SUMMARY
Gut microbial induction of host immune maturation exemplifies host-microbe mutualism. We colonized germ-free (GF) mice with mouse microbiota (MMb) or human microbiota (HMb) to determine whether small intestinal immune maturation depends on a coevolved host-specific microbiota. Gut bacterial numbers and phylum abundance were similar in MMb and HMb mice, but bacterial species differed, especially the Firmicutes. HMb mouse intestines had low levels of CD4+ and CD8+ T cells, few proliferating T cells, few dendritic cells, and low antimicrobial peptide expression–all characteristics of GF mice. Rat microbiota also failed to fully expand intestinal T cell numbers in mice. Colonizing GF or HMb mice with mouse-segmented filamentous bacteria (SFB) partially restored T cell numbers, suggesting that SFB and other MMb organisms are required for full immune maturation in mice. Importantly, MMb conferred better protection against Salmonella infection than HMb. A host-specific microbiota appears to be critical for a healthy immune system.
INTRODUCTION
Mutually beneficial host-microbe interactions shaped by eons of coevolution take place in all orders of life. In humans, the nutrient-rich intestinal environment is inhabited by up to 100 trillion microbes, the vast majority of which are nonpathogenic bacteria essential to human health. Recognition of the importance of microbes to human physiology has led to studies aimed at defining what bacterial species or genes compose a healthy human microbiota (HMb) (Arumugam et al., 2011; Turnbaugh et al., 2007). Identification of microbes on the basis of small subunit (16S) ribosomal RNA (rRNA) gene sequences has substantially elucidated the gut microbiota’s composition. Analyses of the gut microbiota of vertebrates, including humans, have shown that Firmicutes and Bacteroidetes predominate among the >80–100 bacterial phyla on Earth (Eckburg et al., 2005). In the phyla represented, abundant species and strains are found. The HMb is similar to the microbiotas of other mammals at the phylum level but distinct at the species and strain levels (Dethlefsen et al., 2007). Despite vast individual variation in species and strains, a person’s gut microbiota more resembles that of other people than that of other mammals (Ley et al., 2008a).
Work in germ-free (GF) mice, which display developmental defects including abnormal nutrient absorption and altered intestinal morphology and motility (Smith et al., 2007), has shown that the gut microbiota is critical for intestinal immune maturation. GF animals have smaller Peyer’s patches (PPs), fewer plasma cells, fewer intraepithelial lymphocytes (IELs), impaired antimicrobial peptide and IgA secretion, and other immunologic deficiencies (Round and Mazmanian, 2009); many deficiencies are corrected by recolonization with a health-associated mouse commensal microbiota. Gut microbiota-stimulated immune maturation maintains gut homeostasis by protecting the host from infections (Duan et al., 2010), injury (Rakoff-Nahoum et al., 2004), and damaging inflammatory responses (Atarashi et al., 2011; Mazmanian et al., 2008). Exclusivity between the host and specific symbiotic bacteria has been studied in invertebrate models. In the squid Euprymna scolopes, Vibrio fischeri is central to tissue development (Koropatnick et al., 2004). In tsetse flies, Wigglesworthia glossinidia enhances host fitness, and flies are sterile in its absence (Pais et al., 2008). In complex mammals, it remains unclear whether health-associated development depends on specific bacterial species exclusive to the host.
Different host species are colonized with different bacterial consortia (Ley et al., 2005). Reciprocal gut microbiota transplantation between zebrafish and mice shows that the host gut habitat selects for certain microbial community structures (Rawls et al., 2006). Diet and host phylogeny are both critical determinants of gut bacterial diversity (Ley et al., 2008b; Ochman et al., 2010). Despite broadened knowledge of factors determining the shape and composition of the gut bacterial community, it is not clear whether the community typically colonizing a given mammalian host species preferentially stimulates a specific program of immune maturation. Have mammals (like invertebrates such as the squid and the fly) coevolved with specific bacterial species uniquely capable of stimulating immune maturation? In other words, is mammalian immune maturation dependent on the mere presence of bacteria, or is a host-specific microbiota required?
To address these questions, we colonized GF mice at birth with a mouse gut microbiota (MMb) or a human gut microbiota (HMb). We studied immune maturation and gut microbiota composition over time by deep pyrosequencing of 16S rRNA genes. MMb and HMb mice share the same major bacterial phyla, but their microbiotas (particularly the Firmicutes) differ significantly at the operational taxonomic unit (OTU) level. MMb and HMb result in remarkably different small intestinal immune systems. Absolute cell numbers and gene transcription in the small intestine indicate that innate and adaptive immune maturation in mice colonized with an HMb (comparable in bacterial abundance to the MMb) resembles that in GF mice. Moreover, HMb mice are more susceptible than MMb mice to gastrointestinal infection. This observation suggests that mammalian hosts have coevolved with a specific consortium of bacterial species that stimulates intestinal immune maturation.
RESULTS
GF Swiss Webster (SW) mice underwent oral gavage with pooled fecal specimens from two healthy humans or with fecal/cecal contents from specific pathogen-free (SPF) SW mice. The two groups of recipient mice were then maintained in separate gnotobiotic isolators. To mimic age-dependent changes in the gut microbiota (Palmer et al., 2007) and the immune system (Chassin et al., 2010; Olszak et al., 2012), we bred mice in the isolator, naturally exposing the offspring to maternal microbes during and after birth (Figure 1A). Both MMb and HMb offspring had a smaller cecum than GF mice, whose cecum is abnormally large (see Figure S1A available online). Body weight did not differ in age-comparable MMb and HMb offspring (Figure S1B).
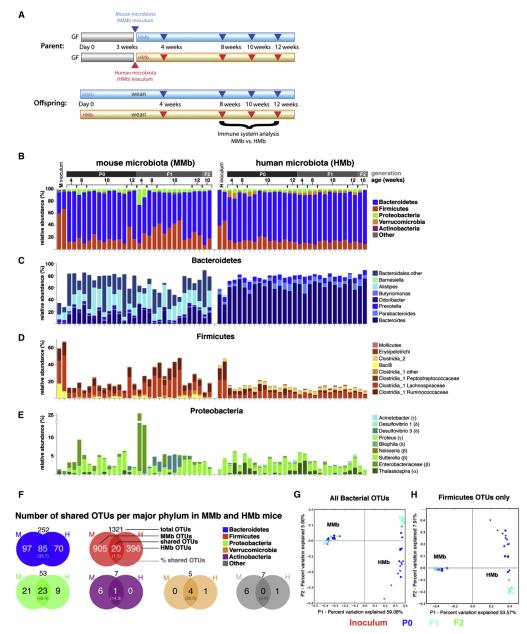
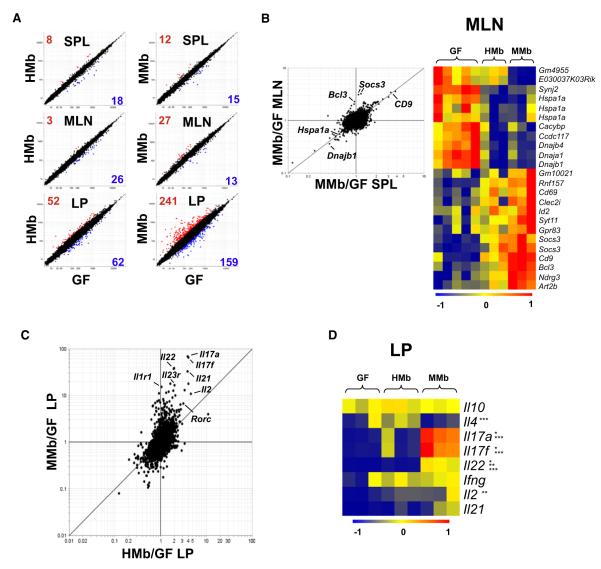
Figure 1. MMb and HMb Mouse Gut Microbiotas Are Similar in Major Bacterial Phyla Abundance with Differences at the OTU Level.
(A) Schematic of colonization model (see text for details) is illustrated. Blue and red arrowheads indicate fecal pellet collection for bacterial 16S rDNA sequencing. Offspring were sacrificed for immune system analysis. (B) Relative abundance of major bacterial phyla in the gut microbiota from MMb and HMb mice is shown. P0, parents; F1, first-generation offspring; F2, secondgeneration offspring. Each bar represents an individual mouse. Apparent differences in the Firmicutes-to-Bacteroidetes ratio between inoculum samples and recipients may have resulted from the observed differential DNA extraction performances of fecal suspensions (high water content) and fecal pellets (low water content). (C–E) Detailed relative abundance of bacterial taxa in the three most abundant major phyla is presented. (F) Number (percentage) of shared OTUs in each major bacterial phylum in MMb and HMb fecal pellets is demonstrated. See also Figures S1D and S1E. (G and H) Gut microbial communities from individual mice, clustered according to principal coordinates analysis of unweighted UniFrac distances, is illustrated. Percentages of variation explained by plotted principal coordinates P1 and P2 are indicated on the x and y axes, respectively.
Fecal Microbiota of MMb and HMb Mice
To monitor the evolution of gut microbial communities in these previously GF mice, we pyrosequenced bar-coded, amplified bacterial 16S rDNA in fecal samples from MMb and HMb parent mice (P0) and their first (F1) and second (F2) offspring generations (Figure 1A). MMb and HMb mice displayed similar relative abundances of major intestinal bacterial phyla, with Bacteroidetes predominating and Firmicutes and Proteobacteria next most abundant (Figure 1B). Quantitative PCR (qPCR) of bacterial 16S rRNA genes and quantitative aerobic and anaerobic culture of bacteria (Figure S1C) showed a similar total bacterial load in MMb and HMb mice.
MMb and HMb mice shared most taxa at the genus level within the Bacteroidetes (e.g., Bacteroides, Parabacteroides, Prevotella, and Alistipes); however, the relative abundance of these taxa drastically differed between MMb and HMb mice, closely resembling the patterns in their respective inocula (Figure 1C). MMb and HMb mice shared many Firmicute classes and families (Figure 1D), with the predominant Lachnospiraceae and Ruminococcaceae accompanied by members of the genera Bacillus in MMb mice and Clostridium 2 in HMb mice. Again, these patterns closely resembled those in the respective inocula. Similarly, MMb and HMb mice shared many genera of Proteobacteria but with different relative abundance resembling the patterns in the respective inocula (Figure 1E). Closer examination of major bacterial phyla at the species (OTU) level revealed striking differences in Firmicutes. Of 1,321 Firmicute-affiliated OTUs, only 20 (1.5%) were shared by MMb and HMb mice. This figure was significantly higher in other major bacterial phyla (Bacteroidetes, 33.7%; Proteobacteria, 43.4%; Figures 1F andS1D; Table S1). MMb and HMb inocula had similar patterns of OTU overlap, with the least overlap (0.8%) in the Firmicutes (Figure S1E). Of OTUs in the mouse inoculum, 7% were not detected in MMb mice; of those in the human inoculum, 30% were not detected in HMb mice. The majority of absent OTUs were low-abundance Firmicute taxa (Tables S2A and S2B). In addition, we found taxa in six fecal specimens from three healthy humans (Dethlefsen and Relman, 2011) that were lacking in HMb mice; most were Firmicutes (Table S2C).
Differences and similarities between microbial communities are revealed by determination of unweighted UniFrac distances, which take into account the presence/absence and evolutionary relatedness of OTUs, and by subsequent principal coordinate analysis. The first principal coordinate (P1), which explained 59% of variance in the data, separated MMb from HMb mouse samples and showed relatively small differences within MMb and HMb mouse samples. The second principal coordinate (P2) showed a high degree of similarity among all MMb mouse samples (including the mouse inoculum); however, the two human inocula had a community distinct from that of HMb mouse samples (Figure 1G). Samples from P0 HMb mice were widely separated along the P2 axis, and an apparently unstable transition state contrasted with the rather stable state in the F1 and F2 generations. Intriguingly, the same pattern was observed when only the Firmicute-affiliated OTUs were taken into account (Figure 1H), a result indicating that members of the Firmicutes are responsible for most observed differences in gut community between MMb and HMb mice and for the apparent instability in the HMb P0 generation. The analyses suggested that MMb and HMb mice have similar total bacterial loads and similar relative abundances of the major bacterial phyla in the gut but differ significantly in bacterial species, especially Firmicutes. The implication is that Firmicutes, in particular, show host specificity.
MMb, but Not HMb, Mice Exhibit Expansion of Adaptive and Innate Intestinal Immune Cells
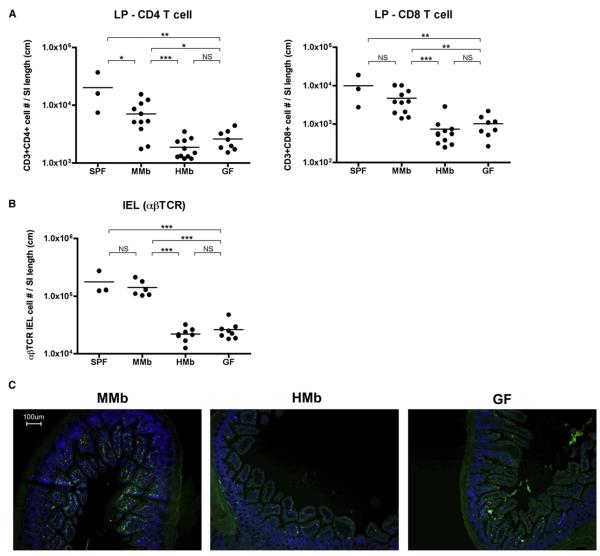
Although MMb colonization of GF mice reverses many intestinal immune abnormalities (Smith et al., 2007), it is not clear whether an HMb can do so. To determine how bacterial species-level differences affect intestinal immune maturation, we measured absolute numbers of T cells in the small intestinal lamina propria (LP). As expected, GF mice had few T cells in the LP, and MMb colonization brought LP T cell numbers closer to those in SPF mice. Despite prolonged exposure to diverse bacteria from birth, HMb offspring were deficient in total numbers and percentages of T cells (CD4+ and CD8+) in the small intestinal LP; this deficiency unexpectedly resembled that in GF mice (Figures 2A and S2A). Next, we examined T cell numbers in the small intestinal IEL compartment; the MMb is known to expand IELs expressing the αβ T cell receptor (αβTCR), but not those expressing the γδTCR (Bandeira et al., 1990). MMb mice had a significantly higher percentage and total number of αβTCR IELs and a higher αβTCR/γδTCR ratio than did HMb and GF mice (Figures 2B, S2A, and S2B). The deficient T cell numbers in HMb mouse small intestines were confirmed by immunohistochemistry (Figure 2C). Similar to their offspring, HMb P0 mice had lower T cell numbers in the IEL and LP compartments than MMb P0 mice (data not shown).
Figure 2. MMb Mice Have More Small Intestinal T Cells Than Do HMb Mice.
(A and B) IELs were extracted from the small intestine; the remaining LP tissue was digested. Absolute numbers of CD3+CD103+TCRβ+ among IELs (B) and CD3+CD4+ and CD3+CD8+ cells from LP (A) were quantitated by flow cytometry and normalized to small intestine length. SPF and GF SW mice were age matched. See also Figures S2A and S2B. *p < 0.05, **p < 0.01, ***p < 0.001. NS, not significant. (C) Sections of small intestine were stained with FITC-conjugated antibody to CD3 (green) and counterstained with DAPI (blue).
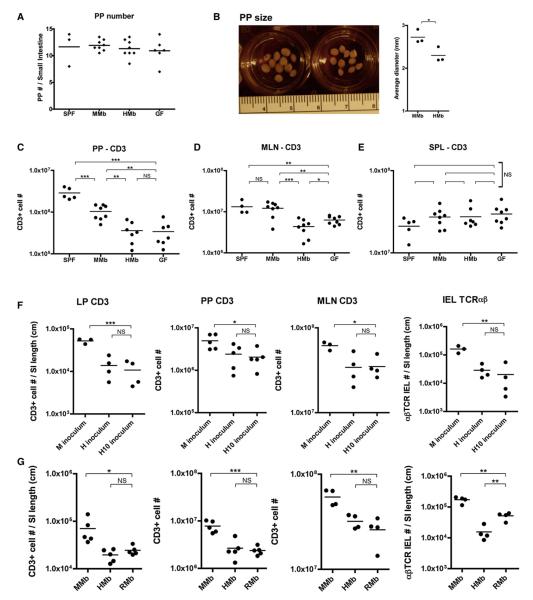
We asked whether the deficiency in T cell numbers in HMb offspring was restricted to intestinal tissue or was broader, involving secondary intestinal lymphoid organs such as PPs and mesenteric lymph nodes (MLNs). PPs sample antigens directly from the gut lumen and are important in initiating gut immune responses. Consistent with reports that PP organogenesis begins in the embryonic stage in the absence of microbes (Eberl et al., 2004), we found no difference in PP numbers along the intestines of SPF, MMb, HMb, and GF mice (Figure 3A). Moreover, the percentages of CD3+ T cells in PPs of MMb and HMb mice were comparable (Figure S3A). Despite the latter similarity, the PPs of MMb mice were visibly larger than those of HMb mice and contained significantly more total CD4+ and CD8+ T cells (Figures 3B, 3C, and S3B). PPs of SPF mice contained more T cells than did PPs of MMb mice; this difference suggested that, upon artificial transfer of MMb to GF mice, some PP-stimulatory mouse bacterial species may have been lost. MLNs, which drain cells from PPs and intestinal tissue, also contained higher percentages and numbers of T cells in MMb mice than in HMb mice (Figures 3D, S3A, and S3C). In contrast to differences found in the small intestine, no difference in large intestinal LP CD3+ and αβTCR IEL numbers was found among SPF, MMb, HMb, and GF mice. However, large intestinal γδTCR IEL numbers were higher in SPF and MMb mice than in HMb and GF mice (Figure S3D). These data suggest that the microbiota regulates T cell populations in the small and large intestines via distinct mechanisms.
Figure 3. The MMb, but Not the HMb or RMb, Expands T Cell Populations in Small Intestinal Tissue and Secondary Gut Lymphoid Organs.
(A–C) PP number (A) and average PP size (B) per small intestine were compared. PPs were mashed, stained for CD3, and subjected to flow cytometry (C). See also Figures S3A and S3B. (D and E) Total T cell numbers in MLNs (D) and spleen (E) are shown. See also Figures S3C–S3G. (F) GF mice (3–4 weeks old) were orally gavaged with the original mouse (M) or human (H) inoculum or with feces pooled from ten additional human donors (H10 inoculum). T cell numbers were measured after 4 weeks of colonization. (G) GF mice were orally gavaged with Sprague-Dawley rat feces and bred in vinyl isolators to obtain RMb offspring. T cell numbers in age-matched MMb, HMb, and RMb offspring were compared. *p < 0.05, **p < 0.01, ***p < 0.001. NS, not significant.
The deficient T cell numbers in HMb mice were limited to the intestine; MMb and HMb offspring did not differ in terms of T cell numbers in the spleen, inguinal lymph nodes, or brachial lymph nodes (Figures 3E and S3E). To assure that the differences in gut T cell numbers between MMb and HMb mice were neither artifactual nor due to inadequate HMb sample numbers, we collected fecal samples from ten additional human donors. GF mice colonized with these fecal samples had lower gut T cell numbers than GF mice colonized with the mouse inoculum (Figure 3F). In all four gut compartments (LP, PPs, MLNs, and IELs), these mice had T cell numbers comparable to those in mice colonized with the original human inoculum.
Dendritic cells (DCs)–CD11c+ innate immune cells–are critical regulators of downstream T cell responses and interact closely with gut bacteria by sampling the intestinal lumen (Kelsall and Rescigno, 2004). DC numbers (defined by CD11chigh expression) in small intestinal LP tissue were similar in MMb and HMb mice (Figure S3F). In contrast, in PPs and MLNs, HMb and GF mice had lower numbers of DCs than did MMb mice (Figure S3F). Antimicrobial peptides constitute another critical arm of innate intestinal immunity, providing protection from bacterial penetration of the gut (Vaishnava et al., 2011). MMb colonization induced gene expression of RegIIIγ, an antimicrobial peptide produced by gut epithelial cells, but HMb was deficient in upregulating RegIIIγ (Figure S3G).
Thus, regardless of exposure to large bacterial numbers with a phylum representation similar to that in MMb mice, HMb mice have very low levels of intestinal T cells, DCs, and antimicrobial peptide expression. These immunologic parameters indicate that the adaptive and innate small intestinal immune system of the HMb mouse–despite the diverse colonizing human bacterial species–is quite similar to that of GF mice, which have no viable gut bacteria at all.
A Rat Microbiota Does Not Increase Intestinal T Cell Numbers in GF Mice
To further evaluate whether intestinal immune maturation depends on host-specific bacterial species, we colonized GF mice with a second foreign microbiota: that from rat gut. Different diets can drastically alter microbial community structure (Turnbaugh et al., 2009b); because mice and rats are fed a similar rodent diet, such effects are minimized in this model. GF mice underwent oral gavage with feces from Sprague-Dawley rats and were bred in a gnotobiotic isolator to obtain offspring colonized at birth with rat microbiota (RMb). Like the HMb, the RMb was not as effective as the MMb in expanding T cell numbers in all four gut compartments examined (Figure 3G). In other words, neither the microbiota from 12 human donors nor that from rat donors restored T cell numbers in the mouse gut. Therefore, certain host-specific bacterial species appear to be required for intestinal immune maturation.
T Cell Proliferation Plays a Role in Expansion of Small Intestinal T Cells and Depends on a Host-Specific Microbiota
Recruitment of CD4+ and CD8αβ+ T cells to the mucosa reportedly is initiated through antigen uptake by antigen-presenting cells (APCs) in intestinal tissue followed by further priming and activation of naive lymphocytes by antigen-loaded APCs in secondary lymphoid organs such as PPs and MLNs. Primed lymphocytes expressing α4β7 and CCR9 in secondary gut lymphoid organs enter the bloodstream and ultimately exit into gut tissue through vessels in the small intestinal LP where MAdCAM and CCL25–ligands of α4β7 and CCR9, respectively–are expressed (Mowat, 2003). Because we found low numbers of T cells in both small intestinal tissue (IEL compartment, LP) and secondary lymphoid organs (PPs, MLNs) of HMb mice, we wondered whether there was a deficiency in T cell activation in the PPs and MLNs of these mice.
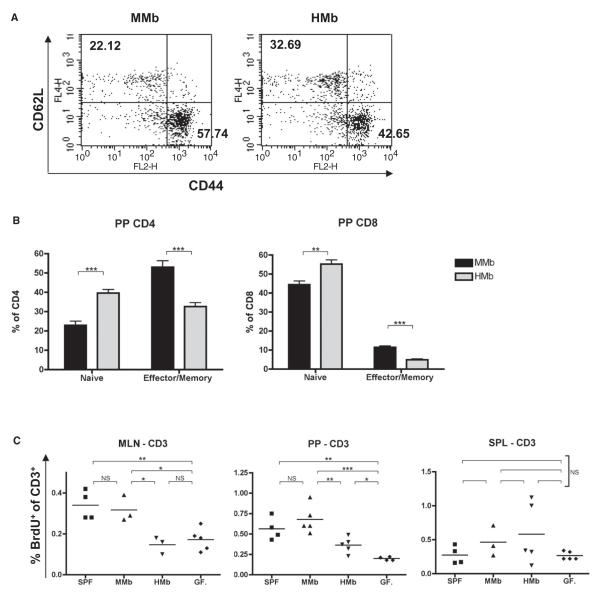
When we compared CD4+ T cells in PPs of HMb and MMb mice, we found a lower frequency of effector/memory cells (CD44hiCD62Llo) but a higher frequency of naive T cells (CD44loCD62Lhi) in HMb mice. Analysis of CD8+ T cells in PPs yielded similar results (Figures 4A and 4B). T cell activation in secondary lymphoid organs can also be assessed by measurement of T cell proliferation frequencies. Mice were injected with bromodeoxyuridine (BrdU), which is incorporated into proliferating cells; the animals were sacrificed 2 hr later, and T cell proliferation frequencies were measured. This time point was chosen to exclude the possibility that BrdU-positive cells were migrating from a distant organ to MLNs and PPs (Rakoff-Nahoum et al., 2004). No differences were evident in the frequency of BrdU-positive T cells in the spleen and peripheral lymph nodes of MMb and HMb mice. However, the frequency of BrdU-positive CD3+ cells in PPs and MLNs was significantly higher in MMb offspring than in HMb offspring. Comparable differences were found in both CD4+ and CD8+ T cell subsets (Figures 4C and S4A). These differences in T cell proliferation were also observed between SPF and GF mice (Figure 4C). Differences in T cell proliferation were confirmed by staining with Ki-67, an antigen associated with proliferating cells (Figure S4B). BrdU-positive T cells in PPs and MLNs, but not in spleen, expressed high and comparable levels of the small-intestine-homing markers CCR9 and α4β7 in both MMb and HMb mice (Figure S4C). This result suggested that proliferating T cells in secondary lymphoid organs (PPs, MLNs) are imprinted to populate small intestinal tissue (IELs, LP). These data are consistent with the hypothesis that lower T cell proliferation in secondary lymphoid organs of HMb mice can contribute to T cell deficiency throughout the small intestine. Intriguingly, T cell proliferation in secondary lymphoid organs depended on colonization with host-specific bacterial species in the intestinal lumen.
Figure 4. Host-Specific Gut Microbiota Induction of T Cell Proliferation in Secondary Gut Lymphoid Organs Leads to Expansion of Small Intestinal T Cells.
(A) Representative flow cytometry plots of CD44hiCD62Llo (effector/memory) and CD44loCD62Lhi (naive) expression on CD3+CD4+ T cells in PPs of MMb and HMb offspring are presented. Numbers indicate cell percentages in the quadrant. (B) Combined data for PP CD3+CD4+ and CD3+CD8+ cells (n = 7) are illustrated. (C) Mice injected with BrdU were sacrificed 2 hr later. CD3+ T cells were stained with FITC-conjugated antibody to BrdU for detection of proliferating cells. See also Figures S4A–S4C. *p < 0.05, **p < 0.01, ***p < 0.001. NS, not significant.
It is certainly possible that immunologic mechanisms besides T cell proliferation contribute to the deficiency in T cell numbers in HMb mice. In PPs, but not in MLNs, we observed a higher rate of T cell apoptosis in HMb and GF mice than in MMb mice; thus, T cell death may play a role in diminishing T cell numbers in some intestinal compartments (Figure S4D). We also addressed whether T cells in HMb mice are deficient in homing to the intestine. DCs from PPs imprint CCR9 and α4β7 on T cells upon activation (Mora et al., 2003), with consequent homing of activated T cells to intestinal tissue where their respective ligands are expressed. MMb and HMb offspring DCs (MLNs, PPs) similarly upregulated CCR9 and α4β7 expression on T cells (Figure S4E). There was no significant difference in MMb and HMb intestinal recruitment of gut-homing T cells (Figure S4F). Analysis of small intestinal tissue consistently indicated no difference between MMb and HMb mice in expression of transcripts MAdCAM and CCL25 (Figure S4G). Although other chemokines or adhesion molecules may contribute to differences in T cell numbers, these data suggest that T cell trafficking to the intestine via CCR9 and α4β7 does not play a major role in differences between gut T cell counts in MMb and HMb mice.
HMb Mice Exhibit a Distinct Intestinal Gene Expression Profile
We addressed whether colonization with a foreign gut microbiota has effects beyond influencing T cell numbers, i.e., whether it also influences T cell phenotype. Using microarray analysis, we examined the transcriptional profiles of purified CD4+ T cells from MMb, HMb, and GF mice. When we compared MMb and HMb mice with GF mice, we found that few genes were differentially expressed in CD4+ T cells from spleen and MLNs, whereas differences were much more extensive in the small intestinal LP (Figure 5A).
Figure 5. Distinct Gene Expression Profile in Small Intestinal T Cells from HMb Mice.
(A) Microarray analysis comparing CD4+ T cell gene expression in GF mice with that in HMb mice (left) and MMb mice (right) is demonstrated. CD4+ T cells were sorted from spleen (SPL), MLNs, and small intestinal LP. Data are mean values from three to five independent experiments. Numbers indicate genes showing a ≥2-fold difference in expression between groups; red numbers indicate overexpression and blue numbers underexpression. (B) Fold change versus fold-change analysis compares MMb mice with GF mice in terms of gene expression in CD4+ T cells sorted from MLNs (y axis) and spleen (x axis) (left). A heatmap (right) shows differentially expressed genes in CD4+ T cells sorted from the MLN. Some genes (Hspa1a, Socs3) were detected with multiple probes. Genes with the highest and lowest transcript levels are red and blue, respectively. See also Figure S5A. (C) Fold change versus fold-change analysis compares gene expression in CD4+ T cells sorted from small intestinal LP of GF mice versus MMb mice (y axis) or HMb mice (x axis). (D) Heatmap shows differential cytokine expression in CD4+ T cells from small intestinal LP. Data are from three independent experiments. See also Figures S5B–S5F. *p < 0.05, MMb versus GF; **p < 0.05, HMb versus GF; ***p < 0.05, HMb versus MMb.
The changes induced in spleen and MLNs were largely shared in both locales, because many genes fall on the diagonal of the FoldChange/FoldChange plot in Figure 5B. In MLNs, we found a number of genes upregulated by both MMb and HMb, including T cell activation genes (CD9, Bcl3, and Socs3); conversely, heat shock transcripts (Hspa1a, dnaja1, dnajb1, dnajb4), which are generally expressed at low levels in normal nonstressed cells (Glover and Lindquist, 1998), exhibited greater expression in MLN CD4+ T cells from GF mice than in those from MMb and HMb mice–perhaps a reflection of the abnormal physiological conditions in GF mice (Figure 5B). In the spleen, some genes (e.g., CD9 and heat shock transcripts) were expressed in a pattern similar to that in MLNs (Figure S5A).
In the small intestinal LP, where transcriptional changes were more extensive, the effects were more profound in MMb mice than in HMb mice. Many of the changes affected the same transcripts (Figure 5C) but were far more extensive in CD4+ T cells from the LP of MMb mice, as denoted by the off-diagonal disposition of most transcripts. Most of these induction events affected cytokine genes, particularly those of the TH17 family, i.e., interleukin-17a (Il17a), Il17f, and Il22. The induction of these cytokine genes was stronger in MMb mice (30- to 80-fold) than in HMb mice (2- to 5-fold). Closer examination of cytokine transcripts (Figure 5D) showed that CD4+ T cells from the LP indeed expressed higher levels of TH17 transcripts in MMb mice, whereas those from the LP of HMb mice expressed higher levels of Il4–indicative of stronger TH2-type differentiation. Interferon γ (Ifnγ), the hallmark of TH1 cells, was induced to the same levels in MMb and HMb mice. Rorc and Rora–transcription factors that control TH17 cell differentiation (Ivanov et al., 2006; Yang et al., 2008)–were more strongly induced in MMb mice than in HMb or GF mice. Furthermore, Gata3–the transcription factor that controls TH2-type differentiation–was detected at higher levels in HMb and GF mice than in MMb mice (Figures S5B and S5C). Thus, colonization with different bacterial species resulted in reciprocal bias in T cell effector phenotypes. These analyses indicated that host specificity of bacteria drastically affects both T cell numbers and phenotypes in the small intestinal LP.
In addition to isolated T cells, we studied gene transcription profiles in ileal tissue. Ilea from MMb mice exhibited enhanced expression of a collection of B cell-specific genes over levels in HMb and GF ilea (Figure S5D). Histologic examination also showed more intestinal IgA+ cells in MMb mice than in HMb mice; in addition to T cell deficiencies, the HMb mouse gut has deficiencies in B cell maturation (Figure S5E). Microarray analysis of ileal tissue showed that certain chemokines (CCL20, CCL28, CXCL9) critical for T cell and DC chemotaxis and activation in the gut (Kunkel et al., 2003) are more strongly induced in MMb mice than in HMb or GF mice. These chemokines are expressed by the epithelium, at the interface between gut bacteria and host (Figure S5F). The host epithelium may be more apt to sense host-specific than foreign commensals.
Segmented Filamentous Bacteria Only Partially Expand Mouse Intestinal T Cell Numbers
Strong induction of TH17 cell-associated gene transcripts in MMb mice led us to assess the presence of segmented filamentous bacteria (SFB) in our mouse colony. SFB are commensal gut bacteria found in mammals such as mice, rats, and chickens (Snel et al., 1995) and are potent inducers of TH17 cells (Gaboriau-Routhiau et al., 2009; Ivanov et al., 2009). Genome sequence analysis showed that SFB lack virulencerelated genes and may depend largely on the host for amino acids and essential nutrients (Prakash et al., 2011; Sczesnak et al., 2011). In an SFB-specific qPCR assay for 16S rDNA in multiple mammalian species, we found that all MMb mice (and the MMb inoculum) carried SFB DNA. In contrast, all HMb mice (and the HMb inoculum) were negative for SFB (Figure 6A). Although there is no DNA sequence-based evidence for SFB in humans, these bacteria are members of the highly host-specific phylum Firmicutes (Figures 1F and S1E), and other Firmicutes may play a role in humans similar to that played by SFB in mice.
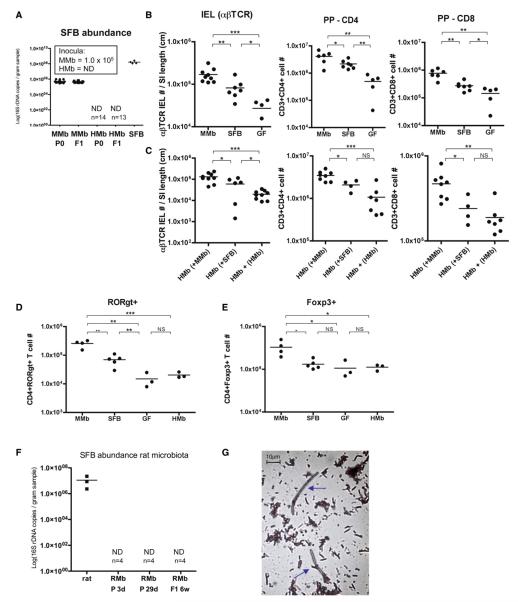
Figure 6. SFB Play a Role in Rescuing Intestinal T Cell Numbers and Exhibit Host Specificity.
(A) Abundance of SFB in MMb, HMb, and SFB-monocolonized mice, measured as SFB-specific 16S rDNA copy numbers by qPCR analysis of fecal pellets, is shown. Inset values indicate number of SFB 16S rDNA copies/ml in inocula. ND, not detected. (B and C) Absolute T cell numbers in IEL (CD3+CD103+TCRb+) and PP (CD3+CD4+ and CD3+CD8+) compartments of MMb, SFB-monocolonized, and GF mice (B) and HMb mice cohoused with MMb or SFB-monocolonized mice for 4 weeks (C) are presented. In (C), as a negative control, HMb mice were cohoused with HMb mice. See also Figures S6A–S6E.
Besides inducing TH17 cells, SFB increase IEL numbers (Umesaki et al., 1999). We tested whether SFB played a role in expansion of IELs in our MMb colony. Although SFB-monocolonized mice had extremely high SFB numbers (Figure 6A), their absolute αβTCR IEL numbers were higher than those in GF mice but significantly lower than those in MMb mice–differences suggesting that SFB alone only partially restored the αβTCR IEL population (Figure 6B). SFB fully expanded absolute numbers of CD8αα-αβTCR IELs–a unique self-reactive population that requires exposure to self-agonists for selection in the thymus (Leishman et al., 2002)–but only partially expanded the CD8αβ-αβTCR IELs, which is generated mostly in secondary lymphoid organs and is primed against nonself-antigens (Figures S6A–S6C). Likewise, in other sites (e.g., PPs and LP), SFB monocolonization partially increased CD4+ and CD8+ T cell numbers but did not fully restore T cell numbers to levels in MMb mice (Figures 6B and S6D). To examine whether the transfer of SFB or microbes from MMb into HMb mice could fully rescue T cell numbers, we separately cohoused HMb mice with either MMb mice or SFB-monocolonized mice. Despite comparable levels of SFB transfer into the two groups (Figure S6E), HMb mice cohoused with MMb mice had more T cells in the IEL compartment, PPs, and LP than HMb mice cohoused with SFB-monocolonized mice (Figures 6C, S6C, and S6D). In addition to SFB, other critical mouse bacterial species may induce immune maturation in mouse gut.
In PPs, SFB appear to play a more prominent role in expanding CD4+ T cells than CD8+ T cells (Figure 6B). Inflammatory CD4+ RORgt+ T cells as well as anti-inflammatory CD4+Foxp3+ T cells are critical in maintaining homeostasis in the gut (Hand and Belkaid, 2010). In contrast to MMb, SFB monocolonization only partially expanded CD4+RORgt+ T cells (Figures 6D andS6F) and failed to increase CD4+Foxp3+ cell numbers in PPs (Figure 6E). The increased percentage of CD4+Foxp3+ cells in PPs of HMb and GF mice (Figure S6G) was due not to an increase in absolute CD4+Foxp3+ cell numbers but rather to a prominent lack of CD4+RORgt+ cells. The failure of SFB and HMb to expand CD4+Foxp3+ populations in PPs (Figure 6E) suggests that mouse-specific bacterial species besides SFB may induce Foxp3 expression.
Rats carry SFB similar in morphology and 16S rRNA sequence to mouse-specific SFB (Klaasen et al., 1993; Snel et al., 1995). To determine whether rat SFB can colonize mice, we sought rat SFB in RMb mice. Fecal pellets from Sprague-Dawley rats that were used to prepare the RMb inoculum were positive for SFB by qPCR (Figure 6F). Using Gram’s stain, we observed organisms with long filamentous structures suggestive of SFB in Sprague-Dawley rat fecal pellets (Figure 6G). However, SFB were not detected in any samples from RMb mice–not even from the parent generation–as soon as 3 days after gavage of the RMb inoculum (Figure 6F).
We conclude that SFB in the mouse gut microbiota play a role in expanding intestinal T cell numbers but do not act alone. We found no sequence-based evidence for SFB in HMb inocula or in our HMb mouse colony. When introduced as part of a complex microbiota, rat SFB did not colonize mice. Therefore, SFB exemplify a host-specific Firmicute lineage. These observations support the hypothesis that the mammalian gut selects for host-specific bacterial species, an effect that in turn strengthens the intestinal immune system.
HMb Mice Are More Susceptible Than MMb Mice to Salmonella Infection
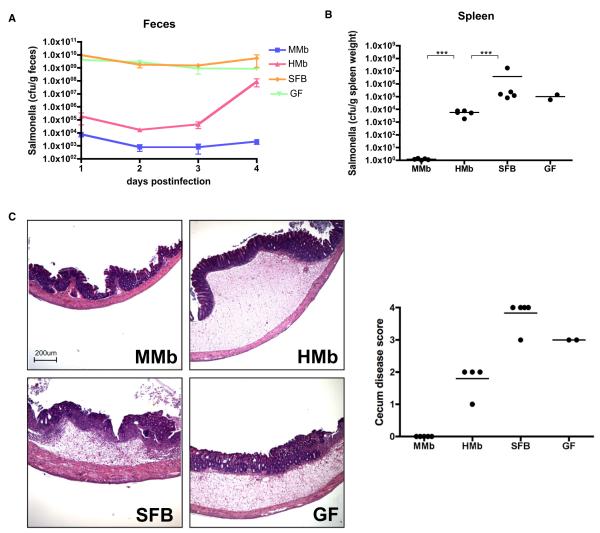
An intact gut microbiota is critical for mucosal protection from bacterial invasion and disease (Ferreira et al., 2011). Furthermore, a high total gut bacterial load alone is insufficient to protect against infection; rather, certain bacterial species correlate with protection (Croswell et al., 2009). Therefore, we orally infected MMb and HMb mice with Salmonella enterica serovar Typhimurium, a zoonotic and clinically relevant pathogen that can be acquired via contaminated food. MMb mice had a significantly lower Salmonella load in feces and less dissemination to the spleen. Interestingly, SFB-monocolonized mice were colonized with high levels of Salmonella similar to GF mice (Figures 7A and 7B). At 4 days after infection, MMb mouse ceca appeared healthy on histology, whereas ceca of HMb, SFB, and GF mice had severe gross pathological changes characterized by thickening of the cecal wall, inflammation, and edema (Figure 7C). Our conclusion that a host-specific microbiota is most effective in defense against an important gastrointestinal disease further supports the hypothesis that hosts may have coevolved with a beneficial host-specific microbiota.
Figure 7. MMb Confers Better Protection against Salmonella enterica Serovar Typhimurium Than HMb.
(A–C) Mice colonized with different microbiotas were orally gavaged with ~1 × 107 salmonellae; the Salmonella load in fecal pellets was measured daily (A). Mice were sacrificed on day 4 after infection, and the Salmonella load in the spleen was measured (B). Cecal sections were stained with hematoxylin and eosin, and disease was scored (C). ***p < 0.001. (D and E) Number of CD3+CD4+ T cells in PPs expressing RORgt+ (D) and Foxp3+ (E), as derived by intracellular staining and flow cytometry, is illustrated. See alsoFigures S6F–S6G. (F) Abundance of SFB in fecal samples from Sprague-Dawley rats and RMb-colonized mice, measured as SFB-specific 16S rDNA copy numbers by qPCR, is shown. Fecal pellets from RMb parents were collected on postgavage days 3 (RMb P 3d) and 29 (RMb P 29d); those from RMb offspring were collected at 6 weeks of age (RMb F1 6w). ND, not detected. (G) Gram-stained Sprague-Dawley rat fecal pellets resuspended in PBS are illustrated. Blue arrows indicate bacteria with long filamentous structures representative of SFB. *p < 0.05, **p < 0.01, ***p < 0.001. NS, not significant.
DISCUSSION
Host-Specific Bacteria Influence Intestinal Immune Maturation
It was reported that short-term colonization of adult GF mice with a human gut microbiota results in a low αβTCR/γδTCR ratio among IELs (Imaoka et al., 2004). Another study documented a predominance of downregulated transcripts in the ilea of mice given human feces (Gaboriau-Routhiau et al., 2009). However, these reports did not clarify whether such phenotypes were due to stalled intestinal immune maturation, an aberrant local effect, or failure to colonize at an early enough age. We analyzed absolute T cell numbers in intestinal compartments of mice naturally colonized with an HMb at birth. Technical advances in high-throughput sequencing permitted deep sampling of 16S rRNA genes and detailed comparison of MMb and HMb after introduction into GF mice–analyses not feasible in the two previous studies.
MMb and HMb mouse gut microbiotas are similar in relative abundances of the major bacterial phyla but have substantial differences at the OTU level, particularly among Firmicutes. Colonization with HMb results in an immature adaptive and innate intestinal immune system, most notably in the small intestine. HMb mice have low numbers of intestinal T cells (partly because of less T cell activation/proliferation) and DCs. HMb mice intestine also displays low-level expression of RegIIIγ, IgA, and various chemokines (CCL20, CCL28, CXCL9) (Figure S7; Table S3). The lack of difference between MMb and HMb mice in large intestinal LP CD3+ and αβTCR IEL numbers suggests that the microbiota regulates the small and large intestinal immune compartments via distinct mechanisms.
Mouse SFB (Firmicutes), but not rat SFB, established colonization in the mouse gut. Rat SFB may not have adhered efficiently to mouse epithelium. In addition, there may exist a host-specific microbial ecology that supports prolonged SFB colonization, especially because SFB may function as an adjuvant for the immunostimulatory functions of other resident gut microbes (Chung and Kasper, 2010). The exclusive partnership of SFB with the host is a proof of concept that the mammalian gut selects for host-specific bacterial species that enhance host immunity. SFB alone partially restored gut T cell numbers but were not fully protective against Salmonella infection–results suggesting that SFB are one but not the only MMb component responsible for immune maturation and protection against infection.
There is redundancy in the gut microbiota in facilitating certain host metabolic functions; i.e., individual HMbs can vary at the bacterial species level but nonetheless share functional genes (Turnbaugh et al., 2009a). A study of GF zebrafish colonized with a MMb showed that a foreign microbiota can partially restore transcription of host genes involved in nutrient absorption and metabolism (Rawls et al., 2006). Despite such bacterial species-level redundancy in stimulating host metabolic functions, our HMb mouse studies demonstrate that the host intestinal immunity maturation depends on a strict set of host-specific bacterial species.
Mechanisms of Intestinal Immune Maturation
We show that host-specific bacterial species induce expansion of intestinal T cells by stimulating T cell activation/proliferation in secondary gut lymphoid organs (PPs and MLNs), with consequent effects on downstream T cell numbers in intestinal tissue (IELs and LP) (Figure S7; Table S3). Many questions remain about the causes of the differences in T cell proliferation. Reduced numbers of DCs in the PPs and MLNs of HMb mice may have caused lower T cell proliferation rates. The MMb, but not the HMb, may induce a cytokine or chemokine milieu effective in activating and recruiting APCs to stimulate downstream T cell proliferation.
We believe that the higher rate of T cell proliferation in MMb mice is predominantly antigen driven. Although the small intestinal epithelium is protected by mucus and antimicrobial peptides (Johansson and Hansson, 2011), certain host-specific microbes may reside near the epithelium and induce antigen uptake from the lumen by stimulating APCs or by modulating the gut epithelial barrier. Compared to HMb, MMb may be more efficient at penetrating the mouse mucus layer or evading certain mouse antimicrobial peptides. Alternatively, certain host-specific microbiotas may regulate epithelial sensitivity in recognizing microbe-associated molecular patterns (MAMPs) through pattern recognition receptors (Eberl and Boneca, 2010). An understanding of these mutually nonexclusive mechanisms potentially employed by the host microbiota could guide efforts to modulate gut immunity in order to improve host health.
The Gut Microbiota, Host Health, and Evolution
HMb mice are more susceptible to Salmonella infection than MMb mice. Because adaptive immunity (T cells) and innate immunity (antimicrobial peptides, DCs) are both critical for defense against Salmonella infection (Salazar-Gonzalez et al., 2006; Vaishnava et al., 2008), the absence of an intact immune system in HMb mice likely played a role in susceptibility to Salmonella infection. In addition, MMb might have been more effective than HMb in physically inhibiting enteropathogen adherence (Heczko et al., 2000). Infectious disease epidemics are among the most powerful selective forces acting on hosts and microbes. We speculate that, throughout evolution, hosts coexisting with bacteria that were well adapted to the host gut environment and capable of enhancing host health survived particular epidemics. Selection against hosts lacking such bacterial species may have promoted the survival and enrichment of beneficial host-specific bacteria.
The Human Microbiome
Metagenomic analysis of the gut microbiota of 124 Europeans suggested that humans share many bacterial species (Qin et al., 2010). Widely shared human-specific bacterial species may not predominate in the gut microbiota, and their prevalence may vary substantially with the individual and perhaps with age. Nonetheless, these microorganisms may be potent stimulators of host immunity. Surveys for SFB-like microbes residing closer to the gut epithelium or for Alcaligenes species within PPs (Obata et al., 2010) may help identify immunomodulatory human microbes.
HMb-colonized mice have been proposed as a useful tool to study human metabolism and disease (Turnbaugh et al., 2009b). Some, but not all, human microbes may be immunostimulatory in a GF mouse monocolonization model. Amplification of one specific microbe may enhance the likelihood of observing a function of that microbe, which otherwise would exert no observable function in a mouse colonized with a complex HMb. For example, the immunomodulatory effects of the human microbe Bacteroides fragilis, which cannot readily colonize the conventionally colonized mouse intestine, were first reported in B. fragilis-monocolonized mice (Mazmanian et al., 2005). Therefore, such GF mouse monocolonization models should be taken into consideration in defining the function of widely shared human microbes.
Revisiting the Hygiene Hypothesis
The revised hygiene hypothesis proposes that exposure to immunomodulatory gut commensals can provide protection from autoimmune diseases (Wills-Karp et al., 2001). Our immunologic analysis of mice colonized with human gut bacteria suggests that exposure to just any gut commensal microbe or its MAMPs (e.g., lipopolysaccharide, cell wall components) is insufficient to induce intestinal immune maturation. Our study rather suggests that only certain host-specific commensals give rise to a mature intestinal immune system. Because the intestinal microbiota can regulate immune responses outside the gut (Wen et al., 2008; Wu et al., 2010), the absence of the “right” gut microbes may conceivably shift the balance toward disease in individuals genetically predisposed to autoimmune diseases.
Heavy processing of food, frequent treatment with antibiotics, and advances in hygiene in industrialized countries may have reduced the stability and transfer of host microbes promoting health. Furthermore, because advances in medicine and technology provide alternative ways to fight disease, humans may be becoming less dependent on their coevolved gut microbiota for health and survival. The current prevalence of autoimmune diseases may be, at least in part, the consequence of increasing vulnerability of the coevolved human-microbe relationship.
EXPERIMENTAL PROCEDURES
Mice and Colonization
SPF and GF SW mice (Taconic Farms) and Sprague-Dawley rats (Harlan) were used. Cecal/fecal contents from ten SPF SW mice served as the MMb inoculum, fresh human feces from two healthy Caucasian adults (one male, one female) as the HMb inoculum, and fecal pellets from four Sprague-Dawley rats as the RMb inoculum. The human donors had not taken antibiotics for 1 year. Fecal contents were diluted (10–1) in prereduced peptone yeast glucose, snap frozen in liquid nitrogen, and stored at –80°C. SW GF mice (3–4 weeks old) received inocula by oral gavage and were then placed in sterile vinyl isolators. Mice were bred in the isolators to obtain offspring and were maintained on an autoclaved NIH-31M rodent diet (Taconic Farms). All procedures with animals were performed according to HMS Office for Research Subject Protection guidelines. Human samples were collected according to Partners Human Research Committee guidelines.
Microbiota Analysis
16S rDNA was amplified from fecal pellets and inoculum samples and subjected to GS-FLX titanium multiplex pyrosequencing (Roche). Reads were filtered and quality trimmed. Phylogenetic assignments were made by clustering reads against a high-quality seed library with Uclust. UniFrac distances were calculated and principal coordinates analyses performed with QIIME software. See Extended Experimental Procedures for detailed analysis and references.
Cell Isolation and Flow Cytometry
PPs were excised from the small intestine, and the remaining tissue was incubated with 1 mM DTT/1 mM EDTA/3% FBS/PBS (30 min, 37°C) for IEL extraction. Residual intestinal tissue was digested in 5% FBS RPMI with 0.15% collagenase II (275 U/mg)/0.05% dispase (1.1 U/mg) (Invitrogen) for 1 hr at 37°C. IELs and LP cells were filtered to minimize mucus contamination. Single-cell suspensions of MLNs, PPs, spleen, and peripheral lymph nodes were prepared by mashing in a cell strainer (70 μm). Cells were stained with fluorophore-conjugated mouse antibodies, and flow cytometry was performed with Fluorospheres/FLOW-COUNT (Beckman Coulter) beads.
Microarray Analysis
CD4+ T cells were isolated from spleen, MLNs, and small intestinal LP of 10- to 14-week-old mice (Feuerer et al., 2010). See also Extended Experimental Procedures.
Salmonella Infections
S. enterica serovar Typhimurium strain SL1344 was grown at 37°C in Luria-Bertani (LB)/streptomycin (200 μg/ml). Mice (8–10 weeks old) were orally gavaged with ~5 × 107 cfu. Fecal samples and spleens were homogenized in PBS and plated on LB/streptomycin (200 μg/ml) agar. Ceca were fixed in Bouin’s solution and stained with hematoxylin and eosin. Slides were scored for inflammation, ulceration, and edema as follows: 0, no disease; 1, mild; 2, moderate; 3, severe; 4, very severe.
Statistical Analysis
All p values were calculated by unpaired/two-tailed t test. In dot plots, each data point represents an individual mouse, and horizontal bars indicate the means.
Supplementary Material
ACKNOWLEDGMENTS
We thank Shakir Edwards for gnotobiotic animal care, Julie B. McCoy for editing, Tom DiCesare for image design, Dr. Rod Bronson for pathology scoring, Jennifer Dinalo for ten human fecal samples, and Les Dethlefsen and Robert Edgar for helpful advice. This work was supported by a Senior Research Award from the Crohn’s & Colitis Foundation of America (to D.L.K.), by the Juvenile Diabetes Research Foundation (to H.C.), by the Danish Council for Independent Research Natural Sciences (to S.J.P.), by National Institutes of Health Grant F32 AI091104 (to N.K.S.), and by Director’s Pioneer Award DP1OD000964 (to D.A.R.). D.A.R. is supported by the Thomas C. and Joan M. Merigan Endowment at Stanford University.
Footnotes
ACCESSION NUMBERS The 16S rDNA sequences have been deposited at GenBank under accession number SRA052958.
SUPPLEMENTAL INFORMATION Supplemental Information includes Extended Experimental Procedures, seven figures, and four tables and can be found with this article online atdoi:10.1016/j.cell.2012.04.037.
REFERENCES
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. MetaHIT Consortium Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandeira A, Mota-Santos T, Itohara S, Degermann S, Heusser C, Tonegawa S, Coutinho A. Localization of gamma/delta T cells to the intestinal epithelium is independent of normal microbial colonization. J. Exp. Med. 1990;172:239–244. doi: 10.1084/jem.172.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassin C, Kocur M, Pott J, Duerr CU, Gütle D, Lotz M, Hornef MW. miR-146a mediates protective innate immune tolerance in the neonate intestine. Cell Host Microbe. 2010;8:358–368. doi: 10.1016/j.chom.2010.09.005. [DOI] [PubMed] [Google Scholar]
- Chung H, Kasper DL. Microbiota-stimulated immune mechanisms to maintain gut homeostasis. Curr. Opin. Immunol. 2010;22:455–460. doi: 10.1016/j.coi.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Croswell A, Amir E, Teggatz P, Barman M, Salzman NH. Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect. Immun. 2009;77:2741–2753. doi: 10.1128/IAI.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. USA. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Chung H, Troy E, Kasper DL. Microbial colonization drives expansion of IL-1 receptor 1-expressing and IL-17-producing gamma/delta T cells. Cell Host Microbe. 2010;7:140–150. doi: 10.1016/j.chom.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl G, Boneca IG. Bacteria and MAMP-induced morphogenesis of the immune system. Curr. Opin. Immunol. 2010;22:448–454. doi: 10.1016/j.coi.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi Y, Littman DR. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 2004;5:64–73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira RB, Gill N, Willing BP, Antunes LC, Russell SL, Croxen MA, Finlay BB. The intestinal microbiota plays a role in Salmonella-induced colitis independent of pathogen colonization. PLoS One. 2011;6:e20338. doi: 10.1371/journal.pone.0020338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerer M, Hill JA, Kretschmer K, von Boehmer H, Mathis D, Benoist C. Genomic definition of multiple ex vivo regulatory T cell subphenotypes. Proc. Natl. Acad. Sci. USA. 2010;107:5919–5924. doi: 10.1073/pnas.1002006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Hand T, Belkaid Y. Microbial control of regulatory and effector T cell responses in the gut. Curr. Opin. Immunol. 2010;22:63–72. doi: 10.1016/j.coi.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heczko U, Abe A, Finlay BB. Segmented filamentous bacteria prevent colonization of enteropathogenic Escherichia coli O103 in rabbits. J. Infect. Dis. 2000;181:1027–1033. doi: 10.1086/315348. [DOI] [PubMed] [Google Scholar]
- Imaoka A, Setoyama H, Takagi A, Matsumoto S, Umesaki Y. Improvement of human faecal flora-associated mouse model for evaluation of the functional foods. J. Appl. Microbiol. 2004;96:656–663. doi: 10.1111/j.1365-2672.2004.02189.x. [DOI] [PubMed] [Google Scholar]
- Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Hansson GC. Microbiology. Keeping bacteria at a distance. Science. 2011;334:182–183. doi: 10.1126/science.1213909. [DOI] [PubMed] [Google Scholar]
- Kelsall BL, Rescigno M. Mucosal dendritic cells in immunity and inflammation. Nat. Immunol. 2004;5:1091–1095. doi: 10.1038/ni1104-1091. [DOI] [PubMed] [Google Scholar]
- Klaasen HL, Koopman JP, Van den Brink ME, Bakker MH, Poelma FG, Beynen AC. Intestinal, segmented, filamentous bacteria in a wide range of vertebrate species. Lab. Anim. 1993;27:141–150. doi: 10.1258/002367793780810441. [DOI] [PubMed] [Google Scholar]
- Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ. Microbial factor-mediated development in a host-bacterial mutualism. Science. 2004;306:1186–1188. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- Kunkel EJ, Campbell DJ, Butcher EC. Chemokines in lymphocyte trafficking and intestinal immunity. Microcirculation. 2003;10:313–323. doi: 10.1038/sj.mn.7800196. [DOI] [PubMed] [Google Scholar]
- Leishman AJ, Gapin L, Capone M, Palmer E, MacDonald HR, Kronenberg M, Cheroutre H. Precursors of functional MHC class I- or class II-restricted CD8alphaalpha(+) T cells are positively selected in the thymus by agonist self-peptides. Immunity. 2002;16:355–364. doi: 10.1016/s1074-7613(02)00284-4. [DOI] [PubMed] [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. Evolution of mammals and their gut microbes. Science. 2008a;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 2008b;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Mora JR, Bono MR, Manjunath N, Weninger W, Cavanagh LL, Rosemblatt M, Von Andrian UH. Selective imprinting of gut-homing T cells by Peyer’s patch dendritic cells. Nature. 2003;424:88–93. doi: 10.1038/nature01726. [DOI] [PubMed] [Google Scholar]
- Mowat AM. Anatomical basis of tolerance and immunity to intestinal antigens. Nat. Rev. Immunol. 2003;3:331–341. doi: 10.1038/nri1057. [DOI] [PubMed] [Google Scholar]
- Obata T, Goto Y, Kunisawa J, Sato S, Sakamoto M, Setoyama H, Matsuki T, Nonaka K, Shibata N, Gohda M, et al. Indigenous opportunistic bacteria inhabit mammalian gut-associated lymphoid tissues and share a mucosal antibody-mediated symbiosis. Proc. Natl. Acad. Sci. USA. 2010;107:7419–7424. doi: 10.1073/pnas.1001061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Worobey M, Kuo CH, Ndjango JB, Peeters M, Hahn BH, Hugenholtz P. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 2010;8:e1000546. doi: 10.1371/journal.pbio.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pais R, Lohs C, Wu Y, Wang J, Aksoy S. The obligate mutualist Wigglesworthia glossinidia influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl. Environ. Microbiol. 2008;74:5965–5974. doi: 10.1128/AEM.00741-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash T, Oshima K, Morita H, Fukuda S, Imaoka A, Kumar N, Sharma VK, Kim SW, Takahashi M, Saitou N, et al. Complete genome sequences of rat and mouse segmented filamentous bacteria, a potent inducer of th17 cell differentiation. Cell Host Microbe. 2011;10:273–284. doi: 10.1016/j.chom.2011.08.007. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Gonzalez RM, Niess JH, Zammit DJ, Ravindran R, Srinivasan A, Maxwell JR, Stoklasek T, Yadav R, Williams IR, Gu X, et al. CCR6-mediated dendritic cell activation of pathogen-specific T cells in Peyer’s patches. Immunity. 2006;24:623–632. doi: 10.1016/j.immuni.2006.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, Huttenhower C, Littman DR, Ivanov II. The genome of th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe. 2011;10:260–272. doi: 10.1016/j.chom.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Snel J, Heinen PP, Blok HJ, Carman RJ, Duncan AJ, Allen PC, Collins MD. Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats, and chickens and proposal of “Candidatus Arthromitus”. Int. J. Syst. Bacteriol. 1995;45:780–782. doi: 10.1099/00207713-45-4-780. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009a;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 2009b;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesaki Y, Setoyama H, Matsumoto S, Imaoka A, Itoh K. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect. Immun. 1999;67:3504–3511. doi: 10.1128/iai.67.7.3504-3511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl. Acad. Sci. USA. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills-Karp M, Santeliz J, Karp CL. The germless theory of allergic disease: revisiting the hygiene hypothesis. Nat. Rev. Immunol. 2001;1:69–75. doi: 10.1038/35095579. [DOI] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.