Abstract
In this study we investigated the dynamics of R-Ras intracellular trafficking and its contributions to the unique roles of R-Ras in membrane ruffling and cell spreading. Wild type and constitutively active R-Ras localized to membranes of both Rab11- and transferrin-positive and -negative vesicles, which trafficked anterograde to the leading edge in migrating cells. H-Ras also co-localized with R-Ras in many of these vesicles in the vicinity of the Golgi, but R-Ras and H-Ras vesicles segregated proximal to the leading edge, in a manner dictated by the C-terminal membrane-targeting sequences. These segregated vesicle trafficking patterns corresponded to distinct modes of targeting to membrane ruffles at the leading edge. Geranylgeranylation was required for membrane anchorage of R-Ras, whereas palmitoylation was required for exit from the Golgi in post-Golgi vesicle membranes and trafficking to the plasma membrane. R-Ras vesicle membranes did not contain phosphatidylinositol (3,4,5)-trisphosphate (PtdIns(3,4,5)P3), whereas R-Ras co-localized with PtdIns(3,4,5)P3 in membrane ruffles. Finally, palmitoylation-deficient R-Ras blocked membrane ruffling, R-Ras/PI3-kinase interaction, enrichment of PtdIns(3,4,5)P3 at the plasma membrane, and R-Ras-dependent cell spreading. Thus, lipid modification of R-Ras dictates its vesicle trafficking, targeting to membrane ruffles, and its unique roles in localizing PtdIns(3,4,5)P3 to ruffles and promoting cell spreading.
Keywords: H-Ras; PI3Kinase; PIP3, vesicle trafficking; R-Ras; cell spreading; geranylgeranyl; membrane ruffles; palmitoyl
Introduction
Among the Ras subfamily of small GTPases, R-Ras is unusual in its ability to promote membrane ruffling,1 enhance cell adhesion2-4 and promote cell spreading and migration.5-7 A close relative of R-Ras, TC21, also promotes cell motility.8 Cell migration is characterized by forward extension of the leading edge plasma membrane in lamellipodia and filopodia as a result of localized actin polymerization, with concomitant retraction of the cell rear.9 These events are precisely controlled and dependent upon the actions of multiple classes of small GTPases, including Ras, Rho, Rab and Arf proteins.10,11 R-Ras promotes spreading and migration of many cell types, distinct from related Ras family GTPases, through multiple mechanisms, such as regulating cell adhesion through integrin receptors and formation of focal adhesions2,3,12-14 and downstream activation of Rac1 and Arf GTPases, phosphatidylinositol 3-kinase (PI3K) and phospholipase Cε (PLCε) and stimulating their effects on actin remodeling, integrin activation and trafficking and membrane protrusion.1,4,6,7,15 At present it is unclear how R-Ras integrates these signaling effects, or selectively engages specific pathways in different cell types.
Forward extensions at the leading edge are made possible in part by incorporation of new membrane material at the migrating front, provided by vesicles delivered from the cell interior. Thus, anterograde vesicle trafficking plays an essential role in forward protrusion and migration.16 Membrane traffic through vesicular transport pathways is tightly coordinated by small GTPases of the Rab and Arf subfamilies.17 As the plasma membrane continuously extends at the migrating front, membrane material is recycled from the leading edge in retrograde membrane ruffles. Vesicular traffic also transports proteins to and from the leading edge (ruffle) of the migrating cell, creating a cycle of anterograde and retrograde movement or proteins and membrane lipids through exocytic and endocytic vesicles.16,18
Subcellular targeting of Ras family small GTPases to endomembranes has garnered much attention as an important mechanism for regulating localized Ras signaling, contributing to distinct cellular functions of Ras isotypes.19-21 Although Ras proteins H-, K-, N- and R-Ras are highly homologous and share nearly identical nucleotide and effector binding domains, their C-termini comprise hypervariable regions (HVRs), which are responsible for isotype-specific subcellular targeting. The HVRs terminate in a so-called CaaX box, consisting of a Cysteine (C) followed by two typically aliphatic residues (aa) and a variable amino acid (X). Ras proteins are initially synthesized as globular, cytoplasmic proteins, but subsequently undergo a series of lipid modifications within the CaaX box and at adjacent residues. The CaaX Cysteine is first subject to isoprenylation—farnesylation (C15) for H-, N- and K-Ras and geranylgeranylation (C20) for R-Ras (K-Ras can also be geranylgeranylated)—followed by proteolytic cleavage removing the aaX sequence and carboxymethylation of the isoprenylated terminal Cysteine. These modifications support interactions of the Ras proteins with ER membranes.22 The HVRs in H-, N- and R-Ras also contain target sites for secondary, reversible acylation, namely S-palmitoylation, whereby palmitate (C16:0) is linked by palmitoyl transferases to an adjacent Cysteine by a thioester linkage (C181 and 184 in H-Ras, C181 in N-Ras and C213 in R-Ras). Palmitoylation of H-, N- and R-Ras occurs at the Golgi, and in the cases of H- and N-Ras, this modification is necessary for sorting into secretory vesicles and subsequent trafficking to the plasma membrane, and depalmitoylation at the plasma membrane completes the cycle by driving H- and N-Ras retrograde recycling to the Golgi.23-27
Palmitoylated H- and N-Ras traffic to the plasma membrane via vesicles which include recycling endosomes (RE), a specialized class of sorting vesicles which facilitate both slow and rapid trafficking and recycling of many proteins including growth factor receptors, integrins and signaling molecules such as small GTPases Rab11 and Arf6.17,28,29 R-Ras has also been localized to RE,30 suggesting a common mode of regulation of trafficking of some Ras proteins by palmitoylation. RE play essential roles in cell spreading and migration, delivering both membrane components, including lipid rafts, as well as signaling proteins to the plasma membrane.28,31-34 Thus, R-Ras trafficking in recycling endosomes may relate to its roles in cell spreading and migration, but this connection has not been well established. However, R-Ras has also been observed in other vesicular compartments, based on co-localization studies with the RE marker Rab11a, which is classically associated with slow recycling pathways.1,35 In this study we investigated the trafficking dynamics of R-Ras in living cells, the contributions of post-translational lipid modifications and downstream effects on cell spreading.
Results
R-Ras traffics to membrane ruffles in recycling vesicles
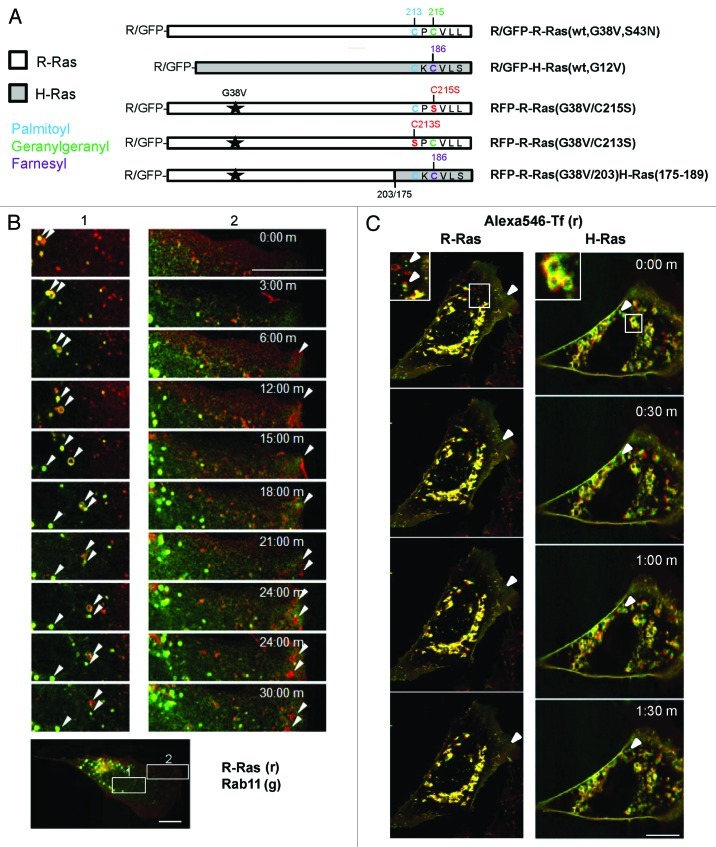
We used N-terminal green (GFP) or red fluorescent protein (RFP) fusions of R-Ras and H-Ras small GTPases (Fig. 1A) coupled with time-lapse confocal microscopy in NIH 3T3 cells to investigate the trafficking dynamics of these proteins in living cells. RFP-R-Ras(G38V), a constitutively active variant of R-Ras,36 was associated with membranes of recycling endosomes (RE) as indicated by co-localization with GFP-Rab11, a RE marker (Fig. 1B), consistent with earlier reports.1,30 Wild type (wt) R-Ras as well as H-Ras also partially co-localized in vesicles with internalized transferrin (Tf), confirming RE targeting for both R-Ras and H-Ras in these cells (Fig. 1C). The R-Ras-containing RE trafficked in anterograde fashion toward the leading edge of migrating cells; however, R-Ras was also in Tf- and Rab11-negative vesicles that also trafficked toward the leading edge (Fig. 1B and C; Vid. S1). R-Ras at the leading edge was enriched in membrane ruffles, as previously described,1 and recycled in retrograde fashion from ruffles into smaller vesicles. However, R-Ras recycling in the ruffles and retrograde vesicles was not associated with Rab11 in our cells (Fig. 1B; Vid. S1). Dominant negative R-Ras(S43N), which cannot bind guanine nucleotide, was localized in puncta dispersed throughout the cell and was completely absent from the plasma membrane (Fig. S1A), as previously observed1; moreover, puncta containing this inactive R-Ras mutant oscillated rapidly in the cytoplasm but did not engage in any apparent directed trafficking (Vid. S2).Thus, active R-Ras traffics to membrane ruffles in part via recycling endosomes and recycles from membrane ruffles at the leading edge via Rab11- and Tf-negative vesicle trafficking.
Figure 1. R-Ras traffics to membrane ruffles in recycling vesicles. (A) R-Ras and H-Ras constructs used in this study, expressed as red or green fluorescent protein (RFP or GFP) fusions. G38V and G12V (starred) are constitutively active mutants of R-Ras and H-Ras, respectively. HVR C-terminal sequences are shown with post-translational modification sites indicated: palmitoyl, blue; geranylgeranyl, green; farnesyl, purple; mutations are shown in red. R-Ras(G38V/203)H-Ras(175–189) is activated R-Ras, residues 1–203, deleted in the HVR and replaced with the farnesyl-specific HVR of H-Ras. R-Ras(S43N) is constitutively inactive. (B) R-Ras and Rab11 trafficking in live, spread cells. RFP-R-Ras(G38V) (red) and GFP-Rab11 (green) were tracked in live NIH 3T3 cells by confocal microscopy. Images were acquired every 30 sec to facilitate vesicle tracking; representative images are shown. Left panels (1) show R-Ras anterograde transport with recycling endosomes. R-Ras transport vesicles included Rab11-containing RE and Rab11-negative puncta. Right panels (2) show retrograde R-Ras transport from membrane ruffles that do not contain Rab11. Arrowheads point to individual R-Ras/Rab11 vesicles tracked across images. The lower panel shows a representative image of the whole cell used for imaging, at lower magnification, with zones 1 and 2 indicated by white boxes. (C) R-Ras and H-Ras (wt) partially traffic in transferrin (Tf)-containing RE. Cells expressing GFP-R-Ras or -H-Ras (green) were labeled with Alexa546-Tf (red) by a pulse-chase scheme (see Materials and Methods) and viewed by live confocal microscopy with images acquired every 30 sec. Areas of co-localization are seen as yellow. Inset areas in white boxes are shown at 2x. Arrowheads point to Tf-negative R-Ras or H-Ras vesicles. Bars, 10 μm.
Endogenous R-Ras and H-Ras partially co-localized with GFP-Rab11 and with GFP-GM130, a cis-Golgi marker,37 as indicated by antibody labeling in fixed cells, but the Ras GTPases were primarily in Rab11-negative tubulo-vesicular structures (Fig. S1B). These structures appeared as a partially connected network and did not resemble the discreet vesicular patterns of fluorescent-tagged Ras or Rab11 in living cells; furthermore vesicles appeared as puncta such that lumens could not be clearly distinguished in this case, suggesting morphological alterations of these structures as a result of fixation. R-Ras has also been reported to be enriched at focal adhesion sites; however, we did not observe characteristic focal adhesion patterns in these cells with RFP-R-Ras, which furthermore did not colocaize with GFP-paxillin, a focal adhesion marker38 (Fig. S1C).
R-Ras and H-Ras segregate into distinct vesicles near the leading edge in a HVR-dependent manner
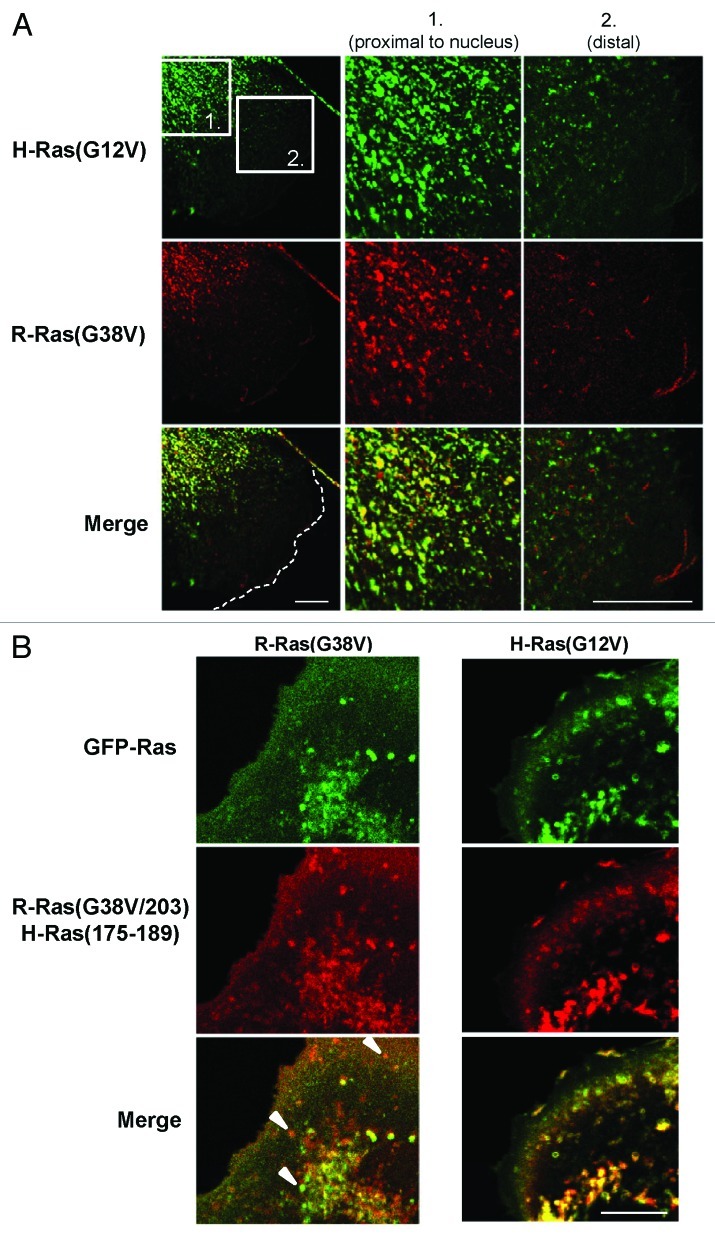
R-Ras is associated with lamellipodia formation, membrane ruffling, cell spreading and cell migration, in a manner which appears to be distinct from H-Ras.1,5-7,15 However, both of these proteins localize to RE, have palmitoylation target sites and share nearly identical effector binding domains.1,25,30,39 We compared the vesicle trafficking patterns of these GTPases by co-expression of GFP and RFP fusions of wild type or mutant H-Ras and R-Ras. As an initial indication of the contributions of vesicle trafficking to the distinct functions of R-Ras and H-Ras, we co-expressed constitutively active GFP-H-Ras(G12V) with RFP-R-Ras(G38V) in NIH3T3 cells and observed the localization of these proteins in spread, fixed cells. As before, vesicular staining in these fixed cells appeared more punctal than in live cells, in which vesicle membrane localization can be more clearly observed. H-Ras and R-Ras co-localized in vesicles proximal to the nucleus and Golgi and also localized in distinct puncta in this region. However, in distal regions of the cell closer to the leading edge, H-Ras and R-Ras segregated completely into distinct vesicle populations (Fig. 2A).
Figure 2. R-Ras and H-Ras segregate into distinct vesicles near the leading edge in a HVR-dependent manner. (A) Cells co-transfected with GFP-H-Ras(G12V) and RFP-R-Ras(G38V) were seeded at low density to allow for random migration, then fixed and imaged by confocal microscopy; merged images are shown at the bottom. The two boxed zones are shown at high magnification. H-Ras and R-Ras co-localized in vesicles proximal to the nucleus (1), but occupied distinct compartments (2) closer to the leading edge of the cell (dotted white line). (B) GFP-R-Ras(G38V) (green) and RFP-R-Ras(G38V/203)H-Ras(175–189) were co-expressed and imaged in live, randomly migrating cells. Merged images are shown at the bottom. White arrowheads point to selected vesicles containing either R-Ras(G38V) or R-Ras(G38V/203)/H-Ras(175–189). Bars, 7.5 μm.
We considered that the segregation of these GTPases into different vesicle populations may have resulted from the divergent membrane targeting domains in the HVRs, which have been shown to be responsible for segregation of these proteins into distinct microdomains at the plasma membrane.40 To test this possibility, we co-expressed a variant of activated R-Ras fused to RFP, R-Ras(G38V/203)/H-Ras(175–189), in which the HVR of R-Ras has been replaced by that of H-Ras41 (Fig. 1A). This mutation is predicted to substitute a geranylgeranylation target site from R-Ras with a farnesylation target site (from H-Ras), while maintaining a palmitoylation site that is common to both. This R-Ras mutant demonstrated H-Ras-like sorting behavior, by which it co-localized with R-Ras only in Golgi-proximal vesicles but segregated from R-Ras near the leading edge [Fig. 2B and Vid. S3; calculated Pearson’s correlation coefficient for R-Ras(G38V/203)/H-Ras(175–189) and R-Ras(G38V) in whole cells, 0.56 ± 0.11]. In contrast, this R-Ras variant co-localized substantially with H-Ras throughout the cell (Fig. 2B; Pearson’s correlation coefficient, 0.77 ± 0.04), and trafficked in vesicles which mostly lacked Rab11 and was only in small membrane ruffles (Vid. S4). Thus, R-Ras and H-Ras segregate into distinct trafficking vesicles toward the leading edge of migrating cells, and the vesicular sorting depends on the HVR sequences.
R-Ras trafficking to ruffles requires the R-Ras HVR and is distinct from H-Ras trafficking
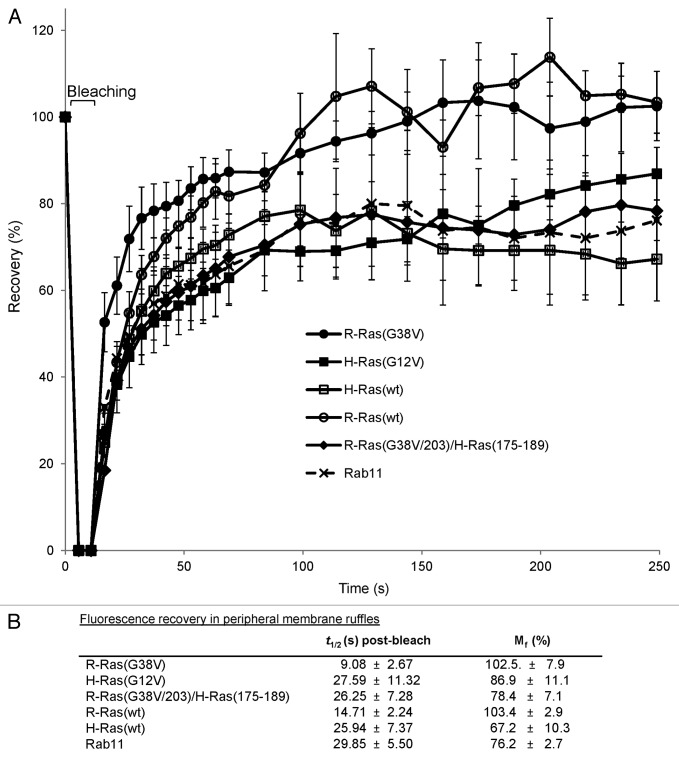
To further understand R-Ras/H-Ras vesicular trafficking toward the leading edge in migrating cells, we measured fluorescence recovery after photobleaching (FRAP) of GFP-R-Ras, -H-Ras and -Rab11 in membrane ruffles at the leading edge in migrating cells. Fluorescence recovery curves showed the expected exponential trend. Interestingly, R-Ras(G38V) fluorescence in ruffles recovered after photobleaching at a faster rate than that of H-Ras(G12V), and bleached fluorescence in membrane ruffles was completely restored for R-Ras but only partially restored for H-Ras in the 250 sec experimental timeframe (Fig. 3;Fig. S2). Furthermore, R-Ras fluorescence recovery was faster than that of Rab11, which more closely resembled the slower trafficking of H-Ras. The slower recovery of H-Ras was primarily due to the farnesyl-specific HVR, as R-Ras(G38V/203)H-Ras(175–189) recovery was much slower, with a lower mobile fraction, than for R-Ras(G38V) and instead resembled the H-Ras recovery profile (Fig. 3). Wild type R-Ras and H-Ras exhibited similar subcellular distributions and FRAP profiles as their constitutively active counterparts. Initial recovery for R-Ras(wt) was slightly slower than for R-Ras(G38V), but in both cases fluorescence completely recovered in ruffles. However, both R-Ras(wt) and H-Ras(wt) showed faster turnover in broad membrane ruffles than the constitutively active mutants, indicated by periodic fluctuations in FRAP of wild type R-Ras and H-Ras in the ruffle zone (Fig. 3). Thus, R-Ras and H-Ras engage in distinct anterograde trafficking patterns toward membrane ruffles at the leading edge in randomly migrating cells, with faster R-Ras trafficking dependent on the R-Ras HVR.
Figure 3. Fast R-Ras trafficking to ruffles requires the R-Ras HVR, and is distinct from slow H-Ras trafficking. (A) Fluorescence recovery after photobleaching (FRAP) was assessed in a ~100 μm2 boxed region at the leading edge of migrating cells expressing GFP-R-Ras, -H-Ras or -Rab11 fusions as indicated. GFP fluorescence intensities were measured for the selected area before (set to a normalized value of 100%) and after bleaching at 5 sec for the first minute, followed by 10 sec intervals for 3 min. Intensities were normalized with an adjacent, non-bleached zone, and are shown as percent recovery ± SD. At least 15 cells per type were measured; representative of three independent experiments. (B) Time of recovery (t1/2) and mobile fraction (Mf) for fluorescence in leading edge membrane ruffles are shown ± S.D, calculated from exponential curve fits with a minimum R2 of 0.95.
R-Ras palmitoylation is required for vesicular localization and for vesicle-mediated R-Ras trafficking to the leading edge
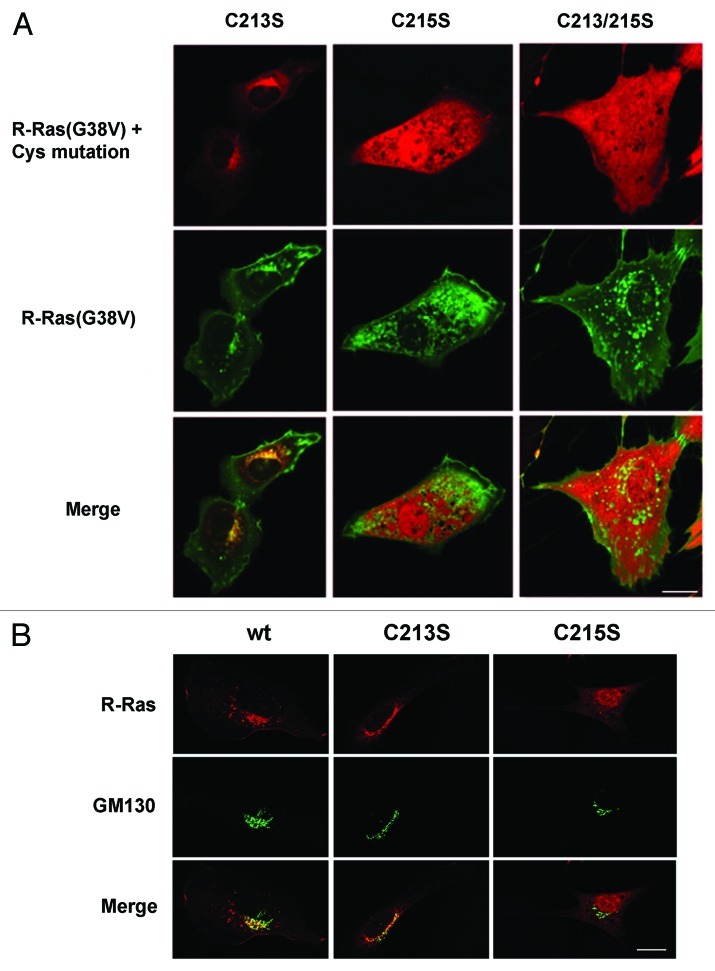
To determine the contributions of lipid modifications to R-Ras localization and trafficking, we mutated each of the two lipid modification sites in the R-Ras HVR (see Fig. 1A). A C215S mutation destroys the geranylgeranylation targeting site, and has been shown by subcellular fractionation techniques to inhibit membrane association.40 Indeed, this variant of constitutively active R-Ras (fused to RFP) did not co-localize with co-expressed R-Ras(G38V), but was diffuse in the cytosol and did not appear to be associated with membranes. The C215S R-Ras variant also localized to the cell nucleus (Fig. 4A). In contrast, RFP-R-Ras(G38V/C213S), mutated at its single palmitoylation target site, appeared associated with Golgi membranes as previously shown,42 where it co-localized with R-Ras(G38V), but was absent from cytosolic vesicles and the plasma membrane. R-Ras(G38V) harboring C213S/C215S double mutation, which ablates both modification sites, was diffuse in the cytosol and in the nucleus, and did not co-localize with R-Ras(G38V) (Fig. 4A). To confirm that the perinuclear sequestration of the C213S mutant reflected Golgi localization, we co-expressed the R-Ras variants with GFP-GM130. Whereas R-Ras co-localized with GM130 and was also present in vesicles and at the plasma membrane, R-Ras harboring the C213S mutation co-localized almost entirely with GM130, confirming Golgi sequestration of this R-Ras mutant (Fig. 4B). The C215S mutant was diffuse in the cytosol and nucleus and did not localize to the Golgi (Fig. 4A and B). Thus, geranylgeranylation of R-Ras is required for R-Ras membrane anchoring and sequestration from the nucleus, whereas palmitoylation of R-Ras is specifically required for exit from the Golgi apparatus into post-Golgi vesicles.
Figure 4. R-Ras palmitoylation is required for localization in vesicles and at the leading edge. (A) Cells co-expressing GFP-R-Ras(G38V) (green) with RFP-R-Ras(G38V) (red) harboring C-terminal site mutations were seeded in chamber slides and imaged live by confocal microscopy. Merged images are shown at the bottom. (B) RFP-R-Ras variants (red) co-expressed with Golgi marker GFP-GM130 (green). R-Ras(wt) partially localized to the Golgi (overlap seen as yellow). The C213S mutant was sequestered in the Golgi, whereas Golgi association was blocked by C215S mutation. Bars, 10 μm.
H-Ras vesicular targeting does not require R-Ras trafficking
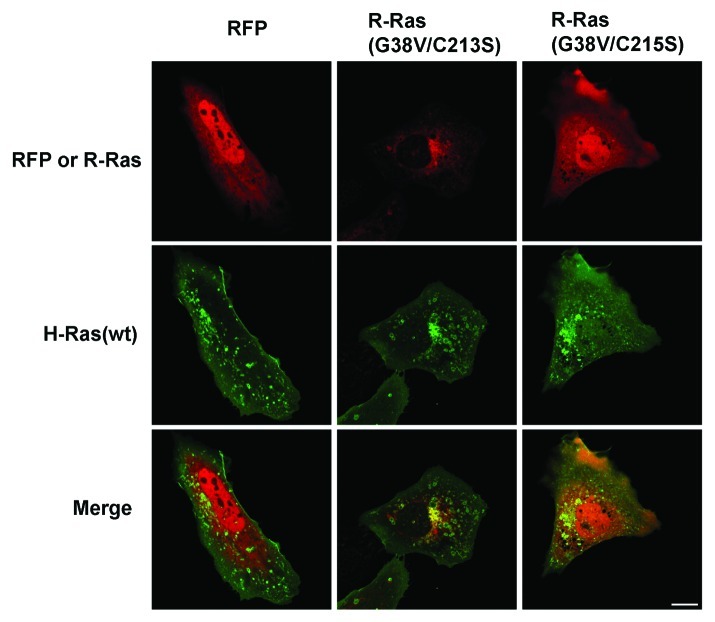
Since both H-Ras and R-Ras traffic from the Golgi into RE, and depalmitoylated R-Ras is sequestered in the Golgi, we considered whether H-Ras anterograde trafficking from the Golgi requires R-Ras. H-Ras(wt) was in perinuclear compartments, presumably Golgi, as well as in vesicles, in the presence of co-expressed R-Ras(G38V/C213S) or R-Ras(G38V/C215S) (Fig. 5). Thus, H-Ras vesicular targeting was not blocked by these R-Ras mutants, indicating that H-Ras is able to localize to trafficking vesicles in cells in which R-Ras is sequestered in the Golgi or displaced from membranes. However, H-Ras was not observed at the plasma membrane in these cells, compared with H-Ras in dorsal ruffles in cells expressing RFP alone. These results suggested that R-Ras mistargeting yields dominant effects on membrane ruffling.
Figure 5. H-Ras vesicular targeting does not require R-Ras trafficking. RFP alone or fused to R-Ras variants as indicated (red) was co-expressed with GFP-H-Ras(wt) (green) and live cells were imaged by confocal microscopy. R-Ras Cysteine mutations altered R-Ras localization and blocked membrane ruffling but did not prevent H-Ras vesicular targeting. Bar, 10 μm.
R-Ras lipid modification is required for membrane ruffling and for R-Ras enhancement of cell spreading
We next investigated the ruffling and spreading capacities of cells transiently transfected with RFP fusions of R-Ras harboring mutations or substitution of the lipid modification sites. Whereas R-Ras(G38V) promoted enhanced membrane ruffling and cell spreading over control as expected,6 activated R-Ras mutated at its palmitoylation or geranylgeranylation site, or at both sites, blocked membrane ruffling (Fig. 6A) and was unable to stimulate spreading (Fig. 6B). R-Ras containing the H-Ras C-terminal domain was still able to support ruffling and spreading, consistent with the ability of this R-Ras variant to traffic in vesicles to membrane ruffles (see also Figs. 2B and 3). Dominant negative R-Ras(S43N) or R-Ras knockdown with a short hairpin RNA (shRNA) each inhibited spreading (and hence, membrane ruffling), confirming the previously noted requirement for R-Ras activity in spreading.6 Thus, palmitoylation and geranylgeranylation at C213 and C215 are both necessary for membrane ruffling and stimulation of cell spreading by activated R-Ras.

Figure 6. Cell spreading and ruffling regulated by R-Ras lipid modification. Cells were transiently transfected with RFP vector alone (RFP), RFP-R-Ras fusions or RFP plus R-Ras shRNA as indicated. After 24 h, cells were seeded onto fibronectin-coated surfaces and membrane ruffling (A) and cell spreading (B) in RFP-positive cells were determined as described in Materials and Methods. (A) Percentages of transfected cells displaying edge ruffles for at least 100 cells per sample are shown + SEM *, p < 0.001. (B) Average cell areas for at least 100 cells per sample are shown + SEM *, p < 0.05; **, p < 0.004; ***, p < 0.003. Data are from three independent experiments each.
R-Ras vesicles are PI(3,4,5)P3-negative, whereas R-Ras co-localizes with PI(3,4,5)P3 in membrane ruffles from which R-Ras is recycled
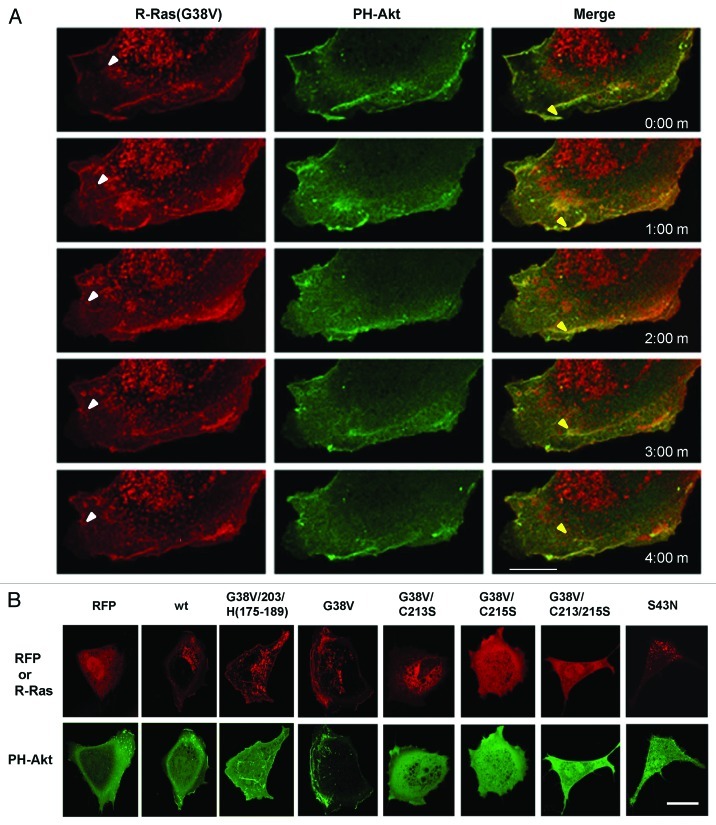
The rapid vesicular trafficking of R-Ras relative to H-Ras to the leading edge plasma membrane and ruffles suggested that delivery of these Ras isotypes to the plasma membrane could localize Ras-specific signaling to this region to regulate cell spreading and migration. We co-transfected cells with RFP-R-Ras(G38V) and GFP fused to the pleckstrin homology domain of the kinase Akt (GFP-PH-Akt), a marker for PI(3,4,5)P3 [PtdIns(3,4,5)P3],43 and hence, a marker for activity of phosphatidylinositol 3-kinase (PI3K), an effector of R-Ras and H-Ras. The dynamics of these proteins in migrating cells were then monitored using time-lapse confocal microscopy. Interestingly, R-Ras co-localized with PH-Akt in the membrane ruffles at the leading edge of the cell, but PH-Akt was absent from R-Ras vesicles trafficking anterograde toward the leading edge (Fig. 7A). PH-Akt remained in ruffles co-localized with R-Ras as the ruffles moved retrograde. In some instances R-Ras-containing vesicles emerged from the ruffles, and these retrograde R-Ras vesicles did not contain PH-Akt (Fig. 7A; Vids. S5A–C).
Figure 7. R-Ras vesicles are PI(3,4,5)P3-negative, whereas R-Ras co-localizes with PI(3,4,5)P3 in membrane ruffles from which R-Ras is recycled. RFP-R-Ras (red) was co-transfected with GFP-PH-Akt (green), as a marker for PI(3,4,5)P3. (A) Dynamics of R-Ras(G38V) and PH-Akt monitored in a migrating cell. Images were acquired every 30 sec; 1 min intervals are shown. R-Ras was in perinuclear vesicles which trafficked toward the plasma membrane (e.g., white arrowheads), and in retrograde membrane ruffles. In contrast, PH-Akt was restricted to the retrograde ruffles, where it co-localized with R-Ras. Both proteins moved in retrograde fashion in the ruffles. In some cases R-Ras recycled from ruffles through vesicular structures (e.g., yellow arrowheads) that lacked PH-Akt. (B) RFP or RFP-R-Ras fusions as indicated, co-expressed with GFP-PH-Akt and imaged in live cells by confocal microscopy. Bars, 7.5 μm.
GFP-PH-Akt was present in some small ruffles in cells expressing RFP alone (Fig. 7B). Cells co-expressing RFP-R-Ras(wt) had broader ruffles than with RFP only, and R-Ras(wt) co-localized with PH-Akt in a manner similar to R-Ras(G38V). In cells expressing R-Ras containing the H-Ras targeting domain, PH-Akt was evident in internal ruffles; however, little if any PH-Akt was present in dorsal membrane ruffles at the cell periphery (Fig. 7B). Interestingly, R-Ras(G38V) mutated at its palmitoylation site (C213S), which sequestered activated R-Ras in the Golgi and prevented ruffling (see Figs. 4 and 6), blocked localization of PH-Akt at the plasma membrane. Geranylgeranyl-deficient R-Ras(G38V/C215S), sequestered in the cytosol, and double mutant C213,215S had similar effects, as did dominant negative R-Ras(S43N) (Fig. 7B).
PtdIns(3,4,5)P3 in ruffles and R-Ras/PI3K interaction require R-Ras palmitoylation and trafficking
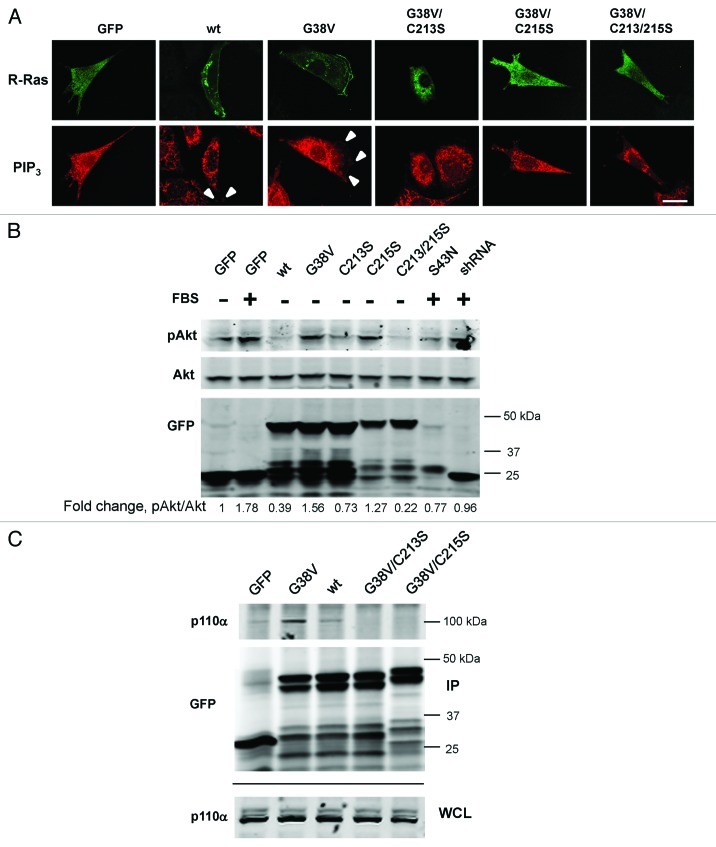
The above results indicated a spatiotemporal correlation of R-Ras anterograde trafficking to the leading edge with PtdIns(3,4,5)P3 generation in membrane ruffles. Combined with our finding that R-Ras palmitoylation is required for its trafficking in vesicles and enrichment in membrane ruffles, we considered whether R-Ras palmitoylation is involved in PI3K activity in ruffles, resulting in the localized production of PtdIns(3,4,5)P3 phospholipids. To investigate this possibility further, we stained fixed cells, transfected with R-Ras variants, using antibodies to PtdIns(3,4,5)P3. The antibody staining patterns revealed PtdIns(3,4,5)P3 localization in vesicular patterns that were not observed with GFP-PH-Akt, indicating potential limitations of the ability of GFP-PH-Akt to label intracellular PtdIns(3,4,5)P3. These results further demonstrate that R-Ras lipid modifications are not required for PtdIns(3,4,5)P3 generation per se. However, PtdIns(3,4,5)P3 vesicles did not co-localize with R-Ras (Fig. 8A). Moreover, we observed PtdIns(3,4,5)P3 staining in peripheral membrane ruffles co-localized with R-Ras(wt and G38V), but we did not detect PtdIns(3,4,5)P3 staining in cells expressing other R-Ras variants. GFP-positive cells co-transfected with R-Ras shRNA were rounded and did not spread (see Fig. 6); hence, these cells also did not ruffle and it was not possible to discern precisely the localization of GFP-PH-Akt or PtdIns(3,4,5)P3 in these cells (results not shown).
Figure 8. PtdIns(3,4,5)P3 in membrane ruffles requires R-Ras lipid modification. (A) GFP-R-Ras fusions as indicated were imaged along with PtdIns(3,4,5)P3 antibody staining in fixed cells. White arrowheads point to PtdIns(3,4,5)P3 in ruffles, co-localized with R-Ras. Bar, 7.5 μm. (B) Cells transfected with the indicated GFP-R-Ras fusions were serum-starved (0.5%) for 2 d and either fed with serum (10%) for 30 min (+ FBS) or kept in starvation medium (-) before being lysed. Lysates were subjected to western blotting with antibodies to GFP, total Akt (Akt) or Akt phosphorylated at Ser473 (pAkt). Densitometric ratios of pAkt:Akt are shown as fold change relative to the starved cell ratio. Representative of three independent experiments. (C) GFP or GFP-R-Ras fusions as indicated were transfected, then immunoprecipitated from cell extracts using GFP antibodies. GFP and R-Ras fusions, and endogenous p110α subunit of PI3K were detected by immunoblotting the immunoprecipitate (IP) or whole cell lysate (WCL) fractions using GFP and p110α-specific antibodies. Representative of four independent experiments.
Next we assessed the ability of R-Ras to stimulate PI3K in serum-starved cells, indicated by phosphorylation of Akt (at Ser473). R-Ras(wt) was a poor stimulator of Akt phosphorylation; however, expression of R-Ras(G38V) induced Akt phosphorylation in starved cells (Fig. 8B). Blockade of R-Ras with dominant negative (S43N) or shRNA partially inhibited serum-induced Akt phosphorylation, demonstrating an important role for R-Ras activation in this process. This stimulatory effect of activated R-Ras was blocked by mutation of the palmitoylation site (C213S), although surprisingly the C215S mutant was able to stimulate Akt phosphorylation in this system. However, the double Cys mutant was not, indicating a selective requirement of R-Ras palmitoylation for inducing Akt phosphorylation. Similarly, R-Ras(G38V) interacted strongly [compared with weaker interaction of R-Ras(wt)] with the p110(α) subunit of PI3K, which is responsible for PtdIns(3,4,5)P3 generation, whereas either Cys mutation (213S or 215S) blocked this interaction (Fig. 8C). Thus, R-Ras interaction with PI3K leading to recruitment of GFP-PH-Akt and localization of PtdIns(3,4,5)P3 at leading edge membrane ruffles requires R-Ras activation and palmitoylation, and R-Ras mutated in the C-terminal lipid modification sites yields dominant negative effects on PtdIns(3,4,5)P3 localization and on ruffle formation.
Discussion
The results from this study describe a mechanism of R-Ras intracellular trafficking and its effects on localized signaling, membrane ruffling and cell spreading. Activated R-Ras associated with Rab11- and Tf-positive and -negative vesicle membranes and trafficked in anterograde fashion, delivering R-Ras into membrane ruffles in migrating cells. Trafficking of R-Ras into ruffles was rapid compared with H-Ras, and R-Ras and H-Ras segregated into distinct vesicle populations approaching the leading edge in a manner dependent on the Ras HVR. Palmitoylation and geranylgeranylation of activated R-Ras were both required for anterograde trafficking, membrane ruffling and cell spreading. Anterograde R-Ras vesicle membranes did not contain PtdIns(3,4,5)P3, whereas activated R-Ras co-localized with PtdIns(3,4,5)P3 in membrane ruffles at the leading edge. Blockade of R-Ras palmitoylation blocked PI3K association and PtdIns(3,4,5)P3 generation at the plasma membrane. Thus, lipid modification of R-Ras, in particular palmitoylation, drives anterograde R-Ras vesicular trafficking leading to localized PtdIns(3,4,5)P3 generation and membrane ruffling at the leading edge, and to cell spreading.
GFP and RFP N-terminal fusions of activated R-Ras co-localized in vesicles with two recycling endosome (RE) markers, transferrin (Tf) and Rab11, and R-Ras was also in Rab11- and Tf-negative vesicles, as previously noted.1,30 These studies described endocytosis of R-Ras from membrane ruffles through Rab11-positive and -negative vesicles; however in our cells these retrograde R-Ras vesicles did not contain Rab11 or Tf. In this study we extended these initial observations to track anterograde R-Ras trafficking from intracellular compartments to membrane ruffles at the plasma membrane in living cells, and we found that R-Ras anterograde trafficking to ruffles also occurred in both Rab11- and Tf-positive and -negative vesicles. This divergence may reflect dynamic sorting within the RE, from which slow trafficking Rab11-dependent vesicles and faster Rab4-dependent vesicles can emerge.35,44 Trafficking driven by both of these Rab GTPases has been implicated in persistent cell migration.45-47 Dominant negative R-Ras was unable to traffic to the plasma membrane, consistent with earlier imaging studies in which inactive R-Ras was absent from the plasma membrane.1 Thus, R-Ras cycles via exocytic and endocytic vesicular trafficking pathways through membrane ruffles. R-Ras has also been observed in focal adhesions in HeLa cells;42 however, in our NIH 3T3 cells seeded on integrin substrates (fibronectin), this was not the case. This difference may be due to cell-type specific effects: R-Ras focal adhesion targeting in HeLa cells was blocked by cholesterol depletion, and membranes of these cells have significantly higher cholesterol:phospholipid ratios than NIH 3T3 cells.48 It is possible that a minimum threshold of membrane cholesterol content is required for focal adhesion targeting of R-Ras and that threshold is not reached in our cells.
Both R-Ras and H-Ras have been observed in RE by different groups,1,25,29,30,49,50 and we found that these two related small GTPases co-localized in perinuclear vesicles which are likely RE. However, proximal to the plasma membrane, R-Ras and H-Ras segregated independently into separate anterograde trafficking pathways. Segregation was dependent upon the different membrane anchors within these GTPases, as activated R-Ras in which the C-terminal membrane-targeting domain is replaced by that of H-Ras,41 co-localized with H-Ras in vesicular compartments throughout the cell, including proximal to the plasma membrane and in ruffles, and segregated from activated R-Ras along separate vesicular trafficking pathways. This R-Ras variant has previously been shown to adopt H-Ras-like plasma membrane microdomain distribution (e.g., displaced from lipid rafts41). Similarly, although both R-Ras and H-Ras were enriched in ruffles, targeting of these small GTPases to ruffles from intracellular compartments occurred on different time scales: R-Ras FRAP in ruffles was significantly faster than that of H-Ras, resulting in more complete recovery over the experimental time frame, even though in both cases recovery was driven by vesicle trafficking to the plasma membrane. Diminished recovery for H-Ras could result from slower anterograde trafficking of H-Ras vesicles (see, e.g., Vid. S3), although other effects such as longer retention times in the Golgi could not be ruled out. Again, replacing geranylgeranylation with farnesylation targeting domain switched R-Ras to H-Ras-like behavior. A switch mutant from farnesylation to geranylgeranylation in H-Ras has been shown to have no effect on retrograde H-Ras trafficking to the Golgi;26 our results suggest that the specific prenylation motifs in H-Ras and R-Ras instead may play an active role in anterograde trafficking.
We note that in our case recovery of H-Ras in ruffles was significantly slower than plasma membrane recovery observed in COS7 cells by Henis et al., using Gaussian-spot FRAP; we attribute these distinctions to known differences in FRAP kinetics using laser scanning confocal FRAP for a large area (~100 μm2, e.g., longer bleaching and recovery times compared with Gaussian-spot FRAP)51 and to differences between rates of lateral diffusion including membrane/cytoplasm exchange, and vesicle delivery to membrane ruffles. Thus, this protracted FRAP timescale in our case allowed us to analyze different rates of transport of Ras proteins into large subcellular structures, namely, membrane ruffles. Indeed, H-Ras fluorescence did recover quickly at the plasma membrane on a timescale matching the lateral diffusion seen previously—to a relatively lower proportion of pre-bleach fluorescence as the initial fluorescence in ruffles is substantially higher than in non-ruffling membranes—but specific recovery in ruffles, apparently as a result of vesicle trafficking, was delayed for H-Ras relative to R-Ras.
The unique trafficking patterns of activated R-Ras, driven by its C-terminal membrane-targeting domains, can be attributed to post-translational lipid modification of R-Ras dictated by the sequence in this domain. Mutation of the geranylgeranylation site (C215S) sequestered activated R-Ras in the cytosol, consistent with earlier biochemical fractionation studies,40 as well as in the nucleus, and this mutant was unable to traffic to the plasma membrane. Nuclear localization may have been due to the inability of this R-Ras variant to attach to extranuclear membranes, which might allow R-Ras to traffic into the nucleus, although the mechanism remains unclear. In contrast, mutation of the palmitoylation site (C213S) did not disrupt Golgi membrane anchorage of activated R-Ras but prevented its delivery from the Golgi into exocytic vesicles,52 in line with Golgi sequestration of a similar mutant (C213A);42 in our cells the palmitoylation mutant was also unable to traffic to ruffles at the plasma membrane. Palmitoylation-dependent trafficking is consistent with increased association of R-Ras with cholesterol/sphingomyelin-rich plasma membrane microdomains by overexpression of the palmitoyl transferase DHHC19, for which R-Ras is a substrate.53 The double Cysteine mutant was also cytosolic and in the nucleus, similar to the single CaaX box C215S mutant, possibly reflecting the general notion that blocking secondary isoprenylation, e.g., geranylgeranylation, for CaaX box proteins such as for the C215S single Cysteine mutant may inhibit palmitoylation as well.54 Together, these results indicate that geranylgeranylation of R-Ras is required for stable membrane anchorage in the cell, whereas palmitoylation at C213 specifies sorting into trafficking vesicles. Interestingly, blockade of H-Ras palmitoylation compromises its activity, possibly due to disruption of favorable conformational effects of palmitate-membrane interactions on H-Ras,55,56 whereas GTP-locked H-Ras undergoes depalmitoylation at a faster rate than inactive H-Ras in cells.57 It is conceivable that GTP-binding of R-Ras is similarly affected by palmitoylation. We note that H-Ras, as well as its close relative N-Ras, are palmitoylated in the Golgi, and are also associated with RE; palmitoylation of those GTPases is necessary for exit from the Golgi in post-Golgi secretory vesicles which associate with RE.25-27 Moreover, although single palmitoylation of H-Ras at C181 provides insufficient membrane anchorage for access to RE, addition of apolar Leucine residues such as a C184L mutation, mimicking a wild type L184 residue in N-Ras, allows for RE localization by enhancing interactions with the membrane.25,58,59 We note that the R-Ras CaaX sequence, CVLL, could potentially support a similar mechanism of enhanced membrane anchorage by monopalmitoylated R-Ras, although this remains to be tested. Our data support the notion that acylation—in the form of S-palmitoylation—combined with increased hydrophobicity due to a second palmitate (H-Ras) or the presence of adjacent Leu resides (N- and R-Ras) together may establish a common motif for H-, R- and N-Ras small GTPases for selective sorting into RE.
A functional role of rapid R-Ras vesicular trafficking to membrane ruffles appears to be to support R-Ras/PI3K interaction in cells and to stimulate the localized production of phosphatidylinositol (3,4,5)P3 [PtdIns(3,4,5)P3] from phosphatidylinositol (3,4)P2 [PtdIns(4,5)P2]. The ability of R-Ras mutated in the geranylgeranylation site to stimulate Akt phosphorylation despite being unable to interact with PI3K was unexpected and the mechanism is unclear. It is conceivable that this R-Ras variant can stimulate PI3K indirectly through unknown cofactors. Nonetheless, this R-Ras mutant did not support PtdIns(3,4,5)P3 or PH-Akt localization at peripheral membrane ruffles. Combined with the findings that either Cys mutant blocked ruffling and was unable to enhance spreading, these data support the notion that R-Ras trafficking and interaction with PI3K are required for R-Ras stimulation of ruffling and PtdIns(3,4,5)P3 localization leading to R-Ras-driven enhanced spreading; however, cells can spread in the absence of these R-Ras signals. PI3K is a major signaling effector of R-Ras; indeed, R-Ras and its paralog, TC21, can potently activate the p110δ isoform of PI3K, whereas H-Ras cannot.60 PI3K has long been associated with cell spreading via lamellipodia formation, and with consequent cell migration, and with membrane ruffling—for example, by stimulating activation of the small GTPase Rac1, which is also downstream of R-Ras.6,15,61-63 More directly, PtdIns(3,4,5)P3 is a hallmark of membrane protrusions induced by localized actin polymerization and may regulate ruffling; however this function may be cell type-specific.64-66 A role for R-Ras activity in membrane ruffling has been noted previously.1 Our studies indicate that R-Ras palmitoylation and its effects on R-Ras vesicular trafficking are responsible for this function of R-Ras, which is further correlated with localized stimulation of lipid phosphorylation in ruffles.
We propose that the ability of R-Ras to activate PI3K at the leading edge plasma membrane and generate membrane ruffles, driven by palmitoylation-dependent vesicular targeting of R-Ras to these regions, is a major mechanism of R-Ras-dependent cell spreading and migration. However, R-Ras has been correlated with cell spreading and migration through multiple pathways3,5-7,15,67; it will be interesting to investigate in more detail how this proposed mechanism contributes to this unique cellular function of the R-Ras small GTPase.
Materials and Methods
Antibodies and reagents
α-R-Ras, α-H-Ras, α-GFP and α-p110α antibodies were purchased from Santa Cruz Biotechnology. Mouse monoclonal α-myc tag antibodies (9E10) were from Millipore. Mouse monoclonal α-PtdIns(3,4,5)P3 antibodies were from MBL International. α-Akt and α-Akt(phosphoSer473) were from Invitrogen. AlexaFluor 546-conjugated transferrin was from Molecular Probes. Fluorophore-conjugated secondary antibodies were from Jackson Immunolabs or LICOR. Restriction endonucleases were obtained from New England Biolabs.
CDNAs
pEGFP-C1 was from Clontech Laboratories. pCS2-mRFP-N1 was a gift of Randall Moon (Addgene plasmid 17143). GFP-H-Ras(wt and G12V) were gifts of K. Svoboda (Addgene plasmids 18662 and 18666, respectively). pEGFP-Rab11a and GFP-GM130 were gifts of Eugene Tkatchenko (University of California San Diego). GFP-paxillin was a gift of Clare Waterman (NHLBI). GFP-PH-Akt was a gift of Eleni Tzima (University of North Carolina). RFP-R-Ras constructs were generated from pEF4-nTAP-R-Ras plasmids68 by subcloning into the pCS2-mRFP-N1 vector using BamH1/Xba1 restriction endonuclease sites. RFP-R-Ras(G38V/203)H-Ras(175–189) and RFP-R-Ras(G38V/C215S) were generated by subcloning from the original FLAG- and myc-tagged R-Ras constructs (gifts of Mark H. Ginsberg, University of California San Diego) into pCS2-mRFP-N1 using EcoR1 sites. RFP-R-Ras(G38V/C213S) was generated by PCR from the RFP-R-Ras(G38V) template using the following primer set: 5′-GGCGGGGGCAGCCCCTGCGTCCTCCTGTAG-3′ and 5′-CTACAGGAGGACGCAGGGGCTGCCCCCGCC-3′. RFP-R-Ras(G38V/C213,215S) was generated by PCR from the RFP-R-Ras(G38V/C215S) template using the following primer set: 5′-GGCGGGGGCAGCCCCAGCGTCCTCCTGTAG-3′ and 5′-CTACAGGAGGACGCTGGGGCTGCCCCCGCC-3′. GFP-R-Ras constructs were made by subcloning from the pCS2-mRFP-N1 vectors into pEGFP-C1 using BamH1/Xba1 restriction endonuclease sites. An R-Ras murine-specific shRNA targeting plasmid was generated in the pSUPER.retro.puro vector (OligoEngine) using the targeting sequence 5′-GCAAGCTCTTCACACAGAT-3′, according to the manufacturer’s instructions.
Cell culture and transfections
NIH 3T3 cells (American Type Culture Collection) were maintained in DME (Cellgro) supplemented with 10% Bovine Calf Serum (BCS), 2 mM L-glutamine, 50 U/mL penicillin, 50 μg/mL streptomycin sulfate, and 1% nonessential amino acids (Sigma-Aldrich) at 37°C in 5% CO2. Cells were transfected with 5–10 μg plasmids/100 mm dish using Lipofectamine 2000 in Opti-MEM (Invitrogen) according to the manufacturer’s instructions. Cells were analyzed 24–48 h after transfection.
Live and fixed cell immunofluorescence microscopy and fluorescence recovery after photobleaching (FRAP)
Glass coverslips or LabTek II 8-chambered microscope slides (Nunc) were incubated with 5 μg/mL plasma fibronectin in 0.1 M NaHCO3 at 4°C overnight; after washing, the coverslips or slides were incubated for 30 min with 1% BSA/PBS that had been heat inactivated by incubation at 80°C for 30 min. Cells were detached with 0.1% trypsin and kept in suspension for 1 h at RT in DME containing 0.2% BSA, and then plated on the coated coverslips for 45 min at 37°C. Non-adherent cells were removed by washing two times with PBS, and in some cases the adherent cells were fixed in 3.7% formaldehyde/PBS for 20 min at RT. Fixed cells were washed and permeabilized with 0.1% Triton X-100/PBS for 5 min, washed with PBS, and incubated with primary antibodies for 1 h at RT, washed, then incubated with fluorophore-conjugated secondary antibodies for 1 h at RT. Coverslips were washed and mounted on slides with ProLong Gold anti-fade reagent (Invitrogen). Alexa546-Transferrin (Al-Tf) recycling experiments were performed in live cells by addition of 100 μg/mL Al-Tf in culture media for 1 h at 4°C. Media was removed, cells were rinsed twice, then ice-cold media was added and the cells were transferred to 37°C. Imaging was started after 10–15 min. Cells in all experiments were imaged using a Leica DM IRE2 microscope with a TCS SL confocal system, using a 63x/1.40 n.a. oil immersion objective and Leica imaging software. Fluorophores used were FITC, Cy3 and Cy5 (Jackson Immunoresearch). Images were captured by line sequential scanning using 4x line averaging and 3x frame averaging at 400 Hz scan speed. For live cell imaging, chamber slides were fitted onto the same microscope stage inside an atmosphere-controlled chamber, maintained at 37°C and 5% CO2. Images were acquired as above using GFP and RFP fluorescence scanning. For FRAP assays, a pre-bleach image was acquired at 10% laser power, after which a selected area was bleached at 100% laser power with two successive bleach scans separated by the minimum scan time of 5 sec, assisted by the microscope software. Post-bleach recovery images were acquired every 5 sec for the first minute, followed by 10 sec intervals for 3 min. Images for these sequences were acquired at 200 Hz scan speed with no line or frame averaging. Intensities were normalized with an adjacent, non-bleached zone. Post-acquisition image processing was performed using ImageJ and Adobe Photoshop. Operations included brightness/contrast adjustment to all pixels in the images, manual tracking of objects across multiple frames, and grouping of images. Fluorescence intensity analysis was performed using Microsoft Excel and FRAP coefficients were calculated from exponential curve fits using Kaleidagraph (Synergy Software). Pearson’s correlation coefficients were calculated using Leica imaging software.
Cell spreading and ruffling
Cells were transiently transfected as above and cultured for 24 h, before being suspended with trypsin and seeded on fibronectin-coated coverslips for 45 min in atmospheric control, then fixed and mounted on slides as above. Coverslips were imaged on an epifluorescence microscope (Nikon E800) fitted with a SPOT charge-coupled device camera (Diagnostic Instruments). The area of RFP-positive cells was measured using ImageJ software.69 Membrane ruffling in RFP-positive cells was scored as percent ruffling cells per field across 5–10 fields per sample, by observers blinded to the identity of the samples.
Immunoprecipitations and western blotting
Cells were transiently transfected and in some cases serum-starved by culturing in DMEM/0.5% serum beginning 24 h after transfection. After 48 h, cell lysates were harvested by scraping in lysis buffer [10 mM Tris-Cl, pH 7.5, 100 mM NaCl, 2 mM MgOAc, 10 μM GTP, 0.5% NP40 plus a cocktail of protease inhibitors (Roche)] and maintained at 4°C. Insoluble material was removed by centrifugation. Fractions of the lysates were separated by SDS-PAGE, followed by western blotting with appropriate antibodies. For immunoprecipitations, supernatants were pre-cleared by incubation with Protein G-coupled sepharose beads (Roche) for 1 h at 4°C. Cleared lysates were incubated with 2 μg of the appropriate antibodies for 16 h at 4°C, followed by antibody capture on protein G-sepharose beads for 1 h. Antibody-bound complexes were precipitated by centrifugation, washed, and separated by SDS-PAGE, followed by western blotting with relevant antibodies.
All experiments in this study were performed at least in triplicate, except where indicated otherwise, and for microscopy results representative images are shown. One-way ANOVA followed by Fisher PLSD analysis was used for all statistical data analysis, using StatView (SAS). A 5% probability was considered significant.
Supplementary Material
Acknowledgments
We thank Bettina Buttaro and Patrick Piggot for assistance with live cell confocal microscopy, and Ankita Patel for technical assistance. This work was supported by NIH grant HL093416 to L.G.
Glossary
Abbreviations:
- PIP3
phosphatidylinositol (3,4,5)-trisphosphate
- PIP2
phosphatidylinositol (4,5)-bisphosphate
- PI3K
phosphatidylinositol 3-kinase
- PLCε
phospholipase Cε
- HVR
hypervariable region
- ER
endoplasmic reticulum
- RE
recycling endosome(s)
- PH
plecktrin homology
- FRAP
fluorescence recovery after photobleaching
- GFP
green fluorescent protein
- RFP
(monomeric) red fluorescent protein
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental material may be found here: www.landesbioscience.com/journals/smallgtpases/article/21084
Footnotes
Previously published online: www.landesbioscience.com/journals/smallgtpases/article/21084
References
- 1.Conklin MW, Ada-Nguema A, Parsons M, Riching KM, Keely PJ. R-Ras regulates beta1-integrin trafficking via effects on membrane ruffling and endocytosis. BMC Cell Biol. 2010;11:14. doi: 10.1186/1471-2121-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sethi T, Ginsberg MH, Downward J, Hughes PE. The small GTP-binding protein R-Ras can influence integrin activation by antagonizing a Ras/Raf-initiated integrin suppression pathway. Mol Biol Cell. 1999;10:1799–809. doi: 10.1091/mbc.10.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwong L, Wozniak MA, Collins AS, Wilson SD, Keely PJ. R-Ras promotes focal adhesion formation through focal adhesion kinase and p130(Cas) by a novel mechanism that differs from integrins. Mol Cell Biol. 2003;23:933–49. doi: 10.1128/MCB.23.3.933-949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole AL, Subbanagounder G, Mukhopadhyay S, Berliner JA, Vora DK. Oxidized phospholipid-induced endothelial cell/monocyte interaction is mediated by a cAMP-dependent R-Ras/PI3-kinase pathway. Arterioscler Thromb Vasc Biol. 2003;23:1384–90. doi: 10.1161/01.ATV.0000081215.45714.71. [DOI] [PubMed] [Google Scholar]
- 5.Holly SP, Larson MK, Parise LV. The unique N-terminus of R-ras is required for Rac activation and precise regulation of cell migration. Mol Biol Cell. 2005;16:2458–69. doi: 10.1091/mbc.E03-12-0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldfinger LE, Ptak C, Jeffery ED, Shabanowitz J, Hunt DF, Ginsberg MH. RLIP76 (RalBP1) is an R-Ras effector that mediates adhesion-dependent Rac activation and cell migration. J Cell Biol. 2006;174:877–88. doi: 10.1083/jcb.200603111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ada-Nguema AS, Xenias H, Hofman JM, Wiggins CH, Sheetz MP, Keely PJ. The small GTPase R-Ras regulates organization of actin and drives membrane protrusions through the activity of PLCepsilon. J Cell Sci. 2006;119:1307–19. doi: 10.1242/jcs.02835. [DOI] [PubMed] [Google Scholar]
- 8.Huang Y, Rangwala F, Fulkerson PC, Ling B, Reed E, Cox AD, et al. Role of TC21/R-Ras2 in enhanced migration of neurofibromin-deficient Schwann cells. Oncogene. 2004;23:368–78. doi: 10.1038/sj.onc.1207075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–9. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 10.Charest PG, Firtel RA. Big roles for small GTPases in the control of directed cell movement. Biochem J. 2007;401:377–90. doi: 10.1042/BJ20061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myers KR, Casanova JE. Regulation of actin cytoskeleton dynamics by Arf-family GTPases. Trends Cell Biol. 2008;18:184–92. doi: 10.1016/j.tcb.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, Vuori K, Wang HG, Reed JC, Ruoslahti E. Integrin activation by R-ras. Cell. 1996;85:61–9. doi: 10.1016/S0092-8674(00)81082-X. [DOI] [PubMed] [Google Scholar]
- 13.Keely PJ, Rusyn EV, Cox AD, Parise LV. R-Ras signals through specific integrin alpha cytoplasmic domains to promote migration and invasion of breast epithelial cells. J Cell Biol. 1999;145:1077–88. doi: 10.1083/jcb.145.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berrier AL, Mastrangelo AM, Downward J, Ginsberg M, LaFlamme SE. Activated R-ras, Rac1, PI 3-kinase and PKCepsilon can each restore cell spreading inhibited by isolated integrin beta1 cytoplasmic domains. J Cell Biol. 2000;151:1549–60. doi: 10.1083/jcb.151.7.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wozniak MA, Kwong L, Chodniewicz D, Klemke RL, Keely PJ. R-Ras controls membrane protrusion and cell migration through the spatial regulation of Rac and Rho. Mol Biol Cell. 2005;16:84–96. doi: 10.1091/mbc.E04-04-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fletcher SJ, Rappoport JZ. Moving forward: polarised trafficking in cell migration. Trends Cell Biol. 2010;20:71–8. doi: 10.1016/j.tcb.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Santiago-Tirado FH, Bretscher A. Membrane-trafficking sorting hubs: cooperation between PI4P and small GTPases at the trans-Golgi network. Trends Cell Biol. 2011;21:515–25. doi: 10.1016/j.tcb.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Guo W. The exocyst complex in exocytosis and cell migration. Protoplasma. 2011 doi: 10.1007/s00709-011-0330-1. In press. [DOI] [PubMed] [Google Scholar]
- 19.Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, et al. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. 2002;4:343–50. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 20.Henis YI, Hancock JF, Prior IA. Ras acylation, compartmentalization and signaling nanoclusters (Review) Mol Membr Biol. 2009;26:80–92. doi: 10.1080/09687680802649582. [Review] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Omerovic J, Prior IA. Compartmentalized signalling: Ras proteins and signalling nanoclusters. FEBS J. 2009;276:1817–25. doi: 10.1111/j.1742-4658.2009.06928.x. [DOI] [PubMed] [Google Scholar]
- 22.Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: post-translational modification of RAS. Nat Rev Mol Cell Biol. 2012;13:39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahearn IM, Tsai FD, Court H, Zhou M, Jennings BC, Ahmed M, et al. FKBP12 binds to acylated H-ras and promotes depalmitoylation. Mol Cell. 2011;41:173–85. doi: 10.1016/j.molcel.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Apolloni A, Prior IA, Lindsay M, Parton RG, Hancock JF. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol Cell Biol. 2000;20:2475–87. doi: 10.1128/MCB.20.7.2475-2487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Misaki R, Morimatsu M, Uemura T, Waguri S, Miyoshi E, Taniguchi N, et al. Palmitoylated Ras proteins traffic through recycling endosomes to the plasma membrane during exocytosis. J Cell Biol. 2010;191:23–9. doi: 10.1083/jcb.200911143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, et al. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307:1746–52. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- 27.Goodwin JS, Drake KR, Rogers C, Wright L, Lippincott-Schwartz J, Philips MR, et al. Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J Cell Biol. 2005;170:261–72. doi: 10.1083/jcb.200502063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Ijzendoorn SC. Recycling endosomes. J Cell Sci. 2006;119:1679–81. doi: 10.1242/jcs.02948. [DOI] [PubMed] [Google Scholar]
- 29.Gomez GA, Daniotti JL. H-Ras dynamically interacts with recycling endosomes in CHO-K1 cells: involvement of Rab5 and Rab11 in the trafficking of H-Ras to this pericentriolar endocytic compartment. J Biol Chem. 2005;280:34997–5010. doi: 10.1074/jbc.M506256200. [DOI] [PubMed] [Google Scholar]
- 30.Takaya A, Kamio T, Masuda M, Mochizuki N, Sawa H, Sato M, et al. R-Ras regulates exocytosis by Rgl2/Rlf-mediated activation of RalA on endosomes. Mol Biol Cell. 2007;18:1850–60. doi: 10.1091/mbc.E06-08-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soldati T, Schliwa M. Powering membrane traffic in endocytosis and recycling. Nat Rev Mol Cell Biol. 2006;7:897–908. doi: 10.1038/nrm2060. [DOI] [PubMed] [Google Scholar]
- 32.Balasubramanian N, Scott DW, Castle JD, Casanova JE, Schwartz MA. Arf6 and microtubules in adhesion-dependent trafficking of lipid rafts. Nat Cell Biol. 2007;9:1381–91. doi: 10.1038/ncb1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Golachowska MR, Hoekstra D, van IJzendoorn SC. Recycling endosomes in apical plasma membrane domain formation and epithelial cell polarity. Trends Cell Biol. 2010;20:618–26. doi: 10.1016/j.tcb.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 34.Hsu VW, Prekeris R. Transport at the recycling endosome. Curr Opin Cell Biol. 2010;22:528–34. doi: 10.1016/j.ceb.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ullrich O, Reinsch S, Urbé S, Zerial M, Parton RG. Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol. 1996;135:913–24. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cox AD, Brtva TR, Lowe DG, Der CJ. R-Ras induces malignant, but not morphologic, transformation of NIH3T3 cells. Oncogene. 1994;9:3281–8. [PubMed] [Google Scholar]
- 37.Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, et al. Characterization of a cis-Golgi matrix protein, GM130. J Cell Biol. 1995;131:1715–26. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, et al. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol. 2004;6:154–61. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 39.Lowe DG, Capon DJ, Delwart E, Sakaguchi AY, Naylor SL, Goeddel DV. Structure of the human and murine R-ras genes, novel genes closely related to ras proto-oncogenes. Cell. 1987;48:137–46. doi: 10.1016/0092-8674(87)90364-3. [DOI] [PubMed] [Google Scholar]
- 40.Oertli B, Han J, Marte BM, Sethi T, Downward J, Ginsberg M, et al. The effector loop and prenylation site of R-Ras are involved in the regulation of integrin function. Oncogene. 2000;19:4961–9. doi: 10.1038/sj.onc.1203876. [In Process Citation] [DOI] [PubMed] [Google Scholar]
- 41.Hansen M, Prior IA, Hughes PE, Oertli B, Chou FL, Willumsen BM, et al. C-terminal sequences in R-Ras are involved in integrin regulation and in plasma membrane microdomain distribution. Biochem Biophys Res Commun. 2003;311:829–38. doi: 10.1016/j.bbrc.2003.10.074. [DOI] [PubMed] [Google Scholar]
- 42.Furuhjelm J, Peränen J. The C-terminal end of R-Ras contains a focal adhesion targeting signal. J Cell Sci. 2003;116:3729–38. doi: 10.1242/jcs.00689. [DOI] [PubMed] [Google Scholar]
- 43.Frech M, Andjelkovic M, Ingley E, Reddy KK, Falck JR, Hemmings BA. High affinity binding of inositol phosphates and phosphoinositides to the pleckstrin homology domain of RAC/protein kinase B and their influence on kinase activity. J Biol Chem. 1997;272:8474–81. doi: 10.1074/jbc.272.13.8474. [DOI] [PubMed] [Google Scholar]
- 44.Yudowski GA, Puthenveedu MA, Henry AG, von Zastrow M. Cargo-mediated regulation of a rapid Rab4-dependent recycling pathway. Mol Biol Cell. 2009;20:2774–84. doi: 10.1091/mbc.E08-08-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White DP, Caswell PT, Norman JC. alpha v beta3 and alpha5beta1 integrin recycling pathways dictate downstream Rho kinase signaling to regulate persistent cell migration. J Cell Biol. 2007;177:515–25. doi: 10.1083/jcb.200609004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mammoto A, Ohtsuka T, Hotta I, Sasaki T, Takai Y. Rab11BP/Rabphilin-11, a downstream target of rab11 small G protein implicated in vesicle recycling. J Biol Chem. 1999;274:25517–24. doi: 10.1074/jbc.274.36.25517. [DOI] [PubMed] [Google Scholar]
- 47.Yoon SO, Shin S, Mercurio AM. Hypoxia stimulates carcinoma invasion by stabilizing microtubules and promoting the Rab11 trafficking of the alpha6beta4 integrin. Cancer Res. 2005;65:2761–9. doi: 10.1158/0008-5472.CAN-04-4122. [DOI] [PubMed] [Google Scholar]
- 48.Kotler-Brajtburg J, Medoff G, Kobayashi GS, Schlessinger D. Sensitivity to amphotericin B and the cholesterol: phospholipid molar ratios of 3T3, L, BHK and HeLa cells. Biochem Pharmacol. 1977;26:705–10. doi: 10.1016/0006-2952(77)90212-X. [DOI] [PubMed] [Google Scholar]
- 49.Porat-Shliom N, Kloog Y, Donaldson JG. A unique platform for H-Ras signaling involving clathrin-independent endocytosis. Mol Biol Cell. 2008;19:765–75. doi: 10.1091/mbc.E07-08-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKay J, Wang X, Ding J, Buss JE, Ambrosio L. H-ras resides on clathrin-independent ARF6 vesicles that harbor little RAF-1, but not on clathrin-dependent endosomes. Biochim Biophys Acta. 2011;1813:298–307. doi: 10.1016/j.bbamcr.2010.11.019. [DOI] [PubMed] [Google Scholar]
- 51.Henis YI, Rotblat B, Kloog Y. FRAP beam-size analysis to measure palmitoylation-dependent membrane association dynamics and microdomain partitioning of Ras proteins. Methods. 2006;40:183–90. doi: 10.1016/j.ymeth.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 52.Woodman PG, Futter CE. Multivesicular bodies: co-ordinated progression to maturity. Curr Opin Cell Biol. 2008;20:408–14. doi: 10.1016/j.ceb.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baumgart F, Corral-Escariz M, Pérez-Gil J, Rodríguez-Crespo I. Palmitoylation of R-Ras by human DHHC19, a palmitoyl transferase with a CaaX box. Biochim Biophys Acta. 2010;1798:592–604. doi: 10.1016/j.bbamem.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 54.Hancock JF, Magee AI, Childs JE, Marshall CJ. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 1989;57:1167–77. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- 55.Cadwallader KA, Paterson H, Macdonald SG, Hancock JF. N-terminally myristoylated Ras proteins require palmitoylation or a polybasic domain for plasma membrane localization. Mol Cell Biol. 1994;14:4722–30. doi: 10.1128/mcb.14.7.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gorfe AA, Hanzal-Bayer M, Abankwa D, Hancock JF, McCammon JA. Structure and dynamics of the full-length lipid-modified H-Ras protein in a 1,2-dimyristoylglycero-3-phosphocholine bilayer. J Med Chem. 2007;50:674–84. doi: 10.1021/jm061053f. [DOI] [PubMed] [Google Scholar]
- 57.Baker TL, Zheng H, Walker J, Coloff JL, Buss JE. Distinct rates of palmitate turnover on membrane-bound cellular and oncogenic H-ras. J Biol Chem. 2003;278:19292–300. doi: 10.1074/jbc.M206956200. [DOI] [PubMed] [Google Scholar]
- 58.Gorfe AA, Pellarin R, Caflisch A. Membrane localization and flexibility of a lipidated ras peptide studied by molecular dynamics simulations. J Am Chem Soc. 2004;126:15277–86. doi: 10.1021/ja046607n. [DOI] [PubMed] [Google Scholar]
- 59.Huster D, Vogel A, Katzka C, Scheidt HA, Binder H, Dante S, et al. Membrane insertion of a lipidated ras peptide studied by FTIR, solid-state NMR, and neutron diffraction spectroscopy. J Am Chem Soc. 2003;125:4070–9. doi: 10.1021/ja0289245. [DOI] [PubMed] [Google Scholar]
- 60.Rodriguez-Viciana P, Sabatier C, McCormick F. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol Cell Biol. 2004;24:4943–54. doi: 10.1128/MCB.24.11.4943-4954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiménez C, Portela RA, Mellado M, Rodríguez-Frade JM, Collard J, Serrano A, et al. Role of the PI3K regulatory subunit in the control of actin organization and cell migration. J Cell Biol. 2000;151:249–62. doi: 10.1083/jcb.151.2.249. [In Process Citation] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Funamoto S, Meili R, Lee S, Parry L, Firtel RA. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell. 2002;109:611–23. doi: 10.1016/S0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- 63.Innocenti M, Frittoli E, Ponzanelli I, Falck JR, Brachmann SM, Di Fiore PP, et al. Phosphoinositide 3-kinase activates Rac by entering in a complex with Eps8, Abi1, and Sos-1. J Cell Biol. 2003;160:17–23. doi: 10.1083/jcb.200206079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chung CY, Firtel RA. Signaling pathways at the leading edge of chemotaxing cells. J Muscle Res Cell Motil. 2002;23:773–9. doi: 10.1023/A:1024479728970. [DOI] [PubMed] [Google Scholar]
- 65.Asano Y, Nagasaki A, Uyeda TQ. Correlated waves of actin filaments and PIP3 in Dictyostelium cells. Cell Motil Cytoskeleton. 2008;65:923–34. doi: 10.1002/cm.20314. [DOI] [PubMed] [Google Scholar]
- 66.Kortholt A, Bolourani P, Rehmann H, Keizer-Gunnink I, Weeks G, Wittinghofer A, et al. A Rap/phosphatidylinositol 3-kinase pathway controls pseudopod formation [corrected] Mol Biol Cell. 2010;21:936–45. doi: 10.1091/mbc.E09-03-0177. [corrected] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nakada M, Niska JA, Tran NL, McDonough WS, Berens ME. EphB2/R-Ras signaling regulates glioma cell adhesion, growth, and invasion. Am J Pathol. 2005;167:565–76. doi: 10.1016/S0002-9440(10)62998-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goldfinger LE, Ptak C, Jeffery ED, Shabanowitz J, Han J, Haling JR, et al. An experimentally derived database of candidate Ras-interacting proteins. J Proteome Res. 2007;6:1806–11. doi: 10.1021/pr060630l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.