Abstract
Biochemical studies with model DNA heteroduplexes have implicated RecJ exonuclease, exonuclease VII, exonuclease I, and exonuclease X in Escherichia coli methyl-directed mismatch correction. However, strains deficient in the four exonucleases display only a modest increase in mutation rate, raising questions concerning involvement of these activities in mismatch repair in vivo. The quadruple mutant deficient in the four exonucleases, as well as the triple mutant deficient in RecJ exonuclease, exonuclease VII, and exonuclease I, grow poorly in the presence of the base analogue 2-aminopurine, and exposure to the base analogue results in filament formation, indicative of induction of SOS DNA damage response. The growth defect and filamentation phenotypes associated with 2-aminopurine exposure are effectively suppressed by null mutations in mutH, mutL, mutS, or uvrD/mutU, which encode activities that act upstream of the four exonucleases in the mechanism for the methyl-directed reaction that has been proposed based on in vitro studies. The quadruple exonuclease mutant is also cold-sensitive, having a severe growth defect at 30°C. This phenotype is suppressed by a uvrD/mutU defect, and partially suppressed by mutH, mutL, or mutS mutations. These observations confirm involvement of the four exonucleases in methyl-directed mismatch repair in vivo and suggest that the low mutability of exonuclease-deficient strains is a consequence of under recovery of mutants due to a reduction in viability and/or chromosome loss associated with activation of the mismatch repair system in the absence of RecJ exonuclease, exonuclease VII, exonuclease I, and exonuclease X.
Methyl-directed mismatch repair ensures the fidelity of Escherichia coli chromosome replication by correcting DNA biosynthetic errors that escape the DNA polymerase proof reading function (1–3). Mismatch correction by this pathway is directed to the daughter strand at the replication fork by virtue of the transient absence of d(GATC) methylation on newly synthesized DNA (4). Repair is initiated by the binding of MutS to the mismatch (5). MutL binds to heteroduplex DNA in a MutS- and ATP-dependent manner (6). Assembly of this ternary complex is sufficient to activate the d(GATC) endonuclease activity of MutH, which cleaves the unmethylated strand of a hemimethylated d(GATC) sequence that can be located on either side of the mispair (7). DNA helicase II (uvrD/mutU product), which also is activated in a MutS- and MutL-dependent manner, enters the helix at the strand break with an orientation bias so that unwinding proceeds back toward the mismatch (8). That portion of the incised strand unwound in this manner is subject to degradation by one of several single-strand exonucleases (refs. 9 and 10; unpublished work), and the ensuing gap is filled by DNA polymerase III holoenzyme, with DNA ligase restoring covalent continuity to the repaired strand (11).
In vitro analysis of the repair reaction has implicated four exonucleases in the repair reaction, with the requirement for an individual activity depending on placement of the strand break 5′ or 3′ to the mismatch. When MutH incision occurs 5′ to the mispair on the unmethylated strand, in vitro excision depends on either exonuclease VII or RecJ exonuclease (9), both of which catalyze 5′ to 3′ hydrolysis of single-stranded DNA (12, 13). Initial biochemical analysis implicated the 3′ to 5′ hydrolytic function of exonuclease I (14) in mismatch repair when the MutH-produced strand break occurs 3′ to the mispair (9, 11). More recent work has demonstrated that the 3′ to 5′ excision activities of exonuclease VII (12), or exonuclease X (15) also can suffice in this regard (unpublished work).
Despite their requirement for in vitro mismatch repair, the involvement of these four exonucleases in the methyl-directed reaction in vivo has been conjectural. In contrast to mutH, mutL, mutS, or uvrD/mutU mutations, which confer a 50- to 100-fold increase in the spontaneous mutation rate, the quadruple mutant lacking RecJ, ExoVII, ExoI, and ExoX displays only a modest 6-fold increase in mutability (unpublished work). Although it is possible that the exonuclease requirements are an in vitro artifact, a plausible alternative is that the low mutability of exonuclease-deficient strains is due to lethal chromosome damage caused by activation of mismatch repair in the absence of exonucleases. By this reasoning, the low mutability of exonuclease-deficient cells is not due to their ability to correct mismatches but rather to a failure to recover chromosomes in which mismatches occur.
In this study we show that the quadruple mutant lacking RecJ, ExoVII, ExoI, and ExoX, as well the triple mutant deficient in RecJ, ExoVII, and ExoI, are characterized by sensitivity to the base analogue 2-aminopurine (2APur). The quadruple mutant also displays a cold-sensitive phenotype and is unable to form colonies at 30°C on rich medium. Because exposure of E. coli to 2APur is known to activate the methyl-directed mismatch repair system (16, 17), and because the phenotypes noted above are suppressed by introduction of mutH, mutL, mutS, or uvrD/mutU mutations, we have concluded that activation of the repair system in the absence of the exonucleases implicated in vitro is associated with a high incidence of cell death and/or chromosome loss.
Experimental Procedures
Bacterial Strains.
Strains used in this work (Table 1) were grown in LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl) on LB plates containing 1.4% agar, or in enriched minimal medium [containing 56/2 salts (18) supplemented with 0.2% vitamin-free casamino acids and 0.2% glucose]. Isogenic strains used for genetic studies were constructed by P1virA-mediated transduction. P1 lysates were prepared and transductions were performed in LB medium supplemented with 10 mM calcium chloride and 0.2% glucose. Transductants were selected on LB agar containing 10 mM sodium citrate supplemented with ampicillin, tetracycline, kanamycin, or chloramphenicol at concentrations of 100 μg/ml, 15 μg/ml, 25 μg/ml, and 15 μg/ml, respectively.
Table 1.
E. coli strains
| Strain | Relevant genotype | Source or derivation |
|---|---|---|
| BT199 | W. Wackernagel | |
| STL4534 | ΔexoX∷npt | S. Lovett collection |
| STL4150 | ΔxonA300∷cat xseA18∷amp recJ284∷Tn10 | S. Lovett collection |
| STL4150-S | STL 4150 mutS∷Tn5 | P1 RK1517 X STL4150 |
| STL4150-L | STL4150 mutL459∷kan | P1 GM4250 X STL4150 |
| STL4150-H | STL4150 mutH471∷kan | P1 GW3773 X STL4150 |
| STL4150-D | STL4150 uvrD254∷Tn5 | P1 STL1526 X STL4150 |
| VB1-VB4 | STL4150 ΔexoX∷npt | P1 STL4534 X STL4150 |
| VB31* | VB3 ΔexoX (Kms) | Kms cured VB3 |
| VB31-S | VB31 mutS∷Tn5 | P1 RK1517 X VB31 |
| VB31-L | VB31 mutL459∷kan | P1 GM4250 X VB31 |
| VB31-H | VB31 mutH471∷kan | P1 GW3773 X VB31 |
| VB31-D | VB31 uvrD254∷Tn5 | P1 STL1526 X VB31 |
| VB101† | VB1 2APurr | |
| VB105† | VB1 2APurr | |
| VB201† | VB2 2APurr | |
| VB301† | VB3 2APurr | |
| VB305† | VB3 2APurr | |
| VB401† | VB4 2APurr | |
| VB701† | STL4150 2APurr | |
| MG1655 | Wild type | M. Berlyn |
| RK1517 | mutS∷Tn5 | R. Kolodner |
| STL1526 | uvrD254∷Tn5 | S. Lovett collection |
| GW3773 | mutH471∷kan | G. Walker |
| GM4250 | mutL459∷kan | M. Marinus |
2APur Sensitivity.
Cultures were grown to about 2 × 108 per ml in LB in the absence of drug. Plates without and with 350 μg/ml 2APur were spotted with 10 μl of 10-fold serial dilutions of cells such that the inocula ranged from 102 to 105 colony-forming units. Plates were examined after 24 h at 37°C. 2APur-resistant derivatives were selected as spontaneous mutants on plates containing 350 μg/ml 2APur after overnight growth in enriched minimal medium containing 400–600 μg/ml 2APur.
Microscopic Observation of Cells.
Cultures were grown in enriched minimal medium in the absence or presence of 2APur (350 μg/ml) or in LB medium for cold-sensitivity study. For measurement of cell lengths, samples of culture were fixed in 1.4% formaldehyde in 56/2 salts, 0.05 M HCl, 0.4% eosinY to immobilize the cells. Treated cells were placed in a Petroff–Hausser counting chamber (Hausser Scientific, Horsham, PA), visualized by phase-contrast microscopy using a Leitz Labor Lux S microscope and photographed onto Kodak TMAX400 film. Cell lengths were determined by using the ruled grid of the counting chamber (50 μm × 50 μm) as internal standard. For observation of nucleoids, cells were stained with 4′,6-diamidino-2-phenylindole according to the fluo-phase method of Hiraga et al. (19) and visualized under oil immersion using combined phase-contrast and fluorescence optics.
Preparation of Extracts and Mismatch Repair Assays.
Extracts were prepared and mismatch repair assays were performed as described (9) except that reactions (20 μl) contained 24 fmol of heteroduplex. Before use for in vitro complementation, exonuclease I, single-strand binding protein (U.S. Biochemical), and DNA helicase II (20) were dialyzed against 20 mM Tris⋅HCl (pH 7.6), 150 mM KCl, 0.5 mM EDTA, 5 mM 2-mercaptoethanol for 6 h at 4°C (two buffer changes) and stored at 4°C. MutS, MutL, and MutH, purified as described (6, 21, 22) were essentially homogeneous. Proteins were diluted as necessary into 20 mM KPO4 (pH 7.4), 50 mM KCl (100 mM KCl in the case of MutS), 1 mM EDTA, 2 mM DTT, 100 μg/ml BSA.
Results
Initial analysis of the in vitro exonuclease dependence of methyl-directed mismatch repair implicated RecJ, ExoVII, ExoI, and an unidentified exonuclease in the reaction (9). The fourth activity recently was identified as ExoX, and extracts prepared from strains deficient in all four exonucleases are devoid of in vitro mismatch repair activity (unpublished work). Nevertheless, such cells display only a small increase in mutation rate. As discussed above, this discrepancy may indicate that the four exonucleases have little, if any, role in cellular mismatch repair.
However, it is also possible that activation of the mismatch repair system in the absence of these exonucleases may be associated with reduced viability or chromosome loss, resulting in failure to recover mutants that otherwise might occur. Precedent for a similar effect has been demonstrated with E. coli dam mutants that are deficient in the DNA methylase that modifies d(GATC) sequences and confers strand-specificity on methyl-directed repair. dam mutants do not grow in the presence of the base analogue 2APur (16, 17), an adenine analogue and weak mutagen that occasionally pairs with cytosine (23). This growth defect, which is suppressed by mutH, mutL, or mutS mutations, has been attributed to activation of the mismatch repair system by mispairs involving 2APur followed by double-strand cleavage at d(GATC) sites, which are unmethylated on both DNA strands in dam mutants (7, 16).
RecJ− ExoVII− ExoI− Triple Mutants and RecJ− ExoVII− ExoI− ExoX− Quadruple Mutants Are Sensitive to 2APur.
To test the possibility that a related phenomenon might be occurring in the exonuclease-deficient backgrounds we examined the 2APur sensitivity of an isogenic set of strains representing all possible combinations of one, two, three, or four null mutations in the genes encoding the exonucleases implicated in the in vitro reaction. All strains were found to be resistant to 2APur (data not shown) with the exception of the triple mutant deficient in RecJ, ExoVII, and ExoI, and several independently constructed quadruple mutants lacking RecJ, ExoVII, ExoI, and ExoX. In contrast to the BT199 parental strain, these triple and quadruple mutants grow poorly in the presence of 2APur (Fig. 1). The exonuclease deficient mutants only grow as microcolonies, although faster growing, 2APur-resistant colonies occasionally were observed (see below). Identical results were obtained for triple and quadruple exonuclease mutants in the prototrophic background of E. coli MG1655 (24) (data not shown).
Figure 1.
Suppression of 2APur sensitivity by inactivation of mismatch repair functions. LB agar plates containing 2APur (350 μg/ml) were spotted with 10-fold serial dilutions of each culture such that spots contained roughly 102–104 colony-forming units as indicated. Plates were photographed after 24 h at 37°C. Strain designations (Table 1) are shown on the left and relevant genotypes on the right. The small variability in cell densities evident is due to variation in the initial culture cell densities. In the absence of drug (data not shown), plating efficiency and colony size were similar for all strains tested.
2APur Sensitivity of Exonuclease-Deficient Strains Is Suppressed by mutS, mutL, mutH, or uvrD/mutU Mutations.
2APur-resistant derivatives were isolated (Experimental Procedures) from STL4150 (RecJ−, ExoVII−, ExoI−) and four independently constructed quadruple mutants (VB1, VB2, VB3 and VB4) deficient in RecJ, ExoVII, ExoI, and ExoX. All of these 2APur-resistant isolates (24 of 24) were characterized by a high rate of mutation to rifampicin resistance, similar to that observed with mutS, mutL, mutH, or uvrD/mutU mutants (data not shown). Accordingly, seven independent, 2APur-resistant isolates were tested for potential MutS, MutL, MutH, or helicase II defects by in vitro complementation assay and Western blotting. As shown in Table 2, in vitro complementation indicated six of these seven isolates to be deficient in MutS or MutL. MutS defects were identified in two (VB101 and VB401) and MutL deficiency in three resistant isolates (VB105, VB201, and VB305) obtained from quadruple exonuclease mutant strains. One 2APur-resistant isolate (VB701) derived from the RecJ− ExoVII− ExoI− triple mutant also was characterized by in vitro complementation and found to be defective in MutL activity. Furthermore, Western blotting indicated absence of the MutS polypeptide in extracts of VB101 and the MutL polypeptide in VB105, VB201, and VB305.
Table 2.
In vitro complementation of mismatch repair defects in 2APurR isolates
| 2APurR cell extract
|
||||||||
|---|---|---|---|---|---|---|---|---|
| None | VB101 | VB105 | VB201 | VB301 | VB305 | VB401 | VB701 | |
| Repair (fmol/hr per 0.1 mg extract) | ||||||||
| Addition | ||||||||
| None | — | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 |
| mutS− extract | <0.5 | <0.5 | 13 | 12 | 5.2 | 12 | <0.5 | 11 |
| mutL− extract | <0.5 | 10 | 0.8 | <0.5 | 9.2 | <0.5 | 7.4 | <0.5 |
| mutH− extract | <0.5 | 13 | 6.6 | 4.9 | 13 | 8.8 | 13 | 6.7 |
| ExoI | — | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 |
| ExoI + MutS | — | 6.7 | n.t. | <0.5 | 4.3 | n.t. | 1.3 | n.t. |
| ExoI + MutL | — | <0.5 | 20 | 7.2 | 1.0 | 21 | <0.5 | 20 |
| ExoI + MutH | — | <0.5 | <0.5 | <0.5 | 1.3 | <0.5 | <0.5 | <0.5 |
| ExoI + SSB | — | 2.5 | n.t. | <0.5 | 10 | n.t. | <0.5 | n.t. |
| ExoI + UvrD | — | <0.5 | <0.5 | <0.5 | 3.2 | <0.5 | <0.5 | <0.5 |
| Western blot | MutS | − | + | + | + | + | + | + |
| MutL | + | − | − | + | − | + | + | |
Extracts of 2APur resistant strains were assayed (Experimental Procedures) for repair of a 3′-G-T heteroduplex (9) in the presence of extracts derived from mutS−, mutL−, or mutH− derivatives of the parental strain BT199 or in the presence of purified proteins as indicated. For extract complementation, assays contained approximately 0.1 mg of each extract tested. For complementation with purified proteins, individual extracts (0.1–0.2 mg) were supplemented with ExoI (10 ng), together with MutS (35 ng), MutL (80 ng), MutH (1 ng), single-strand binding protein (400 ng), or UvrD (DNA helicase II, 20 ng) as shown. ExoI restores mismatch repair directed by a 3′ strand signal to E. coli extracts deficient in RecJ, ExoVII, ExoI, and ExoX (unpublished work). No detectable repair was observed in VB101 or VB401 extracts supplemented with MutS in the absence of ExoI, or in VB105, VB201, VB305, or VB701 extracts supplemented with MutL in the absence of the exonuclease. Despite the efficient complementation of VB401 extracts by extracts deficient in MutH or MutL, restoration of repair by addition of ExoI and purified MutS was consistently low, although other purified proteins did not increase repair to detectable levels. Immunological blot analysis was performed on extract samples using polyclonal serum against MutS, MutL, or DNA helicase II by a method described previously (44). Normal levels of the helicase II polypeptide were present in all extracts. n.t., not tested.
The remaining isolate VB301, which was derived from the VB3 quadruple exonuclease mutant background, did not behave in a simple manner. VB301 extracts complemented extracts derived from mutS, mutL, mutH, or uvrD/mutU mutants to restore in vitro repair. Supplementation of VB301 extracts with purified MutL, MutH, or DNA helicase II in the presence of exogenous ExoI resulted in small, but significant, increases in basal repair activity. Addition of MutS and ExoI resulted in intermediate levels of repair, whereas addition of single-stranded DNA binding protein (SSB) and ExoI yielded near normal levels of activity. These observations indicate a complex defect, possibly due to a mutS or ssb mutation. Although the function of SSB in the in vitro mismatch repair reaction has not been delineated (11), the protein probably participates in both the excision and repair DNA synthesis steps of the reaction.
In a second approach, null alleles of mutS, mutL, mutH, or uvrD were introduced into STL4150 and VB31 (Table 1). As shown in Fig. 1, defects in each of these loci effectively suppress the 2APur sensitivities of the RecJ− ExoVII− ExoI− triple mutant and the RecJ− ExoVII− ExoI− ExoX− quadruple mutant.
Filamentation Associated with 2APur Sensitivity Is Suppressed by mutH, mutL, mutS, and uvrD/mutU Mutations.
DNA damage in E. coli often elicits the SOS DNA damage response, one characteristic of which is filamentation due to continued cell elongation in the absence of division (25). In view of the 2APur sensitivity of the triple and quadruple exonuclease mutants, the morphologies of wild-type and exonuclease-deficient strains were examined to assess the effects of exposure to the base analogue.
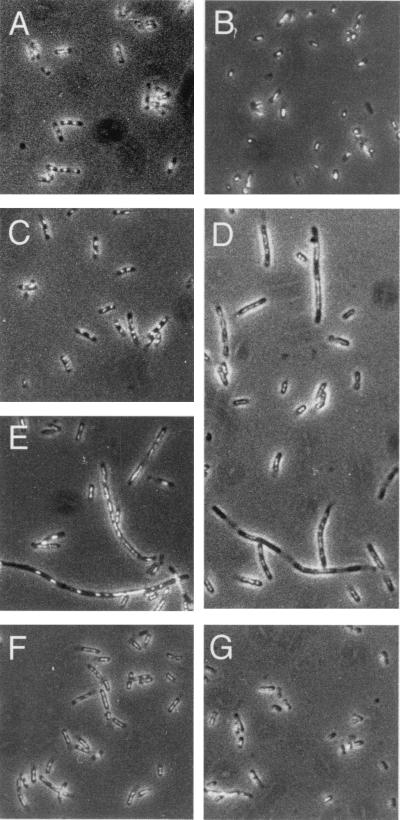
At 37°C in the absence of 2APur, wild-type parental cells (BT199) appeared as short rods of median length of 3.2 μm, with the largest cells ≈5 μm in length (Table 3). Similar morphologies were observed for the RecJ− ExoVII− ExoI− triple mutant (STL4150) and the RecJ− ExoVII− ExoI− ExoX− quadruple (VB31): STL4150 had a median cell length of 3.2 μm, identical to that of BT199, although VB31 was significantly smaller with median length of 1.8 μm. Fluorescence microscopy after 4′,6-diamidino-2-phenylindole staining (19) revealed the presence of one or two evenly spaced nucleoids within these cells without regard to genotype (Fig. 2).
Table 3.
Cell length distributions in the presence or absence of 2APur
| Strain | 2APur | Cell
lengths (μm)
|
|||||
|---|---|---|---|---|---|---|---|
| >5 μm | Range | Median | Avg. | SD | N | ||
| BT199 | − | 0% | 1.4–5.0 | 3.2 | 3.3 | 0.8 | 45 |
| + | 7% | 1.8–18 | 3.2 | 3.8 | 2.4 | 110 | |
| STL4150 | − | 2% | 1.4–5.1 | 3.2 | 3.1 | 0.7 | 51 |
| + | 43% | 2.1–49 | 4.6 | 7.5 | 7.2 | 147 | |
| STL4150-S | − | 4% | 2.1–8.6 | 3.2 | 3.4 | 1.1 | 102 |
| + | 12% | 1.8–16 | 3.6 | 4.3 | 2.6 | 72 | |
| VB31 | − | 0% | 1.1–4.3 | 1.8 | 2.1 | 0.8 | 60 |
| + | 36% | 1.8–46 | 4.3 | 6.6 | 6.9 | 140 | |
| VB31-H | − | 0% | 1.5–3.9 | 2.9 | 2.9 | 0.6 | 52 |
| + | 8% | 1.9–7.8 | 2.9 | 3.1 | 1.2 | 50 | |
| VB31-L | − | 3% | 1.4–11 | 2.9 | 3.0 | 1.4 | 150 |
| + | 3% | 1.1–10 | 2.9 | 2.8 | 1.3 | 90 | |
Cultures were grown and cell lengths were determined as described in Experimental Procedures. N, cells measured. Graphical presentation of some of these data is available in Fig. 4, which is published as supplemental material on the PNAS web site, www.pnas.org.
Figure 2.
Cell and nucleoid morphologies associated with response to 2APur. Samples were removed from cultures grown overnight at 37°C in enriched minimal medium in the presence (350 μg/ml) or absence of 2APur and fixed on microscope slides. After staining with 4′,6-diamidino-2-phenylindole, cells were visualized and photographed by using phase-contrast and fluorescence optics as described in Experimental Procedures. All photographs are shown at the same magnification (×1,250). (A) BT199 no 2APur; (B) VB31 no 2APur; (C) BT199 plus 2APur; (D) VB31 plus 2APur; (E) STL4150 plus 2APur; (F) STL4150mutS plus 2APur; (G) VB31mutL plus 2APur.
After exposure to 2APur, 90% of BT199 wild-type cells appeared morphologically normal, but 7% were extended in length, with filaments up to 18 μm in length observed. Filamentation was much more pronounced with the triple and quadruple exonuclease mutants STL4150 and VB31 (Table 3). After exposure to 2APur, about 40% of the mutant cells in each case were greater than 5 μm in length, with filaments as long as 49 μm observed. These elongated cells did not appear to be septated and contained multiple 4′,6-diamidino-2-phenylindole-positive elements that were not uniformly distributed within the elongated cells (Fig. 2). In contrast to STL4150 and VB31, the morphologies of the mut derivatives of the triple and quadruple exonuclease-deficient strains after 2APur exposure were similar to those observed with wild-type BT199 cells (Table 3 and Fig. 2).
As noted above, the 1.8-μm median size of RecJ− ExoVII− ExoI− ExoX− VB31 cells in the absence of 2APur is significantly less than the 3.2-μm median length of wild-type cells. Introduction of a mutH or mutL null mutation also appeared to suppress this effect (Table 3).
The RecJ− ExoVII− ExoI− ExoX− Quadruple Mutant Is Cold-Sensitive for Growth on Rich Media.
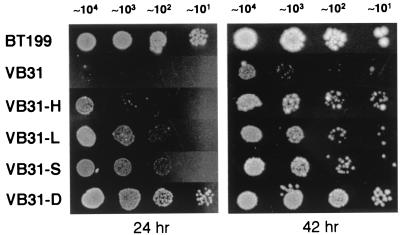
In addition to the 2APur-sensitive phenotype, RecJ− ExoVII− ExoI− ExoX− quadruple mutants are also cold-sensitive, an effect illustrated with strain VB31 in Fig. 3. When growth on LB plates was examined at 30°C by spot test, VB31 colonies were barely visible after 24 h, a phenotype that appeared to be due to reduced colony forming ability as well as very slow growth of those colonies that did become visible. Growth rates in LB broth at 30°C also were reduced as monitored either by A600 or by measurement of viable titer (data not shown). The cold-sensitive phenotype is media-dependent and was not observed with minimal glucose medium or minimal glucose medium supplemented with casamino acids. This phenotype is restricted to the quadruple mutant and was not observed with the triple mutant STL4150 deficient in RecJ, ExoVII, and ExoI.
Figure 3.
Cold sensitivity of a RecJ− ExoVII− ExoI− ExoX− strain and suppression by mismatch repair defects. Two independent cultures of the strains shown (Table 1) were grown overnight in LB at 37°C, and 10-fold serial dilutions from each were prepared and spotted onto LB agar plates. One set of plates was photographed after incubation at 30°C for 24 h and the second after incubation for 42 h. Although not shown, growth at 37°C was comparable for all strains and was similar to that shown here for BT199. The approximate number of colony-forming units is indicated above each spot.
The cold-sensitive growth of the RecJ− ExoVII− ExoI− ExoX− quadruple mutant VB31 was suppressed by introduction of null alleles of mutH, mutL, mutS, or uvrD/mutU. However, as shown in Fig. 3, these repair defects were not equally effective in this regard as judged by qualitative assessment of rates of colony growth at 30°C. When compared with identical dilutions spot tested for growth at 37°C, the uvrD/mutU null allele was the most effective suppressor of the quadruple mutant cold sensitivity, resulting in colony growth characteristics similar to those of BT199 wild-type cells. Colonies of VB31 harboring a mutS or mutL defect were consistently found to grow somewhat more slowly than VB31 uvrD/mutU, and colonies of the VB31 derivative with a mutH null allele came up even more slowly than those with a mutS or mutL mutation. Furthermore, in contrast to the filamentation observed upon exposure of the quadruple exonuclease mutant to 2APur, extensive filamentation was not observed after overnight incubation of RecJ− ExoVII− ExoI− ExoX− VB31 at 30°C. The nature of this complex phenotype will be considered below.
Discussion
The initial evidence implicating ExoI, RecJ, and ExoVII in methyl-directed mismatch repair was based strictly on in vitro studies using model heteroduplexes (9, 11). Although these experiments also indicated involvement of a fourth activity, the in vivo requirements for ExoI, RecJ, and ExoVII in mismatch repair have been a matter of debate due to the insignificant or limited increases in mutability associated with defects in one or more of these activities (9, 26, 27). We have reassessed this question with the identification of ExoX (15), the fourth activity implicated in the in vitro reaction (unpublished work). Extracts prepared from cells lacking all four exonucleases are devoid of mismatch repair as judged by in vitro assay.
Strains deficient in the three exonucleases originally implicated in vitro mismatch repair, as well as the quadruple mutant deficient in ExoI, RecJ, ExoVII, and ExoX, grow poorly in the presence of 2APur. Of the four possible triple mutant combinations, only strains deficient in ExoI, RecJ, and ExoVII display 2APur sensitivity. This effect is associated with filamentation of exonuclease-deficient cells and depends on the action of MutH, MutL, MutS, and DNA helicase II. Because MutH is activated in a mismatch-, MutS- and MutL-dependent manner and because the only known function of activated MutH is its endonucleolytic activity at hemimethylated or unmethylated d(GATC) sites (7), these findings implicate the four exonucleases in the methyl-directed repair of 2APur lesions in newly synthesized DNA. The observation that the triple and quadruple mutants are equally sensitive to 2APur suggests that the triple mutant deficient in ExoI, RecJ, and ExoVII is seriously crippled in its ability to process DNA biosynthetic errors.
Although we were unable to distinguish the triple and quadruple mutants based on 2APur sensitivity, these strains are distinguishable by virtue of the cold-sensitive phenotype of strains deficient in ExoI, RecJ, ExoVII, and ExoX. While the studies reported here were performed in a BT199 background (Table 1), identical results with respect to 2APur and cold sensitivity were obtained in those instances tested in the wild-type E. coli background of MG1655 (24). The cold sensitivity of the quadruple exonuclease mutant was suppressed by uvrD/mutU mutS, mutL, or mutH null mutations. However, as noted above, these repair defects were not equally effective in this regard, indicating that this phenotype is complex. The observation that uvrD/mutU is the most efficient suppressor suggests that a significant component of cold sensitivity is independent of the action of mutS, mutL, and mutH gene products; in addition to its role in mismatch repair, the uvrD/mutU gene product also participates in nucleotide excision repair, a function that also might contribute to the cold-sensitive phenotype. Nevertheless, cold sensitivity was suppressed by mutS, mutL, or mutH mutations, indicating that the products of these repair genes contribute to the growth defect of ExoI− RecJ− ExoVII− ExoX− strains at 30°C. In view of its known molecular functions described above, we infer that the MutH-dependent component of cold sensitivity is due to a failure to properly process spontaneous DNA biosynthetic errors at 30°C under conditions of exonuclease deficiency.
The finding that mutS and mutL mutations are better suppressors than is a mutH defect implies that MutH-independent events also are involved in the cold-sensitive phenotype. Inasmuch as MutS and MutL have been implicated in the MutH-independent processing of recombination intermediates (28, 29) and certain types of DNA damage (30, 31), it is possible that cold sensitivity of the quadruple exonuclease mutant also may reflect a recombination or damage repair function of MutS and MutL. However, it is important to note that correction of a significant fraction of DNA biosynthetic errors has been postulated to be MutH independent. Careful evaluation of mutation rates as a function of d(GATC) site density and mutH−, mutL−, and mutS− defects has led to the suggestion that 50–70% of DNA biosynthetic errors are repaired by a mechanism that requires MutS and MutL, but is independent of MutH and d(GATC) sequences (32). Because the function of MutH and d(GATC) sites in strand-specific mismatch repair is simply provision of a strand discontinuity that serves as the real signal that directs repair (11, 33), discontinuities that occur during the normal course of DNA biosynthesis also may suffice to direct repair to the newly synthesized strand (32, 34).
The suppression of 2APur and cold sensitivity by mutH, mutS, mutL, and uvrD/mutU mutations is of interest in terms of the mechanism for methyl-directed mismatch repair that has been deduced from in vitro experiments (8–10). In this scheme, MutS, MutL, MutH, and DNA helicase II all act upstream of the postulated involvement of RecJ, ExoVII, ExoI, and ExoX. We therefore suggest that mismatch-, MutS-, and MutL-dependent activation of the MutH endonuclease and unwinding by DNA helicase II at the ensuing strand discontinuity is associated with a substantial probability of chromosome loss and/or cell death when these events occur in the absence of RecJ, ExoVII, ExoI, and ExoX. This explanation accounts for the observations described here as well as the limited increase in mutability associated with deficiency of these four activities.
As discussed above, we attribute a component of the cold sensitivity of quadruple exonuclease mutants to activation of the mismatch repair system by spontaneous DNA biosynthetic errors in the absence of the four hydrolytic activities, but the temperature-dependent nature of this effect is unclear. One possibility is that DNA polymerase errors may be more common at 30°C than at 37°C, a feasible explanation because removal of 3′-terminal mismatches by the DNA polymerase III editing exonuclease is strongly temperature-dependent (35). In fact, the rate limiting step in exonuclease editing is a conformational transition that may correspond to local melting of the helix in the vicinity of the primer terminus (36). Poor growth of the quadruple mutant at 30°C also might be indicative of a temperature-dependent ability of E. coli to cope with DNA lesions produced upon activation of the mismatch repair system in the absence of the required exonucleases. According to this model, rescue of a chromosome harboring an abortive repair intermediate, perhaps by recombination, would be more efficient at 37°C than at 30°C. A third possibility attributes this effect to an active process that leads to chromosome inactivation upon repair failure, a process that would be more efficient at the lower temperature.
The redundancy of exonuclease involvement in mismatch repair does not appear to be restricted to E. coli because a similar redundancy has been suggested in the yeast system. Deficiency of the 5′ to 3′ exonuclease EXOI causes a weak mutator phenotype in Schizosaccharomyces pombe and Saccharomyces cerevisiae that is epistatic to msh2 defects, and the EXOI protein has been shown to interact with MSH2 (37, 38). A subset of the mutations that occur in yeast rad27 mutants also are observed under conditions of mismatch repair deficiency, and it has been postulated that the RAD27 5′ to 3′ exonuclease also may function in mismatch correction (39, 40). Because in vitro analysis of mismatch repair in higher cells has indicated a bidirectional excision mechanism similar to that of E. coli (41), involvement of 3′ to 5′ hydrolytic activities in the eukaryotic reaction also is anticipated. The editing exonucleases of DNA polymerases δ and ɛ have been suggested as possible candidates in this regard based on the concerted enhancement of mutation rates in yeast double mutants that are deficient in EXOI and also harbor a mutation that inactivates the editing function of polymerase δ or ɛ (42). However, this interpretation has been questioned with the demonstration that the enhanced mutability of a polymerase δ editing exonuclease mutant depends on S-phase checkpoint activation (40). Consequently, the identity of the putative 3′ to 5′ hydrolytic activities involved in eukaryotic mismatch repair remains an open question.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants GM23719 (to P.M.), GM43889 (to S.T.L.), and T32 GM07122 (to M.V.). P.M. is an Investigator of the Howard Hughes Medical Institute.
Abbreviation
- 2APur
2-aminopurine
References
- 1.Modrich P. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 2.Radman M, Matic I, Halliday J A, Taddei F. Philos Trans R Soc London B. 1995;347:97–103. doi: 10.1098/rstb.1995.0015. [DOI] [PubMed] [Google Scholar]
- 3.Modrich P, Lahue R. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 4.Meselson M. In: Recombination of the Genetic Material. Low K B, editor. San Diego: Academic; 1988. pp. 91–113. [Google Scholar]
- 5.Su S-S, Lahue R S, Au K G, Modrich P. J Biol Chem. 1988;263:6829–6835. [PubMed] [Google Scholar]
- 6.Grilley M, Welsh K M, Su S-S, Modrich P. J Biol Chem. 1989;264:1000–1004. [PubMed] [Google Scholar]
- 7.Au K G, Welsh K, Modrich P. J Biol Chem. 1992;267:12142–12148. [PubMed] [Google Scholar]
- 8.Dao V, Modrich P. J Biol Chem. 1998;273:9202–9207. doi: 10.1074/jbc.273.15.9202. [DOI] [PubMed] [Google Scholar]
- 9.Cooper D L, Lahue R S, Modrich P. J Biol Chem. 1993;268:11823–11829. [PubMed] [Google Scholar]
- 10.Grilley M, Griffith J, Modrich P. J Biol Chem. 1993;268:11830–11837. [PubMed] [Google Scholar]
- 11.Lahue R S, Au K G, Modrich P. Science. 1989;245:160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- 12.Chase J W, Richardson C C. J Biol Chem. 1974;249:4553–4561. [PubMed] [Google Scholar]
- 13.Lovett S T, Kolodner R D. Proc Natl Acad Sci USA. 1989;86:2627–2631. doi: 10.1073/pnas.86.8.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehman I R, Nussbaum A L. J Biol Chem. 1964;239:2628–2636. [PubMed] [Google Scholar]
- 15.Viswanathan M, Lovett S T. J Biol Chem. 1999;274:30094–30100. doi: 10.1074/jbc.274.42.30094. [DOI] [PubMed] [Google Scholar]
- 16.Glickman B W, Radman M. Proc Natl Acad Sci USA. 1980;77:1063–1067. doi: 10.1073/pnas.77.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGraw B R, Marinus M G. Mol Gen Genet. 1980;178:309–315. doi: 10.1007/BF00270477. [DOI] [PubMed] [Google Scholar]
- 18.Willetts M S, Clark A J, Low B. J Bacteriol. 1969;97:244–249. doi: 10.1128/jb.97.1.244-249.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiraga S, Niki H, Ogura T, Ichinose C, Mori H, Ezaki B, Jaffé A. J Bacteriol. 1989;171:1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Runyon G T, Wong I, Lohman T M. Biochemistry. 1993;32:602–612. doi: 10.1021/bi00053a028. [DOI] [PubMed] [Google Scholar]
- 21.Su S-S, Modrich P. Proc Natl Acad Sci USA. 1986;83:5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welsh K M, Lu A-L, Clark S, Modrich P. J Biol Chem. 1987;262:15624–15629. [PubMed] [Google Scholar]
- 23.Ronen A. Mutat Res. 1980;75:1–47. doi: 10.1016/0165-1110(80)90026-3. [DOI] [PubMed] [Google Scholar]
- 24.Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 25.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol. Press; 1995. [Google Scholar]
- 26.Harris R S, Ross K J, Lombardo M J, Rosenberg S M. J Bacteriol. 1998;180:989–993. doi: 10.1128/jb.180.4.989-993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viswanathan M, Lovett S T. Genetics. 1998;149:7–16. doi: 10.1093/genetics/149.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rayssiguier C, Thaler D S, Radman M. Nature (London) 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 29.Petit M-A, Dimpfl J, Radman M, Echols H. Genetics. 1991;129:327–332. doi: 10.1093/genetics/129.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellon I, Champe G N. Proc Natl Acad Sci USA. 1996;93:1292–1297. doi: 10.1073/pnas.93.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lieb M. J Bacteriol. 1987;169:5241–5246. doi: 10.1128/jb.169.11.5241-5246.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Claverys J P, Mejean V. Mol Gen Genet. 1988;214:574–578. doi: 10.1007/BF00330497. [DOI] [PubMed] [Google Scholar]
- 33.Längle-Rouault F, Maenhaut M G, Radman M. EMBO J. 1987;6:1121–1127. doi: 10.1002/j.1460-2075.1987.tb04867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holmes J, Clark S, Modrich P. Proc Natl Acad Sci USA. 1990;87:5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenowitz S, Kwack S, Goodman M F, O'Donnell M, Echols H. J Biol Chem. 1991;266:7888–7892. [PubMed] [Google Scholar]
- 36.Bloom L B, Otto M R, Eritja R, Reha-Krantz L J, Goodman M F, Beechem J M. Biochemistry. 1994;33:7576–7586. doi: 10.1021/bi00190a010. [DOI] [PubMed] [Google Scholar]
- 37.Szankasi P, Smith G R. Science. 1995;267:1166–1169. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- 38.Tishkoff D X, Boerger A L, Bertrand P, Filosi N, Gaida G M, Kane M F, Kolodner R D. Proc Natl Acad Sci USA. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson R E, Kovvali G K, Prakash L, Prakash S. Curr Genet. 1998;34:21–29. doi: 10.1007/s002940050362. [DOI] [PubMed] [Google Scholar]
- 40.Datta A, Schmeits J L, Amin N S, Lau P J, Myung K, Kolodner R D. Mol Cell. 2000;6:593–603. doi: 10.1016/s1097-2765(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 41.Fang W-H, Modrich P. J Biol Chem. 1993;268:11838–11844. [PubMed] [Google Scholar]
- 42.Tran H T, Gordenin D A, Resnick M A. Mol Cell Biol. 1999;19:2000–2007. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kristensen C S, Eberl L, Sanchez-Romero J M, Givskov M, Molin S, De Lorenzo V. J Bacteriol. 1995;177:52–68. doi: 10.1128/jb.177.1.52-58.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spampinato C, Modrich P. J Biol Chem. 1999;275:9863–9869. doi: 10.1074/jbc.275.13.9863. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.