogen and dissolved in protein lysis buffer containing 2% SDS, 60 mM Tris-HCl, pH 8.5, 2.5% glycerol, 0.13 mM EDTA, and 1× complete protease inhibitor (Roche). Total root protein (50 to 80 µg) of each sample was loaded onto 4 to 12% Bis-Tris SDS-PAGE gels (NuPAGE System) and transferred to polyvinylidene difluoride membranes. The membrane was blocked with 1% BSA in 1× PBS solution with 0.2% Tween 20 (PBST), pH 7.2) at room temperature for more than 30 min and hybridized with primary antibody in 1× PBST for 1 to 2 h. The membrane was washed three times with 1× PBST for 5 min followed by hybridization with the horseradish peroxidase–conjugated secondary antibody in 1× PBST for 1 h. After three washes in 1× PBST for 5 min and a rinse with distilled water, chemiluminescent substrates for signal detection were applied. Polyclonal rabbit PHO1 antibody was raised against an N-terminal fragment of PHO1 corresponding to the amino acid residues 28 to 46 (KQIKKIKTSRKPKPASHYP) and affinity purified. Polyclonal rabbit antibody against PIP1;1/2/3 was purchased from Agrisera. The protein transferred onto membranes was visualized with naphthol blue black staining solution (0.1% naphthol blue black, 10% methanol, and 2% acetic acid).
Fluorescence Microscopy
Confocal microscopy images were taken using a Zeiss LSM 510 META NLO DuoScan with objectives LCI Plan-Neofluar ×63/1.3 Imm and Plan-Apochromat ×100/1.4 oil. Excitation/emission wavelengths were 458 nm/465 to 510 nm for CFP and TFP, 514 nm/520 to 550 nm for YFP, 514 nm/630 to 700 nm for propidium iodide, and 561 nm/575 to 630 nm for RFP and mCherry. The cell wall was stained with propidium iodide (1 µg/mL).
Chemical Treatments
CHX (200 mM; Sigma-Aldrich), MG132 (50 mM; Calbiochem), and E-64d (50 mM; Cayman) stock solutions were prepared in DMSO. The final working concentration used in liquid +Pi medium was specified according to the experiment. For control experiments, a 0.1% DMSO solution was used.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: PHO1, At3g23430; and PHO2, At2g33770.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Pi Concentrations of 20-d-Old pho2 Suppressors.
Supplemental Figure 2. Segregation Analysis of F2 Progeny Resulting from Genetic Crosses between s1 and pho2.
Supplemental Figure 3. The Shoot Pi Concentrations of PHO1-Complemented pho2 Suppressors.
Supplemental Figure 4. The Shoot Pi Concentrations of Heterozygous pho1-2/+ Relative to Wild-Type and pho1-2 Plants.
Supplemental Figure 5. Protein Stability of PHO1 Variants.
Supplemental Figure 6. Complementation Tests of pho2 and pho1-2 Mutants by Mutated PHO2 and PHO1-YFP, Respectively.
Supplemental Figure 7. Complementation of pho2 by Fluorescent Protein–Tagged PHO2.
Supplemental Figure 8. The Effect of E-64d and MG132 on the Expression of PHO1.
Supplemental Table 1. Oligonucleotides Used for Plasmid Constructs and Quantitative RT-PCR Analysis.
Supplementary Material
Acknowledgments
This work was supported by Academia Sinica, Taiwan (Grant 98-CDA-L11 and postdoctoral fellowship to T.-Y.L.) and by the National Science Council of the Republic of China (Grant NSC100-2321-B-001-005). We thank the Plant Tech Core Facility at the Agricultural Biotechnology Research Center of Academia Sinica for assistance in protoplast transformations and Shu-Chen Shen from the Scientific Instrument Center of Academia Sinica for assistance in the confocal microscopy analysis. We thank Yu-Ying Hsu for analyzing the F1 progeny of crosses between the wild type and pho1-2, Ching-Mei Sun for assistance with the PHO2 variant complementation experiment, Ya-Ni Chen for generating the PHO1-YFP construct, Tzu-Yun Chang for testing PHO1 antibody, and AndreAna Peña for English editing.
AUTHOR CONTRIBUTIONS
T.-Y.L., T.-K.H., and T.-J.C. designed and analyzed the experiments. T.-Y.L., T.-K.H., C.-Y.T., Y.-S.L., S.-I.L., W.-Y.L., and J.-W.C. performed the experiments. T.-Y.L. and T.-J.C. wrote the article.
Glossary
- Pi
inorganic phosphate
- PM
plasma membrane
- ER
endoplasmic reticulum
- UBC
ubiquitin-conjugating
- CHX
cycloheximide
- EM
endomembrane
- YFP
yellow fluorescent protein
- BiFC
bimolecular fluorescence complementation
- MVB
multivesicular body
- PBST
PBS solution with 0.2% Tween 20
- CFP
cyan fluorescent protein
- RFP
red fluorescent protein
References
- Arai M., Mitsuke H., Ikeda M., Xia J.-X., Kikuchi T., Satake M., Shimizu T. (2004). ConPred II: a consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Res. 32 (Web Server issue): W390–W393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpat A., Magliano P., Wege S., Rouached H., Stefanovic A., Poirier Y. (March 26, 2012). Functional expression of PHO1 to the Golgi and trans-Golgi network and its role in Pi export. Plant J. http://dx.doi.org/10.1111/j.1365-313X.2012.05004.x. [DOI] [PubMed] [Google Scholar]
- Aung K., Lin S.I., Wu C.C., Huang Y.T., Su C.L., Chiou T.J. (2006). pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 141: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon M., Zelazny E., Robert S., Conéjéro G., Curie C., Friml J., Vert G. (2011). Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. USA 108: E450–E458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R., Datt Pant B., Stitt M., Scheible W.R. (2006). PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke T., Pohl C., Pyrowolakis G., Jentsch S. (2004). Dual role of BRUCE as an antiapoptotic IAP and a chimeric E2/E3 ubiquitin ligase. Mol. Cell 14: 801–811 [DOI] [PubMed] [Google Scholar]
- Bayle V., Arrighi J.-F., Creff A., Nespoulous C., Vialaret J., Rossignol M., Gonzalez E., Paz-Ares J., Nussaume L. (2011). Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 23: 1523–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J.S., Weissman A.M. (1998). Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu. Rev. Cell Dev. Biol. 14: 19–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Liu Y., Ni J., Wang Y., Bai Y., Shi J.Gan J., Wu Z., Wu P. (2011). OsPHF1 regulates the plasma membrane localization of low- and high-affinity Pi transporters and determines Pi uptake and translocation in rice. Plant Physiol. 157: 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-F., Li L.-Q., Xu Q., Kong Y.-H., Wang H., Wu W.-H. (2009). The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell 21: 3554–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T.J., Aung K., Lin S.I., Wu C.C., Chiang S.F., Su C.L. (2006). Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18: 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T.-J., Lin S.-I. (2011). Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62: 185–206 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E., Randall P.J. (1995). Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol. 107: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B., Rengel Z., Delhaize E. (1998). Uptake and translocation of phosphate by pho2 mutant and wild-type seedlings of Arabidopsis thaliana. Planta 205: 251–256 [DOI] [PubMed] [Google Scholar]
- Drew M.C., Saker L.R. (1978). Nutrient supply and the growth of the seminal root system in barley. J. Exp. Bot. 29: 435–451 [Google Scholar]
- Drew M.C., Saker L.R. (1984). Uptake and long-distance transport of phosphate, potassium and chloride in relation to internal ion concentrations in barley: Evidence of non-allosteric regulation. Planta 160: 500–507 [DOI] [PubMed] [Google Scholar]
- Duttweiler H.M. (1996). A highly sensitive and non-lethal beta-galactosidase plate assay for yeast. Trends Genet. 12: 340–341 [DOI] [PubMed] [Google Scholar]
- Eguez L., Chung Y.-S., Kuchibhatla A., Paidhungat M., Garrett S. (2004). Yeast Mn2+ transporter, Smf1p, is regulated by ubiquitin-dependent vacuolar protein sorting. Genetics 167: 107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fader C.M., Colombo M.I. (2009). Autophagy and multivesicular bodies: Two closely related partners. Cell Death Differ. 16: 70–78 [DOI] [PubMed] [Google Scholar]
- Gaymard F., Pilot G., Lacombe B., Bouchez D., Bruneau D., Boucherez J., Michaux-Ferrière N., Thibaud J.-B., Sentenac H. (1998). Identification and disruption of a plant shaker-like outward channel involved in K+ release into the xylem sap. Cell 94: 647–655 [DOI] [PubMed] [Google Scholar]
- Geldner N., Dénervaud-Tendon V., Hyman D.L., Mayer U., Stierhof Y.-D., Chory J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Hyman D.L., Wang X., Schumacher K., Chory J. (2007). Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 21: 1598–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González E., Solano R., Rubio V., Leyva A., Paz-Ares J. (2005). PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 17: 3500–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe M., Xu J., Möbius W., Ueda T., Nakano A., Geuze H.J., Rook M.B., Scheres B. (2003). Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr. Biol. 13: 1378–1387 [DOI] [PubMed] [Google Scholar]
- Grefen C., Donald N., Hashimoto K., Kudla J., Schumacher K., Blatt M.R. (2010). A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64: 355–365 [DOI] [PubMed] [Google Scholar]
- Hao Y., Sekine K., Kawabata A., Nakamura H., Ishioka T., Ohata H., Katayama R., Hashimoto C., Zhang X., Noda T., Tsuruo T., Naito M. (2004). Apollon ubiquitinates SMAC and caspase-9, and has an essential cytoprotection function. Nat. Cell Biol. 6: 849–860 [DOI] [PubMed] [Google Scholar]
- Hamburger D., Rezzonico E., MacDonald-Comber Petétot J., Somerville C., Poirier Y. (2002). Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14: 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hürlimann H.C., Pinson B., Stadler-Waibel M., Zeeman S.C., Freimoser F.M. (2009). The SPX domain of the yeast low-affinity phosphate transporter Pho90 regulates transport activity. EMBO Rep. 10: 1003–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Suzuki T., Hattori M., Yoshimoto K., Ohsumi Y., Moriyasu Y. (2006). AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. 47: 1641–1652 [DOI] [PubMed] [Google Scholar]
- Karimi M., De Meyer B., Hilson P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kataoka T., Hayashi N., Yamaya T., Takahashi H. (2004). Root-to-shoot transport of sulfate in Arabidopsis. Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol. 136: 4198–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D. (2009). The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 37: 937–953 [DOI] [PubMed] [Google Scholar]
- Kraft E., Stone S.L., Ma L., Su N., Gao Y., Lau O.-S., Deng X.-W., Callis J. (2005). Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 139: 1597–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxmi A., Pan J., Morsy M., Chen R. (2008). Light plays an essential role in intracellular distribution of auxin efflux carrier PIN2 in Arabidopsis thaliana. PLoS ONE 3: e1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.-H., Kuo H.-F., Canivenc G., Lin C.-S., Lepetit M., Hsu P.-K., Tillard P., Lin H.-L., Wang Y.-Y., Tsai C.-B., Gojon A., Tsay Y.-F. (2008a). Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20: 2514–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.-I., Chiang S.-F., Lin W.-Y., Chen J.-W., Tseng C.-Y., Wu P.-C., Chiou T.-J. (2008b). Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol. 147: 732–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J., Thibaud M.-C., Bechtold N., Raghothama K., Nussaume L. (2004). Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol. Biol. 55: 727–741 [DOI] [PubMed] [Google Scholar]
- Moriyasu Y., Hattori M., Jauh G.-Y., Rogers J.C. (2003). Alpha tonoplast intrinsic protein is specifically associated with vacuole membrane involved in an autophagic process. Plant Cell Physiol. 44: 795–802 [DOI] [PubMed] [Google Scholar]
- Muchhal U.S., Pardo J.M., Raghothama K.G. (1996). Phosphate transporters from the higher plant Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93: 10519–10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge S.R., Rae A.L., Diatloff E., Smith F.W. (2002). Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 31: 341–353 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D., Riezman H. (2007). Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315: 201–205 [DOI] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Pant B.D., Buhtz A., Kehr J., Scheible W.R. (2008). MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 53: 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiter E., Montanini B., Gobert A., Pedas P., Husted S., Maathuis F.J.M., Blaudez D., Chalot M., Sanders D. (2007). A secretory pathway-localized cation diffusion facilitator confers plant manganese tolerance. Proc. Natl. Acad. Sci. USA 104: 8532–8537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman M.G., Wildes R.A., Schaefer N., Wellfare D. (1977). Effect of azetidine 2-carboxylic acid on ion uptake and ion release to the xylem of excised barley roots. Plant Physiol. 60: 240–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y., Bucher M. (2002). Phosphate transport and homeostasis in Arabidopsis. The Arabidopsis Book 1: e0024, doi/10.1199/tab.0024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y., Thoma S., Somerville C., Schiefelbein J. (1991). Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol. 97: 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch C., Bucher M. (2002). Molecular mechanisms of phosphate transport in plants. Planta 216: 23–37 [DOI] [PubMed] [Google Scholar]
- Risinger A.L., Kaiser C.A. (2008). Different ubiquitin signals act at the Golgi and plasma membrane to direct GAP1 trafficking. Mol. Biol. Cell 19: 2962–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S.W. (2001). Kiss and run mechanism in exocytosis. J. Membr. Biol. 181: 67–76 [PubMed] [Google Scholar]
- Shin H., Shin H.S., Dewbre G.R., Harrison M.J. (2004). Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 39: 629–642 [DOI] [PubMed] [Google Scholar]
- Stefanovic A., Arpat A.B., Bligny R., Gout E., Vidoudez C., Bensimon M., Poirier Y. (2011). Over-expression of PHO1 in Arabidopsis leaves reveals its role in mediating phosphate efflux. Plant J. 66: 689–699 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Watanabe-Takahashi A., Smith F.W., Blake-Kalff M., Hawkesford M.J., Saito K. (2000). The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J. 23: 171–182 [DOI] [PubMed] [Google Scholar]
- Takano J., Miwa K., Yuan L., von Wirén N., Fujiwara T. (2005). Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc. Natl. Acad. Sci. USA 102: 12276–12281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J., Noguchi K., Yasumori M., Kobayashi M., Gajdos Z., Miwa K., Hayashi H., Yoneyama T., Fujiwara T. (2002). Arabidopsis boron transporter for xylem loading. Nature 420: 337–340 [DOI] [PubMed] [Google Scholar]
- Vembar S.S., Brodsky J.L. (2008). One step at a time: Endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9: 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wang Y., Ribot C., Rezzonico E., Poirier Y. (2004). Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol. 135: 400–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F.-H., Shen S.-C., Lee L.-Y., Lee S.-H., Chan M.-T., Lin C.-S. (2009). Tape-Arabidopsis sandwich — A simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Fuji K., Shimada T., Nishimura M., Hara-Nishimura I. (2005). Endosomal proteases facilitate the fusion of endosomes with vacuoles at the final step of the endocytotic pathway. Plant J. 41: 888–898 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.