Mitogen-activated protein kinase kinase kinase MEKK2 functions as a positive regulator of plant immunity mediated by the nucleotide binding–Leu-rich repeat Resistance protein SUMM2. MEKK2 interacts with MPK4 and is negatively regulated by the MEKK1-MKK1/MKK2-MPK4 kinase cascade.
Abstract
In Arabidopsis thaliana, the MEKK1-MKK1/MKK2-MPK4 mitogen-activated protein (MAP) kinase cascade represses cell death and immune responses. In mekk1, mkk1 mkk2, and mpk4 mutants, programmed cell death and defense responses are constitutively activated, but the mechanism by which MEKK1, MKK1/MKK2, and MPK4 negatively regulate cell death and immunity was unknown. From a screen for suppressors of mkk1 mkk2, we found that mutations in suppressor of mkk1 mkk2 1 (summ1) suppress the cell death and defense responses not only in mkk1 mkk2 but also in mekk1 and mpk4. SUMM1 encodes the MAP kinase kinase kinase MEKK2. It interacts with MPK4 and is phosphorylated by MPK4 in vitro. Overexpression of SUMM1 activates cell death and defense responses that are dependent on the nucleotide binding–leucine-rich repeat protein SUMM2. Taken together, our data suggest that the MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates MEKK2 and activation of MEKK2 triggers SUMM2-mediated immune responses.
INTRODUCTION
Plants use a large repertoire of immune receptors to sense attacks by microbial pathogens and trigger downstream defense responses. One type of immune receptor recognizes conserved microbial components collectively known as pathogen-associated molecular patterns (PAMPs) or microbe-associated molecular patterns (Boller and Felix, 2009). PAMP receptors are usually transmembrane receptor-like kinases (RLKs) or receptor-like proteins that directly interact with PAMPs. The other type of intracellular immune receptors known as Resistance (R) proteins recognizes effector molecules secreted by pathogens (Eitas and Dangl, 2010). This recognition can be either direct or indirect. Most R proteins belong to the nucleotide binding domain and leucine-rich repeat–containing (NB-LRR) protein family.
In plant defense responses, mitogen-activated protein kinase (MAPK) cascades play important roles in transducing signals from upstream receptors to the downstream targets (Pitzschke et al., 2009a). A MAPK cascade is a signaling module usually consisting of a MAP kinase kinase kinase (MAPKKK), a MAP kinase kinase, and a MAPK. Activation of MAPKKKs by upstream signals results in sequential phosphorylation of their downstream MAPKKs and MAPKs. Perception of PAMP signals by PAMP receptors leads to the activation of at least two MAPK cascades composed of MEKK1-MKK4/MKK5-MPK3/MPK6 or MEKK1-MKK1/MKK2-MPK4 (Boller and Felix, 2009).
Without challenges from pathogens, plant defense responses mediated by different plant immune receptors have to be kept under tight control to prevent autoimmunity. Mutants with autoactivated immune responses often exhibit dwarf morphology, accumulate high levels of the defense hormone salicylic acid (SA), and constitutively express immunity marker genes, such as PR1 and PR2. Gain-of-function mutations in immune receptors can lead to autoimmunity, and loss-of-function mutations in negative regulators that prevent autoimmune responses can also result in constitutive activation of defense responses. For example, a number of mutations in NB-LRR–type R genes, such as snc1, ssi4, slh1, Rx, uni-1D, and chs3-3D, have been found to constitutively activate downstream defense responses (Bendahmane et al., 2002; Shirano et al., 2002; Zhang et al., 2003a; Noutoshi et al., 2005; Igari et al., 2008; Bi et al., 2011). Gain-of-function mutations in the RLK SNC4 and receptor-like protein SNC2 also activate downstream defense responses (Bi et al., 2010; Zhang et al., 2010). Additionally, recessive mutations in SRFR1 and CPR1 lead to increased accumulation of R proteins SNC1 and RPS2 and constitutive activation of R protein–mediated immune responses, suggesting that negative regulation of R protein accumulation is important for preventing autoimmunity (Kim et al., 2010; Li et al., 2010; Cheng et al., 2011; Gou et al., 2012). Furthermore, loss of function of the RLK BIR1 causes activation of immunity mediated by another RLK SOBIR1 (Gao et al., 2009).
About a decade ago, it was reported that knocking out Arabidopsis thaliana MPK4 results in constitutive defense responses (Petersen et al., 2000). The mpk4 mutant plants are dwarf, accumulate high levels of SA, and exhibit enhanced pathogen resistance. The dwarf phenotype of mpk4 can be partially suppressed by silencing MAP kinase 4 substrate1 (MKS1) (Andreasson et al., 2005). Later studies showed that mekk1 single mutants and mkk1 mkk2 double mutants also exhibit similar phenotypes like mpk4 (Ichimura et al., 2006; Nakagami et al., 2006; Suarez-Rodriguez et al., 2007; Gao et al., 2008; Qiu et al., 2008; Pitzschke et al., 2009b). Analysis of mekk1 sid2 and mkk1 mkk2 sid2 mutant plants revealed that the elevated SA levels contribute very little to the mutant phenotypes in mekk1 and mkk1 mkk2 (Ichimura et al., 2006; Qiu et al., 2008).
MKK1 and MKK2 interact with MEKK1 and MPK4 in vivo, and activation of MPK4 by flg22 requires MEKK1 as well as MKK1 and MKK2, suggesting that MEKK1, MKK1/MKK2, and MPK4 form a MAPK cascade to negatively regulate plant immune responses. However, the mechanism on how this kinase cascade regulates plant immunity is unclear. In this study, we show that the autoimmunity phenotypes in mpk4, mekk1, and mkk1 mkk2 mutant plants are caused by activation of defense responses mediated by SUMM1 (for SUPPRESSOR OF mkk1 mkk2 1). SUMM1 encodes the MAPKKK MEKK2, which is directly targeted by MPK4.
RESULTS
Identification and Characterization of summ1 Mutants
To understand the mechanism of how the MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates plant immunity, a screen for suppressors of mkk1 mkk2 was performed. Seeds of mkk1-1 mkk2-1 were obtained by growing the mutant plants at 28°C, as high temperature partially suppresses the mutant phenotype of mkk1 mkk2 (Gao et al., 2008). The mkk1-1 mkk2-1 seeds were subsequently mutagenized with ethyl methanesulfonate (EMS), and the M1 plants were grown at 28°C to maturity. To identify mutants that suppress the seedling lethality phenotypes of mkk1 mkk2, M2 plants were grown on soil at 23°C. A total of ∼50 summ mutants were identified. Two alleles of summ1, summ1-1 and summ1-2, were characterized in detail.
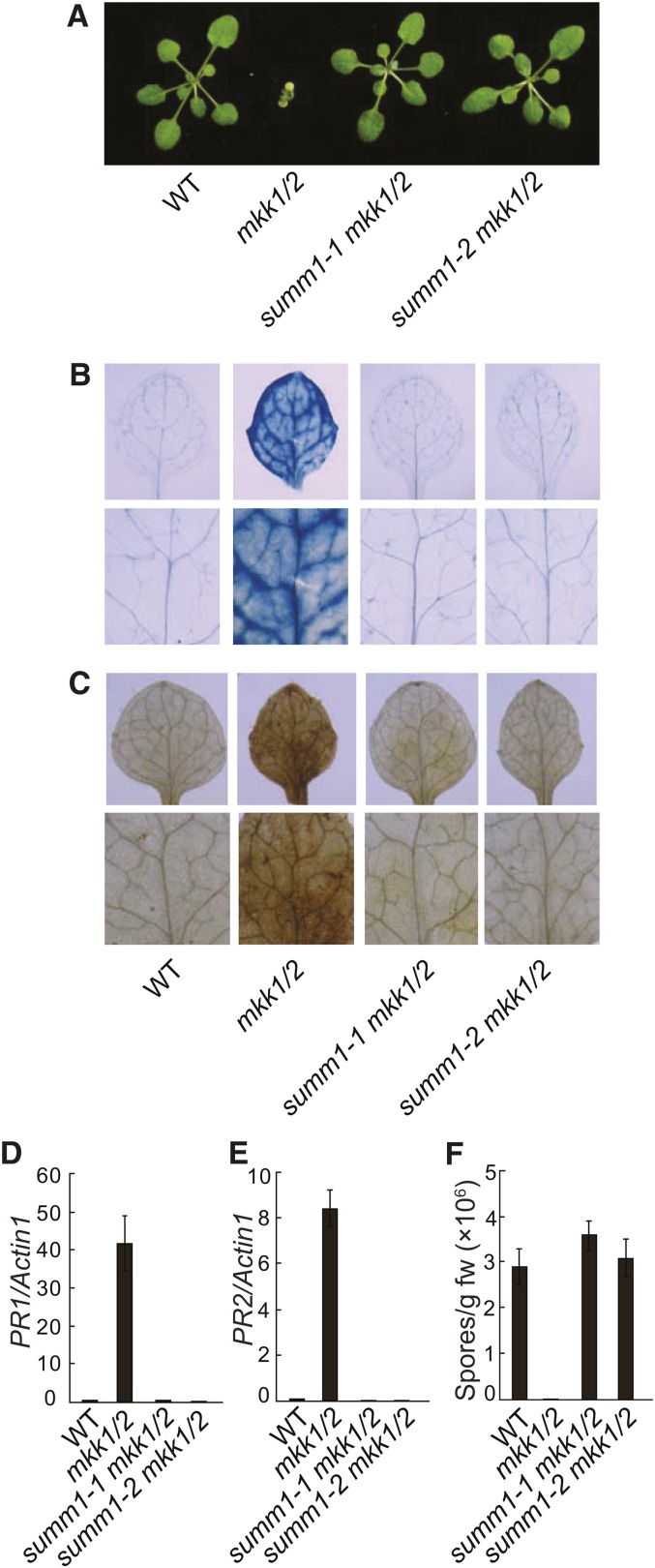
As shown in Figure 1A, summ1-1 mkk1 mkk2 and summ1-2 mkk1 mkk2 exhibited wild type–like morphology. Trypan blue staining revealed that the extensive cell death observed in the mkk1 mkk2 double mutant was completely suppressed in summ1-1 mkk1 mkk2 and summ1-2 mkk1 mkk2 (Figure 1B). 3,3′-diaminobenzidine (DAB) staining showed that accumulation of hydrogen peroxide (H2O2) in mkk1 mkk2 was also blocked in the triple mutants (Figure 1C). In mkk1 mkk2, defense responses are constitutively activated. As shown in Figures 1D and 1E, constitutive expression of defense marker genes PR1 and PR2 was also completely suppressed in the summ1-1 mkk1 mkk2 and summ1-2 mkk1 mkk2 triple mutants.
Figure 1.
Suppression of mkk1 mkk2 Mutant Phenotypes by summ1-1 and summ1-2.
(A) Morphology of the wild type (WT), mkk1 mkk2 (mkk1/2), summ1-1 mkk1 mkk2, and summ1-2 mkk1 mkk2. The photograph shows 4-week-old soil-grown plants.
(B) and (C) Trypan blue (B) and DAB (C) staining of wild-type, mkk1 mkk2 (mkk1/2), summ1-1 mkk1 mkk2, and summ1-2 mkk1 mkk2 seedlings.
(D) and (E) PR1 (D) and PR2 (E) expression in wild-type, mkk1 mkk2 (mkk1/2), summ1-1 mkk1 mkk2, and summ1-2 mkk1 mkk2 seedlings. Values were normalized to the expression of ACTIN1. Error bars represent ±sd of three replicates.
(F) Growth of H.a. Noco2 on the wild type, mkk1 mkk2 (mkk1/2), summ1-1 mkk1 mkk2, and summ1-2 mkk1 mkk2. Three-week-old seedlings were sprayed with H.a. Noco2 spores (5 × 104 spores/mL). Infections were scored 7 d after inoculation by counting the number of resuspended conidia spores per gram of leaf samples. Error bars represent ±sd of three replicates. This experiment was repeated three times with similar results.
To determine whether pathogen resistance in mkk1 mkk2 is affected by the summ1 mutations, seedlings of summ1-1 mkk1 mkk2 and summ1-2 mkk1 mkk2 were challenged with the virulent oomycete pathogen Hyaloperonospora arabidopsidis (H.a.) Noco2. While H.a. Noco2 failed to grow on the mkk1 mkk2 double mutant, growth of the pathogen on summ1-1 mkk1 mkk2 and summ1-2 mkk1 mkk2 plants was comparable to that on the wild type, suggesting that constitutive pathogen resistance in mkk1 mkk2 was suppressed by the summ1 mutations (Figure 1F).
Positional Cloning of SUMM1
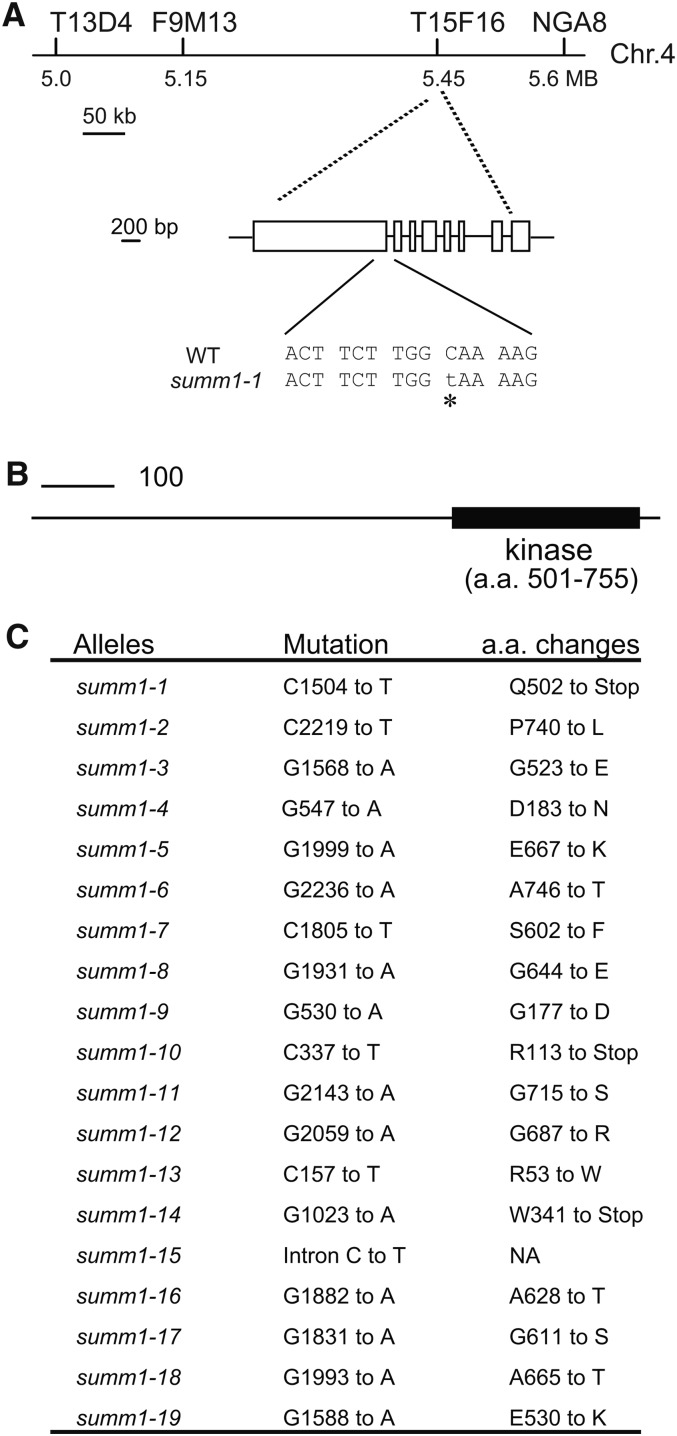
To map the summ1-1 mutation, summ1-1 mkk1 mkk2 (in the Columbia [Col] background) was crossed with Landsberg erecta (Ler). F2 plants homozygous for mkk1 mkk2 were selected for linkage analysis. Crude mapping using 48 such plants revealed that summ1-1 was flanked by markers T13D4 and T15F16 on chromosome 4 (Figure 2A). Further mapping using progeny of plants that were homozygous for mkk1 mkk2 but heterozygous for summ1 indicated that the summ1-1 mutation is between markers F9M13 and T15F16, a region of ∼300 kb. We reasoned that SUMM1 may be induced by pathogen infections. To identify the summ1-1 mutation, candidate genes in this region whose expression is induced by pathogen infections were identified using the microarray database at The Arabidopsis Information Resource and sequenced. Sequencing of candidate genes revealed a C-to-T mutation in the coding region of At4g08480. The mutation introduced an early stop codon in At4g08480, which encodes the MAPKKK MEKK2, also known as MAPKKK9 (Figure 2A).
Figure 2.
Map-Based Cloning of SUMM1.
(A) Map position and the mutation in summ1-1. WT, the wild type.
(B) Protein structure of SUMM1/MEKK2. a.a., amino acids.
(C) Mutations identified in the summ1 alleles and the consequences of mutations to SUMM1/MEKK2 protein.
Sequence analysis of At4g08480 in summ1-2 revealed that it also contains a mutation in MEKK2. This mutation changes Pro-740 to Leu. To determine whether other summ mutants also contain mutations in At4g08480, At4g08480 was amplified from the other 48 mutants by PCR and sequenced. Seventeen additional summ mutants were found to contain mutations in At4g08480 (Figures 2C; see Supplemental Figure 1 online). Most of the mutations result in amino acid changes in the kinase domain. These results suggest that SUMM1 is At4g08480.
Suppression of Cell Death and Defense Responses of mpk4 by summ1-1
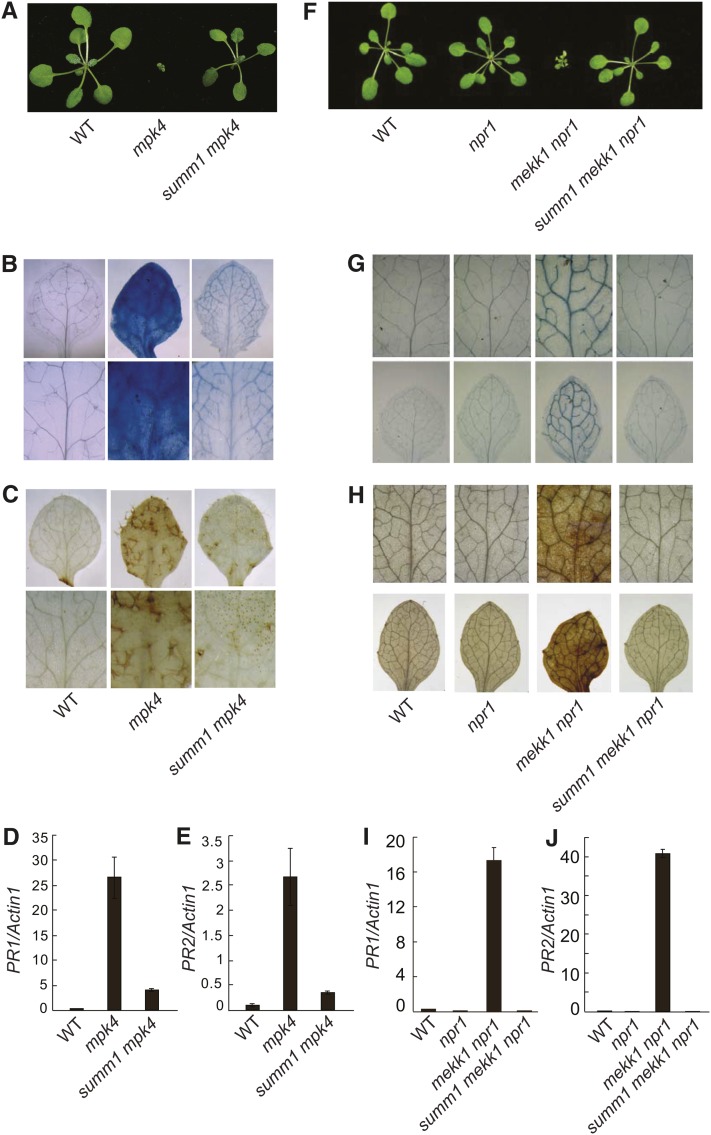
MPK4 has previously been shown to function downstream of MKK1 and MKK2 (Gao et al., 2008; Qiu et al., 2008). Like the mkk1 mkk2 double mutant, mpk4 mutants in Col background are also seedling lethal. To determine whether the mutant phenotypes in mpk4-3 can be suppressed by summ1-1, the summ1-1 mpk4-3 double mutant was created by crossing mpk4-3 (in Col) and summ1-1 mkk1 mkk2. As shown in Figure 3A, the dwarf morphology of mpk4-3 was completely suppressed by summ1-1, and no obvious cell death was observed in the double mutant.
Figure 3.
Suppression of mpk4-3 and mekk1-4 Mutant Phenotypes by summ1 Mutants.
(A) Morphology of the wild type (WT), mpk4-3 (mpk4), and summ1-1 mpk4-3 (summ1 mpk4). The photograph shows 4-week-old soil grown plants.
(B) and (C) Trypan blue (B) and DAB (C) staining of wild-type, mpk4-3, and summ1-1 mpk4-3 seedlings.
(D) and (E) PR1 (D) and PR2 (E) expression in wild-type, mpk4-3, and summ1-1 mpk4-3 seedlings. Values were normalized to the expression of ACTIN1. Error bars represent ±sd of three replicates.
(F) Morphology of the wild type, npr1-1 (npr1), mekk1-4 npr1-1 (mekk1 npr1), and summ1-3 mekk1-4 npr1-1 (summ1 mekk1 npr1). The photograph was taken on 4-week-old soil-grown plants.
(G) and (H) Trypan blue (G) and DAB (H) staining of wild-type, npr1-1, mekk1-4 npr1-1, and summ1-3 mekk1-4 npr1-1 seedlings.
(I) and (J) PR1 (I) and PR2 (J) expression in the wild type, npr1-1, mekk1-4 npr1-1, and summ1-3 mekk1-4 npr1-1. Values were normalized to the expression of ACTIN1. Error bars represent ±sd of three replicates.
Trypan blue staining showed that the massive cell death observed in mpk4-3 was largely suppressed in the summ1-1 mpk4-3 double mutant (Figure 3B). However, there is still some staining in the double mutant, suggesting the existence of microscopic cell death in summ1-1 mpk4-3. DAB staining showed that the H2O2 level in summ1-1 mpk4-3 was much lower than that in mpk4-3 but still modestly higher than in the wild type (Figure 3C). Analysis of PR gene expression indicated that the expression of PR1 and PR2 in summ1-1 mpk4-3 is dramatically reduced compared to those in mpk4-3, but still higher than the wild type (Figures 3D and 3E). These data suggest that the activation of cell death and defense responses in mpk4 is mainly dependent on SUMM1.
Suppression of Cell Death and Defense Responses in mekk1 by summ1-3
MEKK1 functions upstream of MKK1 and MKK2, and mutations in MEKK1 result in similar mutant phenotypes as the mkk1 mkk2 double mutant (Ichimura et al., 2006; Nakagami et al., 2006; Suarez-Rodriguez et al., 2007). We therefore speculated that mekk1 activates SUMM1-dependent defense responses, and a double mutant of mekk1 and summ1 would help us to test our hypothesis. Because SUMM1 and MEKK1 are closely linked and the distance between the two genes is only ∼12 kb, it is not feasible to obtain the summ1 mekk1 double mutant by crossing summ1 and mekk1. To address this problem, we mutagenized mekk1-4 npr1-1 (Gao et al., 2008) with EMS and screened for mutants that suppressed the seedling lethality phenotype of the mutant. The mutants were subsequently sequenced to determine whether they contained mutations in SUMM1.
One of the mutants that completely suppressed the morphological phenotypes of mekk1-4 npr1-1 was found to contain a mutation in SUMM1 (Figure 3F). The mutation is the same as the summ1-3 mutation identified from the suppressor screen of mkk1 mkk2 (Figure 2C). When a construct expressing SUMM1 with a C-terminal 3xFLAG tag under its own promoter was transformed into summ1-3 mekk1-4 npr1-1, all transgenic plants displayed a seedling-lethal phenotype, suggesting that suppression of the seedling-lethal phenotype of mekk1-4 npr1-1 was caused by the summ1-3 mutation. Trypan blue staining showed that cell death in mekk1-4 npr1-1 was suppressed by summ1-3 (Figure 3G). DAB staining indicated that the elevated H2O2 level in mekk1-4 npr1-1 was reduced to the wild-type level in summ1-3 mekk1-4 npr1-1 (Figure 3H). Analysis of defense gene expression showed that expression of PR1 (Figure 3I) and PR2 (Figure 3J) in summ1-3 mekk1-4 npr1-1 was also comparable to that in wild-type plants. These data indicate that summ1-3 completely suppress the cell death and constitutive defense responses in mekk1-4 npr1-1.
flg22-Induced PAMP Responses Are Not Affected in summ1-1
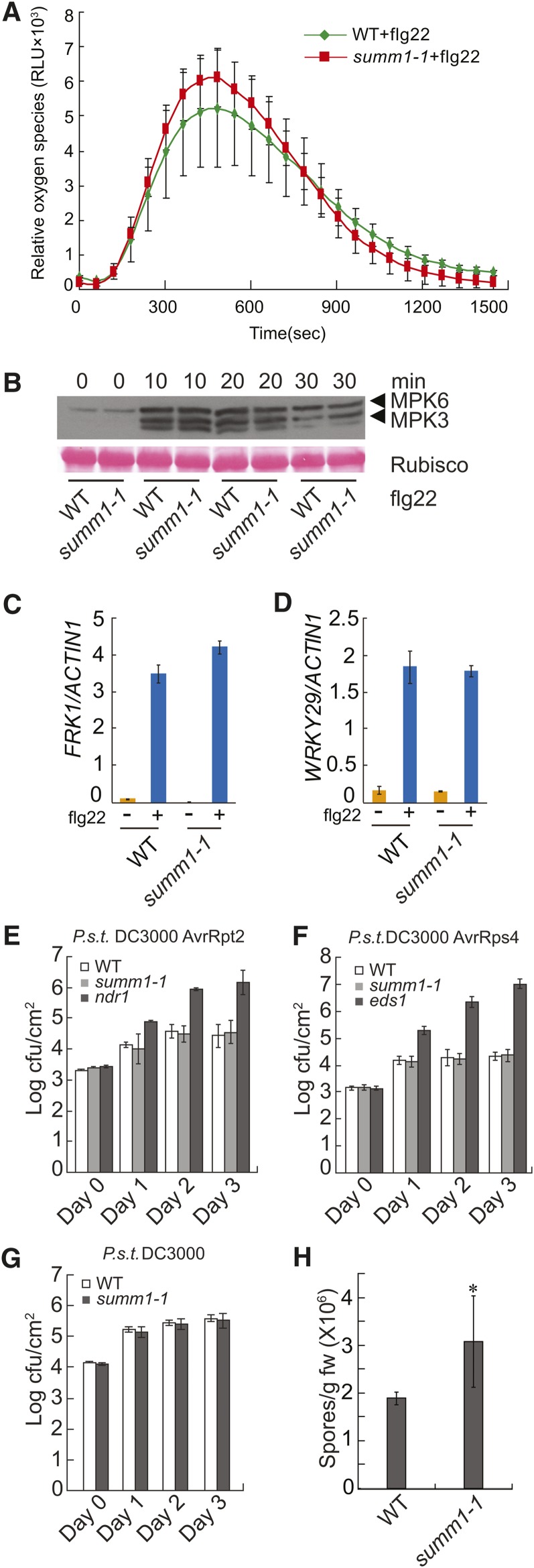
MEKK1 was previously shown to be required for PAMP-induced activation of MPK4 (Ichimura et al., 2006; Nakagami et al., 2006; Suarez-Rodriguez et al., 2007). Because SUMM1/MEKK2 shares high sequence similarity with MEKK1, we tested whether SUMM1 is required for flg22-induced reactive oxygen species (ROS) production, activation of MAPKs, and upregulation of FRK1 and WRKY29. As shown in Figure 4A, induction of ROS by flg22 was comparable in the wild type and summ1-1. Activation of MPK3 and MPK6 was not affected in summ1-1 either (Figure 4B). Real-time RT-PCR showed that induction of FRK1 and WRKY29 was also comparable in the wild type and summ1-1 (Figures 4C and 4D). These data indicate that flg22-induced PAMP responses are intact in summ1-1.
Figure 4.
Analysis of Different Immune Responses in summ1-1.
(A) flg22-induced oxidative burst in summ1-1. Leaf slices were treated with 1 μM flg22 before ROS was measured. Error bars represent the sd of 12 independent samples. RLU, relative luminescence units; WT, wild type.
(B) flg22-induced MAPKs activation. Two-week-old seedlings grown on half-strength MS medium were treated with 10 μM of flg22. Samples were collected at 0, 10, 20, and 30 min and analyzed by immunoblots using an anti-Erk antibody (Cell Signaling; #4370S).
(C) and (D) Real-time RT-PCR analysis of the induction of FRK1 (C) and WRKY29 (D) in the wild type and summ1-1 by flg22. Two-week-old seedlings grown on half-strength MS plates were sprayed with 10 μM of flg22 4 h before samples were taken. Error bars represent ±sd of three replicates.
(E) to (G) Growth of P.s.t. DC3000 avrRpt2 (E), P.s.t. DC3000 avrRps4 (F), and P.s.t. DC3000 (G) on the indicated genotypes. Five-week-old plants grown under short-day conditions were infiltrated with P.s.t. DC3000 at a concentration of OD600 = 0.002 and P.s.t. DC3000 avrRpt2 and P.s.t. DC3000 avrRps4 at a concentration of OD600 = 0.001. Samples were taken at 0 h (Day 0), 24 h (Day 1), 48 h (Day 2), and 72 h (Day 3) after inoculation, respectively. Error bars represent ±sd of six replicates.
(H) Growth of H.a. Noco2 on the wild type and summ1-1. Four-week-old plants were sprayed with spores of H.a. Noco2 at a concentration of 50,000 spores/mL. Error bars represent sd of three replicates. *P < 0.05, statistical difference from the wild type.
All experiments in this figure were independently repeated three times with similar results.
[See online article for color version of this figure.]
SUMM1 Is Not Required for Resistance Mediated by RPS2 and RPS4
Next, we tested whether resistance mediated by R genes such as RPS2 and RPS4 is affected in summ1-1. As shown in Figures 4E and 4F, growth of Pseudomonas syringae pv tomato (P.s.t.) DC3000 carrying avrRpt2 or avrRps4 was comparable in the wild type and summ1-1, suggesting that resistance mediated by RPS2 and RPS4 was not affected by summ1-1 and SUMM1 is not a general defense regulator downstream of plant immune receptors. We further tested whether SUMM1 is required for basal resistance against virulent pathogens. As shown in Figure 4G, growth of the virulent P.s.t. DC3000 on wild-type and summ1-1 plants was similar. Interestingly, when summ1-1 was challenged with H.a. Noco2, it supported significantly higher growth of the oomycete pathogen (Figure 4H), suggesting that summ1-1 affects basal resistance against H.a. Noco2.
Overexpression of SUMM1 Activates Cell Death and Defense Responses
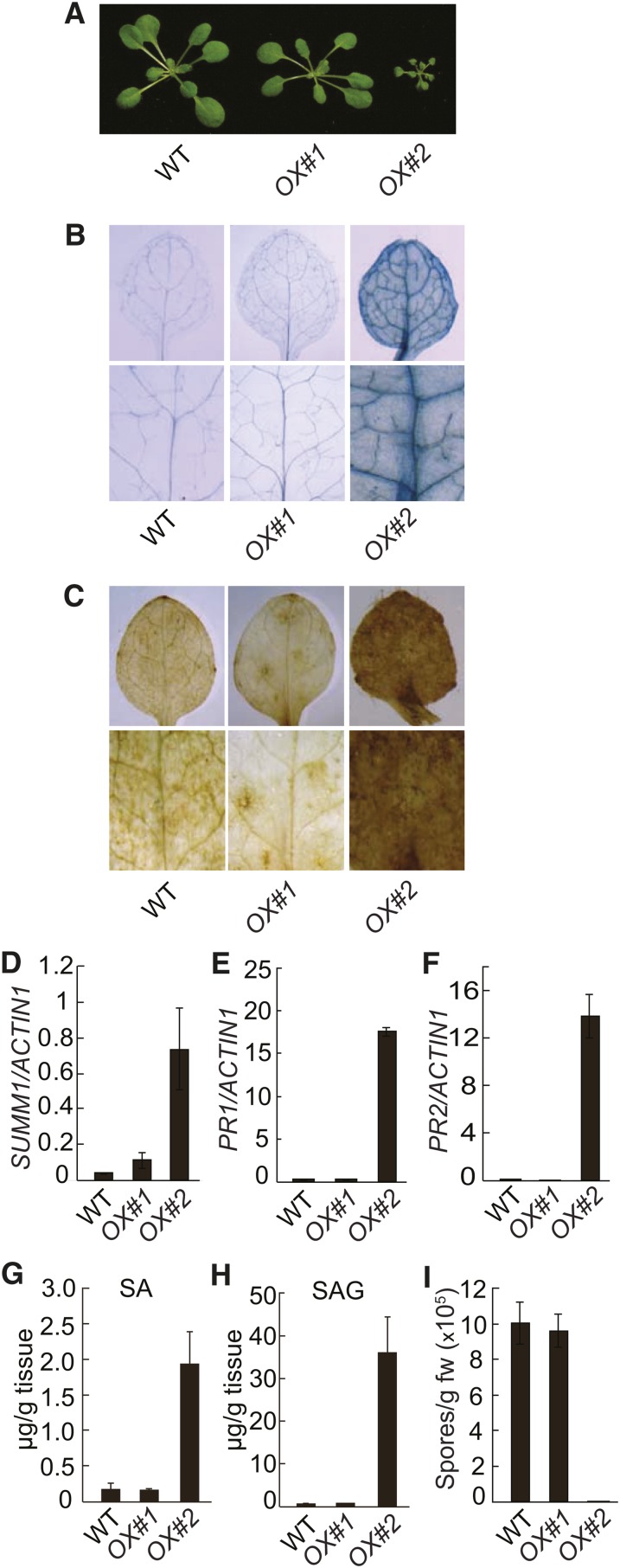
When SUMM1 with a C-terminal 3xFLAG-tag was expressed in wild-type plants under its own promoter, about half of T1 transgenic plants exhibited dwarf morphology, suggesting that defense responses might be activated in these plants. Two representative SUMM1-FLAG lines with SUMM1-FLAG expressed at different levels were characterized in detail. As shown in Figure 5A, line #2 was much smaller than the wild type and line #1. Trypan blue staining showed that there was extensive cell death in line #2 (Figure 5B). Line #2 also accumulated high levels of H2O2 (Figure 5C) compared with the wild type and line #1. Real-time RT-PCR showed that the expression of SUMM1 in line #2 was about 10-fold higher than in line #1 (Figure 5D). In addition, both PR1 (Figure 5E) and PR2 (Figure 5F) were constitutively expressed in line #2. Analysis of SA levels showed that both free and total SA accumulated more in line #2 compared with the wild type and line #1 (Figures 5G and 5H). Furthermore, line #2 exhibited strongly enhanced resistance to H.a. Noco2 (Figure 5I). These data suggest that overexpression of SUMM1 leads to constitutive activation of cell death and defense responses.
Figure 5.
Overexpression of SUMM1 Leads to Activation of Cell Death and Defense Responses.
(A) Morphology of the wild type (WT) and SUMM1-FLAG transgenic lines OX#1 and OX#2. The photograph shows soil-grown plants ∼4 weeks after planting.
(B) and (C) Trypan blue (B) and DAB (C) staining of the seedlings of the wild type and the SUMM1-FLAG transgenic lines OX#1 and OX#2.
(D) Real-time RT-PCR analysis of SUMM1 expression in the wild type and the SUMM1-FLAG transgenic lines OX#1 and OX#2. Error bars represent ±sd of three replicates.
(E) and (F) PR1 (E) and PR2 (F) expression in the wild type and the SUMM1-FLAG transgenic lines. Error bars represent ±sd of three replicates.
(G) and (H) Free SA (G) and SAG (H) levels in the wild type and the SUMM1-FLAG transgenic lines. This experiment was repeated twice with similar results. Error bars represent ±sd of four replicates.
(I) Growth of H.a. Noco2 on the wild type and the SUMM1-FLAG transgenic lines. Inoculation of the pathogen and scoring of the infection were performed as shown in Figure 1F. Error bars represent ±sd of three replicates. This experiment was repeated three times with similar results. fw, fresh weight.
SUMM1/MEKK2 Interacts with MPK4
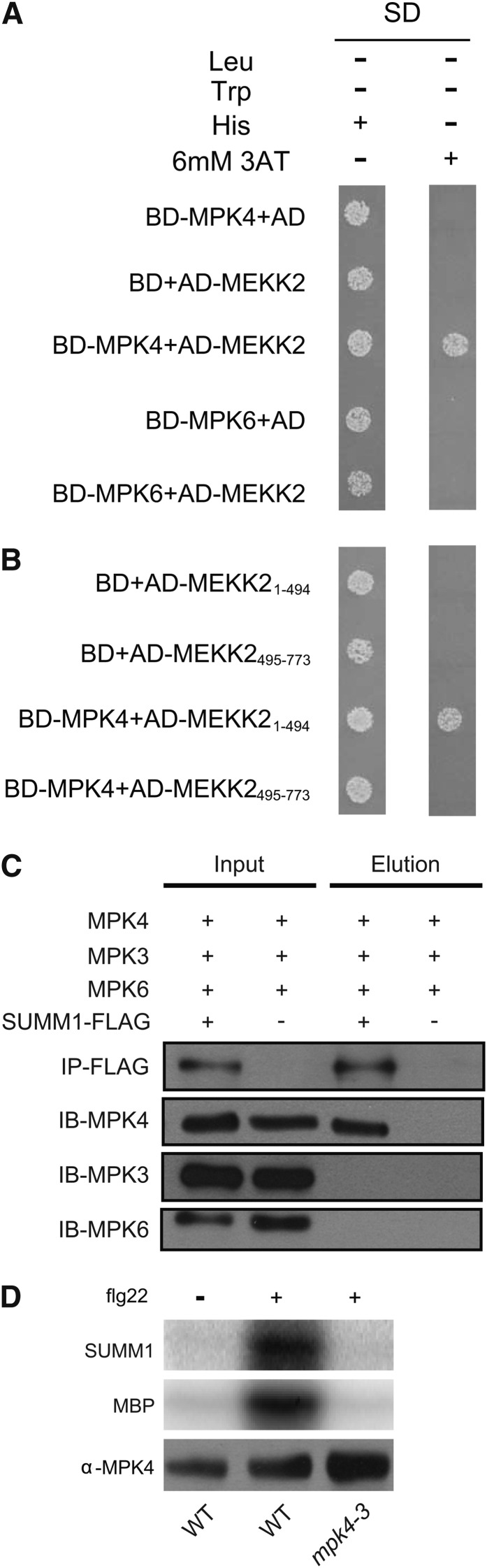
The epistatic relationship between SUMM1 and all members of the MEKK1-MKK1/MKK2-MPK4 MAPK cascade suggests that MPK4 might be a negative regulator of SUMM1. However, whether MPK4 and SUMM1 physically interact with each other is unclear. Thus, we first tested whether SUMM1/MEKK2 and MPK4 interact with each other in yeast two-hybrid assays. As shown in Figure 6A, MEKK2 interacted with MPK4 but not MPK6 or the empty vector. MEKK2 contains two distinct domains. The N-terminal domain is quite divergent from other MEKKs and its function is unknown. The C terminus contains the kinase domain, which is highly conserved in MEKKs. To determine which region of MEKK2 interacts with MPK4, we expressed the N-terminal part of MEKK2 and the C-terminal kinase domain separately. As shown in Figure 6B, the N-terminal but not the C-terminal domain of MEKK2 interacted with MPK4 in the yeast two-hybrid assay, suggesting that the N terminus of MEKK2 contains the interface of interaction with MPK4.
Figure 6.
MPK4 Interacts with MEKK2 and Phosphorylates the N Terminus of MEKK2.
(A) Yeast two-hybrid analysis of the interaction between MPK4 and MEKK2.
(B) Yeast two-hybrid analysis of the interaction between MPK4 and the N-terminal (MEKK21-494) and C-terminal (MEKK2495-773) domains of MEKK2.
(C) Co-IP of MPK4 with MEKK2-3xFLAG in total proteins extracts from SUMM1-3xFLAG transgenic plants. Total protein extracts were subjected to immunoprecipitation with anti-FLAG Sepharose beads. Crude lysates (left panel, Input) and immunoprecipitated proteins (right panels, Elution) were detected with anti-FLAG, anti-MPK4, anti-MPK3, and anti-MPK6 antibodies, respectively. Wild-type plants without the SUMM1-3xFLAG transgene were used as a negative control. This experiment was repeated three times with similar results.
(D) Phosphorylation of the N terminus of SUMM1/MEKK2 by MPK4. MPK4 was immunoprecipitated from the wild type (WT) and mpk4-3. After incubation with [γ-32P]ATP and the immunoprecipitated MPK4 or mpk4-3 mutant protein in protein kinase buffer, the E. coli–expressed N-terminal domain of SUMM1 was separated on 10% SDS-PAGE. The autoradiograph of the gel is shown in the top panel, and immunoblot analysis of MPK4 levels is shown in the bottom panel. This experiment was repeated four times with similar results.
To test whether MEKK2 and MPK4 associate with each other in vivo, we performed coimmunoprecipitation (co-IP) analysis using transgenic plants expressing the MEKK2-3xFLAG fusion protein under its own promoter. As shown in Figure 6C, MPK4 coimmunoprecipitated with 3xFLAG-tagged MEKK2 from total protein extracts of the transgenic plants but not from the protein extract of wild-type plants without the transgene. Immunoblot analysis of the immunoprecipitated proteins using anti-MPK3 and anti-MPK6 antibodies showed that MPK3 and MPK6 were not coimmunoprecipitated with MEKK2. These data suggest that MEKK2 interacts with MPK4 but not MPK3 and MPK6 in planta.
Phosphorylation of the N-Terminal Domain of SUMM1/MEKK2 by MPK4
The direct physical interaction between MPK4 and MEKK2/SUMM1 prompted us to test whether MEKK2 is a substrate for MPK4. To test whether MPK4 can phosphorylate MEKK2, the N-terminal domain of MEKK2 (MEKK21-500) with a 6×His-tag was expressed in Escherichia coli and purified using Ni2+-nitrilotriacetate chromatography. MPK4 was purified from wild-type or mpk4-3 mutant plants by immunoprecipitation using anti-MPK4 antibodies. In vitro kinase assays were subsequently performed using the MEKK21-500 and MPK4 proteins. As shown in Figure 6D, MEKK21-500 was phosphorylated by MPK4 from the flg22-treated wild-type plants but not the mutant protein from mpk4-3 plants, suggesting that MEKK2 is indeed a substrate of MPK4.
To identify the amino acid(s) in MEKK21-500 that was phosphorylated by MPK4, MPK4-treated E. coli–expressed MEKK21-500 was analyzed by mass spectrometry. As shown in Table 1, Ser-365 was phosphorylated in the protein. When we analyzed MEKK2-3xFLAG immunoprecipitated from transgenic plants expressing the fusion protein, Ser-365 was also found to be phosphorylated in vivo (Table 1), suggesting that Ser-365 in MEKK2 is most likely a target site of MPK4. Several additional phosphorylation sites were also identified in the MEKK2-3xFLAG protein (Table 1). It remains to be determined which kinase(s) is responsible for the phosphorylation of these sites.
Table 1. Phosphopeptides from MEKK2.
| Assay | Amino Acid No. | Peptide Sequence |
|---|---|---|
| In vitro | 362–375 | GVTpSPVLNLRPTDK |
| In vivo | 77–96 | SNpSSENKIPNEDISVSTSSR |
| 150–156 | pSLDFPNR | |
| 362–375 | GVTpSPVLNLRPTDK | |
| 362–386 | GVTpSPVLNLRPTDKEVVDSGTVENR | |
| 757–772 | RPLPSSGSGSTpSPLIR |
Phosphopeptides were identified by mass spectrometry analysis as previously described (Tang et al., 2009). A lowercase p indicates phosphorylation of the Ser residue that follows.
Root Development Defects in mpk4-3 Are Not Suppressed by summ1-1
In addition to negatively regulating plant defense responses, MPK4 is also important for cytokinesis and plant development (Beck et al., 2010; Kosetsu et al., 2010; Takahashi et al., 2010). The mpk4-2 mutant exhibits increased root width and has abnormal root hairs. To test whether summ1-1 suppresses the root development phenotypes of mpk4, we measured the root width in mpk4-3 and summ1-1 mpk4-3 and found that both mpk4-3 and summ1-1 mpk4-3 have increased root width compared with the wild type and summ1-1 (see Supplemental Figure 2A online). In addition, abnormal root hairs were also observed in both mpk4-3 and summ1-1 mpk4-3 but not in the wild type and summ1-1 (see Supplemental Figure 2B online), suggesting that summ1-1 cannot suppress the developmental phenotypes of mpk4 roots.
We also measured the root width and examined the morphology of root hairs in mekk1-4 npr1-1, summ1-3 mekk1-4 npr1-1, mkk1 mkk2, and summ1-1 mkk1 mkk2. As shown in Supplemental Figures 3A to 3D online, the root width of these mutants is comparable to that in wild-type plants and root hairs developed normally in the mutant plants. We also analyzed the roots of the SUMM1-FLAG transgenic line OX#2. No difference was observed between wild-type and the transgenic plants in root width and root hair morphology (see Supplemental Figures 3E and 3F online). These data suggest that MEKK1, MKK1/MKK2, and SUMM1 do not function in Arabidopsis root development as MPK4 does.
SUMM1/MEKK2 Functions Upstream of SUMM2
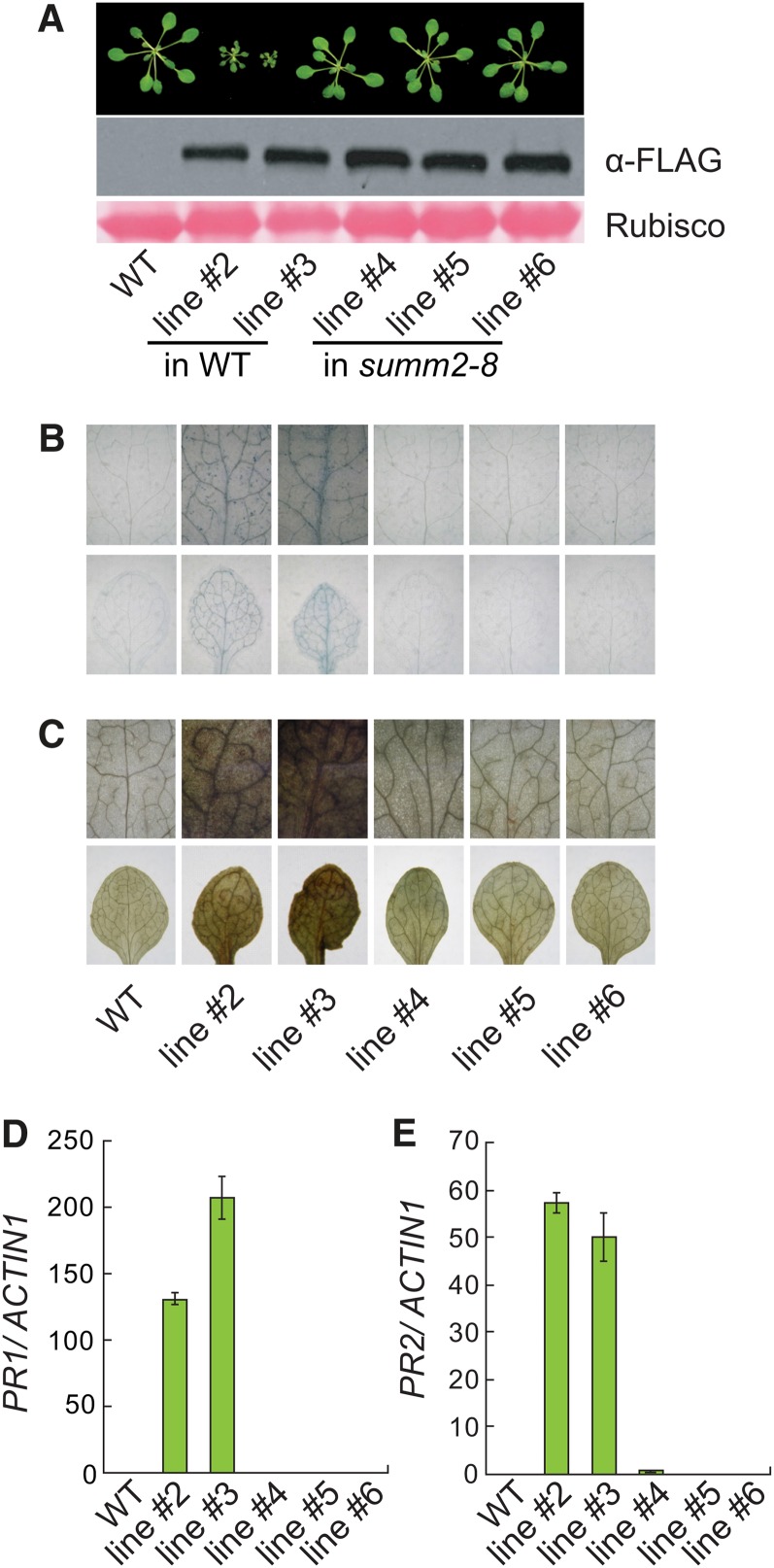
Our study on another suppressor mutant of mkk1 mkk2, summ2-1, revealed that SUMM2 encodes a coiled-coil NB-LRR R protein whose activity is negatively regulated by the MEKK1, MKK1/MKK2, and MPK4 kinase cascade (Zhang et al., 2012). To determine whether SUMM2 is required for activation of defense responses by overexpression of SUMM1, we transformed summ2-8, a T-DNA knockout mutant of SUMM2, with the construct expressing SUMM1-FLAG fusion protein under its own promoter. None of the transgenic plants exhibited a dwarf phenotype.
Three transgenic lines in summ2-8 background and two transgenic lines in wild-type background with similar SUMM1-FLAG protein levels were analyzed further. As shown in Figure 7A, the transgenic lines in summ2-8 background displayed wild-type morphology, whereas the transgenic lines in wild-type background exhibited dwarf morphology. Trypan blue staining and DAB staining revealed extensive cell death (Figure 7B) and high level of H2O2 (Figure 7C) in the transgenic lines in wild-type background but not in transgenic lines in summ2-8 background. Analysis of the expression of PR1 (Figure 7D) and PR2 (Figure 7E) showed that they were also constitutively expressed in the transgenic lines in wild-type background but not in transgenic lines in summ2-8 background. These data suggest that SUMM1 functions upstream of SUMM2 to regulate plant defense responses.
Figure 7.
SUMM2 Is Required for Defense Responses Activated by SUMM1.
(A) Morphology of ∼3-week-old SUMM1-FLAG transgenic lines and SUMM1-FLAG expression levels. Line #2 and #3 are two representative SUMMI1-FLAG transgenic lines from the transformation of wild-type plants. Line #4, #5, and #6 are three representative SUMM1-FLAG transgenic lines from the transformation of summ2-8 plants. Rubisco, ribulose-1,5-bis-phosphate carboxylase/oxygenase; WT, the wild type.
(B) and (C) Trypan blue staining (B) and DAB staining (C) of the true leaves of indicated genotypes.
(D) and (E) Expression levels of PR1 (D) and PR2 (E) as determined by quantitative PCR. Values were normalized relative to the expression of ACTIN1. Error bars represent sd of three measurements.
DISCUSSION
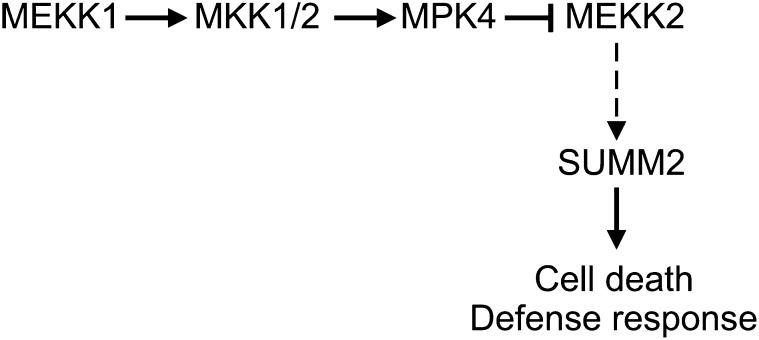
Our analysis of suppressor mutants of mkk1 mkk2 identified MEKK2 as a positive regulator of plant immunity. Not only do mutations in MEKK2 suppress the dwarf phenotype and the constitutive defense responses in mekk1, mkk1 mkk2, and mpk4 mutant plants, overexpression of MEKK2 is also sufficient to activate defense responses mediated by the NB-LRR R protein SUMM2. Our data suggest that the MEKK1, MKK1/MKK2, and MPK4 kinase cascade negatively regulates MEKK2 and MEKK2 functions as a positive regulator of SUMM2-mediated plant immunity (Figure 8).
Figure 8.
A Working Model for Repression of MEKK2-Mediated Immunity by the MEKK1-MKK1/2-MPK4 MAPK Cascade.
MEKK2 functions as a positive regulator of the NB-LRR R protein SUMM2, and its activity is negatively regulated by the MEKK1-MKK1/2-MPK4 kinase cascade. As shown by the dashed arrow, the mechanism by which MEKK2 activates SUMM2 remains to be determined.
Although mekk1, mkk1 mkk2, and mpk4 mutants exhibit similar morphology, mekk1 knockout mutants and mpk4 alleles in Col background are more severely dwarfed than mkk1 mkk2 (Gao et al., 2008), suggesting that functions of MKK1 and MKK2 can be partially compensated for by another MKK with overlapping functions. Probably due to the more severe phenotypes in mpk4-3, suppression of cell death, accumulation of H2O2, and upregulation of PR genes in summ1-1, mpk4-3 is not as complete as that in the summ1-1 mkk1 mkk2 triple mutant. The incomplete suppression of defense responses in mpk4-3 by summ1-1 suggests that MPK4 regulates additional defense components in addition to MEKK2. Previously, MKS1 was also identified as a substrate of MPK4 (Andreasson et al., 2005). Silencing MKS1 partially suppresses the mpk4 mutant morphology. It is possible that MEKK2 and MKS1 function independently and the residual cell death and defense responses in summ1-1 mpk4-3 are results of activation of MKS1-mediated defense responses.
Both yeast two-hybrid and co-IP analyses showed that MPK4 interacts with MEKK2. In addition, MPK4 can phosphorylate MEKK2 in vitro. These data suggest that MEKK2 is a direct substrate of MPK4 and MPK4 most likely suppresses defense responses through inactivating MEKK2 by phosphorylation. Since MAPKKKs usually function upstream of MAPKs, it is unexpected that MEKK2 serves as a target of MPK4. Previously it was shown that MEKK1 interacts with MPK4 in yeast two-hybrid assays (Ichimura et al., 1998). It is unclear whether MPK4 negatively regulates the activity of MEKK1 by phosphorylation and whether phosphorylation of MAPKKKs by their downstream MAPKs is used as a mechanism of negative feedback regulation of MAPK cascades.
MEKK2 contains an N-terminal domain with unknown functions and a C-terminal kinase domain. The N-terminal domain interacts with MPK4 and can be phosphorylated by MPK4 in vitro, indicating that it may have a regulatory function. Most of the mekk2 mutations found to suppress the mkk1 mkk2 mutant phenotypes are located in the kinase domain, suggesting that the kinase domain of MEKK2 is important for activation of defense responses. It remains to be determined whether MEKK2 functions like traditional MAPKKKs, which transduce signals through downstream MAPKKs and MAPKs.
In addition to its function in plant defense, MPK4 also plays an important role in regulating cytokinesis (Beck et al., 2010; Kosetsu et al., 2010). Due to defect in cytokinesis, mpk4 mutants also exhibit various root development phenotypes. Interestingly, summ1-1 suppresses the autoimmune phenotypes but not the root development phenotypes of mpk4-3, suggesting that the constitutive defense responses and defects in root development can be uncoupled. This is also supported by findings that overexpressing SUMM1 results in cell death and activation of defense responses, but not defects in root development. Similarly, the mpk4-1 mutant in Ler background displays constitutive defense responses but has no visible defects in cytokinesis and root development (Kosetsu et al., 2010). Our results suggest that MPK4 is a multifunctional MAPK regulating at least two separate biological processes. On one hand, it functions together with ANP2/ANP3 and MKK6 to regulate microtubule organization and cytokinesis. On the other hand, it forms a kinase cascade together with MEKK1 and MKK1/MKK2 to negatively regulate MEKK2-mediated defense responses.
Studies on bacterial effectors revealed that HopAI1 inactivates MAPKs by removing the phosphate group from phosphothreonine through a unique phosphothreonine lyase activity to suppress PAMP responses (Zhang et al., 2007), whereas HopF2 suppresses PAMP-mediated immunity by inhibiting MAPKKs (Wang et al., 2010), suggesting that targeting MAPK cascades downstream of plant immune receptors plays important roles in bacterial virulence. Since MEKK2 functions upstream of the NB-LRR R protein SUMM2, it is likely that MEKK2 evolved to sense the attack of the MEKK1-MKK1/MKK2-MPK4 kinase cascade by microbial pathogens. Disruption of the activity of MEKK1, MKK1/MKK2, or MPK4 leads to activation of MEKK2, which triggers SUMM2-mediated immune responses.
One important question is how MEKK2 activates SUMM2-mediated defense responses. We were not able to detect any direct interaction between MEKK2 and SUMM2. One possibility is that MEKK2 functions as a component of a MAPK cascade and activates defense responses through its downstream MAP kinase kinase and MAPK. Alternatively, MEKK2 may not function as a traditional MAPKKK. In this scenario, it may directly activate SUMM2-mediated defense responses by phosphorylation of its target protein(s), which is recognized by SUMM2. Future studies on other summ mutants may help us identify genes that function between MEKK2 and SUMM2 and lead to better understanding about how MEKK2 regulates plant immune responses.
METHODS
Plant Materials, Mutant Screen, and Characterization
Mutants mkk1-1 mkk2-1 (mkk1 mkk2), mpk4-3, mekk1-4 npr1-1, and summ2-8 were reported previously (Gao et al., 2008; Zhang et al., 2012). The summ1-1 single mutant was obtained by backcrossing summ1-1 mkk1 mkk2 to Col wild-type plants. The summ1-1 mpk4-3 double mutant was obtained by crossing summ1-1 mkk1 mkk2 with mpk4-3. The summ1-3 mekk1-4 npr1-1 triple mutant was identified from an EMS-mutagenized mutant population of mekk1-4 npr1-1.
For gene expression analysis, RNA was purified from 2-week-old seedlings grown on half-strength Murashige and Skoog (MS) plates. Reverse transcription was performed using the M-MLV RTase cDNA synthesis kit from Takara. Real-time PCR was performed on the cDNA reverse transcribed from three independent RNA samples using Takara SYBR Premix Ex Taq II. Primers used for real-time PCR analysis of PR1, PR2, and Actin1 were described previously (Zhang et al., 2003b). The primers used for real-time PCR analysis of MEKK2 are RT-MEKK2-F and RT-MEKK2-R (see Supplemental Table 1 online). All experiments on gene expression analysis were repeated at least three times. For cell death and H2O2 analysis, 12-d-old seedlings grown on half-strength MS plates were stained with trypan blue and DAB according to procedures previously described (Parker et al., 1996; Thordal-Christensen et al., 1997). ROS were measured using a luminol-dependent assay (Trujillo et al., 2008). H a. Noco2 infection was performed by spraying 2-week-old seedlings with spore suspensions in H2O at a concentration of 50,000 spores per mL and scored as previously described (Bi et al., 2010). SA was extracted and measured using HPLC as previously described (Li et al., 1999).
Map-Based Cloning of summ1-1
The markers used for mapping were designed using the Monsanto Arabidopsis polymorphism and Ler sequence collections (Jander et al., 2002). All primer sequences are listed in Supplemental Table 1 online. T13D4 and NGA8 are based on Indel polymorphisms. F9M13 and T15F16 are based on single nucleotide polymorphisms. For marker F9M13, primers F9M13-F and F9M13-Col-R were used to detect the presence of the Col allele, and primers F9M13-F and F9M13-Ler-R were used to detect the presence of the Ler allele. For marker T15F16, primers T15F16-F and T15F16-Col-R were used to detect the presence of the Col allele, and primers T15F16-F and T15F16-Ler-R were used to detect the presence of the Ler allele.
For testing whether the summ1-3 mutation is responsible for the suppression of mekk1-4 mutant phenotypes in summ1-3 mekk1-4 npr1-1, the promoter region of SUMM1 was amplified by PCR using primers SUMM1-Promoter-F and SUMM1-Promoter-R, and the coding region of SUMM1 was amplified by PCR using primers SUMM1-F and SUMM1-R from wild-type genomic DNA. The two PCR fragments were sequentially cloned into a modified pCAMBIA1305 vector to obtain pCAMBIA1305-SUMM1-FLAG for expressing the MEKK2-3xFLAG fusion protein under its own promoter. pCAMBIA1305-SUMM1-FLAG was transformed into summ1-3 mekk1-4 npr1-1 and wild-type plants by the floral dip method (Clough and Bent, 1998).
Yeast Two-Hybrid Assays
To create the MPK4 and MPK6 bait plasmids, MPK4 cDNA was amplified by primers MPK4-F and MPK4-R, whereas MPK6 cDNA was amplified using MPK6-F and MPK6-R and cloned into pBI880. The MEKK2 cDNA was amplified using primers MEKK2-pBI881-F and MEKK2-pBI881-R and cloned into the prey vector pBI881. The N-terminal fragment of MEKK2 was amplified using primers MEKK2-pBI881-F and MEKK2-N-pBI881-R, whereas the C-terminal fragment of MEKK2 was amplified using primers MEKK2-C-pBI881-F and MEKK2-pBI881-R. Both fragments were cloned into pBI881. All primer sequences are listed in Supplemental Table 1 online. For yeast two-hybrid assays, bait and prey plasmids were cotransformed into yeast strain Y1348. Yeast strains containing the bait and prey plasmids were cultured in SD-Trp-Leu liquid medium overnight and diluted to OD600 = 0.005 using double distilled water. Ten microliters of the diluted culture was plated on SD-Trp-Leu-His with 6 mM 3-amino-1,2,4-triazole and SD-Trp-Leu dropout plates.
Co-IP
For the co-IP experiment, 2-week-old-seedlings of Col-0 and MEKK2-3×FLAG transgenic plants on half-strength MS plates were used. About 0.9 g tissue was ground in liquid nitrogen and suspended in 0.9 mL grinding buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 150 mM NaCl, 0.1% Nonidet P-40, 1 mM PMSF, and 1× Protease Inhibitor Cocktail from Roche). The samples were spun at 21,000g for 10 min at 4°C followed by incubation with 40 μL of protein G beads (GE Healthcare; 17-0618-01) for 30 min with rotation. After the beads were removed by centrifugation, 40 μL anti-FLAG M2 agarose (Sigma-Aldrich; 087K6001) was added to supernatant and incubated at 4°C for 2 h with rotation. The beads were spun down and washed three times using grinding buffer. For elution, the beads were incubated with 100 μg/mL FLAG peptide at 4°C with rotation for 25 min. The supernatant was collected by centrifugation, and the immunoprecipitated proteins were detected by immunoblotting. The primary antibodies used for immunoblotting were mouse anti-FLAG monoclonal antibody (Sigma-Aldrich) and rabbit anti-MPK4, anti-MPK3, and anti-MPK6 (Sigma-Aldrich).
MPK4 Kinase Assays
To express the N-terminal domain of MEKK2 (amino acids 1 to 500) with a 6×His-tag, the cDNA fragment of MEKK2 was amplified using primers MEKK2-N-pET24c-F and MEKK2-N-pET24c-R (see Supplemental Table 1 online) and cloned into pET-24c. The plasmid was transformed into Escherichia coli strain BL21 for expressing the protein and the protein was purified using Ni2+-nitrilotriacetate chromatography.
To isolate MPK4 protein for kinase assays, 12-d-old seedlings of the wild type and mpk4-3 were sprayed with 10 μM flg22. About 0.5 g of tissue from each sample was harvested in liquid nitrogen 10 min later. One milliliter of extraction buffer (50 mM HEPES 7.4, 50 mM NaCl, 10 mM EDTA, 0.1%Triton X-100, 1 mM Na3VO4, 1 mM NaF, 1 mM DTT, 1 mM PMSF, and 1 mM Protease Inhibitor) was added to resuspend the sample. After centrifugation, the supernatant was collected and incubated with 2 μL anti-MPK4 antibodies (Sigma-Aldrich) for 1 h with constant rotation at 4°C. Next, 20 μL of Protein A beads was added into each sample and incubated for another 3 h at 4°C. After centrifugation, the beads were washed with 1 mL of extraction buffer three times and with 1 mL of kinase buffer (50 mM HEPES 7.4, 10 mM MgCl2, 10 mM MnCl2, 1 mM DTT, and 10 μM ATP) once. The beads were spun down and resuspended in 15 μL of kinase buffer.
For the kinase assay, 9 μL MPK4 was incubated with ∼1 μg of MEKK21-500 protein or 0.5 μg of MBP, 0.5 μL ATP (200 μM), 10 μCi [γ-32P]ATP, and kinase reaction buffer (50 mM HEPES, pH 7.4, 10 mM MgCl2, 10 mM MnCl2, 1 mM DTT, and 10 μM ATP) in a total volume of 15 μL at 30°C for 30 min. The reaction was ended by adding SDS loading buffer. After separation by SDS-PAGE, phosphorylation of MEKK21-500aa and MBP was detected by autoradiography.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At4g08480 (SUMM1), AT4G01370 (MPK4), At2g14610 (PR1), At3g57260 (PR2), and At2g37620 (Actin1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Morphological Phenotypes of the Wild Type, mkk1-1 mkk2-1, and Different summ1 Mutant Alleles.
Supplemental Figure 2. Development Defects in the Roots of mpk4-3 Are Not Suppressed by summ1-1.
Supplemental Figure 3. MEKK1, MKK1/MKK2, and SUMM1 Are Not Required for Root Development.
Supplemental Table 1. Primer Sequences.
Supplementary Material
Acknowledgments
We thank Virginia Woloshen for critical reading of the article. We are grateful for financial support from the Chinese Ministry of Science and Technology (Grants 2010CB835302 and 2011CB100700) and the Natural Sciences and Engineering Research Council of Canada.
AUTHOR CONTRIBUTIONS
Q.K., M.G., and Y.Z. designed the research. Q.K., N.Q., M.G., Z.Z., X.D., F.Y., Y.L., and O.X.D. performed research. S.C., X.L., and Y.Z. analyzed data. Q.K., X.L., and Y.Z. wrote the article.
Glossary
- PAMP
pathogen-associated molecular pattern
- RLK
receptor-like kinase
- NB-LRR
nucleotide binding–leucine-rich repeat
- MAPK
mitogen-activated protein kinase
- MAPKKK
MAP kinase kinase kinase
- SA
salicylic acid
- EMS
ethyl methanesulfonate
- DAB
to be defined
- H2O2
hydrogen peroxide
- Col
Columbia
- Ler
Landsberg erecta
- co-IP
coimmunoprecipitation
- MS
Murashige and Skoog
References
- Andreasson E., et al. (2005). The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24: 2579–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M., Komis G., Müller J., Menzel D., Samaj J. (2010). Arabidopsis homologs of nucleus- and phragmoplast-localized kinase 2 and 3 and mitogen-activated protein kinase 4 are essential for microtubule organization. Plant Cell 22: 755–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A., Farnham G., Moffett P., Baulcombe D.C. (2002). Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 32: 195–204 [DOI] [PubMed] [Google Scholar]
- Bi D., Cheng Y.T., Li X., Zhang Y. (2010). Activation of plant immune responses by a gain-of-function mutation in an atypical receptor-like kinase. Plant Physiol. 153: 1771–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi D., Johnson K., Huang Y., Zhu Z., Li X., Zhang Y. (2011). Mutations in an atypical TIR-NB-LRR-LIM resistance protein confer autoimmunity. Front. Plant Sci. 2: 71. [DOI] [PMC free article] [PubMed]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Cheng Y.T., Li Y., Huang S., Huang Y., Dong X., Zhang Y., Li X. (2011). Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc. Natl. Acad. Sci. USA 108: 14694–14699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Eitas T.K., Dangl J.L. (2010). NB-LRR proteins: Pairs, pieces, perception, partners, and pathways. Curr. Opin. Plant Biol. 13: 472–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Liu J., Bi D., Zhang Z., Cheng F., Chen S., Zhang Y. (2008). MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18: 1190–1198 [DOI] [PubMed] [Google Scholar]
- Gao M., Wang X., Wang D., Xu F., Ding X., Zhang Z., Bi D., Cheng Y.T., Chen S., Li X., Zhang Y. (2009). Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6: 34–44 [DOI] [PubMed] [Google Scholar]
- Gou M., Shi Z., Zhu Y., Bao Z., Wang G., Hua J. (2012). The F-box protein CPR1/CPR30 negatively regulates R protein SNC1 accumulation. Plant J. 69: 411–420 [DOI] [PubMed] [Google Scholar]
- Ichimura K., Casais C., Peck S.C., Shinozaki K., Shirasu K. (2006). MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J. Biol. Chem. 281: 36969–36976 [DOI] [PubMed] [Google Scholar]
- Ichimura K., Mizoguchi T., Irie K., Morris P., Giraudat J., Matsumoto K., Shinozaki K. (1998). Isolation of ATMEKK1 (a MAP kinase kinase kinase)-interacting proteins and analysis of a MAP kinase cascade in Arabidopsis. Biochem. Biophys. Res. Commun. 253: 532–543 [DOI] [PubMed] [Google Scholar]
- Igari K., Endo S., Hibara K., Aida M., Sakakibara H., Kawasaki T., Tasaka M. (2008). Constitutive activation of a CC-NB-LRR protein alters morphogenesis through the cytokinin pathway in Arabidopsis. Plant J. 55: 14–27 [DOI] [PubMed] [Google Scholar]
- Jander G., Norris S.R., Rounsley S.D., Bush D.F., Levin I.M., Last R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Gao F., Bhattacharjee S., Adiasor J.A., Nam J.C., Gassmann W. (2010). The Arabidopsis resistance-like gene SNC1 is activated by mutations in SRFR1 and contributes to resistance to the bacterial effector AvrRps4. PLoS Pathog. 6: e1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosetsu K., Matsunaga S., Nakagami H., Colcombet J., Sasabe M., Soyano T., Takahashi Y., Hirt H., Machida Y. (2010). The MAP kinase MPK4 is required for cytokinesis in Arabidopsis thaliana. Plant Cell 22: 3778–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang Y., Clarke J.D., Li Y., Dong X. (1999). Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98: 329–339 [DOI] [PubMed] [Google Scholar]
- Li Y., Li S., Bi D., Cheng Y.T., Li X., Zhang Y. (2010). SRFR1 negatively regulates plant NB-LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog. 6: e1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H., Soukupová H., Schikora A., Zárský V., Hirt H. (2006). A Mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J. Biol. Chem. 281: 38697–38704 [DOI] [PubMed] [Google Scholar]
- Noutoshi Y., Ito T., Seki M., Nakashita H., Yoshida S., Marco Y., Shirasu K., Shinozaki K. (2005). A single amino acid insertion in the WRKY domain of the Arabidopsis TIR-NBS-LRR-WRKY-type disease resistance protein SLH1 (sensitive to low humidity 1) causes activation of defense responses and hypersensitive cell death. Plant J. 43: 873–888 [DOI] [PubMed] [Google Scholar]
- Parker J.E., Holub E.B., Frost L.N., Falk A., Gunn N.D., Daniels M.J. (1996). Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8: 2033–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Pitzschke A., Djamei A., Bitton F., Hirt H. (2009b). A major role of the MEKK1-MKK1/2-MPK4 pathway in ROS signalling. Mol. Plant 2: 120–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A., Schikora A., Hirt H. (2009a). MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 12: 421–426 [DOI] [PubMed] [Google Scholar]
- Qiu J.L., Zhou L., Yun B.W., Nielsen H.B., Fiil B.K., Petersen K., Mackinlay J., Loake G.J., Mundy J., Morris P.C. (2008). Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 148: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirano Y., Kachroo P., Shah J., Klessig D.F. (2002). A gain-of-function mutation in an Arabidopsis Toll Interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14: 3149–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Rodriguez M.C., Adams-Phillips L., Liu Y., Wang H., Su S.H., Jester P.J., Zhang S., Bent A.F., Krysan P.J. (2007). MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 143: 661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Soyano T., Kosetsu K., Sasabe M., Machida Y. (2010). HINKEL kinesin, ANP MAPKKKs and MKK6/ANQ MAPKK, which phosphorylates and activates MPK4 MAPK, constitute a pathway that is required for cytokinesis in Arabidopsis thaliana. Plant Cell Physiol. 51: 1766–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.T., Li S., Long C., Cha J., Huang G., Li L., Chen S., Liu Y. (2009). Setting the pace of the Neurospora circadian clock by multiple independent FRQ phosphorylation events. Proc. Natl. Acad. Sci. USA 106: 10722–10727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H., Zhang Z., Wei Y., Collinge D.B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11: 1187–1194 [Google Scholar]
- Trujillo M., Ichimura K., Casais C., Shirasu K. (2008). Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr. Biol. 18: 1396–1401 [DOI] [PubMed] [Google Scholar]
- Wang Y., Li J., Hou S., Wang X., Li Y., Ren D., Chen S., Tang X., Zhou J.M. (2010). A Pseudomonas syringae ADP-ribosyltransferase inhibits Arabidopsis mitogen-activated protein kinase kinases. Plant Cell 22: 2033–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., et al. (2007). A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1: 175–185 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Goritschnig S., Dong X., Li X. (2003a). A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15: 2636–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Tessaro M.J., Lassner M., Li X. (2003b). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15: 2647–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yang Y., Fang B., Gannon P., Ding P., Li X., Zhang Y. (2010). Arabidopsis snc2-1D activates receptor-like protein-mediated immunity transduced through WRKY70. Plant Cell 22: 3153–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Wu Y., Gao M., Zhang J., Kong Q., Liu Y., Ba H., Zhou J., Zhang Y. (2012). Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11: 253–263. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.