Abstract
We discovered recently that the Drosophila Ral GTPase regulates Notch signaling and thereby affects cell patterning in the eye. Although Ral functions in the ligand signaling cells, Ral does not stimulate ligand signaling directly. Rather, in cells that express both Notch receptor and ligand, Ral activity promotes a cell to become the signaler by inhibiting Notch receptor activation in that cell. Moreover, Ral inhibits a particular pathway of Notch activation—receptor activation that occurs independent of ligand binding. In this Commentary, we discuss the phenomenon of ligand-independent Notch receptor activation and how this event might be regulated by Ral.
Keywords: Drosophila eye, Notch signaling, Ral, endosome, presenilin cleavage
Introduction
Ral proteins are Ras-like GTPases whose known roles are mainly in regulation of membrane trafficking.1 The best-characterized Ral effectors are RalBP1 (aka RLIP76)2,3 and the exocyst complex proteins Sec5 and Exo84.4 Through RalBP1, Ral coordinates endocytosis and mitochondrial fission with mitosis.5-8 Through the exocyst, Ral regulates many cellular processes including secretion,4 cytokinesis,9 cytoskeletal remodeling,10,11 membrane remodeling,12 and apoptosis.13,14
There are two separate Ral genes in vertebrates, RalA and RalB, which produce proteins comparable in amino acid sequence (81% identical and 89% similar) and organization of functional units.1,15 RalA and RalB have different cellular functions through RalA-specific and RalB-specific RalGEFs.9 Drosophila has a single Ral gene and a single Ral protein that resembles RalA more closely than RalB in overall amino acid sequence, but has amino acids specific to both vertebrate Ral proteins.10
Most of what we know about the cellular functions of Ral and its effectors is from studies in vertebrate cell culture. Genetic and biochemical studies in Drosophila suggest that Ral function is largely conserved between flies and vertebrates.16 Moreover, Drosophila provides a multicellular context in which to understand the significance of Ral activity. Before Drosophila Ral loss-of-function mutants were generated, experiments were performed with transgenic flies that overexpressed wild-type, constitutively active, or dominant negative Ral proteins. This work implicated Ral as a regulator of the cytoskeleton to effect cell shape.10,17 Subsequent analysis of Ral loss-of-function mutants showed that Ral, through regulation of Jak/Stat signaling, controls the fate and survival of so-called polar cells in the ovary that control oocyte polarization.18 Another study using sec5 mutants showed that through the exocyst effector, Ral regulates JNK signaling and p38 MAPK to prevent apoptosis in sensory organ precursor cells.13 Although there has not yet been a genetic analysis of RalBP1 effector function, Drosophila Ral does bind in vitro to RalBP1 as well to Sec5.16
We recently discovered a specific role for Drosophila Ral in Notch signaling.19 We identified Ral mutants originally in a screen for mutations that modify the eye defects caused by overexpression of Drosophila Epsin.20 Epsin is an endocytic protein that is required specifically for Notch signaling in Drosophila,21,22 C. elegans,23 and vertebrates.24 Epsin is required in signal-sending cells in order for ligand to activate the Notch receptor in adjacent signal-receiving cells.21-23 The Notch receptor and its ligands (Delta and Serrate in Drosophila) are transmembrane proteins.25 Notch is unusual in its mechanism of activation; ligand binding leads to cleavage of the Notch receptor intracellular domain (NICD), which travels to the nucleus and derepresses transcription of target genes.25 Most evidence suggests that Epsin is required after ligand/receptor binding to internalize ligand into the signal-sending cells thereby exerting mechanical force on the receptor that leads to its activation.26,27
We determined that we identified Ral mutants in our screen because Ral is a negative regulator of Notch signaling.19 We do not understand the mechanism by which Epsin overexpression disrupts eye development and so we cannot say precisely why loss of Ral activity enhances the eye defects. Nevertheless, we determined that loss of Ral activity results in Notch receptor activation independent of ligand binding.19 The main evidence for this lies in four observations. First, the specific defects in Ral mutant eyes were typical effects of too much Notch activity. Second, Ral interacts genetically with several Notch pathway genes in a manner consistent with the idea that Ral inhibits Notch activation. For example, Notch overexpression worsens the eye defects caused by loss of Ral activity. Third, we observed that overexpression of a hy brid Notch protein with its extracellular domain replaced by that of a different receptor had the same effect of worsening the defects in Ral mutant eyes. As the hybrid Notch protein cannot bind Notch ligand, this result suggests that in the absence of Ral Notch might be activated without binding ligand. Finally, we observed that this is indeed the case. In each overexpressed Notch protein NICD was fused to Gal4. Activation of the Notch/Gal4 receptor allows the cleaved NICD/Gal4 fragment to enter the nucleus and activate a UAS-gfp reporter transgene. We detected GFP in animals that overexpressed either normal or hybrid Notch/Gal4 and only in Ral mutants.
We wanted to determine whether or not the idea that Ral inhibits ligand-independent Notch activation made sense in a developmental context. One of the defects in Ral mutant eyes typical of Notch pathway dysfunction was the symmetry of two photoreceptor cells, R3 and R4. These two photoreceptors are normally positioned asymmetrically, and as the molecular mechanism by which Notch signaling mediates R3/R4 specification is understood at least in part, we decided to investigate a possible role for Ral in this process. The ~800 identical ommatidia (or facets) of the fly eye each has eight photoreceptors (R1-R8) arranged in a central trapezoid, with R3 at its apex (Fig. 1). The difference between R3 and R4 is established early during eye development in a five-cell ommatidial precluster that includes adjacent R3/R4 precursor cells. The R3/R4 precursor closest to the dorsal/ventral midline (equator) always becomes R3, and the other cell of the pair becomes R4. R3/R4 specification depends on Frizzled and Notch signaling (Fig. 1).28-30 Although each R3/R4 precursor expresses both Notch and the ligand Delta, Notch activation is asymmetric in R3/R4. Because it is closer to the equator, the pre-R3 cell experiences more Frizzled activation and thus activates Notch in the adjacent pre-R4 cell. Frizzled activation is thought to result in asymmetric Notch activation in pre-R4 through either or both of two pathways. Frizzled signaling leads to elevated transcription in pre-R3 of the ligand Delta and also the Ubiquitin-ligase Neuralized.28,31 Neuralized ubiquitylates Delta, a prerequisite for Delta endocytosis by pre-R3 and activation of Notch in pre-R4.27 Alternatively or in addition, activated Frizzled relocalizes in a complex with a cortical protein called Disheveled to the R3/R4 interface where the complex prevents Notch receptor activation directly in pre-R3.30,32
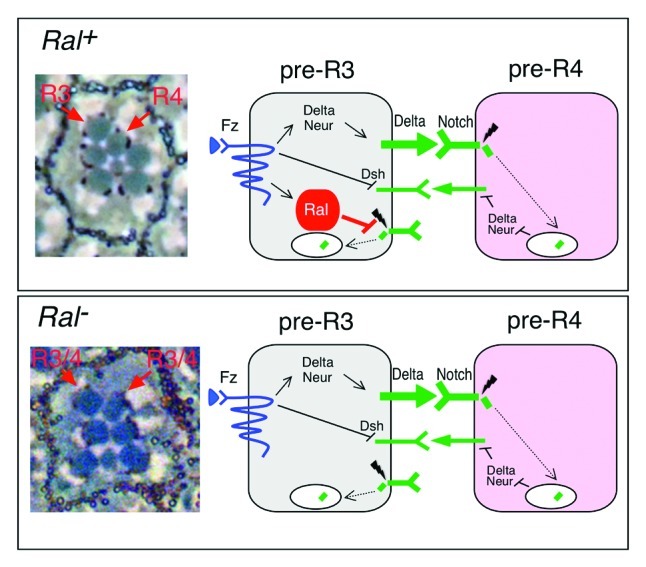
Figure 1. Model for Ral function in R3/R4 specification. At left are light microscope images of a single ommatidium. At right are diagrams of the signaling events that distinguish R3 and R4. The top box is wild type (Ral+). Photoreceptors in the wild-type adult Drosophila eye (top) are arranged in a trapezoid with R3 at the apex. The asymmetric arrangement of the originally equivalent R3/R4 pair is due to pre-R3 experiencing more Frizzled (Fz) activation than does pre-R4. This difference in Fz activity is amplified by subsequent Notch activation in pre-R4.28-30 Fz activation results in increased transcription of Delta and neuralized (neur)28-31 and also relocalization of a Fz/Dsh (Disheveled) complex that represses Notch receptor at the pre-R3 plasma membrane that faces pre-R4.32 Thus pre-R3 becomes the Delta signaling cell and Notch is activated in pre-R4. Ral defines a unique pathway for insuring that pre-R3 becomes the Delta signaling cell.19Ral transcription also depends on Fz, and Ral activity prevents activation of Notch receptor that is not bound to ligand.19 Loss of Ral activity (bottom) results in symmetric R3/R4 pairs and ligand-independent Notch activation.19
We found that Ral defines a distinct pathway by which R3 is biased to become the Delta signaling cell.19 In response to Frizzled activation, elevated Ral activity in R3 prevents ligand-independent Notch activation in pre-R3, thereby helping to tip the scales in favor of pre-R3 becoming the signaling cell. The main evidence for this idea is 2-fold. First, we analyzed mosaic eyes in which one of the R3/R4 pair was Ral+ and the other was Ral-. We found that when pre-R3 was Ral-, the R3/R4 pair was often symmetrical or reversed and Notch was often activated in pre-R3 or else weakly in both pre-R3 and pre-R4. In contrast, loss of Ral activity in R4 had no effect. Second, Ral transcription in R3/R4 precursors was dependent on Frizzled and elevated in R3 relative to R4.
It should be noted that the effects of Ral on R3/R4 asymmetry while consistent were weak; most ommatidia in Ral- eyes had normally asymmetric R3/R4 pairs.19 We used three different Ral loss-of-function mutations in these experiments, none of which are null alleles. Therefore, one possible explanation for the preponderance of ommatidia in Ral mutants with normally differentiated R3/R4 pairs is the presence of residual Ral activity in the R3 cells. Alternatively, the fact that Ral defines just one of several independent parallel pathways for biasing pre-R3 to become the signaling cell could explain why loss of Ral activity has a limited effect on R3/R4 specification. We are presently generating a Ral null allele that should allow us to distinguish between these two alternatives.
The big question that remains is: how does Ral block ligand-independent activation of Notch? We will discuss below the phenomenon of ligand-independent Notch activation and speculate about how Ral might inhibit it.
How is Notch Activated Without Binding Ligand?
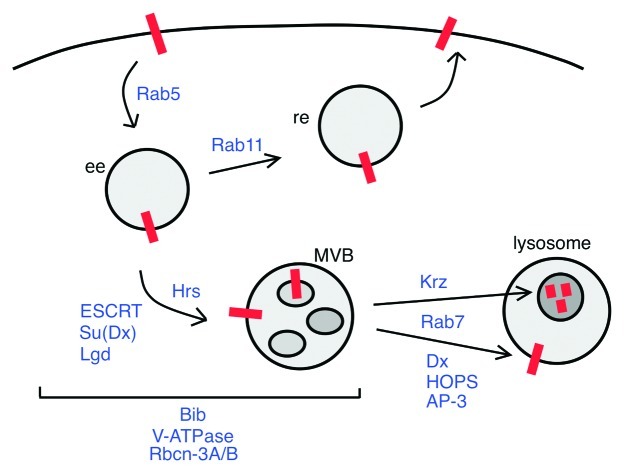
Notch receptor not bound to ligand undergoes endocytosis and endosomal trafficking continually.26,33 Internalized Notch in an early endosome may be recycled to the plasma membrane or routed to the lysosome for degradation (Fig. 2). One way in which ligand-independent Notch activation is thought to occur is as an accident when the flow of Notch through the endosomal pathway is blocked.26 This idea comes from analysis of mutants in which genes required for efficient routing of Notch to the lysosome are inactive. In Drosophila, these genes include Suppressor of Deltex (Su(Dx)),34 which encodes a Ubiquitin-ligase thought to ubiquitylate Notch, genes for ESCRT complex proteins,35-39 which sort ubiquitylated cargos into intralumenal vesicles of multivesicular bodies (MVBs), lethal giant discs (lgd),40-42 which encodes a cytosolic lipid-binding protein, and Kurtz (Krz),43 a gene for a β-arrestin required to route Notch to the lysosome. When Notch is activated by ligand binding, the Notch extracellular domain is cleaved by a metalloprotease, which allows the subsequent γ-secretase cleavage that releases NICD.25 Cleavage of ligand-activated Notch by γ-secretase may occur in endosomes normally, although this point is controversial.26,33,35,44 Nevertheless, ligand-independent Notch activation in mutants where endosomal flow is blocked requires γ-secretase.35,42 It is thought that accumulation of Notch in the acidic environment of endosomes may alter Notch protein conformation and thereby allow γ-secretase cleavage of NICD without ligand binding.26 This idea is supported by the observation that ligand-independent Notch signaling also occurs in mutants where endosome acidity is increased. These mutants are in the gene for an aquaporin called Bigbrain (Bib),26,33 a gene for a subunit of the vacuolar proton pump V-ATPase,45 and genes for Rbcn-3A and Rbcn-3B, two V-ATPase regulators.45 In addition, γ-secretase activity is enhanced by an acidic environment,46 which also could play a role in endosomal activation of unliganded Notch.
Figure 2. Endosomal trafficking of unliganded Notch receptor. A diagram of the known endosomal routes of the Notch receptor that was not activated by ligand binding. The precise step in the endocytic pathway where the Ubiquitin ligases Su(Dx) and Dx function is not clear. Their locations in the diagram are meant to indicate the process for which each is required. See text for explanation of diagram. ee = early endosome, re = recycling endosome, Hrs protein is required to deliver proteins to MVBs. Other abbreviations are defined in the text.
Ligand-independent Notch activation may also occur normally in the lysosomal membrane. A pathway antagonistic to Krz and that requires the Ubiquitin-ligase Deltex (Dx), the HOPS (homotyic fusion and vacuole protein sorting) complex and the endocytic adaptor complex AP-3 is thought to route Notch from MVBs to the lysosomal membrane.47 How low-level Notch signaling from the lysosome might affect cell function is unclear.
How Might Ral Prevent Ligand-independent Notch Activation?
The observation that a hybrid Notch receptor lacking the normal extracellular domain is activated specifically in Ral mutants indicates that Ral functions at or downstream of NICD cleavage by γ-secretase.19 Given our present understanding of ligand-independent Notch activation, Ral might regulate endosome pH or endosomal trafficking of unliganded Notch. Immunofluorescence experiments with Ral antibodies showed that in developing Drosophila eyes most Ral resides in intracellular puncta.19 Although this observation tempts us to speculate that Ral is endosomal, we have not yet been able to identify the Ral-positive puncta, which are basal in photoreceptor cells. We asked whether or not the Ral puncta colocalize with Rab5, Rab7, or Rab11, markers for early, late, and recycling endosomes, respectively.48 YFP-Rab549 expressed in the developing eye using the Gal4/UAS expression system is punctate like Ral but mainly apical (Fig. 3A). There are Rab5 puncta that are as basal as Ral but they colocalize with Ral very little (Fig. 3A-A”’). Rab7-GFP50 expressed using Gal4/UAS is apical and there is essentially no overlap between Rab7-GFP and Ral along the apical/basal axis of photoreceptor cells (Fig. 3B-B”’). Similarly, Rab11 detected with an antibody51 is in puncta that are more apical than those containing Ral (Fig. 3C-C”’).
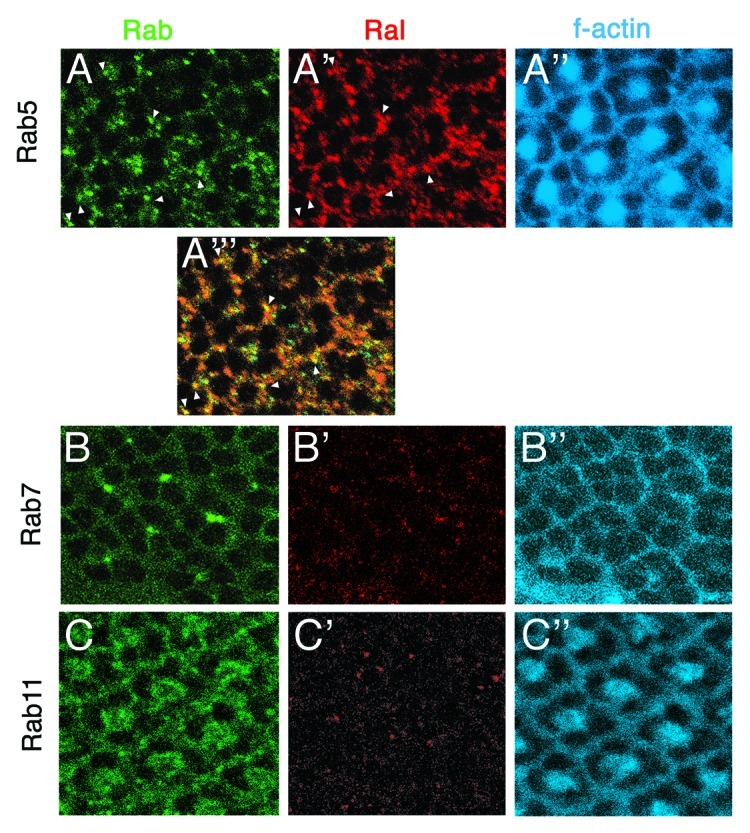
Figure 3. Ral does not colocalize with Rab5, Rab7, nor Rab11. Confocal microscope images of part of three developing eyes (third instar larval eye discs) are shown. All three discs were labeled with anti-Ral49 and phalloidin (binds f-actin at the plasma membrane). (A-A”’) A YFP-Rab5 expressing disc is shown. The genotype is ey-gal4, GMR-gal4/ UAS-yfp-rab5. A”’ is a merge of A and A’. The white arrows indicate the few puncta that are simultaneously YFP-Rab5-positive and Ral-positive. The focal plane of this disc image is more basal than that of the other two discs shown below. (B-B”) A Rab7-GFP expressing disc is shown. The genotype is Actin5C-gal4, UAS-rab7-gfp. At the apical focal plane at which there are Rab7-GFP-positive puncta (B), Ral puncta are absent (B’). (C-C”) A wild-type disc also labeled with anti-Rab11 is shown. At the apical focal plane at which Rab11 resides, no Ral is detected (C’).
The analysis of Ral-positive puncta above is only preliminary and the results should be taken in that light. Yet, the results are surprising as they suggest that the majority of Ral is not present in early or late endosomes defined by Rab5 or Rab7, nor in the recycling endosomes defined by Rab11. Additional experiments of this type with other markers may help to identify the Ral-positive vesicles. Also, it would be useful to examine the Ral puncta in mutants where endocytosis in general is inhibited or specific endosomal pathways are blocked. Another aspect of this analysis that must remain in mind is that the Ral-positive puncta represent only the location of the majority of Ral protein at any given time. This may or may not reflect the Ral protein involved in repression of ligand-independent Notch signaling.
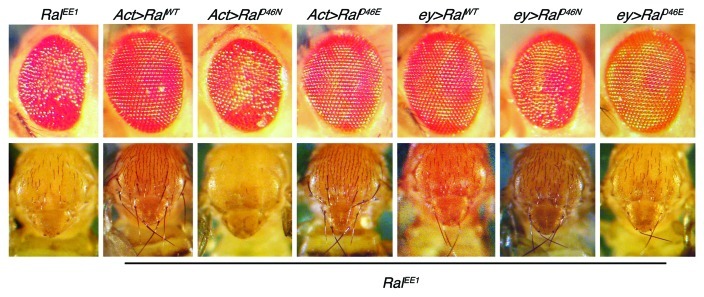
Another means to uncover the critical function of Ral in the eye is to determine which effector is involved. One experiment we did along these lines was to ask if Drosophila Ral proteins with mutations that specifically inhibit binding of two effectors, RalBP1 (RalD46N) or the exocyst protein Sec5 (RalD46E), could substitute for endogenous Ral in the fly eye. Ral proteins with these identical mutations have been characterized biochemically using vertebrate RalA.3,4 We expect that they would have the same effect in Drosophila Ral because fly Ral binds RalBP1 and Sec5,16 and because the amino acid sequence of the effector binding region in Drosophila Ral and vertebrate Ral A are identical. Normal Ral (RalWT), RalD46N or RalD46E were expressed in flies under Gal4 control. When expressed using the eyeless-gal4 driver, either RalWT or RalD46E (binds RalBP1) rescued the eye defects of RalEE1 mutants to wild type (Fig. 4). In contrast, RalD46N (binds exocyst) rescued the eye defects only slightly. Similar results were obtained in the eye with the Actin5C-gal4 driver (Fig. 4). These results suggest that in the eye, Ral works through RalBP1, consistent with a role in endocytosis. One caveat, however, to this interpretation is that the difference in rescuing activity between RalD46E and RalD46N could potentially be explained by RalD46E protein accumulating to higher levels than RalD46N. For example, different levels of transcription could result from position effects, or the two mutant Ral proteins could be differentially stable. While this explanation for the results is possible, we think it is unlikely. First, we know that RalD46N is stably produced in the fly because when their expression is driven by eyeless-gal4, RalD46E and RalD46N each rescue the notal bristle defects of RalEE1 completely (Fig. 4). Second, when RalD46N expression is driven by either eyeless-gal4 or Actin5C-gal4, a similar small amount of rescue activity is observed in the eye. The Actin5C-gal4 driver is much more active than eyeless-gal4, and so if the small amount of activity of RalD46N were due to low expression, more rescue of the eye defects would be expected with Actin5C-gal4. We saw evidence that Actin5C-gal4 is likely driving high levels of RalD46N expression in our flies. When expressed with eyeless-gal4, RalD46N rescues the bristle loss in the notum (Fig. 4). In contrast, when overexpressed with Actin5C-gal4, RalD46N not only fails to rescue the bristle development defect, it makes it worse presumably by acting in a dominant negative fashion.
Figure 4. Rescue of RalEE1 eye organization and notal bristle defects by wild-type and mutant Ral transgenes. Adult eyes and nota of flies of RalEE1 flies with no transgene (leftmost), or that express the indicated Ral transgenes using the Gal4/UAS system are shown. Act is the Actin5C-gal4 driver and ey is the eyeless-gal4 driver. UAS-RalWT has been described.17 Details of how we constructed UAS-RalD46N and UAS-RalD46E are available upon request.
Although the tentative conclusion from these preliminary experiments is that in its role in Notch signaling Ral may function through RalBP1, to be more secure in this interpretation, careful quantitation of protein levels in the different tissues is necessary. In addition, genetic studies using mutations in the effectors genes could be illuminating.
Conclusions
There is presently some evidence suggestive of an endosomal function for Ral in the Notch pathway. Ral inhibits ligand-independent Notch activation, all known forms of which take place in endosomes. Moreover, Ral localizes mainly to intracellular puncta suggestive of endosomes. Also, the failure of RalD46N and the ability of RalD46E to function in the eye suggests that the key Ral effector for Notch signaling is not the exocyst and may be RalBP1 which functions in endocytosis. The role of RalBP1 in endocytosis, however, is poorly defined. Future experiments are needed to identify the Ral puncta and determine if they are associated with Ral function in Notch signaling, and to identify the key Ral effector and how it works. It will also be informative to observe the effect of Ral mutation, if any, on endosome pH and morphology. The discovery of the Ral pathway in pre-R3 revealed that ligand-independent Notch activation is regulated to achieve specific developmental outcomes. It seems likely that more surprises await unraveling the mechanism by which Ral blocks Notch activation.
Acknowledgments
Work in the authors’ laboratory is supported by NIHCD grant #RO1-HD30680 to J.A.F. We are grateful to B. Cohen for anti-Rab11, K. Sawamoto, M. Gonzalez-Gaitan, and M. Scott for flies, and Chang Hoon Lee for help with constructing the Ral variants.
Footnotes
Previously published online: www.landesbioscience.com/journals/smallgtpases/article/19802
References
- 1.van Dam EM, Robinson PJ. Ral: mediator of membrane trafficking. Int J Biochem Cell Biol. 2006;38:1841–7. doi: 10.1016/j.biocel.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Jullien-Flores V, Dorseuil O, Romero F, Letourneur F, Saragosti S, Berger R, et al. Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J Biol Chem. 1995;270:22473–7. doi: 10.1074/jbc.270.38.22473. [DOI] [PubMed] [Google Scholar]
- 3.Bauer B, Mirey G, Vetter IR, García-Ranea JA, Valencia A, Wittinghofer A, et al. Effector recognition by the small GTP-binding proteins Ras and Ral. J Biol Chem. 1999;274:17763–70. doi: 10.1074/jbc.274.25.17763. [DOI] [PubMed] [Google Scholar]
- 4.Moskalenko S, Henry DO, Rosse C, Mirey G, Camonis JH, White MA. The exocyst is a Ral effector complex. Nat Cell Biol. 2002;4:66–72. doi: 10.1038/ncb728. [DOI] [PubMed] [Google Scholar]
- 5.Jullien-Flores V, Mahé Y, Mirey G, Leprince C, Meunier-Bisceuil B, Sorkin A, et al. RLIP76, an effector of the GTPase Ral, interacts with the AP2 complex: involvement of the Ral pathway in receptor endocytosis. J Cell Sci. 2000;113:2837–44. doi: 10.1242/jcs.113.16.2837. [DOI] [PubMed] [Google Scholar]
- 6.Rossé C, L’Hoste S, Offner N, Picard A, Camonis J. RLIP, an effector of the Ral GTPases, is a platform for Cdk1 to phosphorylate epsin during the switch off of endocytosis in mitosis. J Biol Chem. 2003;278:30597–604. doi: 10.1074/jbc.M302191200. [DOI] [PubMed] [Google Scholar]
- 7.Kashatus DF, Lim K-H, Brady DC, Pershing NLK, Cox AD, Counter CM. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat Cell Biol. 2011;13:1108–15. doi: 10.1038/ncb2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakashima S, Morinaka K, Koyama S, Ikeda M, Kishida M, Okawa K, et al. Small G protein Ral and its downstream molecules regulate endocytosis of EGF and insulin receptors. EMBO J. 1999;18:3629–42. doi: 10.1093/emboj/18.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cascone I, Selimoglu R, Ozdemir C, Del Nery E, Yeaman C, White M, et al. Distinct roles of RalA and RalB in the progression of cytokinesis are supported by distinct RalGEFs. EMBO J. 2008;27:2375–87. doi: 10.1038/emboj.2008.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawamoto K, Yamada C, Kishida S, Hirota Y, Taguchi A, Kikuchi A, et al. Ectopic expression of constitutively activated Ral GTPase inhibits cell shape changes during Drosophila eye development. Oncogene. 1999;18:1967–74. doi: 10.1038/sj.onc.1202522. [DOI] [PubMed] [Google Scholar]
- 11.Sugihara K, Asano S, Tanaka K, Iwamatsu A, Okawa K, Ohta Y. The exocyst complex binds the small GTPase RalA to mediate filopodia formation. Nat Cell Biol. 2002;4:73–8. doi: 10.1038/ncb720. [DOI] [PubMed] [Google Scholar]
- 12.Hase K, Kimura S, Takatsu H, Ohmae M, Kawano S, Kitamura H, et al. M-Sec promotes membrane nanotube formation by interacting with Ral and the exocyst complex. Nat Cell Biol. 2009;11:1427–32. doi: 10.1038/ncb1990. [DOI] [PubMed] [Google Scholar]
- 13.Balakireva M, Rossé C, Langevin J, Chien YC, Gho M, Gonzy-Treboul G, et al. The Ral/exocyst effector complex counters c-Jun N-terminal kinase-dependent apoptosis in Drosophila melanogaster. Mol Cell Biol. 2006;26:8953–63. doi: 10.1128/MCB.00506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chien Y, Kim S, Bumeister R, Loo Y-M, Kwon SW, Johnson CL, et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127:157–70. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 15.Feig LA. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 2003;13:419–25. doi: 10.1016/S0962-8924(03)00152-1. [DOI] [PubMed] [Google Scholar]
- 16.Mirey G, Balakireva M, L’Hoste S, Rossé C, Voegeling S, Camonis J. A Ral guanine exchange factor-Ral pathway is conserved in Drosophila melanogaster and sheds new light on the connectivity of the Ral, Ras, and Rap pathways. Mol Cell Biol. 2003;23:1112–24. doi: 10.1128/MCB.23.3.1112-1124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawamoto K, Winge P, Koyama S, Hirota Y, Yamada C, Miyao S, et al. The Drosophila Ral GTPase regulates developmental cell shape changes through the Jun NH(2)-terminal kinase pathway. J Cell Biol. 1999;146:361–72. doi: 10.1083/jcb.146.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghiglione C, Devergne O, Cerezo D, Noselli S. Drosophila RalA is essential for the maintenance of Jak/Stat signalling in ovarian follicles. EMBO Rep. 2008;9:676–82. doi: 10.1038/embor.2008.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho B, Fischer JA. Ral GTPase promotes asymmetric Notch activation in the Drosophila eye in response to Frizzled/PCP signaling by repressing ligand-independent receptor activation. Development. 2011;138:1349–59. doi: 10.1242/dev.056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eun SH, Lea K, Overstreet E, Stevens S, Lee J-H, Fischer JA. Identification of genes that interact with Drosophila liquid facets. Genetics. 2007;175:1163–74. doi: 10.1534/genetics.106.067959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Overstreet E, Fitch E, Fischer JA. Fat facets and Liquid facets promote Delta endocytosis and Delta signaling in the signaling cells. Development. 2004;131:5355–66. doi: 10.1242/dev.01434. [DOI] [PubMed] [Google Scholar]
- 22.Wang W, Struhl G. Drosophila Epsin mediates a select endocytic pathway that DSL ligands must enter to activate Notch. Development. 2004;131:5367–80. doi: 10.1242/dev.01413. [DOI] [PubMed] [Google Scholar]
- 23.Tian X, Hansen D, Schedl T, Skeath JB. Epsin potentiates Notch pathway activity in Drosophila and C. elegans. Development. 2004;131:5807–15. doi: 10.1242/dev.01459. [DOI] [PubMed] [Google Scholar]
- 24.Chen H, Ko G, Zatti A, Di Giacomo G, Liu L, Raiteri E, et al. Embryonic arrest at midgestation and disruption of Notch signaling produced by the absence of both epsin 1 and epsin 2 in mice. Proc Natl Acad Sci U S A. 2009;106:13838–43. doi: 10.1073/pnas.0907008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–89. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 26.Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Dev Cell. 2009;16:633–47. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Weinmaster G, Fischer JA. Notch ligand ubiquitylation: what is it good for? Dev Cell. 2011;21:134–44. doi: 10.1016/j.devcel.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fanto M, Mlodzik M. Asymmetric Notch activation specifies photoreceptors R3 and R4 and planar polarity in the Drosophila eye. Nature. 1999;397:523–6. doi: 10.1038/17389. [DOI] [PubMed] [Google Scholar]
- 29.Cooper MTD, Bray SJ. Frizzled regulation of Notch signalling polarizes cell fate in the Drosophila eye. Nature. 1999;397:526–30. doi: 10.1038/17395. [DOI] [PubMed] [Google Scholar]
- 30.Tomlinson A, Struhl G. Decoding vectorial information from a gradient: sequential roles of the receptors Frizzled and Notch in establishing planar polarity in the Drosophila eye. Development. 1999;126:5725–38. doi: 10.1242/dev.126.24.5725. [DOI] [PubMed] [Google Scholar]
- 31.del Alamo D, Mlodzik M. Frizzled/PCP-dependent asymmetric neuralized expression determines R3/R4 fates in the Drosophila eye. Dev Cell. 2006;11:887–94. doi: 10.1016/j.devcel.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 32.Strutt D, Johnson R, Cooper K, Bray S. Asymmetric localization of frizzled and the determination of notch-dependent cell fate in the Drosophila eye. Curr Biol. 2002;12:813–24. doi: 10.1016/S0960-9822(02)00841-2. [DOI] [PubMed] [Google Scholar]
- 33.Fortini ME, Bilder D. Endocytic regulation of Notch signaling. Curr Opin Genet Dev. 2009;19:323–8. doi: 10.1016/j.gde.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkin MB, Carbery AM, Fostier M, Aslam H, Mazaleyrat SL, Higgs J, et al. Regulation of notch endosomal sorting and signaling by Drosophila Nedd4 family proteins. Curr Biol. 2004;14:2237–44. doi: 10.1016/j.cub.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 35.Vaccari T, Lu H, Kanwar R, Fortini ME, Bilder D. Endosomal entry regulates Notch receptor activation in Drosophila melanogaster. J Cell Biol. 2008;180:755–62. doi: 10.1083/jcb.200708127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moberg KH, Schelble S, Burdick SK, Hariharan IK. Mutations in erupted, the Drosophila ortholog of mammalian tumor susceptibility gene 101, elicit non-cell-autonomous overgrowth. Dev Cell. 2005;9:699–710. doi: 10.1016/j.devcel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 37.Thompson BJ, Mathieu J, Sung H-H, Loeser E, Rørth P, Cohen SM. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev Cell. 2005;9:711–20. doi: 10.1016/j.devcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 38.Vaccari T, Bilder D. The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating notch trafficking. Dev Cell. 2005;9:687–98. doi: 10.1016/j.devcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 39.Herz HM, Chen Z, Scherr H, Lackey M, Bolduc C, Bergmann A. vps25 mosaics display non-autonomous cell survival and overgrowth, and autonomous apoptosis. Development. 2006;133:1871–80. doi: 10.1242/dev.02356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Childress JL, Acar M, Tao C, Halder G. Lethal giant discs, a novel C2-domain protein, restricts notch activation during endocytosis. Curr Biol. 2006;16:2228–33. doi: 10.1016/j.cub.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallagher CM, Knoblich JA. The conserved c2 domain protein lethal (2) giant discs regulates protein trafficking in Drosophila. Dev Cell. 2006;11:641–53. doi: 10.1016/j.devcel.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 42.Jaekel R, Klein T. The Drosophila Notch inhibitor and tumor suppressor gene lethal (2) giant discs encodes a conserved regulator of endosomal trafficking. Dev Cell. 2006;11:655–69. doi: 10.1016/j.devcel.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 43.Mukherjee A, Veraksa A, Bauer A, Rosse C, Camonis J, Artavanis-Tsakonas S. Regulation of Notch signalling by non-visual β-arrestin. Nat Cell Biol. 2005;7:1191–201. doi: 10.1038/ncb1327. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto S, Charng W-L, Bellen HJ. Endocytosis and intracellular trafficking of Notch and its ligands. Curr Top Dev Biol. 2010;92:165–200. doi: 10.1016/S0070-2153(10)92005-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan Y, Denef N, Schüpbach T. The vacuolar proton pump, V-ATPase, is required for notch signaling and endosomal trafficking in Drosophila. Dev Cell. 2009;17:387–402. doi: 10.1016/j.devcel.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pasternak SH, Bagshaw RD, Guiral M, Zhang S, Ackerley CA, Pak BJ, et al. Presenilin-1, nicastrin, amyloid precursor protein, and γ-secretase activity are co-localized in the lysosomal membrane. J Biol Chem. 2003;278:26687–94. doi: 10.1074/jbc.M304009200. [DOI] [PubMed] [Google Scholar]
- 47.Wilkin M, Tongngok P, Gensch N, Clemence S, Motoki M, Yamada K, et al. Drosophila HOPS and AP-3 complex genes are required for a Deltex-regulated activation of notch in the endosomal trafficking pathway. Dev Cell. 2008;15:762–72. doi: 10.1016/j.devcel.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 48.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–17. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 49.Zhang J, Schulze KL, Hiesinger PR, Suyama K, Wang S, Fish M, et al. Thirty-one flavors of Drosophila rab proteins. Genetics. 2007;176:1307–22. doi: 10.1534/genetics.106.066761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Entchev EV, Schwabedissen A, González-Gaitán M. Gradient formation of the TGF-beta homolog Dpp. Cell. 2000;103:981–91. doi: 10.1016/S0092-8674(00)00200-2. [DOI] [PubMed] [Google Scholar]
- 51.Bogard N, Lan L, Xu J, Cohen RS. Rab11 maintains connections between germline stem cells and niche cells in the Drosophila ovary. Development. 2007;134:3413–8. doi: 10.1242/dev.008466. [DOI] [PubMed] [Google Scholar]