Abstract
Members of the Ras superfamily of small guanosine triphosphatases (GTPases) function as key nodes within signaling networks in a remarkable range of cellular processes, including cell proliferation, differentiation, growth, cell-cell adhesion and apoptosis. We recently described a novel role for the Ras-like small GTPases Rap1 and Ral in regulating cortical polarity and spindle orientation during asymmetric neuroblast division in Drosophila. The participation of these proteins in promoting cell polarization seems to be a common theme throughout evolution.
Keywords: Asymmetric cell division, Ral, Rap1, cell polarity, small GTPases
Introduction
Small GTPases act as binary molecular switches, cycling between the GDP-bound inactive and GTP-bound active states. This cycling is regulated by guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs), the former substituting GDP for GTP and the latter promoting GTPase activity.1,2 Five major branches of the Ras superfamily of small GTPases have been distinguished on the basis of sequence and functional similarities: Ras, Rho, Rab, Ran and Arf.2 These proteins are crucial regulators of many biological processes and as such, they represent a very intriguing group from which we still have plenty to learn. The small GTPases Rap1 and Ral belong to the Ras family branch, which also includes the Ras GTPase, the founding member of the whole superfamily.3,4
Rap1 and Ral Small GTPases
A signaling circuit centered on Ral (Ras/Rap1-RalGEF-Ral) has proven to be highly conserved between flies and mammals, although some differences in the organization of this network have been found. For example, in mammals Ras acts directly on the Ral-GEF without the participation of Rap1, whereas in flies it is Rap1 that linearly activates the Ral-GEF called Rgl, in turn activating Ral. Ras does interact with this pathway in Drosophila, although not in a linear way.5 Both the Rap1 and Ral GTPases perform crucial functions in physiological and pathological conditions. Rap1 plays a key and evolutionary conserved role in regulating morphogenesis, integrin and cadherin-mediated cell-cell adhesion as well as junction formation. In addition, Rap1 has central functions in signal transduction.5-13 The functions of Ral have remained more elusive for a long time, although we now have some important clues as to the roles it fulfils. For example, over the past years it has been unveiled a key function of Ral as regulator of the exocyst, a complex of proteins involved in the sorting and delivery of secretory vesicles to the plasma membrane.14 Ral proteins also regulate cell morphology, they participate in the JNK and Jak/Stat-dependent apoptotic pathways and they have been associated with the initiation and maintenance of Ras-dependent carcinogenesis in humans.15-19
Asymmetric Cell Division: An Intricate Regulatory Protein Network
Asymmetric cell division is an essential process during development, cancer and stem cell biology.20,21 Asymmetric cell divisions are necessary to produce two distinct daughter cells, one that retains the self-renewal capacity of the mother cell and another that is committed to entering a program of differentiation. Asymmetric cell division has been extensively studied in Drosophila neuroblasts (NBs), the neural stem cells of the central nervous system (CNS). NBs delaminate from the neuroectoderm, inheriting the apico-basal polarity of the neuroectodermal cells. This axis of cell polarity along which the mitotic spindle aligns is fundamental for the asymmetry of the division and, hence, must be tightly regulated.22 Intrinsic cues, mostly polarized at the apical cortex of the NB, regulate the orientation of the spindle along the apico-basal axis. Among the proteins that provide the intrinsic cues crucial to establish this cortical polarity are the highly conserved partitioning defective proteins Par6 and Par3, called Bazooka (Baz) in Drosophila, Cdc42, as well as the atypical protein kinase C (aPKC). Baz/Par3 binds to a protein called Inscuteable, which associates the Par complex with Partner of Inscuteable (Pins) that thereafter orchestrates the orientation of the spindle.20-23 Two main pathways have been shown to modulate this process. One of these is activated by the Aurora-A kinase, which phosphorylates Pins promoting the recruitment of the Discs large (Dlg)/Khc-73 complex. The Khc-73 motor protein in this complex binds to the spindle microtubules, anchoring the spindle to the apical cortex.24 The second pathway involves additional partners of Pins such as the heterotrimeric subunit Gαi that is directly attached to the plasma membrane, the PDZ protein Canoe (Cno, AF-6/Afadin in vertebrates) and Mushroom body defect (Mud, NuMA in vertebrates). Mud may associate with the Lys1/Dynein/Dynactin complex, which is capable of generating pulling forces on the spindle pole microtubules.24,25 The tight coupling of apical proteins and spindle positioning is fundamental for the asymmetric localization of cell-fate determinants to the basal pole of the NB, such as Numb and Prospero (Pros). In this way, these determinants will be delivered exclusively to the smaller daughter cell, the ganglion mother cell (GMC), which will then activate a specific program of differentiation.20,21,23
A Novel Role for the Rap1-Rgl-Ral Signaling Network in Asymmetric Cell Division
Despite the complexity of the protein network involved in regulating asymmetric cell division, it seems that this network may become even more tangled.26 We recently found that in Drosophila, the Rap1-Rgl-Ral signaling network fulfils a novel role in the regulation of cortical polarity and spindle orientation during asymmetric cell division.27 Rap1 is present in NBs, where it is slightly enriched at the apical pole. The apical proteins Par6 and aPKC form a complex in vivo with Rap1 contributing to regulate its distribution. In Rap1 mutants the localization of some apical proteins and the orientation of the spindle are both impaired and, consequently, the localization of cell-fate determinants such as Numb and Pros is disturbed. Very similar phenotypes are detected in Rgl and Ral mutants indicating that the whole Rap1-Rgl-Ral signaling network modulates the process. Rap1 is a known partner of the apical protein Cno, which forms a complex with Pins and acts upstream of Mud during asymmetric NB division.25,28 Rap1 binds to the Ras-associating domains of Cno/AF-6 in flies and mammals, a region that interacts with different Ras-like GTPases.28-31 The distribution of both Cno and Mud is modified in Rap1 mutants and this might explain the mis-orientation of the mitotic spindle observed in those mutants. However, it is intriguing that the Pins and Gαi apical crescents are not affected in Rap1 mutants. This suggests that Rap1 belongs to a third pathway, besides the Pins centered ones, that contributes through Cno and Mud, and probably additional unknown effectors, to regulate the spindle orientation. In fact, we observe stronger spindle misorientation phenotypes in Rgl pins double mutants, which suggests that there is certain synergism between the different pathways involved in orientating the mitotic axis. The defects in cortical polarity in Rap1 mutants also include mislocalization of the aPKC and Baz/Par3 apical proteins, although to a much lesser extent than the misplacement of the Cno and Mud proteins. For this reason, we propose that the Rap1-Rgl-Ral signaling network is not the main driving force initially required to establish the distribution of the Par complex. Rather, it is this complex that facilitates the polarization of the Rap1 pathway to the apical pole of the NB. Then, Rap1-Rgl-Ral signaling may to some extent contribute to stabilize the Par complex through a positive feedback loop, which would explain the defects on aPKC and Baz/Par3 localization in Rap1 mutants. Interestingly, Rap1B also has a dual relationship with Par proteins in cultured mammalian neurons, acting both upstream and downstream of them (see below).
Rap1 and Ral Small GTPases: Establishing Cell Polarity in Little Buddies and Bigger Mates
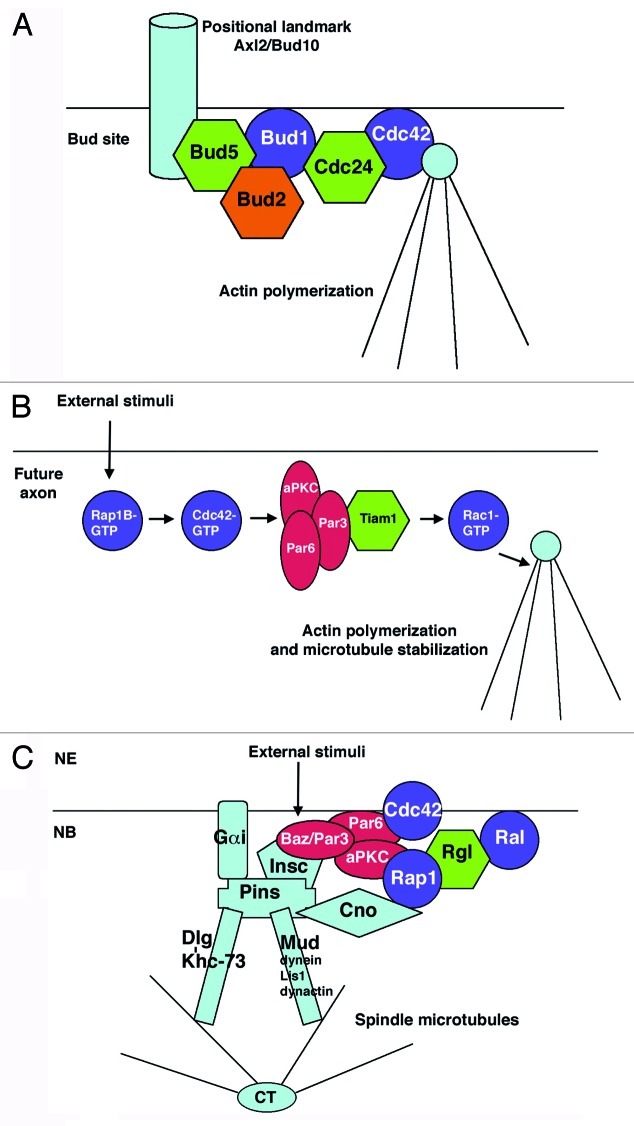
Cell polarity is obviously fundamental to generating asymmetric cell divisions. Intriguingly, other biological processes that require cell polarization are also regulated by Rap1 and Ral GTPases. For example, in the budding yeast Saccharomyces cerevisiae, the Rap1 ortholog Bud1/Rsr1 and its GEF/GAP regulators, Bud5/Bud2, are involved in bud-site selection and, consequently, in determining cell polarity. Bud1 selects the specific sites for growth by recruiting Cdc24, the GEF for the GTPase Cdc42, which promotes cytoskeleton assembly at the bud site. The Bud5 GEF also binds positional landmarks, such as the transmembrane glycoprotein Axl2/Bud1032,33 (Fig. 1A).
Figure 1. Rap1 participates in the establishment of cell polarity in different contexts. (A) The bud site formation in the budding yeast Saccharomyces cerevisiae. (B) Axon specification in cultured mammalian neurons. External stimuli, such as growth factors, may promote the activation of Rap1B in the neurite that will become the axon (C) The asymmetric division of neural stem cells in Drosophila. External unknown stimuli, coming from the neuroectoderm (NE), contribute to the establishment of cortical polarity in the underlying neuroblast (NB). Rap1 and other small GTPases are represented in purple; GEFs and GAPs regulators appear in green and orange, respectively. Polarity proteins of the partitioning defective complex appear in red (see text for more details).
Rap1 also fulfils a key role in establishing neuronal polarity and axonogenesis in vertebrates. The accumulation of Rap1B-GTP in a particular neurite activates Cdc42 and the Par complex, promoting the activation of Rac1 in this neurite and driving the development of an axon. External stimuli, such as growth factors, may contribute to the initial activation of Rap1B. Additionally, an E3 ubiquitin ligase called Smurf2 targets the inactive Rap1B-GDP for degradation in the neurites that are destined to become dendrites. This process also depends on another activity of Par3, which recruits Smurf2 to the growth cones by binding the motor protein Kif3A. Hence, Par3 seems to play a dual role, upstream of Rap1B by recruitment of Smurf2 to the growth cones allowing Rap1B restriction to a single neurite, and downstream of Rap1B in a complex with aPKC and Par6 to subsequently activate Rac1 in the developing axon34-37 (Fig. 1B).
In Drosophila, the Rap1/DE-Cadherin signaling pathway is thought to regulate polarized niche formation and stem cell anchoring.13 Moreover, Ral has very recently been shown to respond to planar cell polarity signals to modulate asymmetric Notch signaling in the Drosophila eye, contributing to the specification of the two initially equivalent R3/R4 photoreceptors.38 Apart from Rap1 and Ral, which have been highlighted here, other small GTPases are also emerging as crucial modulators of cell polarity.39 Cooperation and cross-talk between small GTPases, their effectors/regulators and polarity proteins should be analyzed in depth in different organisms, tissues and contexts in the next years. These interactions will unveil links with both extrinsic signals (i.e growth factors) and intrinsic cues (i.e., actin and microtubule cytoskeleton), fundamental to achieve cell polarization. This is certainly an engaging issue, whose ongoing analysis will enlighten the mechanisms underlying many biological processes in the near future.
Acknowledgments
I would like to thank anonymous reviewers for helpful comments and suggestions. Work in our lab is supported by Grants from the Spanish Government BFU2009–08833 and CONSOLIDER-INGENIO 2010 CSD2007–00023.
Footnotes
Previously published online: www.landesbioscience.com/journals/smallgtpases/article/19631
References
- 1.van Dam TJ, Bos JL, Snel B. Evolution of the Ras-like small GTPases and their regulators. Small Gtpases. 2011;2:4–16. doi: 10.4161/sgtp.2.1.15113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–6. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 3.Gloerich M, Bos JL. Regulating Rap small G-proteins in time and space. Trends Cell Biol. 2011;21:615–23. doi: 10.1016/j.tcb.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Neel NF, Martin TD, Stratford JK, Zand TP, Reiner DJ, Der CJ. The RalGEF-Ral Effector Signaling Network: The Road Less Traveled for Anti-Ras Drug Discovery. Genes Cancer. 2011;2:275–87. doi: 10.1177/1947601911407329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mirey G, Balakireva M, L’Hoste S, Rossé C, Voegeling S, Camonis J. A Ral guanine exchange factor-Ral pathway is conserved in Drosophila melanogaster and sheds new light on the connectivity of the Ral, Ras, and Rap pathways. Mol Cell Biol. 2003;23:1112–24. doi: 10.1128/MCB.23.3.1112-1124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asha H, de Ruiter ND, Wang MG, Hariharan IK. The Rap1 GTPase functions as a regulator of morphogenesis in vivo. EMBO J. 1999;18:605–15. doi: 10.1093/emboj/18.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bos JL, de Rooij J, Reedquist KA. Rap1 signalling: adhering to new models. Nat Rev Mol Cell Biol. 2001;2:369–77. doi: 10.1038/35073073. [DOI] [PubMed] [Google Scholar]
- 8.Caron E. Cellular functions of the Rap1 GTP-binding protein: a pattern emerges. J Cell Sci. 2003;116:435–40. doi: 10.1242/jcs.00238. [DOI] [PubMed] [Google Scholar]
- 9.Knox AL, Brown NH. Rap1 GTPase regulation of adherens junction positioning and cell adhesion. Science. 2002;295:1285–8. doi: 10.1126/science.1067549. [DOI] [PubMed] [Google Scholar]
- 10.Kooistra MR, Dubé N, Bos JL. Rap1: a key regulator in cell-cell junction formation. J Cell Sci. 2007;120:17–22. doi: 10.1242/jcs.03306. [DOI] [PubMed] [Google Scholar]
- 11.O’Keefe DD, Gonzalez-Niño E, Burnett M, Dylla L, Lambeth SM, Licon E, et al. Rap1 maintains adhesion between cells to affect Egfr signaling and planar cell polarity in Drosophila. Dev Biol. 2009;333:143–60. doi: 10.1016/j.ydbio.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price LS, Hajdo-Milasinovic A, Zhao J, Zwartkruis FJ, Collard JG, Bos JL. Rap1 regulates E-cadherin-mediated cell-cell adhesion. J Biol Chem. 2004;279:35127–32. doi: 10.1074/jbc.M404917200. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Singh SR, Zheng Z, Oh SW, Chen X, Edwards K, et al. Rap-GEF signaling controls stem cell anchoring to their niche through regulating DE-cadherin-mediated cell adhesion in the Drosophila testis. Dev Cell. 2006;10:117–26. doi: 10.1016/j.devcel.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Moskalenko S, Henry DO, Rosse C, Mirey G, Camonis JH, White MA. The exocyst is a Ral effector complex. Nat Cell Biol. 2002;4:66–72. doi: 10.1038/ncb728. [DOI] [PubMed] [Google Scholar]
- 15.González-García A, Pritchard CA, Paterson HF, Mavria G, Stamp G, Marshall CJ. RalGDS is required for tumor formation in a model of skin carcinogenesis. Cancer Cell. 2005;7:219–26. doi: 10.1016/j.ccr.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 16.Balakireva M, Rossé C, Langevin J, Chien YC, Gho M, Gonzy-Treboul G, et al. The Ral/exocyst effector complex counters c-Jun N-terminal kinase-dependent apoptosis in Drosophila melanogaster. Mol Cell Biol. 2006;26:8953–63. doi: 10.1128/MCB.00506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghiglione C, Devergne O, Cerezo D, Noselli S. Drosophila RalA is essential for the maintenance of Jak/Stat signalling in ovarian follicles. EMBO Rep. 2008;9:676–82. doi: 10.1038/embor.2008.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camonis JH, White MA. Ral GTPases: corrupting the exocyst in cancer cells. Trends Cell Biol. 2005;15:327–32. doi: 10.1016/j.tcb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Feig LA. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 2003;13:419–25. doi: 10.1016/S0962-8924(03)00152-1. [DOI] [PubMed] [Google Scholar]
- 20.Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–87. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- 21.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–97. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–74. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- 23.Chia W, Somers WG, Wang H. Drosophila neuroblast asymmetric divisions: cell cycle regulators, asymmetric protein localization, and tumorigenesis. J Cell Biol. 2008;180:267–72. doi: 10.1083/jcb.200708159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston CA, Hirono K, Prehoda KE, Doe CQ. Identification of an Aurora-A/PinsLINKER/Dlg spindle orientation pathway using induced cell polarity in S2 cells. Cell. 2009;138:1150–63. doi: 10.1016/j.cell.2009.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Speicher S, Fischer A, Knoblich J, Carmena A. The PDZ protein Canoe regulates the asymmetric division of Drosophila neuroblasts and muscle progenitors. Curr Biol. 2008;18:831–7. doi: 10.1016/j.cub.2008.04.072. [DOI] [PubMed] [Google Scholar]
- 26.Carmena A. Signaling networks during development: the case of asymmetric cell division in the Drosophila nervous system. Dev Biol. 2008;321:1–17. doi: 10.1016/j.ydbio.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 27.Carmena A, Makarova A, Speicher S. The Rap1-Rgl-Ral signaling network regulates neuroblast cortical polarity and spindle orientation. J Cell Biol. 2011;195:553–62. doi: 10.1083/jcb.201108112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boettner B, Harjes P, Ishimaru S, Heke M, Fan HQ, Qin Y, et al. The AF-6 homolog canoe acts as a Rap1 effector during dorsal closure of the Drosophila embryo. Genetics. 2003;165:159–69. doi: 10.1093/genetics/165.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boettner B, Govek EE, Cross J, Van Aelst L. The junctional multidomain protein AF-6 is a binding partner of the Rap1A GTPase and associates with the actin cytoskeletal regulator profilin. Proc Natl Acad Sci U S A. 2000;97:9064–9. doi: 10.1073/pnas.97.16.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuriyama M, Harada N, Kuroda S, Yamamoto T, Nakafuku M, Iwamatsu A, et al. Identification of AF-6 and canoe as putative targets for Ras. J Biol Chem. 1996;271:607–10. doi: 10.1074/jbc.271.2.607. [DOI] [PubMed] [Google Scholar]
- 31.Wee B, Johnston CA, Prehoda KE, Doe CQ. Canoe binds RanGTP to promote Pins(TPR)/Mud-mediated spindle orientation. J Cell Biol. 2011;195:369–76. doi: 10.1083/jcb.201102130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gulli MP, Peter M. Temporal and spatial regulation of Rho-type guanine-nucleotide exchange factors: the yeast perspective. Genes Dev. 2001;15:365–79. doi: 10.1101/gad.876901. [DOI] [PubMed] [Google Scholar]
- 33.Kang PJ, Sanson A, Lee B, Park HOA. A GDP/GTP exchange factor involved in linking a spatial landmark to cell polarity. Science. 2001;292:1376–8. doi: 10.1126/science.1060360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishimura T, Kato K, Yamaguchi T, Fukata Y, Ohno S, Kaibuchi K. Role of the PAR-3-KIF3 complex in the establishment of neuronal polarity. Nat Cell Biol. 2004;6:328–34. doi: 10.1038/ncb1118. [DOI] [PubMed] [Google Scholar]
- 35.Schwamborn JC, Khazaei MR, Püschel AW. The interaction of mPar3 with the ubiquitin ligase Smurf2 is required for the establishment of neuronal polarity. J Biol Chem. 2007;282:35259–68. doi: 10.1074/jbc.M703438200. [DOI] [PubMed] [Google Scholar]
- 36.Schwamborn JC, Müller M, Becker AH, Püschel AW. Ubiquitination of the GTPase Rap1B by the ubiquitin ligase Smurf2 is required for the establishment of neuronal polarity. EMBO J. 2007;26:1410–22. doi: 10.1038/sj.emboj.7601580. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Schwamborn JC, Püschel AW. The sequential activity of the GTPases Rap1B and Cdc42 determines neuronal polarity. Nat Neurosci. 2004;7:923–9. doi: 10.1038/nn1295. [DOI] [PubMed] [Google Scholar]
- 38.Cho B, Fischer JA. Ral GTPase promotes asymmetric Notch activation in the Drosophila eye in response to Frizzled/PCP signaling by repressing ligand-independent receptor activation. Development. 2011;138:1349–59. doi: 10.1242/dev.056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iden S, Collard JG. Crosstalk between small GTPases and polarity proteins in cell polarization. Nat Rev Mol Cell Biol. 2008;9:846–59. doi: 10.1038/nrm2521. [DOI] [PubMed] [Google Scholar]