Abstract
In species of the frog genus Xenopus, lens regeneration occurs through a process of transdifferentiation, in which cornea epithelial cells presumably undergo dedifferentiation and subsequently redifferentiate to form a new lens. Experimental studies have shown that the retina provides the key signal(s) required to trigger this process once the original lens is removed. A previous study showed that addition of an exogenous Fibroblast Growth Factor (i.e., FGF1 protein) could initiate transdifferentiation of cornea epithelial cells in culture. To determine the role of FGF signaling in X. laevis lens regeneration, we have examined the presence of specific FGFs and their receptors (FGFRs) during this process and evaluated the necessity of FGFR signaling. RT-PCR analyses reveal that a number of FGF family members are expressed in cornea epithelium and retinal tissues both before and during the process of lens regeneration. Of these, FGF1, FGF8, and FGF9 are expressed principally in retinal tissue and not in the cornea epithelium. Hence, these ligands could represent key signaling factors originating from the retina that trigger regeneration. The results of experiments using an in vitro eye culture system and an FGFR inhibitor (SU5402) suggest that FGFR signaling is required for lens regeneration in Xenopus.
Keywords: Xenopus laevis, Xenopus, FGF, FGFR, eye, lens, cornea, retina, regeneration, transdifferentiation
Introduction
The fibroblast growth factors (FGFs), formerly known as “heparin-binding growth factors,” are a family of growth factors with high affinity for heparin sulfate proteoglycans (reviewed in Ornitz and Itoh, 2001). In mammalian systems, there are a total of 22 FGFs, numbered FGF1–23, as FGF15/19 represents a single FGF initially discovered in different species (mouse FGF15 and human FGF19; Ornitz and Itoh, 2001; Robinson 2006). These FGFs have been divided into seven subfamilies, each classified by sequence homology (Itoh and Ornitz, 2004; Itoh, 2007; see Table 1). Among the members of the FGF family, significant differences among FGFs concerning localization and function have been found between various tissues (Xu et al., 1999; Ford-Perriss et al., 2001; Bottcher and Niehrs, 2005; Dailey et al., 2005; Thisse and Thisse, 2005; Robinson, 2006; Itoh, 2007; Lea et al., 2009). However, the members of specific FGF subfamilies generally have common FGFR receptor specificities, suggesting similar downstream effects may be elicited by members of each subfamily (Ornitz et al., 1996; Zhang et al., 2006).
Table 1.
List of known vertebrate FGF family members and corresponding FGFR interactions
| Subfamily | FGF | FGFR specificity |
|---|---|---|
| FGF1 | FGF1 | 1b, 1c, 2b, 2c, 3b, 3c, 4 |
| FGF2 | 1b, 1c, 2c, 3c, 4 | |
|
| ||
| FGF4 | FGF4 | 1c, 2c, 3c, 4 |
| FGF5 | 1c, 2c | |
| FGF6 | 1c, 2c, 4 | |
|
| ||
| FGF7 | FGF3 | 1b, 2b |
| FGF7 | 2b, 4 | |
| FGF10 | 1b, 2b | |
| FGF22 | 1b, 2b | |
|
| ||
| FGF8 | FGF8 | 1c, 2c, 3b, 3c, 4 |
| FGF17 | 1c, 2c, 3c, 4 | |
| FGF18 | 2c, 3c, 4 | |
|
| ||
| FGF9 | FGF9 | 2c, 3b, 3c |
| FGF16 | 2c, 3c | |
| FGF20 | 1c, 2c, 3b, 3c, 4 | |
| FGF11 | FGF11 | No FGFRs |
| FGF12 | No FGFRs | |
| FGF13 | No FGFRs | |
| FGF14 | No FGFRs | |
|
| ||
| FGF 19 | FGF15/19 | 1c, 2b, 2c, 3b, 3c, 4 |
| FGF21 | 1b, 1c, 2b, 2c, 3b, 3c, 4 | |
| FGF23 | 1c, 2b, 2c, 3b, 3c, 4 | |
Vertebrate FGF family members are listed by subfamily, and interacting FGFR isoforms (i.e. b, c) for each FGF are listed as determined by Ornitz et al. (1996) and Zhang et al. (2006). Members of the intracellular FGF subfamily do not activate any known FGFR. FGFs highlighted in bold indicate those that have been identified in X. laevis and that have been investigated in this study.
There are a total of four FGF receptors (FGFRs), and each has multiple isoforms. The most commonly made distinction in FGFR isoforms are the IIIb and IIIc isoforms, differing by alternative splicing of a pair of exons, and possessing different FGF affinities (Groth and Lardelli, 2002). Known FGF/FGFR interactions are summarized in Table 1. Each receptor is activated by binding FGF and heparin, resulting in the formation of FGFR homodimers and their subsequent activation via autophosphorylation (Mohammadi et al., 2005). This interaction initiates various downstream signaling cascades, consisting of the phospholipase C-gamma (PLCγ) pathway, the phosphoinositide 3-kinase (PI3K) pathway, and the mitogen-activated protein kinase (MAPK) pathway (reviewed in Schlessinger, 2000; Dailey et al., 2005; Mason, 2007; Dorey and Amaya, 2010). There is some evidence of downstream signaling differences between different FGFRs, such as the difference in activation levels of the MAPK pathway between FGFR1 and FGFR4 as determined using Xenopus animal cap assays (Umbhauer et al., 2000). It is thought that cellular competence and timing of expression may ultimately be responsible for the different downstream effects of FGFRs (Dailey et al., 2005; Thisse and Thisse, 2005; Branney et al., 2009).
It is well established that some vertebrates are able to regenerate parts of the eye including the lens (Henry, 2003; Tsonis and Del Rio-Tsonis, 2004; Filoni, 2008; Henry et al., 2008; Henry and Tsonis, 2010). Lens regeneration is restricted among vertebrates, generally limited to some urodeles and anurans of the genus Xenopus (Henry, 2003; Tsonis et al., 2004; Henry et al. 2008; Henry and Tsonis, 2010). The latter include X. laevis (Freeman, 1963), X. tropicalis (Henry and Elkins, 2001), and X. borealis (Filoni et al., 2006). Lens regeneration in the pre-metamorphic frog tadpole occurs by cornea-lens transdifferentiation. In this process, the outer cornea epithelium forms a thickening and subsequently a lens vesicle that develops into a mature lens (Freeman 1963). Thus, in Xenopus, both the embryonic lens and the regenerated lens originate from head ectodermal tissues. In contrast, urodeles regenerate the lens by Wolffian regeneration, a process whereby the dorsal pigmented iris epithelium transdifferentiates to form new lens cells (Tsonis et al. 2004; Call et al. 2005). Unlike the lens, the dorsal pigmented iris originates from neuroectoderm (Davis-Silberman and Ashery-Padan, 2008). There has been some evidence correlating FGF pathway function with urodele lens regeneration (McDevitt et al. 1997; Del Rio-Tsonis et al., 1998; Hayashi et al. 2004). Specifically, FGF2 appears to be necessary for lens regeneration in the newt (McDevitt et al. 1997; Hayashi et al. 2004), and FGFR3 expression and FGFR activation in general are correlated with Wolffian lens regeneration (McDevitt et al. 1997; Del Rio-Tsonis et al., 1998).
In X. laevis lens regeneration, the involvement of FGF signaling has been implicated to a lesser extent. In one study, it was shown that the addition of FGF1 protein (formerly referred to as “aFGF” or “acidic FGF”) to isolated cultured corneas would trigger lens cell differentiation. (Bosco et al., 1997b). Specifically, Bosco et al. (1997b) showed that the addition of FGF1 enabled cultured outer corneas to undergo transdifferentiation into lentoids containing lens fibers, whereas these cultures in media alone do not transdifferentiate. In addition, a later study demonstrated a correlation between FGFR2 protein expression and lens regeneration competent ectoderm (Arresta et al., 2005). In this study, Arresta et al. (2005) established that only those ectodermal tissues known to be competent to transdifferentiate into lenses were labeled by an antibody specific to FGFR2 IIIc protein, also known as the bek isoform of FGFR2, suggesting that FGFR2 may play a role in Xenopus lens regeneration.
Currently, we do not know exactly which FGFs and FGFRs are expressed in Xenopus larval eye tissues, and the requirement of FGFR signaling has not been shown in the context of cornea-lens transdifferentiation in the larval eye. To examine these questions, we have characterized the expression of FGFs and FGFRs during lens regeneration, and further, using a pharmacological inhibitor of FGFRs (SU5402), our experiments suggest the necessity of FGFR function in lens regeneration in X. laevis.
Materials and Methods
Xenopus laevis larvae
Adult pigmented X. laevis were obtained from Nasco (Fort Atkinson, WI). Fertilized eggs were prepared and larvae were reared to stages 48–51, as previously described (Henry and Grainger 1987; Schaefer et al. 1999). Larvae were staged according to Nieuwkoop and Faber (1956). All animal care was carried out as approved by the University of Illinois IACUC (protocol #08192).
RT-PCR analysis
Control corneas and retinas were collected from X. laevis larvae at stages 48–51 using fine iridectomy scissors. To generate transdifferentiating tissues, lenses were removed from the right eyes of X. laevis larvae at stages 48–51, as described previously (Schaefer et al. 1999). Transdifferentiating corneas and retinas were collected 1-, 3-, 5-, and 7- days after lentectomy.
Total RNA was extracted using TRIzol reagent (following the manufacturer's directions, Invitrogen, Carlsbad, CA) and RNA concentrations were measured using a NanoDrop, ND-1000 Spectrophotometer (ThermoScientific, Wilmington, DE). First strand cDNA was synthesized from 10ng total RNA using Superscript II (Invitrogen). As a positive control, stage 37–38 embryonic total RNA was used to generate first strand cDNA. Oligonucleotides were designed from established X. laevis sequences in the NCBI database (http://www.ncbi.nlm.nih.gov/nucleotide/). For those X. laevis FGF sequences that were unavailable, oligonucleotides were designed from putative X. tropicalis FGF sequences identified in the JGI genome project (http://genome.jgi-psf.org/Xentr4/Xentr4.home.html; Appendix Table 1). The amplified region of FGFRs was limited to the transmembrane domain to include both isoforms of each FGFR and to exclude the secreted forms of FGFRs (Hanneken et al. 1994; Groth and Lardelli, 2002). PCR reactions were performed using Taq polymerase (New England BioLabs, Ipswich, MA), amplified for 35 cycles. Each reaction was repeated two to five times to verify results. PCR products were confirmed by sequencing (Biotechnology Center, Urbana, IL).
in vitro eye culture
In preparation for in vitro eye cultures, stage 47–49 larvae were treated with 100U/mL Penicillin and 100μg/mL Streptomycin (Mediatech, Manassas, VA) in 1/20× Normal Amphibian Media (NAM, see Slack 1984) for three days before surgery. Larvae were anesthetized and ultimately euthanized by the addition of MS 222 (1:3000; Sigma-Aldrich, St. Louis, MO) and all surgeries were performed in this solution. This treatment helped reduce the level of bacterial contamination in the cultures of the isolated eye tissues. Using good sterile technique we found that 90% of the cultures remained free of any bacterial contamination for the duration of these cultures. Any cultures that became contaminated with bacteria were discarded.
Modified L-15 tissue culture media was formulated, as described by Kay and Peng (1991), using 61% L-15 powder (Invitrogen), 100U/mL Penicillin and 100μg/mL Streptomycin (Mediatech), and 10% fetal bovine serum (Invitrogen) diluted with sterilized deionized water. Various amounts of SU5402 (diluted from a 10mg/mL stock in DMSO; Calbiochem, San Diego, CA) were added to the modified L-15 media to assay lens regeneration. Control cultures included an equivalent final concentration of DMSO (0.25%) in modified L-15 media, corresponding to the concentration of DMSO used for the maximal dose of SU5402.
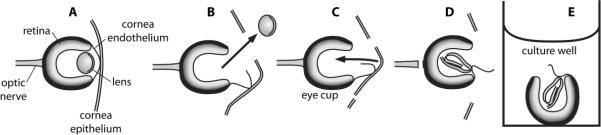
In vitro eye culture was used to assess lens regeneration in a similar manner as previously described (Bosco et al. 1993). Steps for preparing the eye culture are illustrated in Figure 1. First, the lens was removed from a given eye using ultrafine iridectomy scissors and number 5 Dumont forceps (Fig. 1A–B). Using iridectomy scissors an incision was made around the eye into the outer cornea epithelium, while maintaining the central attachment between the inner cornea endothelium and outer cornea epithelium. An incision was then made in the inner cornea endothelium and the lens was removed with forceps. The outer cornea was tucked into the opening of the vitreous chamber of the enucleated eye (Fig. 1C–D). The entire eye was excised from the tadpole by cutting the optic nerve and muscle attachments, and washed three times in modified L-15 culture media before being transferred to a 96-well plate (100μL culture medium per well; Fig. 1E). Each eye was cultured separately, and fresh culture medium was changed daily. Eyes were fixed in 3.7% formaldehyde (Sigma-Aldrich) in modified L-15 media six days after surgery, which is ample time for the formation of lens cells (Henry and Mittleman, 1995).
Figure 1. in vitro eye culture.
in vitro eye culture system used to assay lens regeneration in stage 47–49 larvae of X. laevis. (A) The larval eye is shown with both the inner cornea and outer cornea intact. (B) The lens is removed following incision of the outer and inner corneas. (C–D) The outer cornea is tucked into the vitreous chamber of the enucleated eye. (E) The eye is excised from the tadpole and cultured in modified L-15 media with or without FGFR inhibitor (SU5402). Structures are as labeled.
Immunohistological analysis
Fixed eyes were embedded in Paraplast Plus (McCormick Scientific, Richmond, IL) and sectioned to 8μm thickness (Walter et al. 2008). To detect lenses in sections, antibody staining was performed with a polyclonal rabbit anti-lens antibody specific for Xenopus lens proteins as described previously (Henry and Grainger 1990). Goat anti-rabbit-rhodamine secondary antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) was used, allowing for the positive detection of red fluorescent lens cells. The identity of each lens was confirmed by morphological inspection and fluorescence detection of antibody localization. Results were pooled from 2–4 repetitions for each concentration of SU5402 tested. Statistical analysis was performed using Fisher's exact test under the one-tailed condition. Comparisons with p values less than 0.05 were considered to be significant.
Results
FGFs are expressed in cornea and retinal tissues
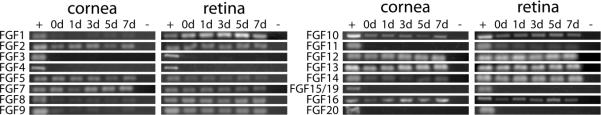
RT-PCR experiments were performed to assess the expression of FGFs in the cornea epithelium and neural retina during various time points prior to and during lens regeneration. mRNA expression within cornea and retinal tissues was evaluated in both control non-regenerating and regenerating eye tissues during four timepoints (1, 3, 5, and 7 days) following lens removal. The presence of mRNA was assessed for the presence of all Xenopus laevis FGF sequences included in the NCBI database (i.e. FGF1, 2, 3, 4, 7, 8, 10, 12, 13, and 20). In addition, RT-PCR primers were designed from available genomic Xenopus tropicalis FGF sequences in the JGI genome database for those not available in the NCBI database. Primers were successfully designed for six additional FGFs in X. laevis (FGF5, 9, 11, 14, 15/19, and 16; Appendix Table 1; amplicon GenBank accession numbers are JF433082, JF433083, JF433084, JF433085, JF433086, and JF433087 respectively). PCR products were verified by sequence analysis. Eleven FGFs (FGF1, 2, 5, 7, 8, 9, 10, 11, 12, 13, 14, and 16; see Fig. 2) were detected in both control cornea tissues and corneas undergoing the process of lens regeneration. Twelve FGFs (FGF1, 2, 5, 7, 8, 9, 10, 11, 12, 13, 14, and16; see Fig. 2) were detected in retinal tissues throughout these timepoints. Though these assays are not quantitative, some potential differences were observed in the level of the amplified PCR products for certain FGFs and at various time points (Fig. 2). Of interest, the expression levels of FGF1, FGF8, and FGF9 mRNA in the cornea were consistently lower than the corresponding levels in the retina.
Figure 2. RT-PCR expression of FGFs in eye tissues.
Expression of FGFs as determined by RT-PCR assays. Total RNA was collected from corneas and retinas of both non-regenerating control and lens-regenerating larvae. Regenerating corneas and retinas were collected at four timepoints (1, 3, 5, and 7 days after lens removal). Note that the bands for FGF1, FGF8, and FGF9 are uniformly less intense in the cornea when compared to the retina, as determined from replicate RT-PCR reactions. 0d denotes non-regenerating control eye tissues; + denotes positive control cDNA derived from mRNA of whole embryos (st. 37–38); - denotes the negative control without addition of template cDNA.
FGFRs are expressed in cornea and retinal tissues
Similarly, RT-PCR experiments were performed to characterize the expression of FGFRs in the cornea and retina during lens regeneration. As shown in Figure 3, only FGFR1, 2, and 3 were expressed in the cornea throughout the period of regeneration examined in this study. In contrast, all four FGFRs were detectable in the retina, though more prominent levels of PCR product were consistently observed for FGFR1 and 4.
Figure 3. RT-PCR expression of FGFRs in eye tissues.
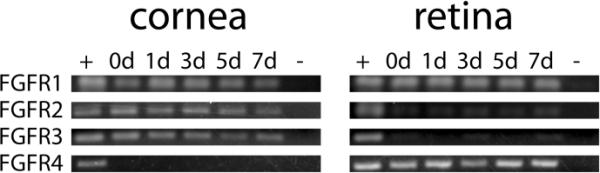
Expression of FGFRs as determined by RT-PCR assays. Total RNA was collected from corneas and retinas of both non-regenerating control and lens-regenerating larvae. Regenerating corneas and retinas were collected at four timepoints (1, 3, 5, and 7 days after lens removal). 0d denotes non-regenerating control eye tissues; + denotes positive control cDNA derived from mRNA of whole embryos (st. 37–38); − denotes the negative control without addition of template cDNA.
SU5402 application inhibits lens regeneration
Increasing dosages of SU5402 (2μM, 5μM, 10μM, and 25μM), a FGFR inhibitor, were tested for their ability to inhibit lens regeneration. Lenses were identified in sectioned eye cultures via anti-lens antibody staining (Fig. 4). Under control conditions (0.25% DMSO in modified L-15 medium), 82% of cultured eyes regenerated a lens (23 out of 28 cases; Fig. 4A–B, 5). On the other hand, application of SU5402 resulted in a dose-dependent inhibition of lens regeneration (Fig. 4C–L, 5). Though there was no inhibition of lens regeneration with 2μM SU5402 (81% regeneration; 13 out of 16 cases examined; p = 0.62), there was almost no regeneration at the higher concentrations tested (10μM and 25μM SU5402). Furthermore, individual cases of lens regeneration at the higher concentrations of SU5402 were small and represented only a preliminary stage of lens regeneration (stage 2, as described in Freeman 1963), in which the developing lens appears only as a thickening of the cornea epithelium (data not shown). The lens regeneration rate was halved at 5μM SU5402 (38% regeneration; 6 out of 16 cases), representing a significant decrease in the lens regeneration rate relative to the control cases (p = 0.0038). There was almost no regeneration upon application of 10μM SU5402 (5.9% regeneration; 1 out of 17 cases; p = 4.5e–7) and 25μM SU5402 (2.5% regeneration; 1 out of 40 cases; p = 2.6e–12). From this data, the IC50 for lens regeneration can be determined to be approximately 5μM SU5402. The cultured eye tissues otherwise all appeared to be healthy and of normal morphology at all doses tested (see Fig. 4).
Figure 4. Sectioned eyes after SU5402 treatment in culture.
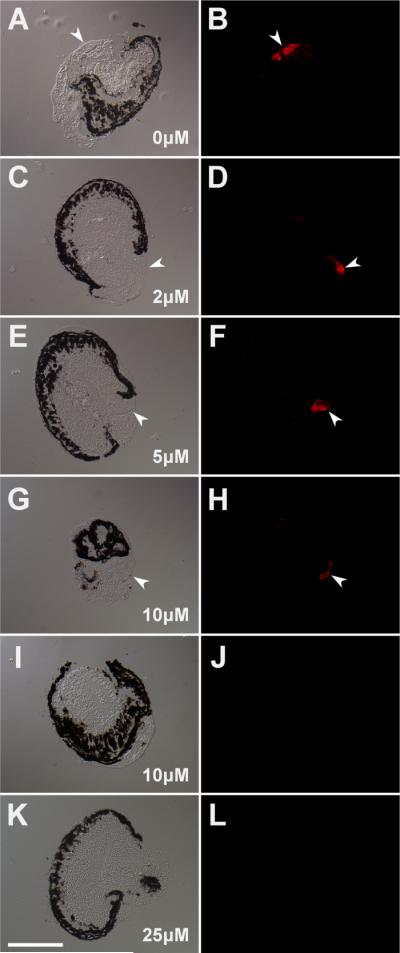
Development of control and SU5402 treated in vitro eye cultures. SU5402 inhibits FGFR function by competitively binding to the FGFR kinase domain. The left column shows sections of representative eyes imaged using differential interference contrast. The right column shows each corresponding fluorescent image illustrating α-lens antibody staining. SU5402 concentrations: (A–B) 0μM control; (C–D) 2μM; (E–F) 5μM; (G–J) 10μM; (K–L) 25μM. The single case of lens regeneration with 10μM SU5402 is shown in G–H. The typical results of non-regenerating cases are shown for 10μM SU5402 (H–I) and 25μM SU5402 (K–L). Arrows point to regenerated lenses; scale bar equals 200μm.
Figure 5. Lens regeneration rates upon application of SU5402.
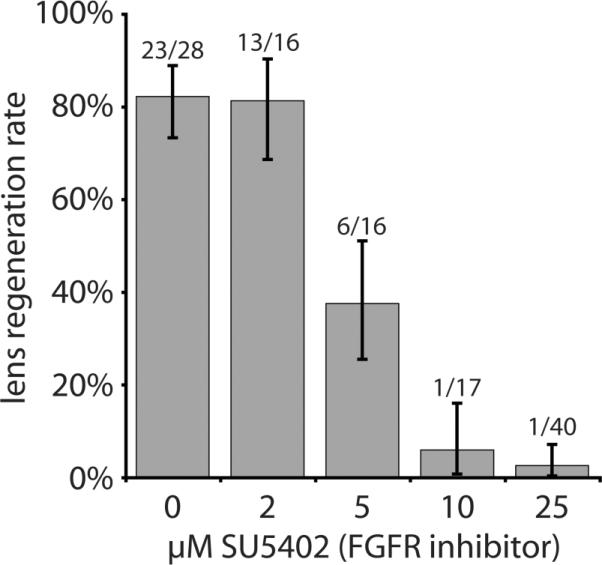
Results of in in vitro eye culture experiments treated with SU5402 to inhibit FGFR function. As shown here, the IC50 for inhibiting lens regeneration is close to 5μM SU5402. Numbers of regenerated lenses and eyes examined are located above each bar; y-axis indicates lens regeneration rate; error bars denote Wilson score intervals in which Z=1.
Discussion
Experimental evidence shows that critical retinal factors trigger Xenopus cornea-lens transdifferentiation or “lens regeneration”. Through the removal of various eye tissues after lentectomy, Filoni et al., (1982) found that the presence of the neural retina was key to inducing lens regeneration during larval stages. In X. laevis, although lens regeneration ability declines as the tadpole ages (Freeman, 1963), cornea epithelia from later stages including adult frogs inserted into the vitreous chamber are still capable of transdifferentiating into lenses in culture (Filoni et al., 1997). Bosco et al. (1993) showed that the presence of neural retina was sufficient to induce lens regeneration in cultured corneas. Further, Bosco et al. (1997a) showed that corneas transdifferentiated into lens after exposure to centrifuged, filtered, retina-conditioned culture medium, thus indicating that some diffusible factor was responsible for inducing lens regeneration. From this evidence, one can hypothesize that a retinal signaling ligand is responsible for inducing lens regeneration, though this has yet to be identified.
As the retina represents the source of key signaling factors required to support lens regeneration, it is possible that one or more of the FGFs detected in the present study could play a role in this process. Normally, the cornea is isolated from the signaling factors provided by the retina via the presence of the lens and the inner cornea endothelium (Freeman, 1963). This ensures that supernumerary lenses are not normally formed in the larval eye (Reeve and Wild, 1978; Bosco et al. 1979; Filoni et al. 1980). Although we are describing expression at the level of transcription, one could argue that key signals involved in lens regeneration should be expressed only by the retinal tissue and not be expressed in the cornea. Likewise, as the receptor for presumptive FGF signaling, the associated FGFRs should be expressed in the cornea during lens regeneration.
Though the RT-PCR analyses reported here are not quantitative, some potential differences in expression between retina and cornea may exist for FGF1, 8, 9, 11, and 14 based on the rather dramatic differences in the intensity of the PCR products detected (Fig. 2). Of these, FGF11 and FGF14 are members of the intracellular FGF subfamily, formerly known as the FGF11 subfamily or the FGF homologous factors, which are expressed in neuronal tissues and do not interact with known FGFRs (Table 1; Olsen et al. 2003; Goldfarb, 2005; Itoh and Ornitz, 2008). Hence, FGF1, FGF8, and FGF9 are plausible candidate FGFs that could represent the key retinal signal(s) that trigger lens regeneration. Other FGFs (FGF2, 5, 7, 10, 12, 13, and 16) were also expressed by retinal tissues and could represent key signals involved in lens regeneration. Of course, this study describes expression at the level of transcription, and it will be important to examine expression at the level of translation in future studies.
From compiling studies in various vertebrates, Robinson (2006) noted that certain members of all seven subfamilies of the FGFs and all four FGFRs (1–4) have been observed in the eye during development or in the adults of various vertebrates. In a recent X. tropicalis study, Lea et al. (2009) showed that FGF1, FGF3, FGF13, FGF14, and FGF20 are expressed during embryonic eye development. Here we have found additional FGFs (FGF2, 5, 7, 8, 9, 10, 11, 12, and 16) that are expressed in the eye during the later larval stage of X. laevis (i.e., stage 48–51) and have not been able to detect FGF3 and FGF20. Furthermore, during embryonic lens development in X. tropicalis, FGFR3 is expressed in the developing lens, FGFR2 is prominently expressed in corneal epithelium, and FGFR1 and FGFR4 are expressed in “cells surrounding the lens” in early tadpoles (Lea et al. 2009). This is similar to our finding that FGFR1 and FGFR4 are prominently expressed in the larval retina, whereas FGFR1, FGFR2, and FGFR3 are expressed in the cornea.
As for lens regeneration, previous research using X. laevis corneal explant cultures has shown that addition of FGF1 protein, previously known as acidic or aFGF, to cornea explants induced the transdifferentiation of these cells into lens cells (Bosco et al. 1997b). In these experiments, the authors established this using in vitro cultured outer cornea epithelium. The corneas transdifferentiated only when FGF1 was added to the serum supplemented L-15 media. Our results agree with the notion that FGF1 may be a signaling ligand for inducing cornea-lens transdifferentiation. In the newt, as opposed to other FGFs (e.g., 1, 4, and 7–10) and various growth factors (i.e., EGF, IGF, and VEGF), FGF2 has the unique ability to trigger the generation of a new lens after injection into the eye chamber (Hayashi et al. 2004). Significantly, we have found that FGF2 is expressed both in the cornea and the retina in X. laevis (Fig. 2). In another study, FGF2 and FGFR3 (also named PFR3, or Pleurodeles homolog of FGF receptor 3) expression specifically seemed to be correlated with Wolffian lens regeneration (McDevitt et al. 1997).
As described above, the 22 known members of the FGF family belong to seven subfamilies based on sequence homology (Itoh and Ornitz, 2004). It is interesting to note that FGF1, FGF8, and FGF9 identified in our study as possible signaling candidates involved in lens regeneration are members of three different FGF subfamilies (Itoh and Ornitz 2004). Two of the three candidates, FGF8 and FGF9, are functionally thought to be ancestral members of their respective subfamilies in the mouse system (Itoh and Ornitz 2008). FGF9 has been shown to interact with only FGFR2 and FGFR3, whereas FGF1 and FGF8 can activate all four FGFRs (Table 1; Ornitz et al. 1996). This may indicate that a combination of FGFRs could be activated during lens regeneration. Here, we have observed the expression of FGFR1, 2, and 3 in the cornea, so activation of these FGFRs could be involved in triggering X. laevis lens regeneration.
As for the role of FGFR in lens regeneration, the bek isoform of FGFR2 was shown to be present in Xenopus epidermis only in regions where the epidermis was capable of transdifferentiating into lenses (Arresta et al. 2005). Specifically, these regions include the cornea epidermis overlying the eye and the pericorneal epidermis immediately surrounding the eye. In that study, the authors utilized an experimental approach to impart lens-forming competence on epidermis not normally competent to undergo transdifferentiation. This consists of implanting eye tissues beneath the target epidermis, based on the protocol of Cannata et al. (2003). If an eye was implanted beneath head epidermis distant from the eye (at stage 46), the head epidermis became competent to transdifferentiate into lenses if challenged later (at stage 53) by implanting this tissue into the vitreous chamber. Arresta et al. (2005) found that this head epidermis also expressed FGFR2 IIIc after exposure to the implanted eye. However, the authors only established a correlation between lens-forming competence and expression of FGFR2, and did not perform any direct tests to see if FGFR2 expression is responsible for establishing this regeneration capability.
There is evidence linking FGF pathway activation with lens development in other vertebrates (reviewed by Robinson, 2006). In particular, regarding FGFR function, expression of the dominant negative FGFR1 in the developing mouse lens placode (using a Pax6 promoter expressing a dominant negative truncated form of FGFR1) inhibited lens cell proliferation and differentiation, thus demonstrating that FGFR activation is necessary for lens development (Faber et al. 2001). Similarly, a conditional knockout of FGFR2 in the mouse lens placode produced very small or absent lenses (Garcia et al. 2005).
The small molecule SU5402 has been shown to inhibit FGFR autophosphorylation by competitively binding the tyrosine kinase domain of FGFR1 (Mohammadi et al. 1997). Due to sequence conservation between the tyrosine kinase domains of FGFRs, SU5402 can inhibit the function of all four FGFRs (Delaune et al. 2004; Grand et al. 2004; Mansukhani et al. 2005). However, SU5402 inhibitory activity is not completely specific to FGFRs, as it has also been shown to inhibit Vascular Endothelial Growth Factor Receptor (VEGFR) tyrosine kinase activity in NIH 3T3 cells, and to a lesser extent, Platelet-Derived Growth Factor Receptor (PDGFR) tyrosine kinase activity (Sun et al. 1999). Past lens regeneration studies in the newt model have investigated the FGFR pathway using SU5402. In the case of Wolffian lens regeneration, Del Rio-Tsonis et al. (1998) found that inhibiting FGFR function by using SU5402 led to inhibition of lens regeneration in that system. More recently, Hayashi et al. (2002) used an in vitro method of culturing newt dorsal iris cell reaggregates and established that FGF2 and FGF4 were able to induce lens formation in these cultures. They then showed that FGFR function was necessary for this phenomenon by inhibiting lens formation by adding SU5402. In our investigation of Xenopus cornea-lens transdifferentiation, SU5402 almost completely inhibited lens regeneration. The observed IC50 for lens regeneration was at 5μM, less than the published IC50 of 10–20μM for FGFR autophosphorylation in NIH 3T3 cells (Mohammadi et al. 1997). This concentration is less than the 20μM concentration found to be effective in the newt by Del Rio-Tsonis et al. (1998) and the 10μM concentration used by Hayashi et al. (2002), mentioned above. Taken together, the evidence suggests that FGFR activation is both necessary (as established in the present study) and sufficient (as established by Bosco et al., 1997b, see also Arresta et al., 2005) for lens regeneration in Xenopus.
Acknowledgments
We would like to thank Kimberly Perry, Paul Hamilton, and Alvin Thomas for comments improving this manuscript. This work was supported by NIH grant EY09844 to JJH.
Appendix
Table 1.
Oligonucleotides used for RT-PCR verification of FGF and FGFR expression
| Gene | Primer sequences (5'-3') | Tm (°C) |
|---|---|---|
|
FGF1 F R |
TCAAGACCACAGAGACAGGG CAAACCAGTTCATGTCTGCG |
58.8 60.3 |
|
FGF2 F R |
AGGCTCTACTGCAAGAACGG TCTCCCATCTTCCTTCATGG |
59.6 60.0 |
|
FGF3 F R |
TTTAGAAATAACCGCCGTGG TGGAACTGTCCGATAAAGGC |
58.9 59.9 |
|
FGF4 F R |
CATCGGGTTTCATATCCAGG TTGATCCATACAGCTTCCCC |
60.2 59.9 |
|
FGF5* F R |
TTTCATCTCCAGATCCACCC GGTGTTGCATGAAGTTTCCC |
59.9 60.4 |
|
FGF7 F R |
AAACGAGGCAATGTGAAAGG CATTGCATGATTTCTTTCCG |
60.1 59.1 |
|
FGF8 F R |
TACACAGCATGTGAGGGAGC TTTCCACGATTAACTTGGCG |
59.9 61.0 |
|
FGF9* F R |
ATGGGACTATCCAAGGGACC CTCTTGCGTTAGCTTTTCCG |
60.0 60.1 |
|
FGF10 F R |
GCACCAAGAAGGAGAATTGC GACGCATAGGTGTTGTAGCC |
59.8 58.3 |
|
FGF11* F R |
TGTCACCTACTCCTCCACCC GCAACTTCACTGAGCTTGGG |
60.0 61.0 |
|
FGF12 F R |
TACACTGTATCGGCAGCAGG CCAATTTCATGCAGTGATGG |
59.9 59.9 |
|
FGF13 F R |
CGAGTGGTGGCTATTCAAGG GTTGAGACCCAAAAACCAGC |
60.7 59.6 |
|
FGF14* F R |
TTGTAATGGGAACCTGGTGG CTGCTGTCGTCCTTTGTTCC |
60.6 60.8 |
|
FGF15/19* F R |
TTGCCATTAAAGGGTATCGC TCCTTGCTTAGGGAGACAGC |
59.9 59.6 |
|
FGF16* F R |
GACTGTACATGGCACAAGGC CTGTTCAGCTTCTTCGACCC |
59.2 60.0 |
|
FGF20 F R |
TTGCTATTGGCCTGGTTAGC GCTACAAAGTATCGCCGTCC |
60.2 59.7 |
|
FGFR1 F R |
TTAAAATGAAGCACCCGTCG CGAGACTCCAGACAACATGG |
61.0 59.2 |
|
FGFR2 F R |
TCTGCATGGTAGTGGTCTGC GATCCTCACGAGTGGAGTGG |
59.9 60.7 |
|
FGFR3 F R |
GTGACCGAGACCAATGAAGG GGTGACCACAATAAGGACGG |
60.5 60.2 |
|
FGFR4 F R |
GAAGATTTCCTTGAGCAAGCC CAGTTTATGGACAGTTGGCG |
60.3 59.2 |
Oligonucleotides used for assaying FGF and FGFR expression. Primer sequences were designed from X laevis FGF and FGFR sequences in the NCBI database where available.
For X. laevis FGFs not in this database, primers were designed from putative genes in the JGIX. tropicalis genome project database (FGFs marked by GenBank accession numbers are provided for each of the respective amplicons in the Results section). Salt adjusted melting temperature (Tm) values are noted for each primer.
Literature Cited
- Arresta E, Bernardini S, Gargioli C, Filoni S, Cannata SM. Lens-forming competence in the epidermis of Xenopus laevis during development. J Exp Zool A Comp Exp Biol. 2005;303A:1–12. doi: 10.1002/jez.a.138. [DOI] [PubMed] [Google Scholar]
- Baird A, Esch F, Mormède P, Ueno N, Ling N, Böhlen P, Ying SY, Wehrenberg WB, Guillemin R. Molecular characterization of fibroblast growth factor: distribution and biological activities in various tissues. Recent Prog.Horm.Res. 1986;42:143–205. doi: 10.1016/b978-0-12-571142-5.50008-2. [DOI] [PubMed] [Google Scholar]
- Bosco L, Filoni S, Cannata S. Relationships between Eye Factors and Lens-forming Transformations in the Cornea and Pericorneal Epidermis of Larval Xenopus laevis. J. Exp. Zool. 1979;209:261–282. doi: 10.1002/jez.1402090208. [DOI] [PubMed] [Google Scholar]
- Bosco L, Testa O, Venturini G, Willems D. Lens fibre transdifferentiation in cultured larval Xenopus laevis outer cornea under the influence of neural retina-conditioned medium. Cellular and Molecular Life Sciences. 1997a;53:921–928. doi: 10.1007/PL00013198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco L, Valle C, Willems D. In Vivo and In Vitro Experimental Analysis of Lens Regeneration in Larval Xenopus laevis. Dev.Growth Differ. 1993;35:257–270. doi: 10.1111/j.1440-169X.1993.00257.x. [DOI] [PubMed] [Google Scholar]
- Bosco L, Venturini G, Willems D. In vitro lens transdifferentiation of Xenopus laevis outer cornea induced by Fibroblast Growth Factor (FGF) Development. 1997b;124:421–428. doi: 10.1242/dev.124.2.421. [DOI] [PubMed] [Google Scholar]
- Bottcher RT, Niehrs C. Fibroblast growth factor signaling during early vertebrate development. Endocr.Rev. 2005;26:63–77. doi: 10.1210/er.2003-0040. [DOI] [PubMed] [Google Scholar]
- Branney PA, Faas L, Steane SE, Pownall ME, Issacs HV. Characterisation of the fibroblast growth factor dependent transcriptome in early development. PLoS One. 2009;4:e4951. doi: 10.1371/journal.pone.0004951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call MK, Grogg MW, Tsonis PA. Eye on regeneration. Anat Rec B New Anat. 2005;287:42–48. doi: 10.1002/ar.b.20084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannata SM, Arresta E, Bernardini S, Gargioli C, Filoni S. Tissue interactions and lens-forming competence in the outer cornea of larval Xenopus laevis. J Exp Zool A Comp Exp Biol. 2003;299:161–171. doi: 10.1002/jez.a.10275. [DOI] [PubMed] [Google Scholar]
- Chamberlain CG, McAvoy JW. Induction of lens fibre differentiation by acidic and basic fibroblast growth factor (FGF) Growth Factors. 1989;1:125–134. doi: 10.3109/08977198909029122. [DOI] [PubMed] [Google Scholar]
- Dailey L, Ambrosetti D, Mansukhani A, Basilico C. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev. 2005;16:233–247. doi: 10.1016/j.cytogfr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Davis-Silberman N, Ashery-Padan R. Iris development in vertebrates; genetic and molecular considerations. Brain Research. 2008;1192:17–28. doi: 10.1016/j.brainres.2007.03.043. [DOI] [PubMed] [Google Scholar]
- Delaune E, Lemaire P, Kodjabachian L. Neural induction in Xenopus requires early FGF signaling in addition to BMP inhibition. Development. 2004;132:299–310. doi: 10.1242/dev.01582. [DOI] [PubMed] [Google Scholar]
- Del Rio-Tsonis K, Trombley MT, McMahon G, Tsonis PA. Regulation of lens regeneration by fibroblast growth factor receptor 1. Dev. Dyn. 1998;213:140–146. doi: 10.1002/(SICI)1097-0177(199809)213:1<140::AID-AJA14>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Dorey K, Amaya E. FGF signaling: diverse roles during early vertebrate embryogenesis. Development. 2010;137:3731–3742. doi: 10.1242/dev.037689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber SC, Dimanlig P, Makarenkova HP, Shirke S, Ko K, Lang RA. Fgf receptor signaling plays a role in lens induction. Development. 2001;128:4425–4438. doi: 10.1242/dev.128.22.4425. [DOI] [PubMed] [Google Scholar]
- Filoni S. Retina and lens regeneration in anuran amphibians. Semin.Cell Dev.Biol. 2008;20:528–534. doi: 10.1016/j.semcdb.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Filoni S, Bernardini S, Cannata S. Experimental analysis of lens-forming capacity in Xenopus borealis larvae. J Exp Zool A Comp Exp Biol. 2006;305:538–550. doi: 10.1002/jez.a.297. [DOI] [PubMed] [Google Scholar]
- Filoni S, Bernardini S, Cannata SM, D'Alessio A. Lens regeneration in larval Xenopus laevis: Experimental analysis of the decline in the regenerative capacity during development. Dev Bio. 1997;187:13–24. doi: 10.1006/dbio.1997.8598. [DOI] [PubMed] [Google Scholar]
- Filoni S, Bosco L, Cioni C. The role of neural retina in lens regeneration from cornea in larval Xenopus laevis. Acta embryologiae et morphologiae experimentalis. 1982;3:15–28. [PubMed] [Google Scholar]
- Filoni S, Bosco L, Paglioni N, Cioni C. Lens Formation From Pericorneal Epidermis in the Presence of the Old Lens in Larval Xenopus laevis. J. Exp. Zool. 1980;211:303–309. doi: 10.1002/jez.1402130103. [DOI] [PubMed] [Google Scholar]
- Ford-Perriss M, Abud H, Murphy M. Fibroblast growth factors in the developing central nervous system. Clin.Exp.Pharmacol.Physiol. 2001;28:493–503. doi: 10.1046/j.1440-1681.2001.03477.x. [DOI] [PubMed] [Google Scholar]
- Freeman G. Lens regeneration from the cornea in Xenopus laevis. J.Exp.Zool. 1963;154:39–65. doi: 10.1002/jez.1401540105. [DOI] [PubMed] [Google Scholar]
- Garcia CM, Kai Y, Zhao H, Ashery-Padan R, Ornitz DM, Robinson ML, Beebe DC. Signaling through FGF Receptor-2 Is Required for Lens Cell Survival and for Withdrawal From the Cell Cycle During Lens Fiber Cell Differentiation. Dev Dyn. 2005;233:516–527. doi: 10.1002/dvdy.20356. [DOI] [PubMed] [Google Scholar]
- Goldfarb M. Fibroblast growth factor homologous factors: evolution, structure, and function. Cytokine Growth Factor Rev. 2005;16:215–220. doi: 10.1016/j.cytogfr.2005.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grand EK, Chase AJ, Heath C, Rahemtulla A, Cross NCP. Targeting FGFR3 in multiple myeloma: inhibition of t(4;14)-positive cells by SU5402 and PD173074. Leukemia. 2004;18:962–966. doi: 10.1038/sj.leu.2403347. [DOI] [PubMed] [Google Scholar]
- Groth C, Lardelli M. The structure and function of vertebrate fibroblast growth factor receptor 1. Int.J.Dev.Biol. 2002;46:393–400. [PubMed] [Google Scholar]
- Hanneken A, Ying W, Ling N, Baird A. Identification of soluble forms of the fibroblast growth factor receptor in blood. Proc. Natl. Acad. Sci. 1994;91:9170–9174. doi: 10.1073/pnas.91.19.9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Mizuno N, Owaribe K, Kuroiwa A, Okamoto M. Regulated lens regeneration from isolated pigmented epithelial cells of newt iris in culture in response to FGF2/4. Differentiation. 2002;70:101–108. doi: 10.1046/j.1432-0436.2002.700205.x. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Mizuno N, Ueda Y, Okamoto M, Kondoh H. FGF2 triggers iris-derived lens regeneration in newt eye. Mech.Dev. 2004;121:519–526. doi: 10.1016/j.mod.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Henry JJ. The cellular and molecular bases of vertebrate lens regeneration. Int.Rev.Cytol. 2003;228:195–265. doi: 10.1016/s0074-7696(03)28005-0. [DOI] [PubMed] [Google Scholar]
- Henry JJ, Wever JA, Vergara MN, Fukui L. Xenopus, an ideal vertebrate system for studies of eye development and regeneration. In: Wittbrodt J, editor. Animal Models for Eye Research. Academic Press; Elsevier: 2008. pp. 57–92. [Google Scholar]
- Henry J, Elkin M. Cornea-lens transdifferentiation in the anuran, Xenopus tropicalis. Dev.Genes Evol. 2001;211:377–387. doi: 10.1007/s004270100163. [DOI] [PubMed] [Google Scholar]
- Henry J, Grainger R. Inductive interactions in the spatial and temporal restriction of lens-forming potential in embryonic ectoderm of Xenopus laevis. Dev.Biol. 1987;124:200–214. doi: 10.1016/0012-1606(87)90472-6. [DOI] [PubMed] [Google Scholar]
- Henry J, Grainger R. Early tissue interactions leading to embryonic lens formation in Xenopus laevis. Dev.Biol. 1990;141:149–163. doi: 10.1016/0012-1606(90)90110-5. [DOI] [PubMed] [Google Scholar]
- Henry J, Mittleman J. The matured eye of Xenopus laevis tadpoles produces factors that elicit a lens-forming response in embryonic ectoderm. Dev. Biol. 1995;171:39–50. doi: 10.1006/dbio.1995.1258. [DOI] [PubMed] [Google Scholar]
- Henry JJ, Tsonis PA. Molecular and cellular aspects of amphibian lens regeneration. Prog Retin Eye Res. 2010;29:543–555. doi: 10.1016/j.preteyeres.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N. The Fgf families in humans, mice, and zebrafish: their evolutional processes and roles in development, metabolism, and disease. Biol.Pharm.Bull. 2007;30:1819–1825. doi: 10.1248/bpb.30.1819. [DOI] [PubMed] [Google Scholar]
- Itoh N, Ornitz DM. Evolution of the Fgf and Fgfr gene families. Trends Genet. 2004;20:563–569. doi: 10.1016/j.tig.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Itoh N, Ornitz D. Functional evolutionary history of the mouse FGF gene family. Dev. Dyn. 2008;237:18–27. doi: 10.1002/dvdy.21388. [DOI] [PubMed] [Google Scholar]
- Kay B, Peng H, editors. Xenopus Laevis: Practical Uses in Cell and Molecular Biology. Academic Press; 1991. [PubMed] [Google Scholar]
- Lea R, Papalopulu N, Amaya E, Dorey K. Temporal and spatial expression of FGF ligands and receptors during Xenopus development. Dev Dyn. 2009;238:1467–1479. doi: 10.1002/dvdy.21913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansukhani A, Ambrosetti D, Holmes G, Cornivelli L, Basilico C. Sox2 induction by FGF and FGFR2 activating mutations inhibits Wnt signaling and osteoblast differentiation. JCB. 2005;168:1065–1076. doi: 10.1083/jcb.200409182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason I. Initiation to end point: the multiple roles of fibroblast growth factors in neural development. Nature Reviews Neuroscience. 2007;8:583–596. doi: 10.1038/nrn2189. [DOI] [PubMed] [Google Scholar]
- McDevitt D, Brahma SK, Courtois Y, Jeanny J-C. Fibroblast growth factor receptors and regeneration of the eye lens. Dev. Dyn. 1997;208:220–226. doi: 10.1002/(SICI)1097-0177(199702)208:2<220::AID-AJA9>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, McMahon G, Sun L, Tang C, Hirth P, Yeh BK, Hubbard SR, Schlessinger J. Structures of the Tyrosine Kinase Domain of Fibroblast Growth Factor Receptor in Complex with Inhibitors. Science. 1997;276:955–960. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, Olsen SK, Ibrahimi OA. Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev. 2005;16:107–137. doi: 10.1016/j.cytogfr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus Laevis (Daudin) North Holland Publishing Company; 1956. [Google Scholar]
- Olsen SK, Garbi M, Zampieri N, Eliseenkova AV, Ornitz DM, Goldfarb M, Mohammadi M. Fibroblast growth factor (FGF) homologous factors share structural but not functional homology with FGFs. J Biol Chem. 2003;278:34226–34236. doi: 10.1074/jbc.M303183200. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2:REVIEWS3005. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Xu J, Colvin JS, Mcewen DG, Macarthur CA, Coulier F, Gao G, Goldfarb M. Receptor Specificity of the Fibroblast Growth Factor Family. J.Biol.Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- Reeve JG, Wild AE. Lens regeneration from cornea of larval Xenopus laevis in the presence of the lens. J. Embryol. Exp. Morph. 1978;48:205–214. [PubMed] [Google Scholar]
- Robinson M. An essential role for FGF receptor signaling in lens development. Semin.Cell Dev.Biol. 2006;17:726–740. doi: 10.1016/j.semcdb.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer JJ, Oliver G, Henry JJ. Conservation of gene expression during embryonic lens formation and cornea-lens transdifferentiation in Xenopus laevis. Dev.Dyn. 1999;215:308–318. doi: 10.1002/(SICI)1097-0177(199908)215:4<308::AID-AJA3>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- Slack JM. Regional biosynthetic markers in the early amphibian embryo. J.Embryol.Exp.Morphol. 1984;80:289–319. [PubMed] [Google Scholar]
- Sun L, Tran N, Liang C, Tang F, Rice A, Schreck R, Waltz K, Shawver LK, McMahon G, Tang C. Design, Synthesis, and Evaluations of Substituted 3-[(3- or 4-Carboxyethylpyrrol-2-yl)methylidenyl]indolin-2-ones as Inhibitors of VEGF, FGF, and PDGF Receptor Tyrosine Kinases. J. Med. Chem. 1999;42:5120–5130. doi: 10.1021/jm9904295. [DOI] [PubMed] [Google Scholar]
- Thisse B, Thisse C. Functions and regulations of fibroblast growth factor signaling during embryonic development. Dev.Biol. 2005;287:390–402. doi: 10.1016/j.ydbio.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Tsonis PA, Del Rio-Tsonis K. Lens and retina regeneration: transdifferentiation, stem cells and clinical applications. Exp.Eye Res. 2004;78:161–172. doi: 10.1016/j.exer.2003.10.022. [DOI] [PubMed] [Google Scholar]
- Tsonis PA, Madhavan M, Tancous EE, Del Rio-Tsonis K. A newt's eye view of lens regeneration. The International journal of developmental biology. 2004;48:975–980. doi: 10.1387/ijdb.041867pt. [DOI] [PubMed] [Google Scholar]
- Umbhauer M, Penzo-Mendez A, Clavilier L, Boucaut J, Riou J. Signaling specificities of fibroblast growth factor receptors in early Xenopus embryo. J.Cell.Sci. 2000;113:2865–2875. doi: 10.1242/jcs.113.16.2865. [DOI] [PubMed] [Google Scholar]
- Walter BE, Perry KJ, Fukui L, Malloch EL, Wever J, Henry JJ. Psf2 plays important roles in normal eye development in Xenopus laevis. Mol.Vis. 2008;14:906–921. [PMC free article] [PubMed] [Google Scholar]
- Xu, Weinstein XM, Li C, Deng CX. Fibroblast growth factor receptors (FGFRs) and their roles in limb development. Cell Tissue Res. 1999;296:33–43. doi: 10.1007/s004410051264. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ibrahami OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J.Biol.Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]