Abstract
Squamous cell carcinoma of the head and neck (SCCHN) is the sixth most common cancer, globally. Previously, we showed that Rap1GAP is a tumor suppressor gene that inhibits tumor growth, but promotes invasion in SCCHN. In this work, we discuss the role of Rap1 and Rap1GAP in SCCHN progression in the context of a microRNA-oncogene-tumor suppressor gene axis, and investigate the role of Rap1GAP in EZH2-mediated invasion. Loss of expression of microRNA-101 in SCCHN leads to upregulation of EZH2, a histone methyltransferase. Overexpression of EZH2 silences Rap1GAP via methylation, thereby promoting activation of its target, Rap1. This microRNA-controlled activation of Rap1, via EZH2-mediated silencing of Rap1GAP, is a novel mechanism of Rap1 regulation. In two independent SCCHN cell lines, downregulation of EZH2 inhibits proliferation and invasion. In both cell lines, stable knockdown of EZH2 (shEZH2) recovers Rap1GAP expression and inhibits proliferation. However, siRNA-mediated knockdown of Rap1GAP in these cells rescues proliferation but not invasion. Thus, EZH2 promotes proliferation and invasion via Rap1GAP-dependent and –independent mechanisms, respectively. Although the studies presented here are in the context of SCCHN, our results may have broader implications, given that Rap1GAP acts as a tumor suppressor in pancreatic cancer, thyroid cancer, and melanoma.
Keywords: EZH2, Rap1GAP, methylation, miR-101, translational
Introduction
Squamous cell carcinoma of the head and neck (SCCHN) is the sixth most common cancer, globally, with approximately 600,000 new cases every year.1 At ~50%, the 5-y survival rate is poorer than breast cancer or melanoma.2 The poor disease-specific survival of SCCHN is due to late detection, which is a consequence of inadequate screening protocols.3 Hence it is important to identify and characterize biomarkers for early detection of SCCHN.
Treatment for SCCHN is selected according to tumor stage; late stage lesions receive more aggressive treatment than early stage lesions, which typically have a better prognosis.3 However, a significant number of patients with Stage I disease die in less than five years.3 Identification of biomarkers that predict the biologic behavior and response to treatment of a lesion are required to facilitate selection of the most effective treatment and to serve as treatment targets. Recent studies suggested that tumor suppressor genes play an important role in development and progression of SCCHN,4,5 supporting their investigation as biomarkers and treatment targets.
Patients with late stage SCCHN are treated with aggressive surgery/radiation or chemotherapy/radiation. The surviving patient must confront challenges that include disfigurement, feeding and speech difficulties, and poor quality of life. Due to minimal improvement in patient survival in four decades, the emphasis moved to dose escalation of radiation and chemotherapy. However, toxic effects limit the extent to which therapy can be intensified. The chemotherapy/radiation protocol led to organ preservation and improved quality of life.6,7 However, ~20–25% of patients do not respond to chemotherapy and are subsequently treated with surgery.7,8 The delay in effective treatment and aggressive residual cells lead to poor patient survival.8 Hence, new treatments must be identified to improve survival.
Rap1 is a member of the ras subfamily of small GTPases that are critical players in the signaling pathways that control cell growth and differentiation.9 Rap1 exhibits close homology to ras in the transformation domains, which include the GTP binding region, effector domain and membrane attachment site.10 Ras is one of the most frequently mutated genes in cancer but ras mutations in SCCHN are rare.4,5,11 The sequence identity in the effector domains of ras and Rap1 suggested either antagonistic or complementary activity in the same signaling cascade.12
Rap1 shuttles between an inactive GDP- and active GTP-bound form. Activation of Rap1 (Rap1-GTP) is mediated by four classes of guanine nucleotide exchange factors (GEFs), including C3G, PDZ-GEF, Epac, and CalDAG. Inactivation of Rap1 is mediated by GTPase activating proteins, including Rap1GAPI and Rap1GAPII, Spa1/SIPA1 and SIPA1L1/E6TP1.13 In SCCHN, Rap1 activation may be induced by cytokines, including galanin, which facilitate tumor progression.14,15 In a recent study, we showed a novel mechanism for sustained activation of Rap1 via downregulation of microRNA-101 (miR-101). Loss of expression of miR-101 upregulates EZH2, which promotes di or tri methylation at lysine 27 of histone H3, resulting in chromatin condensation as well as promoter hypermethylation, thereby silencing Rap1GAP.16 In the work presented here, we will discuss the role of Rap1 and Rap1GAP in SCCHN progression and how a tumor suppressor-oncogene-tumor suppressor gene axis promotes proliferation and invasion via Rap1GAP-dependent and –independent mechanisms, respectively.
Active, GTP-bound Rap1 is Upregulated in SCCHN
Although Rap1 is a ubiquitously expressed protein, till recently its function in epithelial cells, from which more than 80% of cancers are derived, was relatively unexplored. In normal epithelium, Rap1 regulates dynamic as well as local processes such as cell adhesion, cell migration, polarity and secretion.17 Rap1 controls the assembly and function of epithelial cell-cell junctions so that individual cells adhere to the extracellular matrix through integrin and cadherin complexes and maintain complex tissues.17
In early studies, we showed that active Rap1 inhibits proliferation in normal oral keratinocytes and inactivation of Rap1 reverses this phenotype.18 However, in malignant keratinocytes, which exhibit uncontrolled proliferation, active Rap1 was highly expressed,19 consistent with disrupted signaling mechanisms in cancer cells. Rap1GAP promotes inactivation of Rap1-GTP by inducing its endogenous GTPase activity. Subsequently, we showed that upregulation of active Rap1 in SCCHN is due to reduced expression of Rap1GAP in SCCHN as well as cytokine-mediated stimulation of Rap1, which induces cell proliferation and survival.14,20
Rap1GAP is a Tumor Suppressor Gene and Promotes Invasion in SCCHN
In SCCHN, a significant number of patients with early stage disease ultimately die of disease, despite appropriate treatment selection.3 These patients would likely benefit from more aggressive initial therapy. However, aggressive surgery and radiation treatment are physically and emotionally debilitating and are not appropriate for all SCCHN. Therefore, the identification of biomarkers that are prognostic of tumor progression and elucidation of the mechanism of action of these proteins will facilitate appropriate treatment selection.
Invasion promotes tumor extension into the underlying structures and spread to distant sites.21 In pre-cancerous oral lesions (epithelial dysplasia), proliferation, destruction of the basement membrane and detachment of epithelial cells with invasion and migration into the underlying tissues are essential for transformation to cancer and spread to other sites (metastasis). Thus, invasion has an essential role in malignant transformation, but the mechanism of regulation is relatively unexplored.21
Studies in SCCHN supported a novel function for Rap1GAP as a tumor suppressor protein.20 Overexpression of Rap1GAP in SCCHN delayed progression through the cell cycle, thereby reducing cell proliferation in vitro and tumor growth in vivo.20 Although these data suggested that upregulation of Rap1GAP expression in SCCHN would be prognostic of slower growing lesions with a favorable prognosis, subsequent studies showed that Rap1GAP promotes invasion.22 In SCCHN, counter-intuitive to its tumor suppressor role, Rap1GAP promotes invasion via upregulation of MMP2 and MMP9, matrix metalloproteinases that facilitate degradation of the extracellular matrix.22
Furthermore, high MMP9 is correlated with poor disease-specific survival in SCCHN.22 These findings from a small group of patients suggested that pre-treatment screening for MMP9 in larger clinical trials may identify those patients with early N-stage lesions (no metastasis to lymph nodes or a single ipsilateral lymph node) to benefit from aggressive treatment.22 This is an area that needs further investigation.
Rap1 May Promote or Inhibit Adhesion and Invasion Depending on the Cell Type
Epithelial cells adhere to adjacent epithelial cells via adherens junctions leading to the sheet-like architecture of epithelium.21 Rap1 has an important role in adhesion and invasion, which are inversely correlated.23-25 In drosophila, adherens junctions are uniformly distributed around the circumference of cells with wild type Rap1, but are clustered on one side of cells with inactivating Rap1 mutations.25 The mutant cell clones randomly invade surrounding tissues, suggesting that wild type Rap1 promotes cell adhesion and inhibits invasion. A recent study in prostate cancer showed that SPA-1, a Rap1-specific GAP that decreases active Rap1, promotes invasion and metastasis.26 This study showed that upregulation of SPA-1 expression was correlated with prostate cancer progression and metastasis.26 Furthermore, upregulation of SPA-1 in prostate cancer cell lines promotes lymph node metastasis. Conversely, overexpression of constitutively active Rap1, which is resistant to SPA-1, reduced invasion and lymph node metastasis. Together, these studies in multiple cell types suggested that active Rap1 promotes cell adhesion and inhibits invasion.
A different study, in prostate cancer cells, showed that inactivation of Rap1A by Rap1GAP inhibited invasion and metastasis.27 In colon cancer cells, Rap1GAP depletion activated Rap1, reduced E-cadherin, disrupted cell-cell junctions and promoted cell-matrix adhesion, which promoted tumor cell dissemination.28
The apparent disparity in effects of Rap1 on cell adhesion/invasion between cell types may be due to its different isoforms. Rap1A and Rap1B share 95% sequence identity, and are encoded by two different genes on chromosomes 1 and 12, respectively.29,30 The two Rap1 isoforms are expressed in the same cell19,31 where their functions may differ. A recent study in endothelial cells showed that Rap1A, but not Rap1B, promotes cell junction formation.31 In SCCHN, Rap1A promotes invasion via β-catenin-induced transcriptional targets.32 Since Rap1GAP, which inhibits both Rap1 isoforms, promotes invasion in SCCHN, it is likely that Rap1B inhibits invasion.
Alternatively, the apparently contradictory effects of Rap1 on invasion/ adhesion in drosophila and colon cancer/SCCHN may be related to the exchange factors, C3G and Epac that activate Rap1 or other critical regulatory proteins. By localizing at intracellular or surface membrane sites, these activating factors direct Rap1 to different effectors.33 For example, the ERK signaling pathway is activated by surface membrane-bound C3G/Rap1 but not by intracellular Epac/Rap1.33 These possibilities are under investigation.
Rap1GAP is Silenced by EZH2
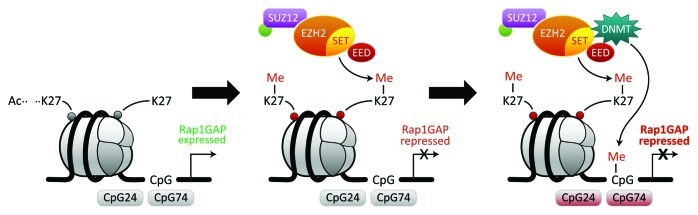
The Rap1GAP studies conclusively showed that therapeutic inhibition of Rap1GAP in SCCHN would not be judicious due to the possibility of inhibiting tumor growth but promoting aggressive tumors by promoting invasion. Therefore, we investigated upstream mechanisms of regulation that would concurrently silence Rap1GAP and other tumor suppressor genes to inhibit multiple phenotypes such as invasion and proliferation. In SCCHN, methylation is an important epigenetic mechanism for silencing tumor suppressor genes.16 Methylation-mediated epigenetic silencing may occur via methylation of lysine residues in histones and/ or via methylation of CpG islands by DNA methyltransferase enzymes (DNMT1, DNMT3A, and DNMT3B) that transfer a methyl group from S-adenosyl-l-methionine to cytosines in CpG dinucleotides in the promoter region of genes.34-36 Enhancer of zeste homolog 2 (EZH2) is a histone methyltransferase that in combination with histone deacetylase promotes cancer development and progression by targeting tumor suppressor genes.37,38 EZH2, a master regulatory gene and catalytic member of the polycomb repressive complex 2 (PRC2), modulates multiple cell functions. EZH2 silences Rap1GAP in SCCHN via trimethylation of histone 3 at lysine 27 (H3K27)16 (Fig. 1). Upregulation of EZH2 also facilitates methylation of the CpG islands in the promoter region of Rap1GAP.16 Consistent with histone deacetylation preceding methylation, histone and promoter hypermethylation of Rap1GAP were shown to be inhibited by suberoylanilide hydroxamic acid (SAHA), a histone deacetylase (HDAC) inhibitor. Also, promoter hypermethylation of Rap1GAP, mediated by DNA methyltransferase (DNMT), was blocked by 5-aza-2’-deoxycytidine (AZA), a DNA methyltransferase inhibitor.16Figure 1 depicts progressive silencing of Rap1GAP as a function of histone and promoter hypermethylation. When histones are acetylated, H3K27 is not methylated and Rap1GAP is expressed (Fig. 1, left panel). The deacetylation of H3K27 leads to di- and tri-methylation of H3K27 by EZH2, and repression of Rap1GAP (Fig. 1, middle panel). EZH2 facilitates DNA methyltransferase-mediated methylation of CpG islands in the promoter region of Rap1GAP thereby further repressing Rap1GAP (Fig. 1, right panel).
Figure 1. EZH2 silences Rap1GAP by histone methylation and by facilitating DNMT-mediated methylation of the promoter region. Interaction of the PRC2 complex with Rap1GAP as a target gene. Rap1GAP is expressed in the absence of methylation at histone (Histone 3 at lysine 27 or H3K27) and promoter hypermethylation (left panel). EZH2, the catalytic component of the PRC2 complex, di- and tri-methylates H3K27 resulting in chromatin compaction, the mark of repression. Consequently, Rap1GAP is silenced (middle panel). CpG islands in the promoter region of Rap1GAP are further methylated by DNA methyl transferase (DNMT) recruited by PRC2 complex. The coordination of these enzymes results in further repression and epigenetic silencing of Rap1GAP (right panel). Methylation and consequent silencing are denoted by red.
EZH2-Induced Proliferation but Not Invasion is Mediated by Suppression of Rap1GAP
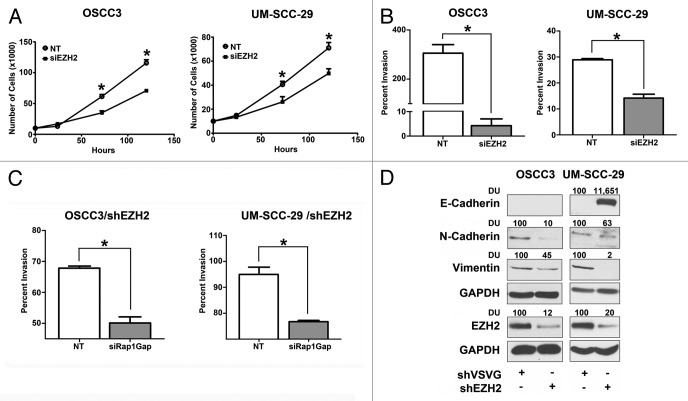
In functional assays, EZH2 promoted proliferation, invasion and survival of immortalized, non-malignant oral keratinocytes.16 Furthermore, overexpression of EZH2 in immortalized keratinocytes suppressed Rap1GAP expression resulting in Rap1 activation. Downregulation of EZH2 in malignant keratinocytes (SCCHN) inhibits proliferation and invasion (Fig. 2A, B), as shown previously in an independent experiment.16 In cells with stable downregulation of EZH2, siRNA-mediated knockdown of Rap1GAP “rescued” the proliferative phenotype of EZH2, establishing that EZH2 promotes proliferation by suppressing Rap1GAP.16 Thus, although EZH2 silences several tumor suppressor genes, suppression of Rap1GAP is required for EZH2-mediated proliferation.16 This emphasizes the critical role of the tumor suppressor Rap1GAP, in SCCHN.
Figure 2. EZH2 promotes proliferation and invasion. OSCC3 and UM-SCC-29 cells were transfected with non-target siRNA or siEZH2 and proliferation (A) and invasion (B) assays were performed (This has been shown in an independent set of experiments previously). (C) OSCC3 and UM-SCC-29 cells stably transduced with shEZH2 were subsequently transfected with non-target siRNA or siRap1GAP. Invasion assays were performed (*p < 0.05). (D) Whole ell lysates from OSCC3 and UM-SCC-29 cells stably transduced with shEZH2 were immunoblotted with E-Cadherin, N-Cadherin, Vimentin and EZH2 antibodies. GAPDH was used as a normalization control. Signal intensity was quantified by densitometric analysis, normalized to GAPDH and then expressed as percent of control.
We previously established that Rap1GAP in SCCHN promotes invasion via MMP9.22 Although EZH2 suppresses Rap1GAP, EZH2 concurrently promotes invasion (Fig. 2B), suggesting that its effects on invasion are independent of Rap1GAP. To investigate this possibility, invasion assays were performed in two independent SCCHN cell lines stably transduced with shEZH2 and transiently transfected with siRap1GAP. Knockdown of Rap1GAP did not “rescue” EZH2-mediated invasion and instead, inhibited invasion in both cell lines (Fig. 2C). These findings independently support our previous study showing that Rap1GAP promotes invasion.22
EZH2-mediated invasion in breast cancer occurs via downregulation of E-cadherin.37 In the studies presented here, an increase in Rap1GAP expression in stable knockdown of EZH2 in SCCHN cell lines (shEZH2) that were characterized elsewhere,16 decreased the mesenchymal markers, vimentin and N-cadherin, in two SCCHN cell lines (Fig. 2D). E-cadherin was not nominated as an EZH2 target in SCCHN16 and may not be directly correlated with the invasive phenotype (2D, left panel). Notably, E-cadherin expression was absent in OSCC3 cells, regardless of invasion. Together, these studies support that EZH2-mediated invasion occurs via a Rap1GAP-independent mechanism and is not dependent on E-cadherin in SCCHN. The mechanism of EZH2-mediated invasion in SCCHN is currently under investigation.
MicroRNA Regulates Rap1 Activation
miR-101, which suppresses EZH2, is downregulated in SCCHN.16 This leads to overexpression of EZH2, which silences Rap1GAP. Consequently, Rap1 is activated in SCCHN. This microRNA-mediated activation of Rap1, via EZH2-mediated silencing of Rap1GAP, is a distinctive mechanism of Rap1 regulation. It is likely that other miRs also regulate Rap1. A potential regulatory miR is miR-138, since a recent study shows that it suppresses EZH2 expression in SCCHN.39 There may be other microRNAs that directly repress Rap1GAP or Rap1 in an EZH2-independent manner, but such a mechanism is yet to be described.
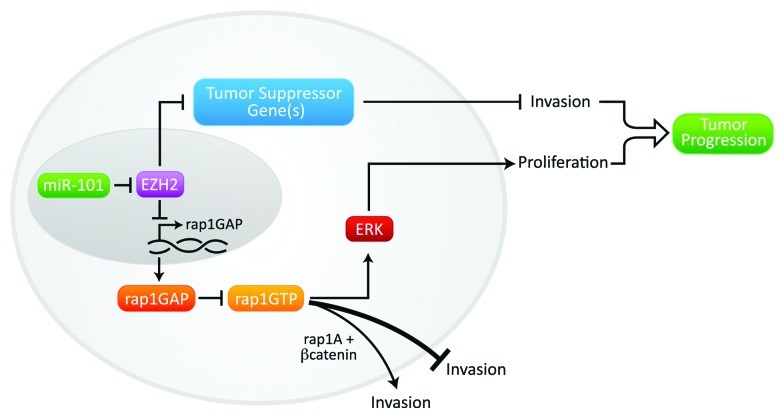
We have summarized our present knowledge about the mir-101-EZH2-Rap1GAP/Rap1 axis in Figure 3. Our findings with Rap1GAP emphasize the need to carefully define the signaling pathways regulated by a putative treatment target, and to correlate functional effects with clinical outcomes prior to developing treatment strategy. Though Rap1GAP inhibits tumor growth, it also promotes invasion in SCCHN. Therefore, therapeutic targeting of Rap1GAP in SCCHN would not be judicious due to the possibility of fostering small but aggressive tumors.
Figure 3. Proposed mechanism. miR-101 expression is reduced in SCCHN thereby contributing to overexpression of EZH2. EZH2-mediated silencing of Rap1GAP promotes proliferation. EZH2 promotes invasion via a Rap1GAP-independent mechanism, via silencing other tumor suppressor gene(s). The effects of Rap1GAP on invasion are likely modulated by the individual Rap1 isoforms or the concurrent expression of critical signaling molecules, such as β-catenin.
The polycomb group protein EZH2, is an attractive target because it regulates multiple genes via chromatin remodeling. It is a histone methyltransferase that interacts with histone deacetylase to silence genes thereby modulating several cell functions. The critical role of EZH2 in SCCHN,16 and correlation of EZH2 expression with poor survival in SCCHN,40 suggest that it is a fascinating treatment target. EZH2 silences multiple tumor suppressor genes in SCCHN by histone methylation and by facilitating DNA methylation.16 3-Deazaneplanocin A (DZNep), an antagonist of EZH2, inhibits H3K27 tri-methylation and downregulates EZH2 expression.41,42 DZNep inhibits tumorigenesis in mice with cell lines from solid tumors, such as glioblastoma, breast cancer and colorectal cancer,42,43 but has not been tested in SCCHN. A recent study showed that a small percentage (5%) of SCCHN may have deletion mutations in EZH2.5 This may have implications for overexpression of Rap1GAP and treatment that targets EZH2.
Conclusion
Our studies in SCCHN indicate that new treatments must be developed since current regimens have not improved survival in several decades. Targeted therapy against single growth factors and their receptors had limited success in SCCHN. This is likely due to simultaneous alteration of multiple genes, or cross-talk between signaling pathways that affect tumor growth, spread and treatment resistance. Delineation of the critical signaling molecules in multiple signaling cascades will facilitate the identification of appropriate treatment targets.
Although the studies described here are in the context of SCCHN, these findings may have broader implications, as tumor suppressor role for Rap1GAP has been established in studies in pancreatic cancer, thyroid cancer and melanoma and support our findings.
Acknowledgments
We thank Dr. Ram-S Mani and Christina S. Scanlon for their thoughtful comments and critical reading of the manuscript.
Financial Support
This work was supported by NIDCR DE018512; DE019513; DE017977 grants (NJD).
Footnotes
Previously published online: www.landesbioscience.com/journals/smallgtpases/article/20413
References
- 1.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 2.Todd R, Donoff RB, Wong DT. The molecular biology of oral carcinogenesis: toward a tumor progression model. J Oral Maxillofac Surg. 1997;55:613–23, discussion 623-5. doi: 10.1016/S0278-2391(97)90495-X. [DOI] [PubMed] [Google Scholar]
- 3.Bsoul SA, Huber MA, Terezhalmy GT. Squamous cell carcinoma of the oral tissues: a comprehensive review for oral healthcare providers. J Contemp Dent Pract. 2005;6:1–16. [PubMed] [Google Scholar]
- 4.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–7. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar B, Cordell KG, Lee JS, Prince ME, Tran HH, Wolf GT, et al. Response to therapy and outcomes in oropharyngeal cancer are associated with biomarkers including human papillomavirus, epidermal growth factor receptor, gender, and smoking. Int J Radiat Oncol Biol Phys. 2007;69(Suppl):S109–11. doi: 10.1016/j.ijrobp.2007.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Urba S, Wolf G, Eisbruch A, Worden F, Lee J, Bradford C, et al. Single-cycle induction chemotherapy selects patients with advanced laryngeal cancer for combined chemoradiation: a new treatment paradigm. J Clin Oncol. 2006;24:593–8. doi: 10.1200/JCO.2005.01.2047. [DOI] [PubMed] [Google Scholar]
- 8.Bauer JA, Kumar B, Cordell KG, Prince ME, Tran HH, Wolf GT, et al. Targeting apoptosis to overcome cisplatin resistance: a translational study in head and neck cancer. Int J Radiat Oncol Biol Phys. 2007;69(Suppl):S106–8. doi: 10.1016/j.ijrobp.2007.05.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bokoch GM. Biology of the Rap proteins, members of the ras superfamily of GTP-binding proteins. Biochem J. 1993;289:17–24. doi: 10.1042/bj2890017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrell F, Torti M, Lapetina EG. Rap proteins: investigating their role in cell function. J Lab Clin Med. 1992;120:533–7. [PubMed] [Google Scholar]
- 11.Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 12.Altschuler DL, Peterson SN, Ostrowski MC, Lapetina EG. Cyclic AMP-dependent activation of Rap1b. J Biol Chem. 1995;270:10373–6. doi: 10.1074/jbc.270.18.10373. [DOI] [PubMed] [Google Scholar]
- 13.Frische EW, Zwartkruis FJ. Rap1, a mercenary among the Ras-like GTPases. Dev Biol. 2010;340:1–9. doi: 10.1016/j.ydbio.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 14.Banerjee R, Henson BS, Russo N, Tsodikov A, D’Silva NJ. Rap1 mediates galanin receptor 2-induced proliferation and survival in squamous cell carcinoma. Cell Signal. 2011;23:1110–8. doi: 10.1016/j.cellsig.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henson BS, Neubig RR, Jang I, Ogawa T, Zhang Z, Carey TE, et al. Galanin receptor 1 has anti-proliferative effects in oral squamous cell carcinoma. J Biol Chem. 2005;280:22564–71. doi: 10.1074/jbc.M414589200. [DOI] [PubMed] [Google Scholar]
- 16.Banerjee R, Mani RS, Russo N, Scanlon CS, Tsodikov A, Jing X, et al. The tumor suppressor gene rap1GAP is silenced by miR-101-mediated EZH2 overexpression in invasive squamous cell carcinoma. Oncogene. 2011;30:4339–49. doi: 10.1038/onc.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gloerich M, Bos JL. Regulating Rap small G-proteins in time and space. Trends Cell Biol. 2011;21:615–23. doi: 10.1016/j.tcb.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 18.D’Silva NJ, Mitra RS, Zhang Z, Kurnit DM, Babcock CR, Polverini PJ, et al. Rap1, a small GTP-binding protein is upregulated during arrest of proliferation in human keratinocytes. J Cell Physiol. 2003;196:532–40. doi: 10.1002/jcp.10331. [DOI] [PubMed] [Google Scholar]
- 19.Mitra RS, Zhang Z, Henson BS, Kurnit DM, Carey TE, D’Silva NJ. Rap1A and rap1B ras-family proteins are prominently expressed in the nucleus of squamous carcinomas: nuclear translocation of GTP-bound active form. Oncogene. 2003;22:6243–56. doi: 10.1038/sj.onc.1206534. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Z, Mitra RS, Henson BS, Datta NS, McCauley LK, Kumar P, et al. Rap1GAP inhibits tumor growth in oropharyngeal squamous cell carcinoma. Am J Pathol. 2006;168:585–96. doi: 10.2353/ajpath.2006.050132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 22.Mitra RS, Goto M, Lee JS, Maldonado D, Taylor JM, Pan Q, et al. Rap1GAP promotes invasion via induction of matrix metalloproteinase 9 secretion, which is associated with poor survival in low N-stage squamous cell carcinoma. Cancer Res. 2008;68:3959–69. doi: 10.1158/0008-5472.CAN-07-2755. [DOI] [PubMed] [Google Scholar]
- 23.Balzac F, Avolio M, Degani S, Kaverina I, Torti M, Silengo L, et al. E-cadherin endocytosis regulates the activity of Rap1: a traffic light GTPase at the crossroads between cadherin and integrin function. J Cell Sci. 2005;118:4765–83. doi: 10.1242/jcs.02584. [DOI] [PubMed] [Google Scholar]
- 24.Hogan C, Serpente N, Cogram P, Hosking CR, Bialucha CU, Feller SM, et al. Rap1 regulates the formation of E-cadherin-based cell-cell contacts. Mol Cell Biol. 2004;24:6690–700. doi: 10.1128/MCB.24.15.6690-6700.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knox AL, Brown NH. Rap1 GTPase regulation of adherens junction positioning and cell adhesion. Science. 2002;295:1285–8. doi: 10.1126/science.1067549. [DOI] [PubMed] [Google Scholar]
- 26.Shimizu Y, Hamazaki Y, Hattori M, Doi K, Terada N, Kobayashi T, et al. SPA-1 controls the invasion and metastasis of human prostate cancer. Cancer Sci. 2011;102:828–36. doi: 10.1111/j.1349-7006.2011.01876.x. [DOI] [PubMed] [Google Scholar]
- 27.Bailey CL, Kelly P, Casey PJ. Activation of Rap1 promotes prostate cancer metastasis. Cancer Res. 2009;69:4962–8. doi: 10.1158/0008-5472.CAN-08-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsygankova OM, Ma C, Tang W, Korch C, Feldman MD, Lv Y, et al. Downregulation of Rap1GAP in human tumor cells alters cell/matrix and cell/cell adhesion. Mol Cell Biol. 2010;30:3262–74. doi: 10.1128/MCB.01345-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rousseau-Merck MF, Pizon V, Tavitian A, Berger R. Chromosome mapping of the human RAS-related RAP1A, RAP1B, and RAP2 genes to chromosomes 1p12----p13, 12q14, and 13q34, respectively. Cytogenet Cell Genet. 1990;53:2–4. doi: 10.1159/000132883. [DOI] [PubMed] [Google Scholar]
- 30.Takai Y, Kaibuchi K, Kikuchi A, Sasaki T, Shirataki H. Regulators of small GTPases. Ciba Found Symp. 1993;176:128–38, discussion 138-46. doi: 10.1002/9780470514450.ch9. [DOI] [PubMed] [Google Scholar]
- 31.Wittchen ES, Hartnett ME. The small GTPase Rap1 is a novel regulator of RPE cell barrier function. Invest Ophthalmol Vis Sci. 2011;52:7455–63. doi: 10.1167/iovs.11-7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goto M, Mitra RS, Liu M, Lee J, Henson BS, Carey T, et al. Rap1 stabilizes beta-catenin and enhances beta-catenin-dependent transcription and invasion in squamous cell carcinoma of the head and neck. Clin Cancer Res. 2010;16:65–76. doi: 10.1158/1078-0432.CCR-09-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Dillon TJ, Pokala V, Mishra S, Labudda K, Hunter B, et al. Rap1-mediated activation of extracellular signal-regulated kinases by cyclic AMP is dependent on the mode of Rap1 activation. Mol Cell Biol. 2006;26:2130–45. doi: 10.1128/MCB.26.6.2130-2145.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herceg Z, Hainaut P. Genetic and epigenetic alterations as biomarkers for cancer detection, diagnosis and prognosis. Mol Oncol. 2007;1:26–41. doi: 10.1016/j.molonc.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kondo T, Asa SL, Ezzat S. Epigenetic dysregulation in thyroid neoplasia. Endocrinol Metab Clin North Am. 2008;37:389–400, ix. doi: 10.1016/j.ecl.2007.12.002. [ix.] [DOI] [PubMed] [Google Scholar]
- 36.McCabe MT, Brandes JC, Vertino PM. Cancer DNA methylation: molecular mechanisms and clinical implications. Clin Cancer Res. 2009;15:3927–37. doi: 10.1158/1078-0432.CCR-08-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao Q, Yu J, Dhanasekaran SM, Kim JH, Mani RS, Tomlins SA, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27:7274–84. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 39.Lu J, He ML, Wang L, Chen Y, Liu X, Dong Q, et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011;71:225–33. doi: 10.1158/0008-5472.CAN-10-1850. [DOI] [PubMed] [Google Scholar]
- 40.Kidani K, Osaki M, Tamura T, Yamaga K, Shomori K, Ryoke K, et al. High expression of EZH2 is associated with tumor proliferation and prognosis in human oral squamous cell carcinomas. Oral Oncol. 2009;45:39–46. doi: 10.1016/j.oraloncology.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 41.Fiskus W, Wang Y, Sreekumar A, Buckley KM, Shi H, Jillella A, et al. Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cells. Blood. 2009;114:2733–43. doi: 10.1182/blood-2009-03-213496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–63. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suvà ML, Riggi N, Janiszewska M, Radovanovic I, Provero P, Stehle JC, et al. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 2009;69:9211–8. doi: 10.1158/0008-5472.CAN-09-1622. [DOI] [PubMed] [Google Scholar]