Abstract
Tc1/mariner elements are able to transpose in species other than the host from which they were isolated. As potential vectors for insertional mutagenesis and transgenesis of the mouse, these cut-and-paste transposons were tested for their ability to transpose in the mouse germ line. First, the levels of activity of several Tc1/mariner elements in mammalian cells were compared; the reconstructed fish transposon Sleeping Beauty (SB) was found to be an order of magnitude more efficient than the other tested transposons. SB then was introduced into the mouse germ line as a two-component system: one transgene for the expression of the transposase in the male germ line and a second transgene carrying a modified transposon. In 20% of the progeny of double transgenic male mice the transposon had jumped from the original chromosomal position into another locus. Analysis of the integration sites shows that these jumps indeed occurred through the action of SB transposase, and that SB has a strong preference for intrachromosomal transposition. Analysis of the excision sites suggests that double-strand breaks in haploid spermatids are repaired via nonhomologous end joining. The SB system may be a powerful tool for transposon mutagenesis of the mouse germ line.
Transposon tagging is a valuable tool for functional genomics in model organisms such as Drosophila (P-element) and Caenorhabditis elegans (Tc1 transposon). In the mouse germ line, however, efficient in vivo transposon tagging has not yet been achieved.
Unlike P-element, members of the Tc1/mariner superfamily of transposable elements do not require host-specific factors for their activity in vitro (1, 2). Indeed, several of these cut-and-paste transposons were shown to be active in a wide variety of species other than their original hosts, including bacteria [Himar1 (3)], malaria mosquito [Minos (4)], zebrafish [Tc3 (5) and Mos1 (6)], chicken [Mos1 (7)], and human cells [Tc1 (8), Sleeping Beauty (SB) (9), Himar1 (10), and Minos (11)]. Although the reconstructed fish transposon SB has been shown be able to jump in mouse embryonic stem (ES) cells (12), and more recently in mouse somatic tissue (13), germ-line transposition—an important and powerful tool—has yet to be demonstrated.
To assess the utility of Tc1/mariner elements for transposon tagging in the mouse germ line, we first compared Tc1, Tc3, Himar1, Mos1, and SB in an in vitro mammalian cell culture assay and determined that SB is most efficient. Based on these results, we introduced SB in the mouse germ line. We show that SB is able to jump with a high efficiency in the mouse germ line, demonstrating its potential utility for genetic applications.
Materials and Methods
Plasmids.
PCR fragments of the ORFs encoding the transposase proteins of Tc1, Tc3, Himar1, and Mos1 were cloned into the Klenow-treated, 3.8-kb NotI fragment of pCMVβ (CLONTECH), resulting in, respectively, pRP1341, pRP1342, pRP1389, and pRP1353. The template plasmids were, respectively, pRP470 (14), pRP716 (15), pMar27fH (2), and pMos1 (16). The mutations in the Tc3, Mos1, and Himar1 transposase ORFs were introduced either by site-directed mutagenesis using mutagenic primers or by a PCR-ligation-PCR method (17). The following cytomegalovirus (CMV) expression vectors were constructed: pRP2301 (Tc3 N225D), pRP2302 (Tc3 V41E N225D), pRP1390 (Mos1 F344L), pRP1398 (Himar1 H267R), pRP1399 (Himar1 Q131R E137K), and pRP2300 (Himar1 Q131R E137K H267R). The primer sequences are available on request.
A simian virus 40 (SV40)-G418 resistance cassette (a blunt-ended 1.6-kb BamHI–EcoRI fragment of pRc/CMV (Invitrogen)) was cloned into Tc1 [into the blunt-ended StyI sites of pRP1212 (18)], resulting in pRP1349, into Tc3 [into the blunt-ended BspEI and NcoI sites of Tc3 in pRP790 (19)], resulting in pRP1351, into Himar1 [the Himar1 transposon was cut out from pMarKan (2) by using NotI and EcoRI and cloned into the SmaI site of pUC19, the SV40-G418 resistance cassette was then cloned into the HincII and BstEII sites], resulting in pRP1347, and into Mos1 [the Mos1 transposon was PCR amplified from pMos1 (16) and cloned into the SmaI site of pUC19; the SV40-G418 resistance cassette was subsequently cloned into the NruI site], resulting in pRP1388. The corresponding SB vectors, pSB10 for CMV-transposase expression, and pT/neomycin (neo) (9) containing the marked transposon, were provided by Zoltán Ivics, Max-Delbrück-Centrum für Molekulare Medizin, Berlin. The T/neo transposon is referred to as SB/neo.
The plasmids for germ-line expression of Tc1 and SB transposase (pRP1321 and pPR1345, respectively) were constructed as follows: a double-stranded oligonucleotide containing restriction sites (XhoI–EcoRI–BamHI–KpnI–NdeI) was cloned into the NdeI and XhoI sites of 304P (a pSP72 vector with a 652-bp fragment of the Prm1 promoter cloned into the BglII and XhoI sites; a kind gift of Stephen O'Gorman, Salk Institute, La Jolla, CA), resulting in the plasmid prm1-NX. A 1-kb EcoRI fragment of pRP1439 (Henri van Luenen and R.H.A.P., unpublished data), containing the Tc1 transposase ORF preceded by a Kozak sequence was cloned into the EcoRI site of prm1-NX, resulting in the vector prm1-Tc1. Then, a 1.2-kb BamHI–KpnI fragment containing the rabbit β-globin 3′ splicing and polyadenylation signal was cloned into the BamHI and KpnI sites, resulting in pRP1321. To construct pRP1345, pRP1321 was partially digested with EcoRI, releasing the fragment containing the Tc1 ORF; the EcoRI sites were filled in, and a 1-kb EcoRI–BamHI fragment of pSB10 (9) containing the SB ORF, was inserted. The transgenes were fragments of the following plasmids: pRP1212 (Tc1/kan) (18) (BamHI–SphI fragment), pRP1321 (HpaI–KpnI fragment), pT/neo (9) (SB/neo) (KpnI-SalI fragment), and pRP1345 (HpaI–KpnI fragment).
Cell Culture and Transfections.
Human HeLa cells and mouse NIH 3T3 cells were cultured in OptiMEM (GIBCO/BRL) supplemented with 5% serum and 50 μg/ml penicillin/streptomycin at 37°C and 5% CO2. The tissue culture transposition assay was performed as described (9).
Generation of Transgenic Mice.
The transgene fragments were cut out of the vectors, purified from agarose gel, and injected into fertilized FVB/N (20) oocytes as described (21). The founder mice were bred to FVB. The presence of the transgenes was determined by PCR or Southern blot analysis of tail tip DNA (22).
DNA Isolation and Analysis.
All cloning techniques were performed as described (23). Genomic DNA was isolated from mouse tail tips by using standard procedures. Briefly, tissue was digested overnight in SDS/proteinase K buffer, precipitated with isopropanol, and resuspended in Tris-EDTA buffer. Aliquots of 10 μg of genomic DNA were digested for Southern analysis and treated as described (22). PCR fragments of the neo gene or of the Prm1 promoter were used as probes for Southern blots.
The sequences flanking SB/neo were isolated by using a vectorette transposon display method (24), using primers T/DR, T/BAL, T/JOBB1, and T/JOBB2 (9). All sequencing was done by using an Applied Biosystems 377 or 3700 DNA analyzer. Sequences of the primers used to analyze the DNA flanking SB/neo, before and after transposition, are available on request. The original locus and the insertion sites were mapped by using the T31 mouse/hamster radiation hybrid panel (Research Genetics, Huntsville, AL) (25). The data were submitted to and analyzed by The Jackson Laboratory Mapping Panels.
RNA Isolation and Reverse Transcriptase (RT)–PCR.
RNA was isolated from tissues by using guanadinium thiocyanate (22). Five micrograms of RNA was treated with DNaseI and used to synthesize cDNA by using avian myeloblastosis virus reverse transcriptase with random hexamers pd(N)6 and oligo(dT)12–18 as primers. A PCR was done on the cDNA by using SB primers.
Results
Efficiencies of Tc1/mariner Elements Compared in Human HeLa Cells.
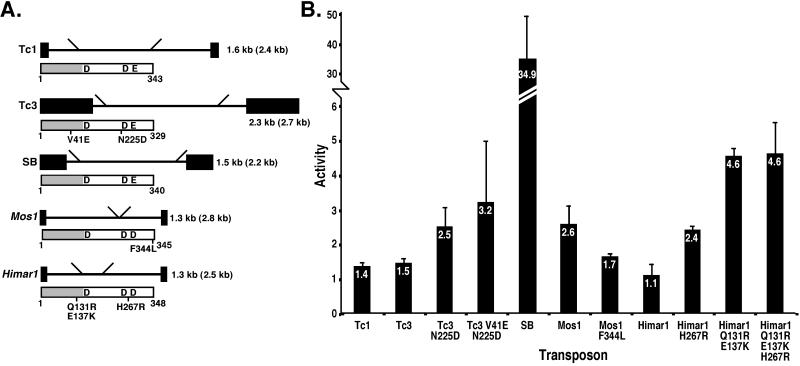
The efficiencies of transposition of the C. elegans transposons Tc1 (26) and Tc3 (27), the fish transposon SB (9), and the insect transposons Himar1 (2), and Mos1 (28) (Fig. 1A) in human HeLa cells were compared. Several naturally occurring polymorphic versions and putative hyperactive versions of the transposase proteins also were tested (indicated in Fig. 1A). The mutations in Tc3 transposase, V41L and N225D, are naturally occurring polymorphisms present in Tc3 elements in the C. elegans strain Bristol N2 (unpublished work). The F344L mutation introduced in Mos1 transposase is one of several amino acid substitutions found in the inactive mariner element peach as compared with Mos1 transposase. This mutation is thought to be primarily responsible for the inactivity of peach (29). The Himar1 transposase mutants H267R and Q131R/E137K were identified in a screen for hyperactive mutants performed in Escherichia coli and are 5- to 50-fold more active in E. coli and in vitro than wild-type Himar1 transposase (3). All transposase ORFs were cloned in identical restriction sites in a CMV expression vector. The corresponding transposons all were disrupted by an SV40-G418-resistance cassette. The transposase expression vector and the corresponding transposon vector were cotransfected into human HeLa cells. After cotransfection the number of G418-resistant colonies was compared with the number obtained after cotransfection of a control expression vector together with the transposon vector. It was shown previously that the increase in the number of resistant colonies is caused by transposase-mediated integration of the transposon (8, 9) and therefore is indicative of the transposition efficiency. Several of the tested transposase mutants have an increased activity compared with their wild-type versions, most notably the Himar1 mutants that were identified as hyperactive in bacteria (3). This result shows that the enhanced activity levels of Himar1 transposase selected in E. coli is not limited to E. coli, but also is apparent in human cells. However, SB is an order of magnitude more efficient than the other Tc1/mariner elements (Fig. 1B). In mouse 3T3 cells, SB also was found to be more active than Tc1 and Tc3 (data not shown).
Figure 1.
Tc1/mariner transposons compared for their activity in HeLa cells. (A) Structure of the transposons tested and of the corresponding transposases (below the transposon). The black boxes represent the terminal inverted repeats. The diagonal lines indicate the positions of the SV40-G418-resistance cassette. The transposon sequence between these lines was deleted. The sizes of the wild-type and, parenthetically, disrupted transposons, are indicated. Each tagged transposon was cloned into the polycloning site in pUC18/19. The gray box in each the transposase protein represents the (predicted) DNA-binding domain, whereas the white part represents the catalytic domain containing the catalytic amino acids DD(34)E or DD(34)D(50). Relevant mutions are indicated beneath each transposon. All five ORFs have comparable numbers of optimal and nonoptimal human codons and are preceded and followed by identical sequences. (B) Activity of Tc1/mariner elements compared in HeLa cells. A vector with the transposon marked by an SV40-G418-resistance cassette was cotransfected with a CMV expression vector with either the ORF of the corresponding transposase or with the ORF of β-galactosidase. G418-resistant colonies were selected and counted. The transposition activity (indicated on the y axis) is the ratio of the number of resistant colonies obtained when the transposase expression vector is cotransfected over the number of resistant colonies when the β-galactosidase expression vector is cotransfected (1 indicates no transposition). The activity indicates an average of between three and nine independent transfections. The error bars indicate SEM.
SB Transposition in the Mouse Germ Line.
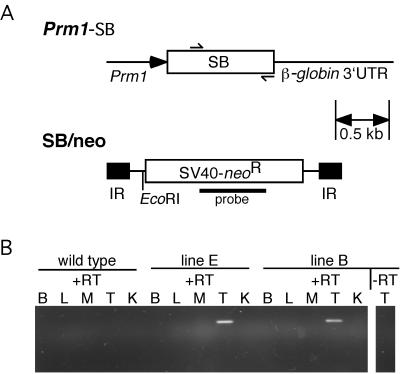
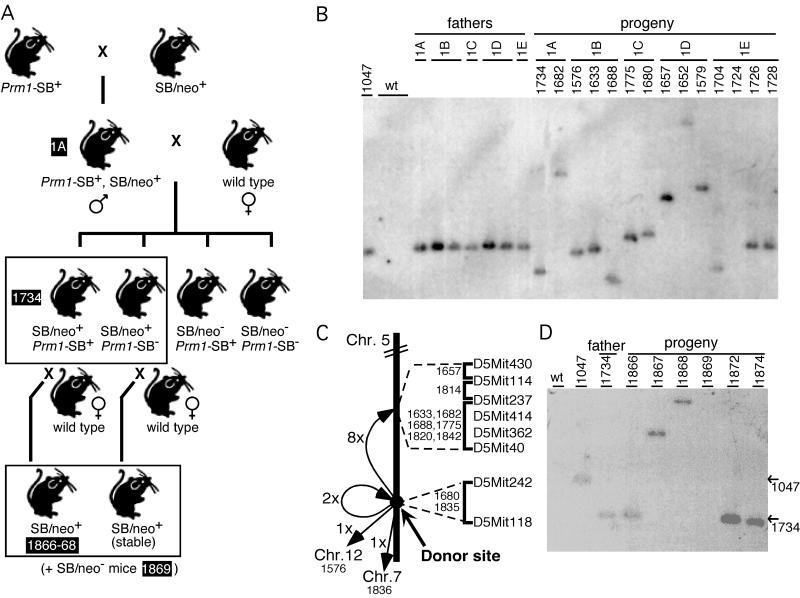
Tc1 and SB were introduced into the mouse germ line. To obtain germ-line transposition in the mouse, a system was devised in which the transposase is expressed in the germ line from a transgene and therefore should mobilize the transposon—present on a second transgene—specifically in the germ line. Transposase expression was driven by the proximal protamine 1 (Prm1) promoter (Fig. 2A), that was shown to drive expression of transgenes during spermiogenesis in haploid round spermatids (30). Both the Prm1-Tc1 (data not shown) and the Prm1-SB transgenic lines show transposase mRNA expression in testis as determined by RT-PCR (Fig. 2B). A second transgene contained the transposon in which the transposase ORF had been disrupted (Fig. 2A). To induce germ-line transposition, double transgenic males were generated for both Tc1 and SB (Fig. 3A). Progeny of these males mated to wild-type females were inspected for germ-line jumps of the transposon. A total of 36 progeny that inherited the Tc1/kan transposon from Tc1/kan+ Prm1-Tc1+ fathers were analyzed for Tc1 transposition by Southern blotting using a transposon probe. In all 36 cases no change in the hybridizing genomic fragment was observed, suggesting that none of these animals was derived from a sperm cell in which a Tc1 transposition event had occurred. However, when 98 SB/neo+ progeny of SB/neo+, Prm1-SB+ males (derived from five independent transposase lines) were analyzed, 20 (20.4%) showed a change in restriction fragment size compared with the fathers (Table 1 and Fig. 3B). This finding suggests that the SB transposon undergoes transposition in the mouse germ line.
Figure 2.
Generation and analysis of the transgenic mice. (A) Structure of the transgenes. The Prm1-SB-β-globin transgene is a fragment of plasmid pRP1345. A fragment of the mouse protamine 1 (Prm1) promoter was cloned in front of the ORF encoding the SB transposase, followed by the rabbit β-globin polyadenylation signal. The SB/neo transgene is a fragment of plasmid pT/neo. It has an SV40-G418R cassette in between the inverted repeats of the transposon. The fragments were injected into different FVB/N oocytes. Five founders (designated as lines A–E) transmitting the Prm1-SB transgene to their offspring were obtained, ranging in copy number from 1 to 5. One line carrying a single copy of the complete SB/neo transposon was obtained (line 1). (B) RT-PCR analysis of the expression of SB transposase mRNA in several tissues, including brain (B), liver (L), skeletal muscle (M), testis (T) and kidney (K) of wild-type and transgenic mice (shown are lines B and E) using primers indicated in A by arrows.
Figure 3.
(A) Mating scheme to induce and detect SB/neo transposition in mouse spermatids. Transposition events that occur in the spermatids of double transgenic males can be detected in their progeny. The progeny are analyzed for the presence of the SB/neo transposon by PCR. SB/neo+ animals (boxed) are further analyzed by PCR for the presence of SB/neo at the original locus and by Southern blotting to detect possible transposition events (shown in B). Males with SB/neo at a new location were further crossed to wild-type mice. SB/neo+ progeny (boxed) were analyzed to detect possible transposition events (shown in D). As an example, the lineage of animal 1734 is indicated (black text boxes). The heterozygous mice with an SB insertion were not homozygosed and had no obvious phenotypes. (B) Southern blot analysis of EcoRI-digested tail tip DNA of Prm1-SB+, SB/neo+ fathers and their offspring. The EcoRI site in SB/neo and the probe, a fragment of the neo-resistance gene, are indicated in Fig. 2A. Animal 1047 is the SB/neo+ founder animal (line 1) and shows the SB/neo transgene at its original locus. This line was crossed to all five Prm1-SB lines, lines A to E. Double transgenic males of lines 1A, 1B, 1C, 1D, and 1E were obtained and crossed with wild-type females. Of their progeny, animal 1724 did not inherit the SB/neo transgene; animals 1726 and 1728 did inherit the SB/neo transgene at its original locus. The remaining progeny represent 11 of 20 animals in which the SB/neo transposon had moved from its original locus to a new site (as the change in fragment size indicates). PCR analysis using a primer flanking the transposon at its original locus and a primer in the transposon confirmed the presence or absence of the transposon at the original locus. (C) Map of mouse chromosome 5 indicating the SB insertion sites. The insertion sites were mapped between the markers indicated, using the T31 mouse/hamster radiation hybrid panel. The distance between the original site on distal chromosome 5 and the region on central chromosome 5 is ≈25 cM. (D) Southern blot analysis of EcoRI-digested tail tip DNA of a Prm1-SB+, SB/neo+ father and his offspring. Animal 1047 is the SB/neo+ founder animal (line 1). Animal 1734 is progeny of a double transgenic male (line 1A), which has SB/neo at a new site and inherited the Prm1-SB transgene. Six progeny of mouse 1734 crossed to a wild-type female are shown: animals 1866, 1872, and 1874 have the transposon at the location of their father, animal 1734. Animal 1869 did not inherit the transposon. In animals 1867 and 1868 new transposition events were detected. PCR analysis confirmed the presence or absence of the transposon at the 1734 locus in the progeny of animal 1734. The positions of the fragments containing SB at its original site in animal 1047, and at the site in animal 1734, are indicated by arrows.
Table 1.
Transposition of the SB/neo transposon in the progeny of Prm1-SB+, SB/neo+ fathers
| SB/neo+Prm1-SB+ line | SB/neo− progeny | SB/neo+ progeny | SB/neo+ progeny with transposed SB/neo |
|---|---|---|---|
| 1A | 22 | 12 | 4 |
| 1B | 37 | 16 | 5 |
| 1C | 17 | 14 | 2 |
| 1D | 20 | 22 | 3 |
| 1E | 27 | 34 | 6 |
| Total | 123 | 98 | 20 (=20.4%) |
One of the characteristics of Tc1/mariner transposition is that the transposon integrates into a TA dinucleotide, and that this TA dinucleotide is duplicated upon integration. To verify that the restriction fragment length polymorphisms observed were indeed caused by transposition mediated by SB transposase, the flanking sequences of SB/neo at 18 insertion sites were determined (Table 2). In all cases, the transposon was flanked by a TA dinucleotide. Furthermore, the sequences flanking the TA dinucleotide were different for all 18 insertions and were different from the sequences flanking SB/neo at its original locus. Sequence analysis of five of these sites before insertion (in a wild-type mouse) revealed the presence of only one TA dinucleotide.
Table 2.
Transposon insertion sites of SB/neo in the mouse genome
| Mouse | Left flank* | SB/neo | Right flank* | Chr† | Linked to† |
|---|---|---|---|---|---|
| Founder | AACACTGATACCCTA | cagt … actg | TAGGGGATCCTCTAG | 5 | D5Mit242 |
| 1576 | CCTAGGGACTAGCTA | cagt … actg | TATAACAGATGTCAA | 12 | D12Mit190 |
| 1579 | cagt … actg | TATCTCATGATGATG | n.d. | ||
| 1633 | CATCTCTCCAATATA | cagt … actg | TAGCTCTAGATGGTC | 5 | D5Mit237 |
| 1657 | cagt … actg | TATGTGAGTTGGGTG | 5 | D5Mit430 | |
| 1680 | cagt … actg | TATATATAGGTATAT | 5 | D5Mit242 | |
| 1682 | GTACCCCCTCACCTA | cagt … actg | TATTAAATCTTTTAA | 5 | D5Mit237 |
| 1688 | CTACAGACAAATATA | cagt … actg | TAGGTTTCACTCTTA | 5 | D5Mit237 |
| 1704 | GATCAGTTA | cagt … actg | n.d. | ||
| 1725 | cagt … actg | TATATC | n.d. | ||
| 1734 | CCTCCGGTAGTACTA | cagt … actg | TATGTCCAATTTTCT | n.d. | |
| 1775 | TATTTATCAGATATA | cagt … actg | TAAGTAAGTCTTCTA | 5 | D5Mit237 |
| 1797 | TATGTTCTTTATGTA | cagt … actg | n.d. | ||
| 1814 | TGCCTAATAAACATA | cagt … actg | TAACACTGCTCCGTG | 5 | D5Mit414 |
| 1818 | cagt … actg | TAGCTCCAGGGCATT | n.d. | ||
| 1820 | AATGGCTAATACATA | cagt … actg | 5 | D5Mit237 | |
| 1835 | GCAGCCCACTACCTA | cagt … actg | TAGTTCTCCCCCTTC | 5 | D5Mit242 |
| 1836 | cagt … actg | TATGTGTAAGAAAAC | 7 | D7Mit161 | |
| 1842 | cagt … actg | TAGGTCTGTTCCATG | 5 | D5Mit362 |
n.d., not determined.
The sequences flanking SB/neo (in capitals) were determined by using a transposon display method (24).
Using the T31 mouse/hamster radiation hybrid panel the insertion sites were mapped. The chromosome and the marker with the highest logarithm of odds score are indicated. Six insertions could not be mapped due to either the lack of sufficient flanking sequence or the presence of multiple copies of the insertion site sequence in the mouse genome. blast (51) searches showed that both in mouse 1579 and in mouse 1734 SB/neo had integrated into an L1 retrotransposon.
The original locus and 12 insertion sites were mapped to the mouse genome (Table 2 and Fig. 3C). In eight of 12 transposition events the transposon had jumped from its original locus on distal chromosome 5 to central chromosome 5; remarkably six of these insertion sites mapped to the same 38.1-cR interval on the chromosome of origin. In two cases, mouse 1680 and 1835, the transposon had jumped close to the original locus on distal chromosome 5. Two instances of interchromosomal transposition, to chromosomes 7 and 12, were observed. We conclude that SB/neo excised from its original chromosomal locus and reintegrated in the mouse genome through bona fide transposition, with a strong preference for reintegration into the chromosome of origin.
Sixteen of the 20 animals in which a transposition event was detected did not have the Prm1-SB transgene, indicating that transposition occurred in the germ cells of the fathers. To prove that the observed transposition events had indeed occurred in the germ line of the mouse, two animals with SB/neo at a new location were further crossed to wild-type mice. Analysis of their progeny showed that these mice stably transmitted the transposon at its new location (data not shown).
To show that SB is able to transpose from an independent chromosomal context other than the original transgene, a Prm1-SB+ male with SB/neo at a new location (1734) was crossed further to a wild-type mouse. Of 10 progeny, six had the SB/neo transposon. Four of these six mice had the transposon at the location of the father, whereas in the remaining two animals the transposon was found at a new location (Fig. 3D).
Double-Strand Break (DSB) Repair After SB Excision in Mouse Spermatids.
After excision Tc1/mariner transposons leave DSBs. In C. elegans DSBs induced by Tc1 transposition are repaired by either nonhomologous end joining (NHEJ), or homologous recombination using the sister chromatid or homologous chromosome as a template (31). After NHEJ, a small footprint is found, consisting of a few base pairs of transposon sequence between the duplicated TA dinucleotide. The footprint arises as a consequence of the staggered cuts that the transposase introduces at the ends of the transposon (32). Analysis of 16 excision sites (Table 3) demonstrates that the DSBs induced by SB transposition in the male germ line of the mouse are repaired through NHEJ. In 12 cases, footprints of one or two base pairs of transposon sequence were found between the TA dinucleotides, similar to the footprints of Tc3 in C. elegans (32). In the other cases small deletions of flanking sequence had occurred. Previously, Luo et al. (12) observed 3-bp footprints after SB transposition in mouse embryonic stem cells, prompting the hypothesis that the SB transposon is excised via 3-bp staggered cuts (12). Our results, however, do not follow this pattern.
Table 3.
DSB repair in haploid spermatids after SB/neo excision
| Mouse | Left flank | Footprint | Right flank | ||
|---|---|---|---|---|---|
| Founder | ACAACACTGATACCC | ta | cagt … actg | ta | GGGGATCCTCTAGAG |
| 1680, 1704, 1797, 1802 | ACAACACTGATACCC | ta | g | ta | GGGGATCCTCTAGAG |
| 1725, 1734, 1814 | ACAACACTGATACCC | ta | c | ta | GGGGATCCTCTAGAG |
| 1576, 1652, 1682 | ACAACACTGATACCC | ta | cg | ta | GGGGATCCTCTAGAG |
| 1633, 1818 | ACAACACTGATACCC | ta | ca | ta | GGGGATCCTCTAGAG |
| 1688 | ACAACACTGATACCC | ta | ga | GGATCCTCTAGAG | |
| 1657 | ACAACACTGATACCC | t | CTAGAG | ||
| 1835 | ACAACACTGATACCC | ta | ct | GGGATCCTCTAGAG | |
| 1836 | ACAACACTGATACC | tcta | GATCCTCTAGAG | ||
The excision sites were sequenced by using primers flanking SB/neo at the original locus. The duplicated TA dinucleotides are indicated in bold; footprints are underlined. Four of 20 sites could not be sequenced, presumably because of deletion of flanking sequences to which the primer anneals.
Nonhomologous end joining is the expected repair pathway as the DSBs are presumably generated and repaired in haploid, round spermatids, when a template for homologous recombination is not present. Our results, in which insertion of an SB element was always accompanied by loss of the element at the donor site, suggest the notion that these events occurred in the haploid stage, when no DSB repair template was present anymore. In later stages, after round spermatids develop into elongated spermatids and finally into spermatozoa, sperm DNA is packed in a highly condensed structure and transcription and presumably also DNA repair (33) cease until fertilization. It is unlikely that the transposition events and repair occurred during these stages. However, it can not be excluded that expression of SB transposase, SB transposition, and DSB repair occur at diploid stages of spermatogenesis. Although most reports have shown that the Prm1 promoter restricts expression of transgenes to round spermatids, low levels of ectopic expression have been reported (34, 35).
Discussion
Mammalian gene function is studied predominantly by gene-driven approaches in the mouse, such as overexpression or loss-of-function mutants. In contrast, forward genetic approaches have been predominant in Drosophila and C. elegans. Forward genetics screens in the mouse primarily rely on N-ethyl-N-nitrosourea as the mutagen. Finding the causal point mutation is difficult and time consuming. Alternatively, large-scale insertional mutagenesis is performed in mouse ES cells (36), which requires the time-consuming generation of mice from ES cells. Therefore, the introduction of an exogenous transposon into the mouse germ line might be valuable tool for genetics in the mouse, especially if a recombinant transposon (e.g., suitable for gene trapping) could be used.
In the past, retroviruses have been used for gene tagging, but with limited success (reviewed in ref. 37). One of the bottlenecks is that the germ line contains many endogenous copies of the retrovirus; another one is that retroviral integration cannot be regulated and therefore requires infection of preimplantation or postimplantation embryos.
In a screen to find the best candidate for transposition in mammalian cells, the fish transposon SB was found to be most efficient. Himar1 and Tc3 are certainly not optimally active: single missense mutations in these transposase proteins could increase the efficiency of the other transposons by several-fold. Therefore it is expected that these transposons can be further mutated to a higher activity. The hyperactive Himar1 mutants were found in a screen performed by Lampe in E. coli (3). We now find that these mutants are also hyperactive in mammalian cells. Therefore, this screen is suitable for selecting mutants (of Tc1, Tc3, Himar1, Mos1) that are more active in mammalian cells. Presumably, the SB transposase also can be mutated to a higher activity.
Still the SB transposon is an order of magnitude more efficient than the most active other transposon in this mammalian system. This difference in efficiency might reflect the vertebrate origin of SB, which might be more favorable for possible stimulating host factors in a mammalian system. Alternatively, the SB transposon might have a higher intrinsic transposition activity than the other transposons.
The frequency of SB transposition in the male germ line was unexpectedly high. Previously, Luo et al. (12) showed that SB is capable of chromosomal transposition in mouse ES cells. However, the frequency of excision in that system is much lower (3.5 × 10−5 excisions/cell per generation) than the frequency of transposition that we find in spermatids (0.2 transposition events/spermatid), although the same donor element was used. Furthermore, the footprint found after repair of the excision site was different in ES cells compared with male spermatids. These differences might be caused by a difference in cell type. Similarly, it was previously was found that the footprints left by Tc3 in zebrafish are different from the footprints found in C. elegans (5).
The percentage of offspring with a SB insertion (20%) is similar to the percentage of retrovirus-infected embryos with a proviral integration in the germ line (2–56% depending on whether postor preimplantation embryos are infected) (38, 39). About 4% of retroviral insertions lead to an obvious mutant phenotype in the mouse homozygous for the insertion (38). Assuming the mutagenicity is similar for SB transposition, with a transposition frequency of 20%, 0.8% of the homozygous progeny of a double transgenic male with a single integrated copy of the transposon will be expected to have a phenotype. With an estimated number of loci of about 35,000 [based on the human genome (40, 41)] the mutation rate is less than 10−6 per locus per gamete derived from a double transgenic male. N-ethyl-N-nitrosourea mutagenesis in the mouse induces mutations at a frequency of about 10−3/locus per gamete (42), 1,000 times higher. Clearly, the current frequency of mutagenesis using SB is not high enough for forward mutagenic screens of recessive loci.
These calculations are based on the assumptions that SB insertion is as mutagenic as retroviral integration and that each locus can be hit. However, both retroviruses and many DNA transposons are known to preferentially integrate into certain sites in the genome. Retroviruses integrate preferentially close to DNaseI-hypersensitive sites (43) or transcriptionally active regions (44). It is not clear whether SB has a similar preference. Luo et al. (12) showed that in three of six transposition events in mouse ES cells SB integrated into the chromosome of origin. We now show that in the mouse germ line SB has a strong preference for transposition within the same chromosome, reminiscent of the target site preference of Ac elements in maize (45).
The utility of the SB system could be increased by generating mice with multiple dispersed copies of the SB transposon on different chromosomes. A single cross then could result in multiple jumps per gamete. Still the main use of SB as a mutagen in the mouse germ line probably will be for specific goals, for enhancer or gene trapping, or as a mutagen in hemizygous strains that carry chromosomal deletions (46–49). In addition, the SB system can be used to study the function of various repair proteins in DSB repair in the germ line of mice by generating DSBs at known locations in the genome.
It recently was shown that SB transposes from a plasmid into the mouse genome in somatic tissues (13). Here we show that SB is capable of high-frequency transposition in the germ line of the mouse; it is the first transposon for which regulated germ-line transposition has been demonstrated in any vertebrate. Because germ-line events are stably transmitted, this system has potential for use in insertional mutagenesis and enhancer trapping.
Acknowledgments
We thank Drs. Zoltán Ivics and Zsuzsanna Izsvák for reagents and advice, Dr. Stephen O'Gorman for providing the protamine promoter, Dr. Hugh Robertson for sharing data before publication, Dr. Paul Krimpenfort and coworkers for oocyte injections and Karen Thijssen, Ton Schrauwers, Caroline Friederich and Angela van Hattum for biotechnical assistance. We thank Drs. Stephen Wicks, René Ketting, and Marcel Tijsterman for discussions. S.E.J.F. is supported by Grant 335-210 from the Netherlands Organization for Scientific Research (NWO).
Abbreviations
- SB
Sleeping Beauty
- neo
neomycin
- DSB
double-strand break
- ES
embryonic stem
- CMV
cytomegalovirus
- SV40
simian virus 40
- RT
reverse transcriptase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Vos J C, de Baere I, Plasterk R H A. Genes Dev. 1996;10:755–761. doi: 10.1101/gad.10.6.755. [DOI] [PubMed] [Google Scholar]
- 2.Lampe D J, Churchill M E A, Robertson H M. EMBO J. 1996;15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 3.Lampe D J, Akerley B J, Rubin E J, Mekalanos J J, Robertson H M. Proc Natl Acad Sci USA. 1999;96:11428–11433. doi: 10.1073/pnas.96.20.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Catteruccia F, Nolan T, Loukeris T G, Blass C, Savakis C, Kafatos F C, Crisanti A. Nature (London) 2000;405:959–962. doi: 10.1038/35016096. [DOI] [PubMed] [Google Scholar]
- 5.Raz E, Van Luenen H G A M, Schaerringer B, Plasterk R H A, Driever W. Curr Biol. 1998;8:82–88. doi: 10.1016/s0960-9822(98)70038-7. [DOI] [PubMed] [Google Scholar]
- 6.Fadool J M, Hartl D, Dowling J E. Proc Natl Acad Sci USA. 1998;95:5182–5186. doi: 10.1073/pnas.95.9.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sherman A, Dawson A, Mather C, Gilhooley H, Li Y, Mitchell R, Finnegan D J, Sang H. Nat Biotechnol. 1998;16:1050–1053. doi: 10.1038/3497. [DOI] [PubMed] [Google Scholar]
- 8.Schouten G J, Van Luenen H G A M, Verra N C V, Valerio D, Plasterk R H A. Nucleic Acids Res. 1998;26:3013–3017. doi: 10.1093/nar/26.12.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivics Z, Hackett P B, Plasterk R H A, Izsvak Z. Cell. 1997;91:1–20. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L, Sankar U, Lampe D J, Robertson H M, Graham F L. Nucleic Acids Res. 1998;26:3687–3693. doi: 10.1093/nar/26.16.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klinakis A G, Zagoraiou L, Vassilatis D K, Savakis C. EMBO Rep. 2000;1:416–421. doi: 10.1093/embo-reports/kvd089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo G, Ivics Z, Izsvak Z, Bradley A. Proc Natl Acad Sci USA. 1998;95:10769–10773. doi: 10.1073/pnas.95.18.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yant S R, Meuse L, Chiu W, Ivics Z, Izsvak Z, Kay M A. Nat Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- 14.Vos J C, Van Luenen H G A M, Plasterk R H A. Genes Dev. 1993;7:1244–1253. doi: 10.1101/gad.7.7a.1244. [DOI] [PubMed] [Google Scholar]
- 15.Van Luenen H G A M, Colloms S D, Plasterk R H A. EMBO J. 1993;12:2513–2520. doi: 10.1002/j.1460-2075.1993.tb05906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medhora M, Maruyama K, Hartl D L. Genetics. 1991;128:311–318. doi: 10.1093/genetics/128.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ali S A, Steinkasserer A. BioTechniques. 1995;18:746–750. [PubMed] [Google Scholar]
- 18.Ketting R F, Fischer S E J, Plasterk R H A. Nucleic Acids Res. 1997;25:4041–4047. doi: 10.1093/nar/25.20.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer S E, van Luenen H G, Plasterk R H. Mol Gen Genet. 1999;262:268–274. doi: 10.1007/pl00008641. [DOI] [PubMed] [Google Scholar]
- 20.Taketo M, Schroeder A C, Mobraaten L E, Gunning K, Hanten G, Fox R F, Roderick T H, Stewart C L, Lilly F, Hansen C T, Overbeek P. Proc Natl Acad Sci USA. 1991;88:2065–2069. doi: 10.1073/pnas.88.6.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 22.Glover D M, Hames B D. In: The Practical Approach Series. Rickwood D, Hames B D, editors. Oxford: IRL Press; 1995. pp. 231–262. [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 24.Wicks S R, de Vries C J, van Luenen H G, Plasterk R H. Dev Biol. 2000;221:295–307. doi: 10.1006/dbio.2000.9686. [DOI] [PubMed] [Google Scholar]
- 25.McCarthy L C, Terrett J, Davis M E, Knights C J, Smith A L, Critcher R, Schmitt K, Hudson J, Spurr N K, Goodfellow P N. Genome Res. 1997;7:1153–1161. doi: 10.1101/gr.7.12.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emmons S W, Yesner L, Ruan K, Katzenberg D. Cell. 1983;32:55–65. doi: 10.1016/0092-8674(83)90496-8. [DOI] [PubMed] [Google Scholar]
- 27.Collins J, Forbes E, Anderson P. Genetics. 1989;121:47–55. doi: 10.1093/genetics/121.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medhora M M, MacPeek A H, Hartl D L. EMBO J. 1988;7:2185–2189. doi: 10.1002/j.1460-2075.1988.tb03057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Capy P, Koga A, David J R, Hartl D L. Genetics. 1992;130:499–506. doi: 10.1093/genetics/130.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braun R E, Peschon J J, Behringer R R, Brinster R L, Palmiter R D. Genes Dev. 1989;3:793–802. doi: 10.1101/gad.3.6.793. [DOI] [PubMed] [Google Scholar]
- 31.Plasterk R H A. EMBO J. 1991;10:1919–1925. doi: 10.1002/j.1460-2075.1991.tb07718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Luenen H G A M, Colloms S D, Plasterk R H A. Cell. 1994;79:293–301. doi: 10.1016/0092-8674(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 33.Van Loon A A, Den Boer P J, Van der Schans G P, Mackenbach P, Grootegoed J A, Baan R A, Lohman P H. Exp Cell Res. 1991;193:303–309. doi: 10.1016/0014-4827(91)90101-y. [DOI] [PubMed] [Google Scholar]
- 34.Behringer R R, Peschon J J, Messing A, Gartside C L, Hauschka S D, Palmiter R D, Brinster R L. Proc Natl Acad Sci USA. 1988;85:2648–2652. doi: 10.1073/pnas.85.8.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Gorman S, Gagenais N A, Qian M, Marchuk Y. Proc Natl Acad Sci USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zambrowicz B P, Friedrich G A, Buxton E C, Lilleberg S L, Person C, Sands A T. Nature (London) 1998;392:608–611. doi: 10.1038/33423. [DOI] [PubMed] [Google Scholar]
- 37.Gridley T. New Biol. 1991;3:1025–1034. [PubMed] [Google Scholar]
- 38.Soriano P, Gridley T, Jaenisch R. Genes Dev. 1987;1:366–375. doi: 10.1101/gad.1.4.366. [DOI] [PubMed] [Google Scholar]
- 39.Soriano P, Jaenisch R. Cell. 1986;46:19–29. doi: 10.1016/0092-8674(86)90856-1. [DOI] [PubMed] [Google Scholar]
- 40.Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature (London) 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 41.Venter J C, Adams M D, Myers E W, Li P W, Mural R J, Sutton G G, Smith H O, Yandell M, Evans C A, Holt R A, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 42.Schimenti J, Bucan M. Genome Res. 1998;8:698–710. doi: 10.1101/gr.8.7.698. [DOI] [PubMed] [Google Scholar]
- 43.Vijaya S, Steffen D L, Robinson H L. J Virol. 1986;60:683–692. doi: 10.1128/jvi.60.2.683-692.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scherdin U, Rhodes K, Breindl M. J Virol. 1990;64:907–912. doi: 10.1128/jvi.64.2.907-912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dooner H K, Belachew A, Burgess D, Harding S, Ralston M, Ralston E. Genetics. 1994;136:261–279. doi: 10.1093/genetics/136.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramirez-Solis R, Liu P, Bradley A. Nature (London) 1995;378:720–724. doi: 10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- 47.You Y, Browning V L, Schimenti J C. Methods. 1997;13:409–421. doi: 10.1006/meth.1997.0547. [DOI] [PubMed] [Google Scholar]
- 48.Su H, Wang X, Bradley A. Nat Genet. 2000;24:92–95. doi: 10.1038/71756. [DOI] [PubMed] [Google Scholar]
- 49.Zheng B, Sage M, Sheppeard E A, Jurecic V, Bradley A. Mol Cell Biol. 2000;20:648–655. doi: 10.1128/mcb.20.2.648-655.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plasterk R H, Izsvak Z, Ivics Z. Trends Genet. 1999;15:326–332. doi: 10.1016/s0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- 51.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]