Plants produce hydrogen peroxide (H2O2) as a stress signal. This study shows that H2O2 promotes the binding of cytosolic, glycolytic enzymes, GAPCs, to the plasma membrane–associated phospholipase PLDδ. The GAPC–PLDδ interaction mediates the plant response to H2O2 and provides a molecular link between stress signaling and the alteration of cellular metabolism and growth in the plant response to drought.
Abstract
Reactive oxygen species (ROS) are produced in plants under various stress conditions and serve as important mediators in plant responses to stresses. Here, we show that the cytosolic glycolytic enzymes glyceraldehyde-3-phosphate dehydrogenases (GAPCs) interact with the plasma membrane–associated phospholipase D (PLDδ) to transduce the ROS hydrogen peroxide (H2O2) signal in Arabidopsis thaliana. Genetic ablation of PLDδ impeded stomatal response to abscisic acid (ABA) and H2O2, placing PLDδ downstream of H2O2 in mediating ABA-induced stomatal closure. To determine the molecular link between H2O2 and PLDδ, GAPC1 and GAPC2 were identified to bind to PLDδ, and the interaction was demonstrated by coprecipitation using proteins expressed in Escherichia coli and yeast, surface plasmon resonance, and bimolecular fluorescence complementation. H2O2 promoted the GAPC–PLDδ interaction and PLDδ activity. Knockout of GAPCs decreased ABA- and H2O2-induced activation of PLD and stomatal sensitivity to ABA. The loss of GAPCs or PLDδ rendered plants less responsive to water deficits than the wild type. The results indicate that the H2O2-promoted interaction of GAPC and PLDδ may provide a direct connection between membrane lipid–based signaling, energy metabolism and growth control in the plant response to ROS and water stress.
INTRODUCTION
Reactive oxygen species (ROS) are produced in plants in response to a wide variety of stresses, including drought, UV irradiation, high light, wounding, ozone, low and high temperatures, and pathogens (Desikan et al., 2001; Apel and Hirt, 2004; Suzuki et al., 2012). ROS were originally viewed as by-products of metabolic pathways, and a high concentration of ROS is toxic to the cells (Apel and Hirt, 2004; Quan et al., 2008; Finkel, 2011). It has now been well documented that ROS are generated as signals that alter various cellular and physiological processes in plant growth and development (Desikan et al., 2001; Apel and Hirt, 2004; Gechevet al., 2006; Shao et al., 2008). Hydrogen peroxide (H2O2) is the major and most stable species of ROS and plays a signaling role in plant response to stresses, such as mediating abscisic acid (ABA)–regulated stomatal closure (Pei et al., 2000; Zhang et al., 2001). H2O2 is thought to affect target protein activities through modification of thiol groups of Cys residues (Hancock et al., 2005). However, it is unclear how such oxidative modification affects a signaling cascade that leads to alteration of cellular function and plant stress responses.
Recent studies indicate that phospholipase D (PLD) and its product phosphatidic acid (PA) play a role in ROS-mediated signaling (Sang et al., 2001; Yamaguchi et al., 2004; Zhang et al., 2009; Lanteri et al., 2011). The Arabidopsis thaliana genome contains 12 PLDs, PLDα(3), β(2), γ(3), δ, ϵv, and ζ(2), and these PLDs exhibit distinguishable biochemical properties and cellular functions. Knockout (KO) of PLDα1 decreases the production of ROS, and addition of PA induces recovery of ROS levels in the PLDα1 mutant (Sang et al., 2001). PA interacts with NADPH oxidase and increases its activity and ROS production (Zhang et al., 2009). PLD and PA are also implicated in promoting the elicitor-induced generation of ROS in suspension rice (Oryza sativa) and tomato (Solanum lycopersicum) cells (Yamaguchi et al., 2004; Lanteri et al., 2011). On the other hand, H2O2-induced activation of PLD enhances elicitor-induced biosynthesis of phytoalexins in rice cells (Yamaguchi et al., 2004). Plasma membrane–associated PLDδ is activated by H2O2, and ablation of it renders Arabidopsis cells more sensitive to H2O2-promoted programmed cell death than the wild type (Wang and Wang, 2001; Zhang et al., 2003, 2005; Wang et al., 2006). These results suggest that whereas PLDα1 promotes the ROS production, PLDδ mediates plant responses to ROS. However, it is unknown how H2O2 activates PLDδ and whether PLDδ is involved in mediating the H2O2 effect in the ABA signaling.
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) catalyzes the conversion of glyceraldehyde-3-phosphate to 1,3-bisphosphoglycerate in the glycolytic pathway, thus functioning to produce energy and supply intermediates for cellular metabolism (Plaxton, 1996). The Arabidopsis genome contains seven phosphorylating GAPDHs, five of which are located in plastids, whereas GAPC1 and GAPC2 are in the cytosol (Rius et al., 2008; Muñoz-Bertomeu et al., 2010). GAPDHs have been implicated in embryo development, pollen development, root growth, and ABA signal transduction (Rius et al., 2006, 2008; Muñoz-Bertomeu et al., 2009, 2010, 2011). The catalytic Cys residues of GAPDH can be oxidized by oxidants such as H2O2, leading to fully or partially reversible inactivation of GAPDH (Hancock et al., 2005; Hara et al., 2005; Holtgrefe et al., 2008). GAPC1 has been suggested to be a H2O2 target potentially involved in mediating ROS response in Arabidopsis (Hancock et al., 2005; Holtgrefe et al., 2008). Here, we show that GAPC1 and GAPC2 bind to PLDδ, that H2O2 promotes the GAPC interaction with PLDδ, and that the interaction mediates plant response to ABA and water deficits.
RESULTS
Ablation of PLDδ Compromises ABA- and H2O2-Induced Stomatal Closure, but Not ABA-Promoted H2O2 Production
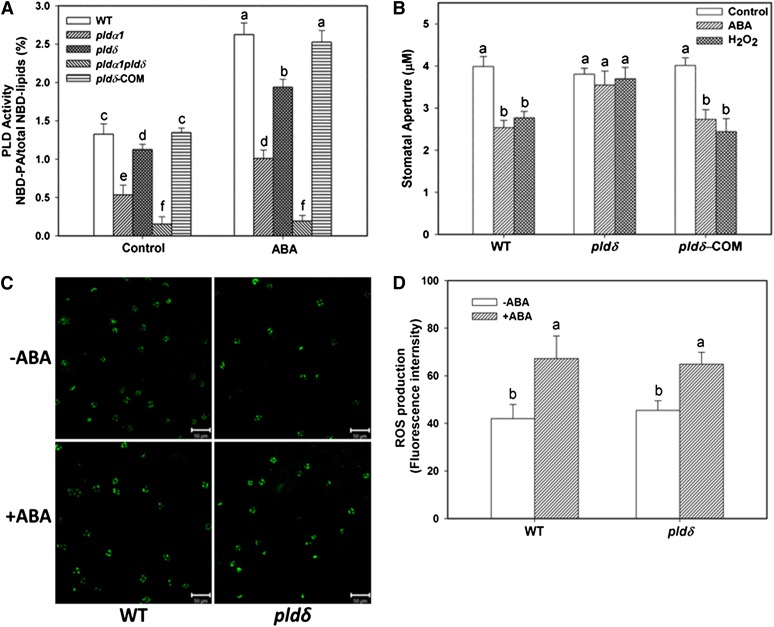
To determine if PLDδ is activated by ABA, we isolated PLDα1 PLDδ double KO pldα1 pldδ (see Supplemental Figure 1 online) and assayed the PLD activity in response to ABA in wild-type, pldα1, pldδ, and pldα1 pldδ using 1-palmitoyl-2-{12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]dodecanoyl}-sn-glycero-3-phosphocholine (NBD-PC)–labeled protoplasts (Figure 1A). The PLDα1 KO mutant was used because PLDα1 was reported to be responsible for a majority of PA produced in response to ABA (Zhang et al., 2009). PA production was increased twofold after wild-type protoplasts were incubated with ABA for 20 min (Figure 1A). The ABA-induced PA production in pldα1 and pldδ was ∼62 and 28% lower, respectively, than in the wild type. No significant PA increase was observed in response to ABA in PLDα1 PLDδ double KO cells (Figure 1A). The results indicate that in addition to PLDα1, PLDδ is also activated by ABA and that PLDα1 and PLDδ together account for virtually all ABA-induced PLD activity, with PLDα1 providing twice as much PA as PLDδ in response to ABA in Arabidopsis.
Figure 1.
Decreased Response of pldδ Plants to H2O2 and ABA.
(A) ABA-induced PA production in leaf protoplasts of pldα1, pldδ, pldα1 pldδ, PLDδ-complementation (COM), and the wild type (WT). Values are means ± se (n = 3).
(B) Stomatal closure induced by 25 µM ABA or 100 µM H2O2. Values are means ± se (n = 50).
(C) Representative image of ROS production in guard cells, visualized by fluorescent dye. +ABA, epidermal peels were loaded with H2DCF-DA for 10 min followed by addition of 25 µM ABA for 5 min; –ABA, no ABA added. Bars = 50 µm.
(D) Quantification of ROS production based on fluorescence intensity (mean pixel intensity). Values are means ± se (n = 50). Columns with different letters are significantly different from each other (ANOVA, P < 0.05).
[See online article for color version of this figure.]
To determine the role of PLDδ in ABA response, we investigated whether the loss of PLDδ alters ABA-promoted stomatal closure and H2O2 production in guard cells. pldδ leaf peels exhibited decreased sensitivity to ABA-promoted stomatal closure (Figure 1B), a response similar to pldα1 (Zhang et al., 2004; Zhang et al., 2009). H2O2 has been shown to induce stomatal closure in pldα1 (Zhang et al., 2009). However, H2O2 failed to induce stomatal closure in pldδ (Figure 1B). Introduction of PLDδ driven by its own promoter into pldδ restored the phenotype for both ABA- and H2O2-induced stomatal closure, indicating that loss of PLDδ is responsible for the ABA and H2O2 response phenotype (Figure 1B). In addition, unlike pldα1, which decreased ABA-promoted H2O2 production (Zhang et al., 2009), KO of PLDδ did not affect the ABA-induced H2O2 production. The basal level of ROS in pldδ and wild-type cells were also similar, as revealed by the fluorescent dye 2′,7′-dichlorofluorescin diacetate (H2DCF-DA) intensity (Figures 1C and 1D). These results indicate that PLDδ is not required for ABA-induced H2O2 production but is involved in stomatal response to ABA and H2O2. The data suggest that PLDδ acts downstream of H2O2 in signaling ABA-induced stomatal closure.
Direct Interaction between GAPC and PLDδ
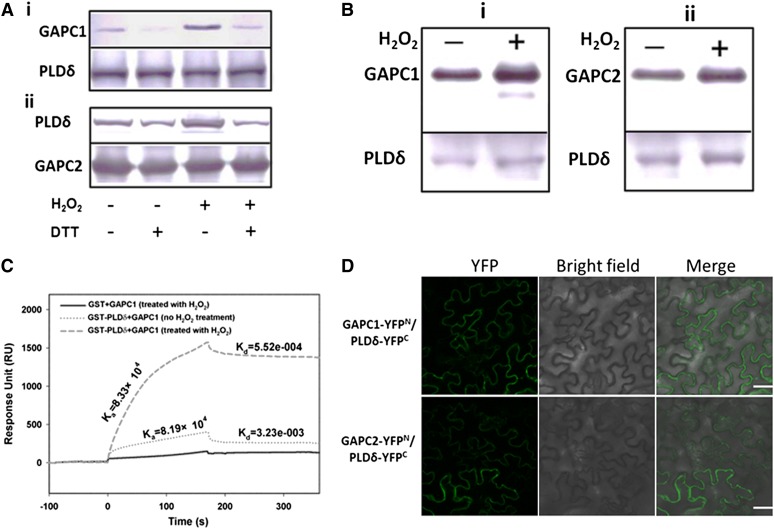
To determine how PLDδ is involved in the H2O2 response, we incubated purified PLDδ with H2O2 and the treatment had no impact on enzyme activity (Zhang et al., 2003). The transcript level of PLDδ was not increased after ABA treatment for 40 min (see Supplemental Figure 2 online). These data indicate that the ABA-induced activation of PLDδ in the early phase is not mediated by increased PLDδ expression or the direct effect of H2O2 on PLDδ. To test whether a protein is involved in the H2O2 activation of PLDδ, we investigated the potential interaction of PLDδ with GAPC, because GAPC was reported as a direct target of H2O2 in Arabidopsis (Hancock et al., 2005; Holtgrefe et al., 2008). His-tagged GAPC1 was expressed in Escherichia coli and incubated with microsomal proteins from Arabidopsis leaves, and immunoblotting with PLDδ antibodies detected PLDδ in the GAPC1 coprecipitate (see Supplemental Figure 3 online). To verify the interaction, we purified His-tagged GAPC1 and GAPC2 proteins expressed in E. coli (see Supplemental Figure 4A online) and used them for reciprocal pulldown with glutathione S-transferase (GST)-PLDδ. GAPC2 pulled down PLDδ. PLDδ also pulled down GAPC1, as indicated by immunoblotting with anti-His or anti-GST antibodies (Figure 2A). In addition, the association of GAPCs and PLDδ was increased in the presence of H2O2 but decreased in the presence of the reducing reagent DTT (Figure 2A). To further validate the interaction, we coexpressed GAPC and PLDδ in yeast (see Supplemental Figure 4B online) and grew the yeast cells with or without H2O2. GAPC1 and GAPC2 were detected in the complex with PLDδ when PLDδ was immunoprecipitated with FLAG antibody. PLDδ also associated with GAPC1 or GAPC2 when GAPCs were immunoprecipitated with cMyc antibody. The presence of H2O2 promoted the interaction between GAPC and PLDδ (Figure 2B). These results indicate that the GAPC–PLDδ interaction is enhanced in an oxidative but weakened in a reducing environment.
Figure 2.
Interaction of GAPC with PLDδ.
(A) Immunoblotting of proteins after coprecipitation using E. coli–expressed GST-PLDδ and His-GAPC1/2, as affected by H2O2 (100 μM) and DTT (100 μM). i, Coprecipitation of His-GAPC1 with GST-PLDδ. GAPC1, immunoblotting of GAPC1 using anti-His antibody for the precipitates; PLDδ, the starting GST-PLDδ used for precipitation. ii, Coprecipitation of GST-PLDδ with His-GAPC2. PLDδ, immunoblotting of PLDδ using anti-GST antibody for the precipitates. GAPC2, the starting His-GAPC2 used for precipitation. DTT was added before the addition of H2O2 when both were applied.
(B) Immunoblotting of coprecipitated GAPC and PLDδ that were coexpressed in yeast grown in the presence or absence of added H2O2 (20 μM). i and ii, Reciprocal pulldown of PLDδ and GAPC1 and GAPC2, respectively. PLDδ was fused with a FLAG tag and GAPC1or GAPC2 with a cMyc tag. GAPC1 or GAPC2 band indicates immunoblotting with cMyc antibody against the sample precipitated with FLAG antibody–conjugated agarose beads. PLDδ band indicates immunoblotting with FLAG antibody against the sample precipitated with cMyc antibody for GAPC1 or GAPC2.
(C) Quantitative SPR analysis of PLDδ binding to GAPC1. GAPC1 (no H2O2 treatment or pretreated with 100 µM H2O2) was first immobilized on the NTA chip followed by injection of GST or GST-PLDδ.
(D) Representative confocal images of BiFC. Green color represents YFP fluorescence, indicating interaction of GAPC with PLDδ. PLDδ-YFPC was cotransformed with GAPC1-YFPN or GAPC2-YFPN into tobacco leaves by infiltration. Bars = 50 µm.
To quantify the interaction between GAPC1 and PLDδ, we used surface plasmon resonance (SPR) to determine the binding kinetics. Purified GAPC1 was immobilized on an nitrilotriacetic acid (NTA) chip followed by injection of purified GST or GST-PLDδ. The representative sensorgram showed an increase in response unit (RU) when GST-PLDδ, but not GST, was injected, indicating that PLDδ interacts with GAPC1 (Figure 2C). When H2O2-treated GAPC1 was used, the GAPC1–PLDδ interaction was enhanced as RU was higher than when GAPC1 was not incubated with H2O2 (Figure 2C). H2O2-treated or untreated GAPC1 displayed comparable association rate constants (Ka = 8.19 × 104 M−1s−1 versus 8.33 × 104 M−1 s−1). However, the dissociation rate constant was lower when GAPC1 was exposed to H2O2 (Kd = 5.52 × 10−4 s−1 versus 3.23 × 10−3 s−1). The maximum specific binding is 1564 RU for H2O2-treated GAPC1 and 286 RU for GAPC1 without H2O2 treatment (Figure 2C). The equilibrium binding constant KD is 6.62 × 10−9 M for GAPC1–PLDδ interaction in the presence of H2O2 and 3.94 × 10−8 M for GAPC1–PLDδ interaction without H2O2. The results indicate that the GAPC1–PLDδ interaction is significantly enhanced by H2O2 and that H2O2 stabilizes the interaction by decreasing dissociation between GAPC1 and PLDδ.
To visualize the GAPC–PLDδ interaction in plant cells, we used bimolecular fluorescence complementation (BiFC) that brings together two yellow fluorescent protein (YFP) fragments fused to two interacting proteins (Walter et al., 2004). GAPC1 or GAPC2 was fused to the N terminus of YFP (GAPC1-YFPN or GAPC2-YFPN), and PLDδ was fused to the C terminus of YFP (PLDδ-YFPC). These constructs were cointroduced into tobacco leaves. No fluorescence was observed when empty vectors YFPN and YFPC were cotransformed or when GAPC-YFPN and PLDδ-YFPC were transformed separately (see Supplemental Figure 5 online). In the positive controls, bZIP63-YFPN and bZIP63-YFPC, the transcription factor, formed dimers and brought YFPN and YFPC together to generate fluorescence in the nucleus (see Supplemental Figure 5 online). GAPC1-YFPN or GAPC2-YFPN coexpressed with PLDδ-YFPC produced fluorescence in the cell, indicating that both GAPCs interacted with PLDδ (Figure 2D).
GAPCs Promote the Activity of PLDδ under Oxidative Conditions
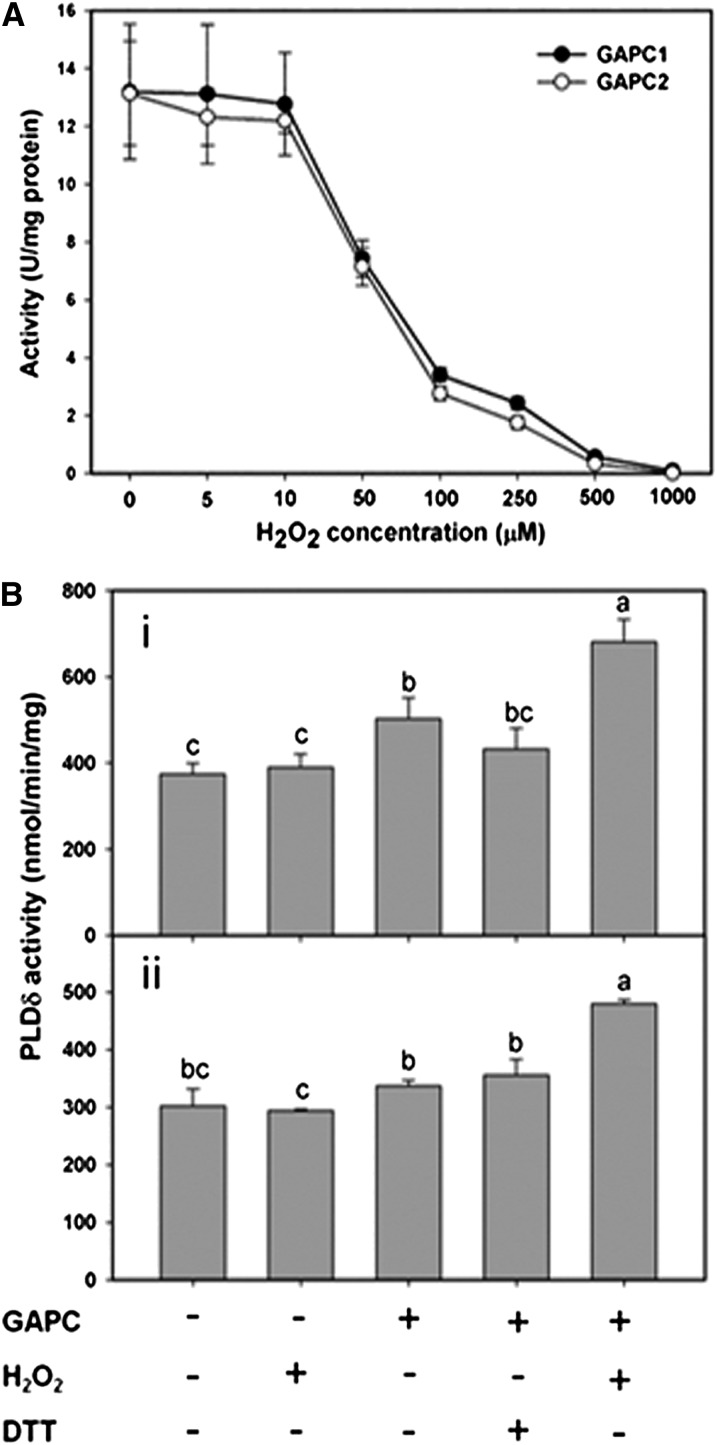
To determine the function of GAPC interaction with PLDδ, we first tested the sensitivity of GAPC1 and GAPC2 purified from E. coli to H2O2. H2O2 inhibited GAPC activity in a dose-dependent manner, and virtually all GAPC1 or GAPC2 activity was inhibited at 500 µM H2O2 (Figure 3A). When different concentrations of DTT were added to GAPCs first, followed by addition of 500 µM H2O2, the loss of GAPC activity was small, showing that H2O2 oxidation of GAPCs can be protected by DTT reduction (see Supplemental Figure 6A online). After incubation with 500 µM H2O2, partial GAPC activity could be recovered by addition of DTT (see Supplemental Figure 6B online).
Figure 3.
Oxidized GAPC Promotes PLDδ Activity.
(A) H2O2 inhibition of GAPC1 and GAPC2 activities.
(B) GAPC promotion of PLDδ activity under oxidative conditions. Equal molar ratios of PLDδ and GAPC proteins were used. PLDδ activity was assayed in the presence of GAPC1 (i) or GAPC2 (ii) under different conditions as indicated; 100 μM DTT or 100 μM H2O2 was used as indicated. Values are means ± se (n = 3). Different letters indicate significant differences (ANOVA, P < 0.05).
Purified PLDδ was then incubated GAPCs with or without H2O2 to determine the effect of H2O2 and GAPC on PLDδ activity. Without GAPC, addition of 100 µM H2O2 did not affect PLDδ activity (Figure 3B), verifying that H2O2 has no direct effect on PLDδ activity. Incubation of PLDδ with GAPC1 and GAPC2 increased PLDδ activity by 34 and 11%, respectively (Figure 3B). However, pretreatment of GAPC1 and GAPC2 with 100 µM H2O2 increased PLDδ activity by 82.1 and 58.9%, respectively (Figure 3B). The data indicate that H2O2 inactivates GAPC but promotes the GAPC binding to PLDδ, and the binding increases PLDδ activity.
GAPC Mediates the H2O2 Activation of PLDδ in the Cell
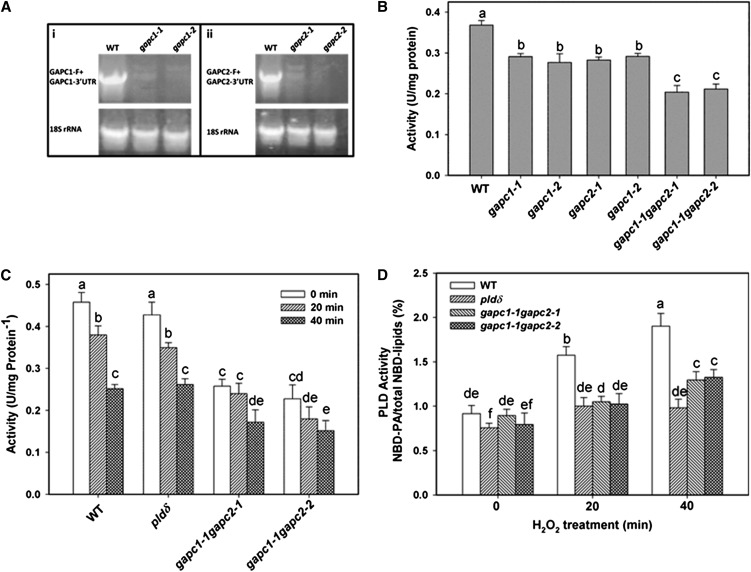
To evaluate whether GAPC affects the activity of PLDδ in living cells, we compared PLD activity in GAPC-KO, PLDδ-KO, and wild-type protoplasts as affected by H2O2. Two homozygous T-DNA insertion KO lines of Arabidopsis were isolated for GAPC1 (gapc1-1, CS328689; gapc1-2, SALK_129091) and for GAPC2 (gapc2-1, SALK_016539; gapc2-2, SALK_070902) (see Supplemental Figure 7 online). The GAPC1 transcript was lost in two GAPC1-KO lines, and GAPC2 transcript was also absent in two GAPC2-KO lines, suggesting that all four GAPC T-DNA lines are null mutants (Figure 4A). We then generated two double KO lines (gapc1-1 gapc2-1 and gapc1-1 gapc2-2) by crossing the single mutants. Two lines of triple KO mutants (gapc1-1 gapc2-1 pldδ and gapc1-1 gapc2-2 pldδ) were also isolated by crossing the GAPC double KO with pldδ. NAD-dependent GAPDH activity was determined in the single and double KO lines of GAPC. The GAPDH activity in leaves was decreased by 21% (gapc1-1), 25% (gapc1-2), 23% (gapc2-1), and 21% (gapc2-2) for GAPC single mutants (Figure 4B). GAPC double KO plants gapc1-1 gapc2-1 and gapc1-1 gapc2-2 had ∼45% decrease in GAPDH activity (Figure 4B). The results indicate that GAPC1 and GAPC2 contribute almost equally to the activity, and together they account for nearly half of NAD-dependent GAPDH activity in Arabidopsis leaves.
Figure 4.
H2O2 Effects on GAPC and PLDδ Activities.
(A) RT-PCR detection of GAPC1 and GAPC2 expression in the leaves of wild-type (WT) and mutant plants. 18S rRNA was a control confirming the synthesis of cDNA.
(B) GAPDH activity in the total protein extracted from the leaves of wild-type and mutant plants.
(C) GAPDH activity using protein extracted from protoplasts after 1 mM H2O2 treatment.
(D) H2O2-promoted PA production in protoplasts. Values are means ± se (n = 3). Different letters mark significant differences from each other (ANOVA, P < 0.05).
To determine if KO of both GAPCs affects PLD activation by H2O2, protoplasts of wild-type, pldδ, and GAPC double mutants were labeled with NBD-PC and treated with H2O2. We first examined how GAPDH activity in protoplasts responded to H2O2. Protoplasts from GAPC double KOs had significantly lower GAPDH activity than the wild type or pldδ (Figure 4C). H2O2 treatments for 20 min had no significant effect on GAPDH activity in the GAPC double KO but decreased GAPDH activity in the wild type and pldδ by 15%. Significant decreases in GAPDH activity occurred in all genotypes after 40 min of H2O2 treatments (Figure 4C). The results indicate that H2O2 inhibits GAPDH activity in the cell and also could mean that the loss of the GAPDH activity in the early phase (20 min) results primarily from H2O2 inhibition of GAPCs.
Without addition of H2O2, the PLD activity, as measured by the formation of PA, in gapc1-1 gapc2-1 and gapc1-1 gapc2-2 was comparable to that of the wild type (Figure 4D). The H2O2 treatment increased PA production nearly twofold after 40 min in the wild type, whereas it increased PA production only 30% in pldδ. The gapc1 gapc2 double KOs and gapc1 gapc2 pldδ triple KOs exhibited similar attenuated PA increase as pldδ in response to H2O2 (Figure 4D). The results indicate that PLDδ is the main PLD responsible for the H2O2 activation of PLD and that GAPCs mediate the H2O2-induced increase of PLDδ activity.
GAPCs Are Involved in ABA-Induced PA Production
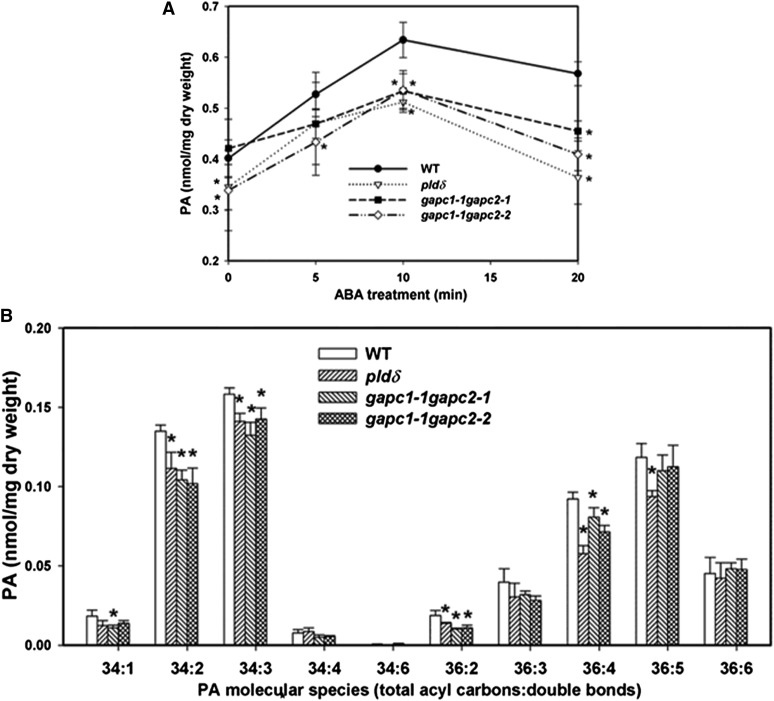
To characterize the effect of GAPC and PLDδ on PA production in response to ABA, we measured the PA levels and composition in 4-week-old Arabidopsis leaves treated with ABA up to 20 min. PA level was induced by ABA in the wild type and reached a plateau at 10 min after ABA treatment. The total PA level was increased in pldδ, gapc1-1 gapc2-1, and gapc1-1 gapc2-2 leaves after ABA treatment (Figure 5A). However, the amount of PA was significantly lower in pldδ, gapc1-1 gapc2-1, and gapc1-1 gapc2-2 than in the wild type at 10 and 20 min after ABA treatment (Figure 5A).
Figure 5.
PA Content of GAPC and PLDδ Mutant Leaves in Response to ABA.
(A) Total PA content of leaves harvested at different times after spraying with ABA (100 µM). WT, the wild type.
(B) PA molecular species in leaves of the wild type and mutants treated with ABA for 10 min. Values are means ± se (n = 5). Asterisks indicate significant difference from the wild type at the same time point of ABA treatment (P < 0.05, t test).
The molecular species of PA in response to ABA at 10 min were analyzed for the wild type, pldδ, gapc1-1 gapc2-1, and gapc1-1 gapc2-2. In wild-type Arabidopsis leaves, 34:2 (16:0/18:2), 34:3 (16:0/18:3), 36:4 (mainly 18:2/18:2), 36:5 (18:2/18:3), and 36:6 (18:3/18:3) are the most abundant PA species (Zhang et al., 2009). The levels of major PA species, including 34:1, 34:2, 34:3, 36:2, 36:4, and 36:5 PA, were significantly decreased in pldδ, and the major overall decrease of total PA level was due to the decrease in 34:2, 34:3, 36:4, and 36:5 PA (Figure 5B). Similarly, the levels of PA species 34:2, 34:3, 36:2 and 36:4 PA were significantly reduced in gapc1-1 gapc2-1 and gapc1-1 gapc2-2 compared with the wild type after 10 min of ABA treatment (Figure 5B). The PA acyl combinations affected by PLDδ and GAPC expression are the molecular species typically derived from hydrolysis of extraplastidic phospholipids (Welti et al., 2002), consistent with the extraplastidic location of these enzymes. The results show that the ablation of either PLDδ or GAPCs decreases the ABA-induced PA production. The attenuation of ABA-induced activation of PLDδ in GAPC double KOs is consistent with the results that GAPCs are required for the activation of PLDδ activity (Figure 4D).
Loss of GAPCs or PLDδ Renders Plants Less Responsive to Water Deficits
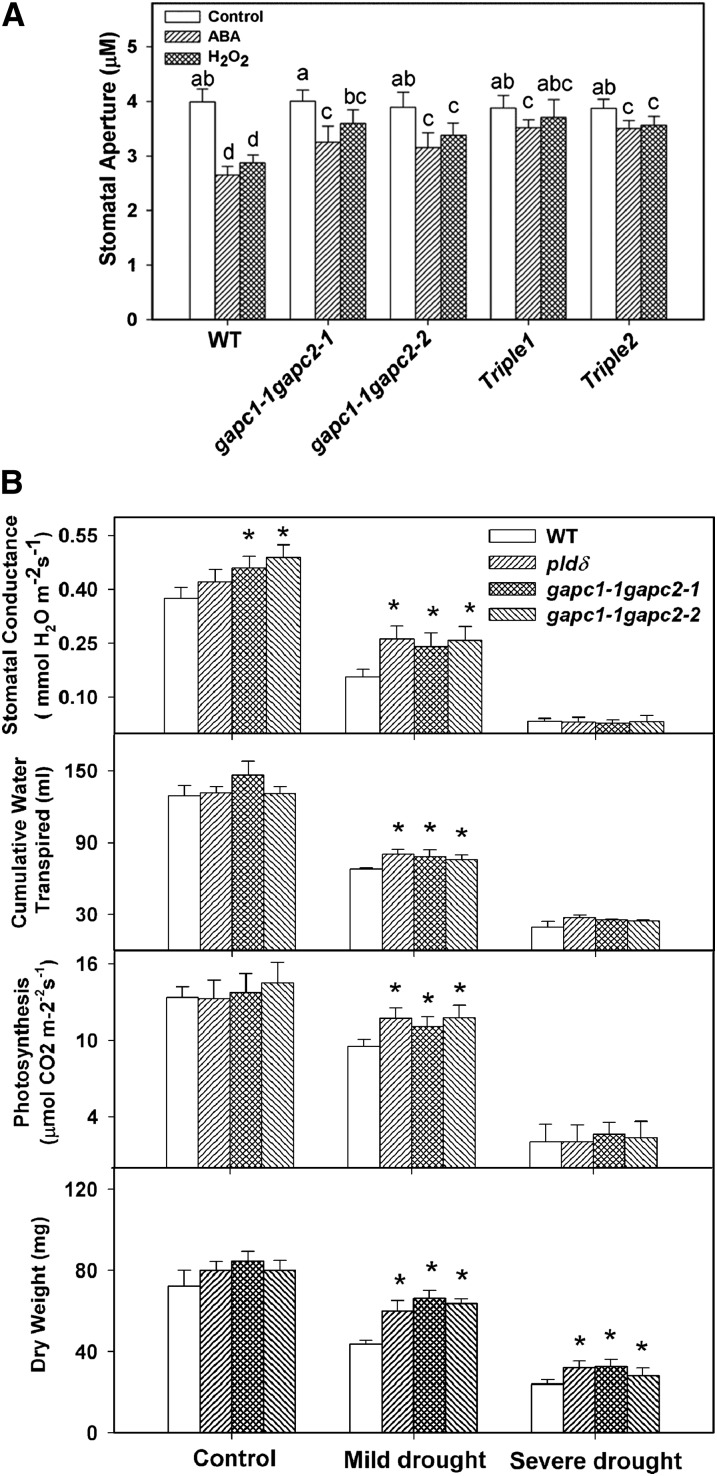
To determine if GAPC–PLDδ interaction is involved in the process of mediating plant response to ROS, we measured stomatal closure in response to ABA and H2O2 in leaves deficient in both GAPCs or GAPC and PLDδ. Stomata of gapc1-1 gapc2-1 and gapc1-1 gapc2-2 were less sensitive to ABA or H2O2, as indicated by greater stomatal aperture in these mutants than that of the wild type after the treatment of ABA or H2O2 (Figure 6A). Two triple mutants (gapc1-1 gapc2-1 pldδ and gapc1-1 gapc2-2 pldδ) were also less sensitive to ABA- and H2O2-promoted stomatal closure (Figure 6A).
Figure 6.
Response of GAPC and PLDδ Mutants to ABA and Water Deficits.
(A) Changes in stomatal aperture after ABA (25 μM) or H2O2 (100 μM) treatment. Values are means ± se (n = 50). Different letters mark significant differences from each other (ANOVA, P < 0.05). WT, the wild type.
(B) Stomatal conductance, cumulative water transpiration, photosynthesis, and dry weight. Asterisks mark significant difference from the wild type under the same growth condition. Values are means ± se (n = 16).
To determine how the effect of GAPCs and PLDδ on ABA and H2O2 signaling impacts plant response to water deficits, we evaluated the effect of GAPCs and PLDδ KOs on Arabidopsis plants grown under three field water capacity (FC) conditions: 100% FC for well-watered control, and 60 and 30% FC for mild and acute drought stress, respectively (see Supplemental Figure 8 online). Under well-watered conditions, pldδ, gapc1-1 gapc2-1, and gapc1-1 gapc2-2 did not show significant difference from the wild type in cumulative water transpired and photosynthetic rate, but gapc1-1 gapc2-1 and gapc1-1 gapc2-2 had higher stomatal conductance than the wild type (Figure 6B). At 60% FC, pldδ, gapc1-1 gapc2-1, and gapc1-1 gapc2-2 displayed higher stomatal conductance, higher cumulative water transpiration, and higher photosynthetic rate than wild-type plants (Figure 6B). At the severe water deficit (30% FC), stomatal conductance was very low in all genotypes, but pldδ, gapc1-1 gapc2-1, and gapc1-1 gapc2-2 mutant lines still exhibited the tendency to have more cumulative water transpiration than the wild type (Figure 6B).
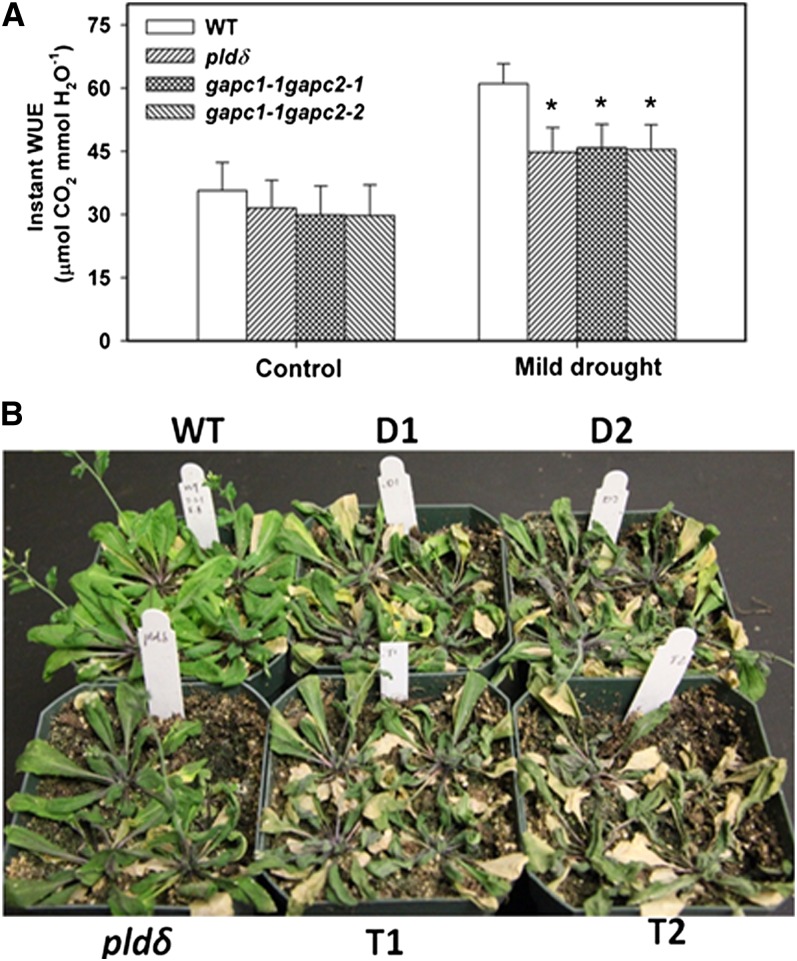
As the FCs decreased, wild-type, pldδ, gapc1-1 gapc2-1, and gapc1-1 gapc2-2 mutants accumulated less biomass, as plant growth was inhibited in response to water deficits. pldδ, gapc1-1 gapc2-1, and gapc1-1 gapc2-2 accumulated more biomass than the wild type under both mild and acute drought conditions. At 60% FC, the three mutants accumulated ∼30% more dry matter than the wild type. The greater biomass in the mutants than the wild type was consistent with higher stomatal conductance and photosynthetic rate. The decreased drought inhibition of plant growth in the mutants suggests that the loss of PLDδ or GAPCs renders plants less responsive to adjusting growth under water deficits. However, the mutants lost much more water and had lower instant water use efficiency (WUE) than the wild type (Figure 7A). When they were grown in separate pots without maintaining FC or watering, the PLDδ and GAPC mutants wilted faster than the wild type (Figure 7B), consistent with the measurements that PLDδ- and GAPC-deficient plants lost more water.
Figure 7.
Increased Water Loss in GAPC-KO and PLDδ-KO Arabidopsis Plants.
(A) Instant WUE of wild-type (WT) and mutant plants under 100 and 60% FC. Arabidopsis seedlings were transplanted to pots and maintained at 100% FC and 60% FC. Instant WUE was calculated as the ratio of the photosynthetic rate to stomatal conductance; measurements were taken after the first 4 d after the onset of required stress. Asterisks indicate significant difference from the wild type. Values are means ± se (n = 16; *P < 0.05, t test).
(B) Increased dehydration of GAPC-KO and PLDδ-KO plants when FC was not maintained. Plants (25 d old) were fully watered and then left unwatered for 16 d when the photograph was taken. D1 and D2 are GAPC1 and 2 double KOs gapc1-1 gapc2-1 and gapc1-1 gapc2-2, respectively. T1 and T2 are GAPC1, GAPC2, and PLDδ triple KOs gapc1-1 gapc2-1 pldδ and gapc1-1 gapc2-2 pldδ, respectively.
[See online article for color version of this figure.]
DISCUSSION
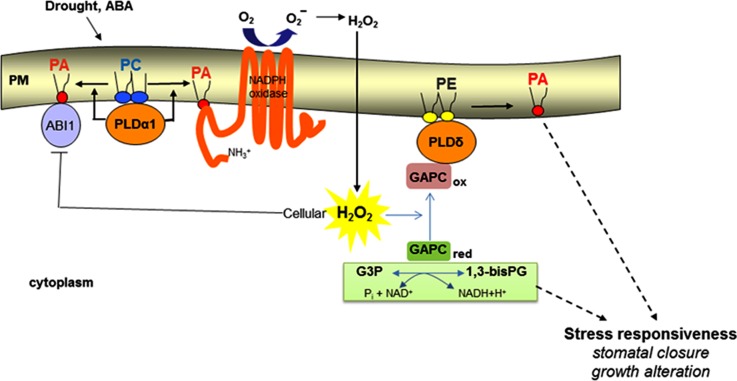
This study demonstrates that PLDδ plays a role in mediating ABA-induced stomatal closure, but it acts in a distinctively different step from PLDα1 in the ABA signaling pathway (Figure 8). PLDα1 promotes NADPH oxidase activity and H2O2 production (Zhang et al., 2009), whereas PLDδ mediates H2O2 response but not H2O2 production. Both PLDα1 and PLDδ are activated in response to ABA to generate PA. This raises the question of whether PA generated by PLDα1 and PLDδ targets the same or different proteins. Our analyses of PLDδ- and PLDα1-deficient mutants show that PLDα1 produces twice as much PA as does PLDδ in response to ABA and that PLDδ is the main PLD responsible for H2O2-stimulated PA production. Also, temporal comparisons of PA formation in these mutants indicate that PLDα1 is activated earlier than PLDδ. In addition, PLDα1 and PLDδ have different subcellular locations and different substrate selectivities with PLDα1 and PLDδ preferring PC and phosphatidylethanolamine, respectively (Figure 8; Wang et al., 2006). It is conceivable that the different magnitude, timing, and location of PA production as affected by PLDα1 and PLDδ will impact their product PA interaction with target proteins. PA has been shown to bind to ABA INSENSITIVE1, NADPH oxidase, and sphingosine kinase. These proteins are involved in the ABA-mediated stomatal closure and targets of PLDα1 (Figure 8) (Zhang et al., 2004; Zhang et al., 2009; Guo et al., 2011). In addition, mitogen-activated protein kinases (MAPKs), which are involved in various cellular processes, such as H2O2-induced cell death and ABA-promoted stomatal closure (Zhang et al., 2003; Zhang et al., 2006; Jammes et al., 2009; Yu et al., 2010), have been implicated as targets of PA. PLDδ-KO cells had a decreased MAPK activity in response to H2O2 (Zhang et al., 2003); thus, MAPKs could be targets regulated by PA involved in PLDδ-mediated stomatal closure.
Figure 8.
A Proposed Model for the Role of PLD/PA in Regulating ROS Production and Response under Water Deficits.
This model depicts only the known targets of PLD/PA in ABA-mediated stomatal closure; other ABA regulators are not included in this model. GAPCox refers to oxidized, catalytically inactive GAPC that interacts with PLDδ and promotes PLDδ activity. GAPCred refers to reduced, active GAPC that converts glyceraldehyde-3-phosphate (G3P) to 1,3-bisphosphoglycerate (1,3-bisPG) with NADH production. PLDα1 uses preferably phosphatidylcholine (PC), whereas PLDδ prefers phosphatidylethanolamine (PE) as substrate. Solid arrows indicate established links, and dashed arrows denote putative links. PM, plasma membrane.
The analyses of GAPC and PLDδ interaction further augment the role of PLDδ and PA in mediating ROS response. This study documented the direct interaction between PLDδ and GAPCs qualitatively and quantitatively using different approaches. H2O2 inhibits GAPC activity by oxidizing the catalytic Cys residues in the enzyme (Hancock et al., 2005). Our results indicate that H2O2 promotes the GAPC interaction with PLDδ by decreasing the dissociation of the GAPC-PLDδ binding. KOs of GAPCs attenuated the ABA- or H2O2-promoted production of PA in the cell, providing in vivo support for the role of GAPCs in the H2O2 activation of PLDδ. It may be noted that the level of H2O2 used in this study is within physiological range reported for Arabidopsis leaves, in which H2O2 levels varied from 60 µM to more than 5 mM under different stress conditions or different assays (Karpinski et al., 1999; Veljovic-Jovanovic et al., 2001; Queval et al., 2008). In our study, GAPC activity in vitro was significantly inhibited at 50 μM H2O2 and almost completely lost at 500 μM H2O2. When H2O2 was applied to protoplasts, we used 1 mM H2O2 to ensure the oxidation of GAPCs because plant cells have a high capacity to degrade H2O2 by several scavenging enzymes.
Plants deficient in GAPCs or PLDδ were less sensitive to ABA-promoted stomatal closure and had higher transpirational water loss than the wild type under drought stress. Without either GAPC or PLDδ, plants are less responsive to drought-induced growth inhibition. These results indicate that GAPC–PLDδ interaction mediates ROS signaling and increases plant responsiveness to water deficits. Under the controlled water deficits with specific FCs maintained, the GAPC- or PLDδ-deficient plants actually accumulated more biomass than the wild type. The data are consistent with the observation that GAPC- or PLDδ-deficient plants have higher stomatal conductance and a higher rate of photosynthesis than the wild type, probably due to more opened stomata to allow more CO2 uptake and increased nutrient transport than the wild type. However, the increase in biomass accumulation was at the expense of increased water use. Indeed, without maintaining a specific soil water level, the GAPC- or PLDδ-deficient plants wilted faster than the wild type when plants were withheld water. Earlier studies showed that KO of PLDδ decreased plant tolerance to severe stresses, such as freezing, UV irradiation, and salt tolerance in Arabidopsis (Katagiri et al., 2001; Zhang et al., 2003; Li et al., 2004; Bargmann et al., 2009). Decreasing growth under water deficits is one of the key strategies for plants to cope with stress and survival. The results indicate that the loss of GAPC or PLDδ compromises plant ability to sense the water stress and to adjust cellular and physiological response accordingly.
The glycolytic enzyme GAPDH occurs in both the cytosol and plastids, and the specific contributions of the two glycolytic pathways to plant metabolism and growth are not well defined (Plaxton, 1996; Muñoz-Bertomeu et al., 2009). Recent studies show that KO of both plastid-localized GAPCps causes severe development and growth defects, including arrested root development, dwarfism, and male sterility in Arabidopsis (Muñoz-Bertomeu et al., 2009, 2010). Genetic ablation of another glycolytic enzyme, phosphoglycerate mutase, also indicates a critical role of glycolysis in stomatal movement, vegetative growth, and pollen production in Arabidopsis (Zhao and Assmann, 2011). By comparison, our study reveals that the KO of both cytosolic GAPCs results in no overt growth defects under normal condition in Arabidopsis. Instead, the GAPC-deficient plants exhibited less growth inhibition than the wild type under drought under the controlled drought conditions. These results suggest that GAPCs are required for plant growth responsiveness to drought, and we propose that the H2O2-promoted interaction of GAPCs with PLDδ is involved in the stress signaling leading to growth inhibition (Figure 8). An alternative hypothesis could be that the decrease in GAPC would alter the flux through carbon metabolism and affect the growth phenotype without GAPC binding to PLDδ. If so, one would expect that under drought stress, the increased H2O2 in plants would inhibit GAPCs, leading to growth inhibition. But this is not the case because GAPC-KO plants display less growth inhibition than the wild type. In addition, GAPC-KO mutants share a similar phenotype as PLDδ-KO; ablation of either GAPCs or PLDδ renders plants less responsive to water deficits, and the drought-induced growth inhibition requires the presence of both PLDδ and GAPCs. Thus, our results are consistent with the proposition that the interaction between GAPCs and PLDδ is involved in mediating H2O2 signals in plant response to water deficits. However, further studies are needed to understand the requirement for and mechanism of the GAPC–PLDδ interaction in modulating plant growth under stress and the metabolic role of GAPCs in plant growth and stress responses.
The identification of GAPC interaction with PLDδ unveils a regulatory function of the carbon metabolic enzymes GAPCs in plants and potentially a molecular node linking stress signaling and metabolic alterations. Some classical metabolic enzymes can have crucial regulatory roles in the cell. For example, hexokinase has been found in the nucleus, where it forms a protein complex mediating glucose signaling in yeast and plant (Ahuatzi et al., 2004; Cho et al., 2006). In animal cells, GAPDH is involved in nonmetabolic processes, including gene transcription, DNA replication, nuclear tRNA export, and DNA repair, and these studies indicate that GAPDH has direct relationship to the pathology of various diseases (Sirover, 1997; Hara et al., 2005; Bae et al., 2006; Harada et al., 2007). Oxidized GAPDH is thought to be translocated to the nucleus to regulate gene expression to initiate apoptotic cell death (Hara et al., 2005; Bae et al., 2006). This study shows that the cytosolic GAPCs interact with the plasma membrane–bound PLDδ and the interaction is promoted by ROS. We propose that the GAPC–PLDδ interaction in response to ROS provides a molecular link between stress signaling and the alteration of cellular metabolism and growth (Figure 8). Further investigations on the specificity, mechanism, and downstream targets of these interactions will provide mechanistic insights to how plants adjust metabolism and growth in response to different stresses.
METHODS
Isolation of KOs and pldδ Complementation
Arabidopsis thaliana (Columbia-0) wild-type and T-DNA insertion lines were obtained from the ABRC at Ohio State University. pldα1 (SALK_053785) was isolated and confirmed previously (Zhang et al., 2004). The homozygous line of pldδ (SALK_023247) was confirmed by PCR. The primers for PCR screening are listed in Supplemental Table 1 online. Four T-DNA lines (gapc1, CS328689, SALK_129091; gapc2, SALK_016539 and SALK_070902) were screened, and the homozygous lines were verified by PCR (see Supplemental Figure 6 online). The open reading frame of GAPC1 and GAPC2 shares 89.7% identity, while the 3′ untranslated regions of both genes are not conserved. Thus, primers in the 3′ untranslated region of GAPC1 and GAPC2 were used to distinguish the GAPC1 and GAPC2 transcripts. To complement pldδ, a genomic sequence including the promoter of PLDδ was cloned (primers listed in see Supplemental Table 1 online) and inserted into binary vector PEC291 for transformations of pldδ.
Plant Growth Conditions and Physiological Analysis
Plants were grown in soil in a growth chamber with cool white light of 160 µmol m−2 s−1 under 12-h-light/12-h-dark and 23°C/19°C cycles. Drought stress was created by a gravimetric approach (Sheshshayee et al., 2005; Peters et al., 2010). Ten-day-old Arabidopsis seedlings were transplanted to pots containing soil saturated to maximum field capacity (100% FC). Soil saturation was achieved by adding a known amount of water based on weight of soil and water holding capacity. The pots were covered with thick polyethylene sheets to prevent evaporation. One set of plants was maintained at 100% FC (control), and the other two sets were maintained at 60% (mild stress) and 30% (acute stress). The pots were weighed every day, and the difference in weight in subsequent days was corrected by adding water to maintain specific FCs. The amount of water added over the experimental period was summed up to give the cumulative water transpired. Stomatal conductance and photosynthetic rate were recorded on fully expanded leaf using a portable gas exchange system (LICOR6400-XT; LiCOR Biosciences). Instant WUE was calculated as ratio of photosynthetic rate to stomatal conductance. Measurements were taken on the first 4 d after the onset of drought stress. At the end of the stress, the shoots were harvested, dried, and weighed.
Stomatal Aperture Measurements
Stomatal aperture was measured using epidermal peels according to a described procedure (Zhang et al., 2004). Briefly, epidermal peels were floated in incubation buffer (10 mM KCl, 0.2 mM CaCl2, 0.1 mM EGTA, and 10 mM MES-KOH, pH 6.15) for 2.5 h under cool white light at 23°C to induce stomatal opening. ABA or H2O2 was applied separately to epidermal peels, which were incubated for 2.5 h under cool white light at 23°C to induce stomatal closure. Stomata were imaged under a microscope with a digital camera and analyzed with ImageJ software (NIH).
ROS Detection
The endogenous ROS levels in guard cells were detected using epidermal peels treated with the dye H2DCF-DA (Sigma-Aldrich) (Zhang et al., 2009). Epidermal peels were floated in incubation buffer for 2 h and then loaded with 50 µM H2DCF-DA (50 mM stock in DMSO) for 10 min, followed by 20 min washing in incubation buffer. The 25 µM ABA was added for desired time of treatment. Epidermal peels were observed with a confocal microscope (Zeiss LSM 510) (green fluorescence: excitation of 488 nm and emission of 525 nm).
GAPC Cloning, Protein Purification, and Activity Assay
The cDNAs of GAPC1 and GAPC2 were amplified and ligated to pET-28a-c(+) vector to produce GAPC1 and GAPC2 with six His residues at the N terminus. The recombinant plasmids were transformed into Escherichia coli BL21(DE3)pLysS. Induction and purification of protein was as described (Guo et al., 2011). Purified proteins were dialyzed in tris-buffered saline buffer with DTT overnight. Dialyzed proteins were centrifuged at 12,000g for 20 min, and protein concentration was determined using the Bradford protein assay. Purified proteins were analyzed by 10% SDS-PAGE, followed by Coomassie Brilliant Blue staining. The prepared proteins were used for activity assay or kept in 50% glycerol at −80°C. NAD-dependent GAPDH activity assay was done using purified bacterially expressed GAPC (2 to 5 μg) or total protein (25 to 50 μg) extracted from Arabidopsis leaves with modification according to the method described previously (Rius et al., 2008).
Protein Coprecipitation and Coimmunoprecipitation Assays
GST-PLDδ construct and expression of PLDδ were described previously (Qin et al., 2002). To pull down GAPC, GST-PLDδ–bound beads (∼15 µg purified proteins) were incubated with total protein extracted from E. coli expressing GAPC1 or GAPC2 at 4°C for 3 h with gentle rotation (Zhao and Wang, 2004). To pull down PLDδ, GAPC-bound agarose beads (∼10 µg purified proteins) were incubated with total protein extracted from E. coli expressing GST-PLDδ at 4°C for 3 h with gentle rotation. The beads were collected and washed three times and subjected to 10% SDS-PAGE followed by immunoblotting. To coexpress PLDδ and GAPC in yeast for coimmunoprecipitation, PLDδ and GAPC1 or GAPC2 were cloned into pESC-HIS vector and transformed into YPH yeast strain (Stratagene). PLDδ and GAPC1 or GAPC2 were coexpressed in yeast after induction by addition of galactose, and the yeasts were grown overnight at 30°C. Then, 20 μM H2O2 was added when oxidative condition was required. Primers used for cloning are listed in Supplemental Table 1 online. Total protein was extracted from harvested yeast and used for coimmunoprecipitation analysis.
SPR Analysis
SPR binding assays were performed as described with some modifications (Guo et al., 2011). The purified proteins were dialyzed in the running buffer (0.01 M HEPES, 0.15 M NaCl, and 50 µM EDTA, pH 7.4) overnight at 4°C and then the proteins were centrifuged at 13,000g to remove insoluble protein before use. For each experiment, the running buffer with 500 µM NiCl2 was injected to saturate the NTA chip with nickel. His-tagged GAPC1 protein (200 nM) was immobilized on a Biacore Sensor Chip NTA via Ni2+/NTA chelation. PLDδ-GAPC1 interaction was monitored as GST-PLDδ (200 nM) was injected in sequence over the surface of the sensor chip. The purified GST protein was used as control. During the evaluation, the sensorgrams from the beginning of association to the end of dissociation for each interaction were analyzed and plotted by SigmaPlot 10.0 (Systat Software, Inc.). Kinetic constants including Bmax, association (kon), and dissociation rate (koff) were analyzed using BIAevaluation software (GE Healthcare).
BiFC
The BiFC vectors were constructed, described, and provided by Walter et al. (2004). GAPC1 or GAPC2 cDNA was cloned into pSPYNE vector (GAPC-YFPN), and PLDδ cDNA was cloned into pSPYCE vector (PLDδ-YFPC). The constructs were transformed into C58C1 Agrobacterium tumefaciens strain and grown to stationary phase. Bacterial cells were collected and resuspended in solution containing 10 mM MES, pH 5.7, 10 mM MgCl2, and 150 mg mL−1 acetosyringone. Three-week-old Nicotiana benthamiana leaves were infiltrated with the mixed bacteria (GAPC-YFPN and PLDδ-YFPN) solutions (Voinnet et al., 2003). YFP fluorescence was examined in tobacco leaves using a Zeiss LSM 510 confocal microscope, with a 488-nm excitation mirror and a 505- to 530-nm filter to record images.
Assaying PLD Activity
For in vivo PLD activity assay, protoplasts prepared from leaves of 4-week-old plants were incubated in 0.5 mg/mL NBD-PC for 80 min on ice (Zhang et al., 2004). To determine PLD activity using fluorescent lipids, as affected by ABA treatment at different time points in vivo, 100 µM ABA was added to the NBD-PC–labeled protoplasts, and 100-μL aliquots (1.5 × 105 for each assay) were transferred to a new tube at the end of each treatment. Then, 0.4 mL hot isopropanol (75°C) was added, and the mixture was incubated for 10 min at 75°C to inactivate PLD. Lipids extraction, separation, and quantification were done according to the procedure as described (Zhang et al., 2004). To test the effect of GAPC on PLDδ, PLDδ activity was assayed using dipalmitoylglycero-3-phospho-[methyl-3H]choline as substrate according to the procedure described previously (Qin et al., 2002).
Electrospray Ionization–Tandem Mass Spectrometry Analysis of Lipid Molecular Species
Lipids were extracted and PA analyzed by electrospray ionization–tandem mass spectrometry (Xiao et al., 2010). Expanded leaves of 4- to 5-week-old plants were sprayed with 100 µM ABA with 0.01% Triton X-100. The leaves were excised and immersed in 3 mL of isopropanol with 0.01% butylated hydroxytoluene (preheated to 75°C) immediately after sampling. The experiment was repeated three times with five replicates of each treatment each time.
Statistical Analysis
Experimental values represent mean values and standard errors. n represents the number of independent samples. P values were calculated with Student\x{2019}s t test (two-tailed) using Microsoft Excel or analysis of variance (ANOVA).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: PLDα1, At3g15730; PLDδ, At4g35790; GAPC1, At3g04120; GAPC2, At1g13440; and UBQ10, At4g05320.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Confirmation of Homozygous T-DNA Insertion PLD Mutants by PCR.
Supplemental Figure 2. Expression Level of PLDδ in Response to ABA.
Supplemental Figure 3. PLDδ-GAPC Association as Identified by GAPC1 Coprecipitation of PLDδ from Microsomal Proteins of Arabidopsis Leaves.
Supplemental Figure 4. Purification and Immunoblotting of PLDδ and GAPCs Produced in E. coli and Yeast.
Supplemental Figure 5. Negative and Positive Control for BiFC.
Supplemental Figure 6. DTT Protection of GAPC Activity.
Supplemental Figure 7. Isolation of GAPC T-DNA Homozygous Lines.
Supplemental Figure 8. Growth Phenotype of the Wild Type and GAPC and PLDδ Mutants under Control and Drought Conditions.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Jörg Kudla for kindly providing the BiFC vectors, Mary Roth for technical assistance, and Ruth Welti for critical reading of the article. This work was supported by National Science Foundation Grant IOS-0818740, by U.S. Department of Energy Grant DE-SC0001295, and by U.S. Department of Agriculture Grant 2007-35318-18393.
AUTHOR CONTRIBUTIONS
L.G. and X.W. designed the research. L.G. performed most experiments. Y.Z. and W.Z. identified pldδ stomatal phenotype. X.P. and S.P.D. performed the interaction and GAPDH activity assays. R.N. performed the physiological study in Figure 6B. L.G. and X.W. analyzed the data and wrote the article.
Glossary
- ROS
reactive oxygen species
- ABA
abscisic acid
- H2O2
hydrogen peroxide
- PLD
phospholipase D
- PA
phosphatidic acid
- KO
knockout
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- NBD-PC
fluorescent-phosphatidylcholine
- GST
glutathione S-transferase
- SPR
surface plasmon resonance
- RU
response unit
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- FC
field water capacity
- WUE
water use efficiency
- MAPK
mitogen-activated protein kinase
- ANOVA
analysis of variance
- H2DCF-DA
2′,7′-dichlorofluorescin diacetate
References
- Ahuatzi D., Herrero P., de la Cera T., Moreno F. (2004). The glucose-regulated nuclear localization of hexokinase 2 in Saccharomyces cerevisiae is Mig1-dependent. J. Biol. Chem. 279: 14440–14446 [DOI] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Bae B.I., Hara M.R., Cascio M.B., Wellington C.L., Hayden M.R., Ross C.A., Ha H.C., Li X.J., Snyder S.H., Sawa A. (2006). Mutant huntingtin: Nuclear translocation and cytotoxicity mediated by GAPDH. Proc. Natl. Acad. Sci. USA 103: 3405–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann B.O., Laxalt A.M., ter Riet B., van Schooten B., Merquiol E., Testerink C., Haring M.A., Bartels D., Munnik T. (2009). Multiple PLDs required for high salinity and water deficit tolerance in plants. Plant Cell Physiol. 50: 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y.H., Yoo S.D., Sheen J. (2006). Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127: 579–589 [DOI] [PubMed] [Google Scholar]
- Desikan R., A-H-Mackerness S., Hancock J.T., Neill S.J. (2001). Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 127: 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T. (2011). Signal transduction by reactive oxygen species. J. Cell Biol. 194: 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev T.S., Van Breusegem F., Stone J.M., Denev I., Laloi C. (2006). Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays 28: 1091–1101 [DOI] [PubMed] [Google Scholar]
- Guo L., Mishra G., Taylor K., Wang X. (2011). Phosphatidic acid binds and stimulates Arabidopsis sphingosine kinases. J. Biol. Chem. 286: 13336–13345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock J.T., Henson D., Nyirenda M., Desikan R., Harrison J., Lewis M., Hughes J., Neill S.J. (2005). Proteomic identification of glyceraldehyde 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxide in Arabidopsis. Plant Physiol. Biochem. 43: 828–835 [DOI] [PubMed] [Google Scholar]
- Hara M.R., et al. (2005). S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 7: 665–674 [DOI] [PubMed] [Google Scholar]
- Harada N., Yasunaga R., Higashimura Y., Yamaji R., Fujimoto K., Moss J., Inui H., Nakano Y. (2007). Glyceraldehyde-3-phosphate dehydrogenase enhances transcriptional activity of androgen receptor in prostate cancer cells. J. Biol. Chem. 282: 22651–22661 [DOI] [PubMed] [Google Scholar]
- Holtgrefe S., Gohlke J., Starmann J., Druce S., Klocke S., Altmann B., Wojtera J., Lindermayr C., Scheibe R. (2008). Regulation of plant cytosolic glyceraldehyde 3-phosphate dehydrogenase isoforms by thiol modifications. Physiol. Plant. 133: 211–228 [DOI] [PubMed] [Google Scholar]
- Jammes F., et al. (2009). MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc. Natl. Acad. Sci. USA 106: 20520–20525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski S., Reynolds H., Karpinska B., Wingsle G., Creissen G., Mullineaux P. (1999). Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 284: 654–657 [DOI] [PubMed] [Google Scholar]
- Katagiri T., Takahashi S., Shinozaki K. (2001). Involvement of a novel Arabidopsis phospholipase D, AtPLDδ, in dehydration-inducible accumulation of phosphatidic acid in stress signalling. Plant J. 26: 595–605 [DOI] [PubMed] [Google Scholar]
- Lanteri M.L., Lamattina L., Laxalt A.M. (2011). Mechanisms of xylanase-induced nitric oxide and phosphatidic acid production in tomato cells. Planta 234: 845–855 [DOI] [PubMed] [Google Scholar]
- Li W., Li M., Zhang W., Welti R., Wang X. (2004). The plasma membrane-bound phospholipase Ddelta enhances freezing tolerance in Arabidopsis thaliana. Nat. Biotechnol. 22: 427–433 [DOI] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J., Bermúdez M.A., Segura J., Ros R. (2011). Arabidopsis plants deficient in plastidial glyceraldehyde-3-phosphate dehydrogenase show alterations in abscisic acid (ABA) signal transduction: Interaction between ABA and primary metabolism. J. Exp. Bot. 62: 1229–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J., Cascales-Miñana B., Irles-Segura A., Mateu I., Nunes-Nesi A., Fernie A.R., Segura J., Ros R. (2010). The plastidial glyceraldehyde-3-phosphate dehydrogenase is critical for viable pollen development in Arabidopsis. Plant Physiol. 152: 1830–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J., Cascales-Miñana B., Mulet J.M., Baroja-Fernández E., Pozueta-Romero J., Kuhn J.M., Segura J., Ros R. (2009). Plastidial glyceraldehyde-3-phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis. Plant Physiol. 151: 541–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z.M., Murata Y., Benning G., Thomine S., Klüsener B., Allen G.J., Grill E., Schroeder J.I. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406: 731–734 [DOI] [PubMed] [Google Scholar]
- Peters C., Li M., Narasimhan R., Roth M., Welti R., Wang X. (2010). Nonspecific phospholipase C NPC4 promotes responses to abscisic acid and tolerance to hyperosmotic stress in Arabidopsis. Plant Cell 22: 2642–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton W.C. (1996). The organization and regulation of plant glycolysis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 185–214 [DOI] [PubMed] [Google Scholar]
- Qin C., Wang C., Wang X. (2002). Kinetic analysis of Arabidopsis phospholipase Ddelta. Substrate preference and mechanism of activation by Ca2+ and phosphatidylinositol 4,5-biphosphate. J. Biol. Chem. 277: 49685–49690 [DOI] [PubMed] [Google Scholar]
- Quan L.J., Zhang B., Shi W.W., Li H.Y. (2008). Hydrogen peroxide in plants: A versatile molecule of the reactive oxygen species network. J. Integr. Plant Biol. 50: 2–18 [DOI] [PubMed] [Google Scholar]
- Queval G., Hager J., Gakière B., Noctor G. (2008). Why are literature data for H2O2 contents so variable? A discussion of potential difficulties in the quantitative assay of leaf extracts. J. Exp. Bot. 59: 135–146 [DOI] [PubMed] [Google Scholar]
- Rius S.P., Casati P., Iglesias A.A., Gomez-Casati D.F. (2006). Characterization of an Arabidopsis thaliana mutant lacking a cytosolic non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase. Plant Mol. Biol. 61: 945–957 [DOI] [PubMed] [Google Scholar]
- Rius S.P., Casati P., Iglesias A.A., Gomez-Casati D.F. (2008). Characterization of Arabidopsis lines deficient in GAPC-1, a cytosolic NAD-dependent glyceraldehyde-3-phosphate dehydrogenase. Plant Physiol. 148: 1655–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y., Cui D., Wang X. (2001). Phospholipase D and phosphatidic acid-mediated generation of superoxide in Arabidopsis. Plant Physiol. 126: 1449–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H.B., Chu L.Y., Shao M.A., Jaleel C.A., Mi H.M. (2008). Higher plant antioxidants and redox signaling under environmental stresses. C. R. Biol. 331: 433–441 [DOI] [PubMed] [Google Scholar]
- Sheshshayee M.S., Bindumadhava H., Ramesh R., Prasad T.G., Lakshminarayana M.R., Udayakumar M. (2005). Oxygen isotope enrichment (delta18O) as a measure of time-averaged transpiration rate. J. Exp. Bot. 56: 3033–3039 [DOI] [PubMed] [Google Scholar]
- Sirover M.A. (1997). Role of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in normal cell function and in cell pathology. J. Cell. Biochem. 66: 133–140 [PubMed] [Google Scholar]
- Suzuki N., Koussevitzky S., Mittler R., Miller G. (2012). ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 35: 259–270 [DOI] [PubMed] [Google Scholar]
- Veljovic-Jovanovic S.D., Pignocchi C., Noctor G., Foyer C.H. (2001). Low ascorbic acid in the vtc-1 mutant of Arabidopsis is associated with decreased growth and intracellular redistribution of the antioxidant system. Plant Physiol. 127: 426–435 [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang C., Wang X. (2001). A novel phospholipase D of Arabidopsis that is activated by oleic acid and associated with the plasma membrane. Plant Physiol. 127: 1102–1112 [PMC free article] [PubMed] [Google Scholar]
- Wang X., Devaiah S.P., Zhang W., Welti R. (2006). Signaling functions of phosphatidic acid. Prog. Lipid Res. 45: 250–278 [DOI] [PubMed] [Google Scholar]
- Welti R., Li W., Li M., Sang Y., Biesiada H., Zhou H.E., Rajashekar C.B., Williams T.D., Wang X. (2002). Profiling membrane lipids in plant stress responses. Role of phospholipase D α in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 277: 31994–32002 [DOI] [PubMed] [Google Scholar]
- Xiao S., Gao W., Chen Q.F., Chan S.W., Zheng S.X., Ma J., Wang M., Welti R., Chye M.L. (2010). Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell 22: 1463–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Tanabe S., Minami E., Shibuya N. (2004). Activation of phospholipase D induced by hydrogen peroxide in suspension-cultured rice cells. Plant Cell Physiol. 45: 1261–1270 [DOI] [PubMed] [Google Scholar]
- Yu L., Nie J., Cao C., Jin Y., Yan M., Wang F., Liu J., Xiao Y., Liang Y., Zhang W. (2010). Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 188: 762–773 [DOI] [PubMed] [Google Scholar]
- Zhang A., Jiang M., Zhang J., Tan M., Hu X. (2006). Mitogen-activated protein kinase is involved in abscisic acid-induced antioxidant defense and acts downstream of reactive oxygen species production in leaves of maize plants. Plant Physiol. 141: 475–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Qin C., Zhao J., Wang X. (2004). Phospholipase D α 1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 101: 9508–9513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Wang C., Qin C., Wood T., Olafsdottir G., Welti R., Wang X. (2003). The oleate-stimulated phospholipase D, PLDdelta, and phosphatidic acid decrease H2O2-induced cell death in Arabidopsis. Plant Cell 15: 2285–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Yu L., Zhang Y., Wang X. (2005). Phospholipase D in the signaling networks of plant response to abscisic acid and reactive oxygen species. Biochim. Biophys. Acta 1736: 1–9 [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhang L., Dong F., Gao J., Galbraith D.W., Song C.P. (2001). Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol. 126: 1438–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhu H., Zhang Q., Li M., Yan M., Wang R., Wang L., Welti R., Zhang W., Wang X. (2009). Phospholipase dalpha1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 21: 2357–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Wang X. (2004). Arabidopsis phospholipase Dalpha1 interacts with the heterotrimeric G-protein α-subunit through a motif analogous to the DRY motif in G-protein-coupled receptors. J. Biol. Chem. 279: 1794–1800 [DOI] [PubMed] [Google Scholar]
- Zhao Z., Assmann S.M. (2011). The glycolytic enzyme, phosphoglycerate mutase, has critical roles in stomatal movement, vegetative growth, and pollen production in Arabidopsis thaliana. J. Exp. Bot. 62: 5179–5189 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.