Abstract
Two inducible temperate bacteriophages ΦS9 and ΦS63 from Clostridium perfringens were sequenced and analyzed. Isometric heads and long non-contractile tails classify ΦS9 and ΦS63 in the Siphoviridae family, and their genomes consist of 39,457 bp (ΦS9) and 33,609 bp (ΦS63) linear dsDNA, respectively. ΦS63 has 3′-overlapping cohesive genome ends, whereas ΦS9 is the first Clostridium phage featuring an experimentally proven terminally redundant and circularly permuted genome. A total of 50 and 43 coding sequences were predicted for ΦS9 and ΦS63, respectively, organized into 6 distinct lifestyle-associated modules typical for temperate Siphoviruses. Putative functions could be assigned to 26 gene products of ΦS9, and to 25 of ΦS63. The ΦS9 attB attachment and insertion site is located in a non-coding region upstream of a putative phosphorylase gene. Interestingly, ΦS63 integrates into the 3′ part of sigK in C. perfringens, and represents the first functional skin-element-like phage described for this genus. With respect to possible effects of lysogeny, we did not obtain evidence that ΦS9 may influence sporulation of a lysogenized host. In contrast, interruption of sigK, a sporulation associated gene in various bacteria, by the ΦS63 prophage insertion is more likely to affect sporulation of its carrier.
Keywords: Clostridium perfringens, prophage, attachment site, sporulation, skin-element
Introduction
Clostridium perfringens is an anaerobic Gram-positive spore-forming rod, frequently isolated from soil, freshwater sediments, sewage and the gastrointestinal tract of both humans and animals. It is the causative agent of food poisoning and gas gangrene in humans, and enteric diseases in these hosts. Fourteen types of toxins are known so far;1 among them, α- (phospholipase C), β-, ε- and ι-toxins are used to classify C. perfringens into five biotypes (A–E). Others include θ-toxin (perfringolysin O), µ-toxin (hyaluronidase), κ-toxin (collagenase), a sporulation-associated (food-poisoning) enterotoxin (CPE), the structure of which was recently solved,2,3 and others.1 The TpeL-toxin, which is produced during sporulation, is another addition to that family.4,5 Phenotypic variations among the isolates, such as different toxins produced and various degrees of symptoms severity could mainly be attributed to a high degree of genomic variability, as evidenced from comparative genomic studies using three complete C. perfringens (biotype A strains) genome sequences (a food poisoning strain S13, a CPE-negative gas gangrene isolate ATCC13124 and a CPE-negative and gas gangrene-causing strain (SM101)).6,7 In addition to the variation in chromosome-encoded toxin/virulence genes, large plasmids with strain-specific genes8,9 were identified, offering insights into a wide range of environmental adaptations and virulence traits.6 Interestingly, no clear explanation regarding the extremely diverse sporulation efficiencies among the isolates could yet be found. Many of the above mentioned genes appear to be located in mobile elements, or are transferred via conjugational processes.6
Bacterial chromosomes contain a significant proportion of prophage sequences, as mobilizable elements. For example, Streptococcus pyogenes features a genome with more than 10% phage-related sequences,10 and in Escherichia coli O157:H7 strain Sakai, prophage elements account for 16% of the total genome.11 These elements are involved in horizontal gene transfer and their characteristics offer insights into evolutionary processes of the host.12 In addition, prophages often encode virulence genes such as toxins, and provide an explanation for various bacterial virulence characteristics among the different strains.10,12 Thus far, only 12 C. perfringens phage sequences are available from public databases. Prophage Φ362613 was the first C. perfringens phage sequence published, later followed by episomal prophage ΦSM101 identified in sequenced C. perfringens genomes.6 Recently, sequences of phages ΦCP39O and ΦCP26F, as well as ΦCP9O, ΦCP13O and ΦCP3O were reported.14,15 The Podovirus ΦCPV1 was described as the smallest C. perfringens phage isolated so far, both in terms of particle dimensions and DNA size.16 Three recently described virulent podoviruses feature slightly bigger genomes of approximately 18 kb.17 Some trials using C. perfringens specific bacteriophages (CPAS-cocktail) to counteract necrotic enterocolitis have been published.18 Phage ΦCP24R was described as a small virulent podovirus featuring an 18.92 kb genome.19
We here report the sequence and analysis of temperate phages ΦS9 (vB_CpeS-PhiS9) and ΦS63 (vB_CpeS-PhiS63), induced from C. perfringens strains S9 and S63, respectively. We determined and compared their physical genome structures and phage integration sites. ΦS9 was previously reported to influence sporulation of C. perfringens,20 which prompted us to investigate the effects of lysogenic conversion of C. perfringens by ΦS9 and ΦS63.
Results
ΦS9 and ΦS63 are Siphoviruses
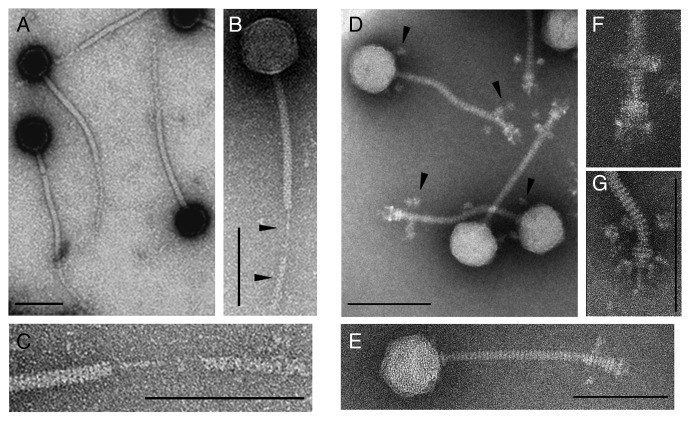
Transmission electron microscopy revealed an icosahedral head (60.4 nm in size) and a long non-contractile tail (Figs. 1A–C) for ΦS9, placing it into the family Siphoviridae in the order of the Caudovirales.21 The tail structure is rather unusual since that it lacks a typical baseplate structure and features a tail fiber cover-like structure (TFC) instead (Fig. 1B, black arrows and 1C). This component was found to quite easily separate from the tail during particle preparation for EM. The tail shaft is 191 nm long and 11 nm wide, and the prominent central tail fiber itself is 46.6 nm long, 3.2 nm wide. Dimension of the TFC is approximately 102 nm long and 10 nm wide (Fig. 1C).
Figure 1. Transmission electron micrographs of negatively stained C. perfringens phage particles. (A–C) ΦS9 and close-up view of the tail fiber cover-like structure. (D–G) ΦS63 and close-up view of the tail adsorption apparatus. Scale bars in images represent 100 nm; scale bars in (F and G) could not be drawn due to size limitations.
Phage ΦS63 also belongs to the Siphoviridae, featuring a 170 nm tail with a diameter of 11 nm and a head of 62 nm diameter (Fig. 1D–G). In contrast to ΦS9, the ΦS63 tail features a classical baseplate structure (Fig. 1F and G). Putative baseplate spikes are visible at the lower end of the base plate (Fig. 1G). It is interesting to note that in all negatively stained ΦS63 particles, very unusual and satellite bubble-like structures are present, arranged around the lower part of the tail just above the base plate, and at the upper portion of the tail just below the head-tail connector (Fig. 1D, black arrows). These structures most likely represent curled tail fibers and/or long whiskers, likely involved in the recognition of and/or interaction with the host cells surface.
Complete nucleotide sequence and genome organization of ΦS9 and ΦS63
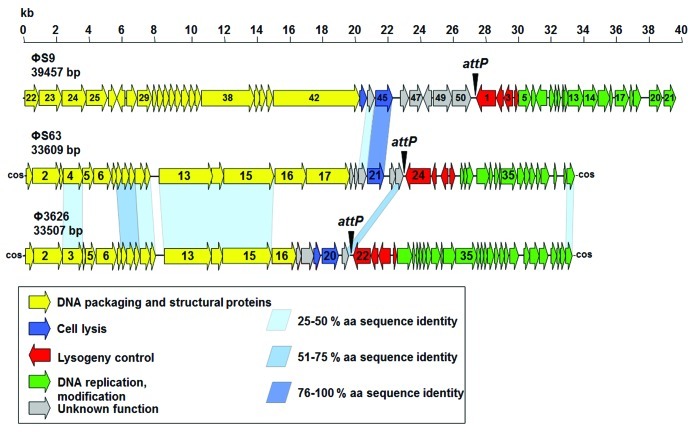
The complete unit genome (not considering possible redundancy of the packaged DNA molecule) of ΦS9 features 39,457 bp, which agrees well with the overall size predicted from restriction analysis (Figs. 2 and 3). The GC content of 28.1 mol% is identical to C. perfringens (28.1–28.4% as determined from sequenced C. perfringens genomes; results not shown). A total of 50 open reading frames with a minimum length of 150 nt were identified in the ΦS9 genome (coding capacity 92.0%) (Table 1), which is organized into distinct functional modules (Fig. 2). One putative tRNAAsn gene was found at nt position 10,367 to 10,439. A putative function could be assigned to 26 gene products.
Figure 2. Genomic maps and alignments of phages ΦS9, ΦS63 and Φ3626. The ΦS9 genome has been reoriented to allow visual alignment. Individual functional modules are indicated by coloring, and significant amino acid sequence identities are indicated by blue shaded bars. Scale represents the nucleotide position in kb. The locus of the attP attachment site is marked by an arrowhead. Gene numbers refer to the annotation provided in Tables S2 and S3.
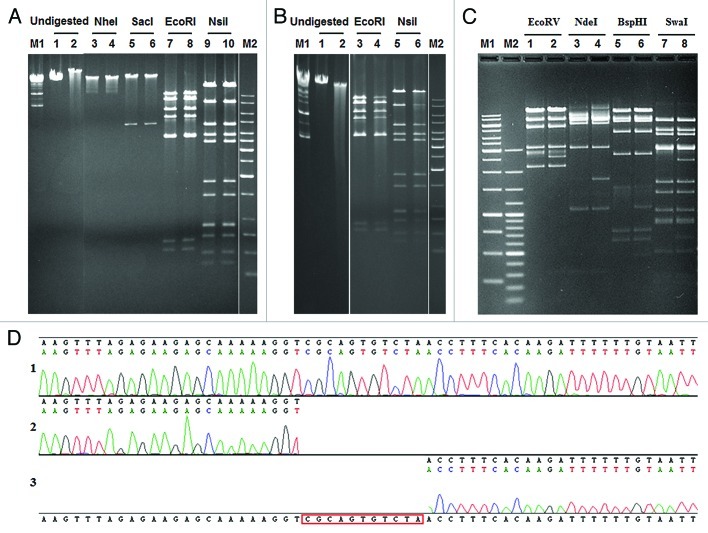
Figure 3. Structure of the ΦS9 and ΦS63 DNA molecules, as determined by gel electrophesis. (A) Restriction enzyme analysis of ΦS9 DNA using different endonucleases as indicated. Samples in lanes 2, 4, 6, 8 and 10 were heated prior to electrophoresis (see Materials and Methods section) (B) Bal31 nuclease treatment of ΦS9 DNA. (1) untreated ΦS9 DNA, (2) treated with Bal31 for 30 min. (3) EcoRI digested DNA, (4) DNA treated with Bal31 prior to digestion with EcoRI. (5) NsiI digested DNA, (6) Bal31 treated DNA and subsequently digested with NsiI. (C) Restriction profile of ΦS63 DNA using different endonucleases as indicated. Samples in lane 2, 4, 6 and 8 have been heated prior to electrophoresis. (D) Determination of single-stranded genome ends (cos-site) of ΦS63 DNA. (1) Sequence of the cos-site region of ΦS63 prophage integrated in C. perfringens. (2) Runoff-sequence of the right end and (3) left end of the ΦS63 DNA molecule, using purified ΦS63 DNA as template. M1, M2: DNA size standards (Fermentas).
Table 1. Synopsis of published Clostridium perfringens phages.
| Name | Family | Genome size (kb) | ORFs | Dimensions (nm) | Genome structure | Integration (attB)/lifestyle |
Reference |
|---|---|---|---|---|---|---|---|
| ΦS9 |
Siphoviridae |
39.46 |
50 |
60 × 190 |
c.p. t.r. |
noncoding, temperate |
This work |
| ΦS63 |
Siphoviridae |
33.61 |
43 |
62 × 170 |
3′ cos |
sigK, temperate |
This work |
| Φ3626 |
Siphoviridae |
33.51 |
50 |
55 × 170 |
3′ cos |
guaA, temperate |
Zimmer et al. 2000 |
| ΦCP9O |
Siphoviridae |
39.59 |
62 |
57 × 100 |
unknown |
n.d. |
Oakley et al. 2011 |
| ΦCP13O |
Siphoviridae |
38.33 |
66 |
57 × 100 |
unknown |
n.d. |
Oakley et al. 2011 |
| ΦCP26F |
Siphoviridae |
39.19 |
62 |
57 × 100 |
c.p., t.r. (putative) |
n.d. |
Seal et al. 2011, Oakley et al. 2011 |
| ΦCP34O |
Siphoviridae |
38.31 |
65 |
57 × 100 |
n.d. |
n.d. |
Oakley et al. 2011 |
| ΦCP39O |
Siphoviridae |
38.75 |
62 |
57 × 100 |
c.p., t.r. (putative) |
n.d. |
Seal et al. 2011, Oakley et al. 2011 |
| ΦSM101 |
Siphoviridae |
38.09 |
54 |
n.d. |
n.d. |
temperate |
Myers et al. 2006 |
| ΦCP24R |
Podoviridae |
18.92 |
22 |
44 |
inverted t.r., possible terminal protein |
none (virulent) |
Morales et al. 2011 |
| ΦCPV1 |
Podoviridae |
16.75 |
22 |
(42 × 23) × 37 |
inverted t.r. (predicted) |
none (virulent) |
Volozhantsev et al. 2011 |
| ΦCPV4 |
Podoviridae |
17.97 |
26 |
40–42 × 35–38 |
inverted t.r. (predicted) |
none (virulent) |
Volozhantsev et al. 2012 |
| ΦZP2 |
Podoviridae |
18.08 |
27 |
40–42 × 35–38 |
inverted t.r. (predicted) |
none (virulent) |
Volozhantsev et al. 2012 |
| ΦCP7R | Podoviridae | 18.40 | 28 | 40–42 × 35–38 | inverted t.r. (predicted) | none (virulent) | Volozhantsev et al. 2012 |
n.d., not determined; c.p., circularly permuted; t.r., terminally redundant.
The ΦS63 unit genome is 33,609 bp in size, which matches very well with PFGE analysis of full-length phage DNA (data not shown), and reflects the physical size of the packaged molecule (see below). It features a GC-content of 27.5 mol%, slightly less than ΦS9 and the Clostridium host strains. A total of 43 open reading frames could be annotated (89.9% of the coding capacity) (Table 1) and a putative function could be assigned to 25. The ΦS63 genome is also organized in a lifestyle specific, modular fashion (Fig. 2).
ΦS9 contains terminally redundant, circularly permuted genomes, and ΦS63 features single-stranded overlapping DNA ends
We determined the genome structure of both phages ΦS9 and ΦS63. Runoff Sanger sequencing reactions with primers complementary to the ends of the ΦS9 single large contig produced sequence complementary to the other end of the contig (data not shown). Ligation of ΦS9 DNA prior to digestion and heat treatment (75°C for 10 min) did not alter restriction patterns (Fig. 3A). In addition, Bal31 exonuclease treatment of ΦS9 DNA followed by EcoRI or NsiI digestion simultaneously decreased the intensity of all restriction fragments over time (Fig. 3B),22,23 and no specific fragment was shortened. These findings clearly indicated that ΦS9 DNA represents a collection of terminally redundant and circularly permuted DNA molecules.
In contrast, when full length ΦS63 DNA was subjected to pulsed field gel electrophoresis, it yielded a pattern of unit-size genomes joined in a concatemeric fashion (data not shown), indicating the presence of self-ligating cohesive (cos) genome ends in these DNA molecules. Heating prior to electrophoresis changed the restriction pattern in a characteristic fashion (Fig. 3C), which also perfectly matched the in silico predictions. Sequencing of a PCR product generated from C. perfringens S63, using primers cos_fw and cos_rev (Table S1), and comparison to sequence generated with the same primer pair using linear ΦS63 DNA yielded the precise structure and sequence of the terminal single-stranded cos site region (Fig. 3D), featuring 3′-overhangs of 11 nt (CGCAGTGTCTA).
Bioinformatic analyses and relationship of ΦS9 and ΦS63 to other phages
Only few similarities were found among ΦS9 and ΦS63, and also to other C. perfringens prophages. The two apparently unrelated viruses feature significant similarities only in the lysogeny control region (integrase and repressor), and the endolysin enzymes. However, proteins of both phages feature several homologies to Siphoviruses of other Firmicutes, such as Listeria, Streptococcus and Bacillus,24,25 as well as to (often cryptic) prophages identified in the genomes of these organisms. Some of the ΦS9 structural genes show sequence homology to Brochothrix phage BL3 (e.g., gp38),25 while some of the early genes feature homologies to Listeria phages A118, A006 and A500.22,24
All predicted gene products encoded by the two phages and putative functional assignments are listed in Tables S2 and S3.
A phylogenetic tree of large terminase subunit amino acid sequences can serve as measure of similarity in DNA packaging strategy and relatedness between phages.26 A tree generated of 109 terminase sequences (Fig. S1) placed ΦSM101 with Φ3626 in close relation to ΦS63 in the branch of 3′ cos-phages. ΦS9 and ΦCP39O cluster in the headful packaging branch and phages ΦCP9O, ΦCP13O, ΦCP26F and ΦCP34O form an own branch in the tree. These findings confirm experimentally evaluated packaging strategies and overall relatedness of the phages. No terminase sequences were available for phages ΦCP7R, ΦCP24R, ΦCPV4 and ΦZP2 and none could be predicted by homology searches.
The ΦS9 attB lies in an intergenic region, whereas ΦS63 inserts into sigK
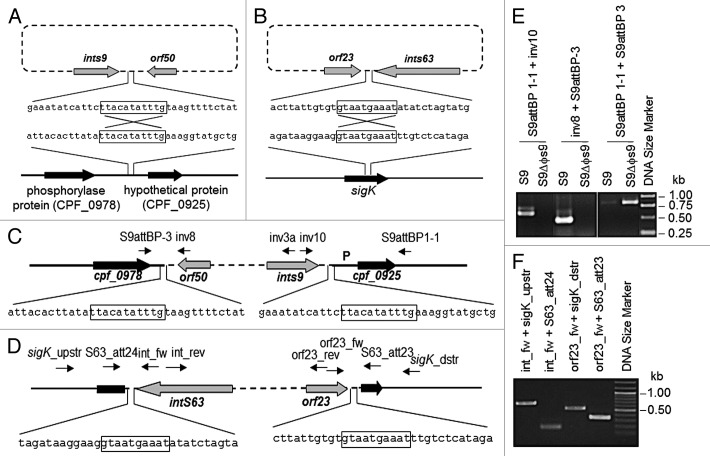
Because genome sequences of C. perfringens strains S9 and S63 have not been available, the insertion sites used by ΦS9 and ΦS63 were identified by inverse PCR from self-ligated C. perfringens S9 or S63 genomic DNA fragments (Fig. 4), and comparison to non-lysogenic host DNA. In the case of ΦS9, the sequence matched a region located next to a different prophage-like element (referred to as Φ13124). Phage ΦS9 integrates into the non-coding intergenic sequence, 541 nt downstream of a gene encoding a putative phosphorylase, and 157 nt upstream of a hypothetical protein. The core sequence of ΦS9 integration is TTACATATTTG (Fig. 4A), which is similar in length to those used by Clostridium phages Φ3626 (12 bp), ΦC2 (11 bp) and ΦCD119 (14 bp).13,27,28
Figure 4. Localization of ΦS9 and ΦS63 attachment sites attB in the C. perfringens genome. (A) Schematic representation of the integration of ΦS9 into C. perfringens genomic DNA (sequence from ATCC 13124). (B) Integration of ΦS63 into C. perfringens (partial genome sequence of S63 determined in this work). (C) Location of ΦS9 in the C. perfringens genome. Core sequence (11 nt) used in recombination is boxed. P, promoter region for cpf_0925 homolog. Primer binding sites are indicated by arrows. (D) Diagram showing the location of ΦS63 in the C. perfringens genome. Core sequence (10 nt) for recombination is boxed. Primer binding sites are indicated by arrows. (E) PCR-based confirmation of the ΦS9 attachment site, using C. perfringens S9 and S9ΔΦS9 genomic DNA as templates, and (F) the ΦS63 attachment site, using C. perfringens S63 genomic DNA as template. Primer binding sites are indicated.
The same approach was used to identify the insertion site for ΦS63 in C. perfringens S63 (Fig. 4B). The GTAATGAAAT 10 nt core of the attB sequence is located at nt position 427 from the the 5′ end and nt 265 from the 3′ end, and the insertion region features significant homology to sigK from C. perfringens S13,7 ATCC 131246 and SM101.6
C. perfringens strain ATCC 13124 harbors two prophages
In the course of our in silico analyses, we identified region 1088991–1128198 (corresponding to CPF_0926–CPF_0977) of strain ATCC131246 as a putative 39,208 bp prophage genome, which was designated Φ13124. Surprisingly, a putative Φ13124 integrase (CPF_0926) was found 100% identical to the ΦS9 integrase. Moreover, the beginning and end of the Φ13124 attP sequence matched the ΦS9 attP, and overall good sequence homologies were found between the two phages. Sequence alignment with ΦS63 indicated another putative prophage sequence in ATCC 13124 (termed Φ13124_2), located on a genomic island6 in between positions 1783746 to 1820131 of the ATCC 13124 genome.6 The putative Φ13124_2 genome is 36,385 bp in size, and this prophage sequence is also flanked by the attachment site used by ΦS63.
Discussion
The high degree of genomic variation and phenotypic diversities among bacteria appears to be mediated by mobile elements such as conjugative plasmids, transposons and insertion elements. Although lysogenic conversion was established for numerous bacterial species including Clostridia, it has not yet been observed for C. perfringens, possibly due to a lack of data regarding temperate C. perfringens phage.
We here describe the two heterogeneous Siphoviridae ΦS9 and ΦS63. Compared with the other studied C. perfringens phages, ΦS9 and ΦS63 feature significantly larger head diameter and tail length. Interestingly, both also feature unusual tail-associated appendices, which probably assume functions comparable to tail fibers and whiskers. ΦS9 possess the second-largest genome of all known C. perfringens phages, and has been shown to represent a collection of terminally redundant and circularly permuted DNA molecules. In contrast, ΦS63 features identical unit-length genomes with cohesive ends, similar to Φ3626.13 Most of the sequence-based similarities exist to proteins of other (putative) prophages infecting members of the Firmicutes, namely Streptococcus, Lactococcus, Bacillus, Staphylococcus, Listeria, Brochothrix and other Clostridium species (Tables S2 and S3). Altogether, these findings clearly indicate horizontal gene transfer among the ancestors of the bacterial host and their mobile genome element, i.e., the prophages. Likewise, the surprisingly few homologies between ΦS9 and ΦS63, and to other known C. perfringens phages can be explained by divergent evolution of these phages from a distant ancestor. Interestingly, ΦS9 and ΦS63 feature a virtually identical endolysin (95.3% amino acid identity), which has most likely been acquired by a more recent horizontal gene exchange. Also, the endolysin of ΦS63 is 98% identical to the murein hydrolase of the episomal C. perfringens phage ΦSM101, strongly suggesting a modular exchange of functional units. Altogether, the significant heterogeneity among C. perfringens phages emphasizes the need for more sequences in order to obtain a better overview of this probably large and diverse group of viruses infecting and interacting with an important pathogen.
Homology searches with ΦS9 sequences identified prophage Φ13124 in the genome of C. perfringens ATCC 13124. Based on significant homology over wide areas of the genome, the two phages appear to have a common origin, and are clearly different from ΦS63 and Φ3626. Similarities of ΦS9 and Φ13124 include almost identical integrases and repressors, tail structural components, the holin-endolysin dual lysis module, and the major capsid protein. Φ13124 is inserted within the largest genomic island (243 kb) of the host bacterium, which also contains genes responsible for iron transport, fucose utilization, and glycolytic activities,6 enabling this strain to exploit various environments.6 Interestingly, it also contains the sporulation-related genes cotJB and cotJC, as well as some putative virulence factors such as a sialidase located near the right arm of Φ13124.6 Whether prophage Φ13124 is able to mobilize these closely positioned genes by either a faulty phage excision or generalized transduction is, however, speculative and needs more investigation.
Another putative prophage Φ13124_2 in the ATCC 13124 genome6 was identified using sequence alignment with the ΦS63 genome, sharing extensive sequence similarity among most of the structural proteins. The lack of homology in lysogeny control or DNA replication proteins suggested a more distinct evolution of these two phage sequences. However, it remains to be determined if Φ13124_2 is a functional virus, in contrast to the frequent occurrence of defective or cryptic remnants of inserted phage.
Lysogenic conversion may result from expression of genes located on an inserted phage genome,10,29 or by integration and disruption of coding sequence.22,27 The putative effect of C. perfringens phage ΦS9 on sporulation of its lysogenized host has been subject of discussion over many years. Stewart and Johnson (1977) claimed that curing of ΦS9 from C. perfringens strain S9 delayed sporulation, while a re-lysogenized strain S9CR restored the sporulation competent phenotype.20 This suggested possible lysogenic conversion of C. perfringens by ΦS9. Unfortunately, we were unable to confirm this hypothesis, i.e., we found no indication for lysogenic conversion of C. perfringens by phage ΦS9. The presence or absence of prophage ΦS9 in the C. perfringens strain S9 genome did not significantly influence the onset of production of heat-resistant spores or the total number of spores produced under experimental conditions similar to those published previously20 (Kim K.-P., unpublished data).
The integration site of ΦS9 is different from the Φ362613 and ΦS63 attachment sites, and lies in an intergenic region upstream of a gene encoding a putative membrane protein of unknown function (homologous to CPF_0925 in ATCC 13124), and downstream of a putative phosphorylase-encoding gene.
In contrast, phage ΦS63 integrates into a B. subtilis sigK-like gene, which encodes a RNA polymerase sigma factor involved in the late stage of spore formation. SigK directs the Stage IV to Stage V transition, i.e., the spore coat formation in the sporulation cascade (reviewed in refs. 30, 31). sigK is encoded on two gene fragments (spoIVCB and spoIIIC) in B. subtilis and is created by splicing and the excision of a sigK intervening sequence (skin element). Interruption of sigK by these prophage-like sequences has been reported not only for B. subtilis (skinBs), but also for C. difficile (skinCd) and C. tetani (skinCt).30,32-34 It should be noted that ΦS63 is the first functional phage reported that inserts into a sigK gene of its host. Also, the presence of a skin element has never been reported in C. perfringens. Phage ΦS63 int is oriented in the opposite direction of sigK, similar to the situation in B. subtilis and C. tetani,35 but different to C. difficile.32 There also seems to be some variability regarding the exact insertion locus; while the integration sites of ΦS63, skinCd and skinCt are at a similar location within the coding sequence, skinBs is located in a different region of sigK.35 It was found that a specific recombinase can excise the B. subtilis skin element from sigK.36 Our findings also demonstrate precise excision, resulting in reconstitution of native sigK (Fig. 4). Altogether, these observations point to an important role of this insertion element for control of sigK function and a potential influence on sporulation. While it was reported, that insertion is not required for sporulation in B. subtilis,33 it is needed in C. difficile. A possible explanation is a missing sigK pro-sequence in C. difficile, which lacks an N-terminal portion that needs to be cleaved in order to activate SigK.32 sigK of strain S63 is not different from other C. perfringens strains (the pro-sequence is present), similar to the situation in B. subtilis.32,33 This would suggest that its interruption might not be strictly required for successful sporulation of the host cell. However, the sequenced C. perfringens strains do not contain a protease SpoIVFB homolog, which is necessary to remove the pro-sequence in B. subtilis.37 A reliable sporulation model for strain S63 is not available, and the precise sporulation phenotype of the ΦS63 sigK integration remains to be elucidated.
Materials and Methods
Bacterial strains and growth
C. perfringens strains used in this study included S9, S13,38 S9ΔΦS9 (cured of the prophage), S63, and ATCC 13124. Strains were anaerobically grown in TGY medium (3%, tryptone peptone; 2%, glucose; 1%, yeast extract; 0.1% cysteine, pH 7.4) at 37°C in a flexible vinyl glove chamber (Coy Laboratories), containing a 95% N2 and 5% H2 atmosphere. Escherichia coli DH5α MCR and XL1-blue MRF` (Invitrogen) were grown in Luria-Bertani medium (LB) (1%, tryptone peptone; 1%, NaCl; 0.5%, yeast extract) at 37°C. If required, media were supplemented with ampicillin (100 μg/ml). or tetracycline (18 µg/ml).
UV induction and preparation of ΦS9 and ΦS63 stocks
To induce temperate phages ΦS9 and ΦS63, C. perfringens S9 and S63 were grown to exponential growth phase, and exposed to UV light (254 nm) in a UVC500 Crosslinker (Amersham) for 4 min at 2 J/cm2. An equal volume of TGY medium was added to the culture, and bacteria were incubated for 2 h at 37°C, followed by centrifugation (14,000 ×g, 5 min) and filter-sterilization (0.2 µm pore size). Serially diluted phage-containing lysates were mixed with C. perfringens strain S13 indicator cells, and plated using soft-agar overlays.39 After overnight incubation, distinct plaques were picked and eluted with SM buffer (50 mM TRIS-HCl (pH 7.5), 100 mM NaCl, 8 mM MgSO4). The procedure was repeated twice. Initial stocks of ΦS9 or ΦS63 were prepared by plating the single plaque eluates onto C. perfringens S13, and elution of the entire soft agar layer with SM buffer. Cell debris was removed by centrifugation, and the phage suspension was filter-sterilized and stored at 4°C.
Propagation and purification of phages
For phage ΦS9, exponentially growing cells of C. perfringens strain S13 in broth culture were infected with ΦS9 at a multiplicity of infection (MOI) of 1, and incubated for 8 h at 37°C. Phage ΦS63 was propagated by the agar overlay method and removed by eluting the phage particles with 4 ml SM-buffer per plate.
Following centrifugation of the lysates at 6,000 ×g for 10 min, 8% (w/v), polyethylene glycol (PEG, MW 8,000) and 0.5 M NaCl were added to the supernatant and incubated overnight at 4°C.40 After centrifugation (10,000 × g, 10 min), the supernatant was removed and precipitated phage particles resuspended in SM buffer, followed by stepped CsCl density gradient centrifugation (76,000 × g, 18 h, L-60 Ultracentrifuge, Beckman) as previously described.23 Finally, virus particles were removed and dialyzed against SM buffer (pore size 50,000 Da, Spectrum) overnight at 4°C.
Electron microscopy
Purified phage particles were negatively stained with either 2% uranyl acetate, or 2% Na-phosphotungstic acid, or 2% ammonium molybdate.41 Samples were observed in a Philips CM100 transmission electron microscope at 100 kV acceleration voltage (FEI Company), equipped with a TVIPS Fastscan CCD camera (Tietz Systems), or in a Tecnai G2 Spirit electron microscope at 120 kV equipped with an EAGLE CCD camera (FEI Company).
Cloning, nucleotide sequencing, and genome analysis
Phage genomic DNA was prepared by proteinase K (Fermentas) treatment of purified phage particles, and subsequent organic extraction as described elsewhere.42 Genomic shotgun libraries of ΦS9 and ΦS63 were constructed as previously described.13,23 Briefly, partial restriction digestion (Tsp509I) (New England Biolabs) or complete digestion with HindIII (Fermentas) or TaqI (Fermentas) were performed, fragments of 1 to 2.5 kb in length were separated on agarose gels (0.8%), eluted using QIAquick Gel Extraction kit (Qiagen), and ligated into pBluescript SK II (-) (Stratagene), followed by transformation into E. coli XL-1 Blue and blue-white screening on agar plates containing ampicillin (100 µg/ml), X-Gal (40 µg/ml) and IPTG (3 mM). Plasmids bearing inserts of the desired size were confirmed by restriction enzyme digestion and the inserts sequenced. Following assembly of the sequences, gaps were closed by primer walking directly on ΦS9 and ΦS63 chromosomal DNA, with the aid of specific primers as sequences became available.
Determination of genome structure
Phage genomic DNA was treated with restriction enzymes as recommended by the manufacturers. The fragments were heat-treated (62°C, 10 min) and separated in 1.0% agarose gels.
For exonuclease treatment, phage genomic DNA was first incubated with Bal31 nuclease (New England Biolabs) (1.5 unit per 1 μg DNA) for 0, 10, 20 and 30 min as directed by the manufacturer, followed by phenol-chloroform extraction and ethanol precipitation.42 Following restriction enzyme digestion, fragment patterns were analyzed electrophoretically.
Identification of attB and attP
Identification of ΦS9 attP was performed as previously described,43 using two divergent primers inv3a and inv10 (Table S1), derived from the 3′ end of the putative ΦS9 integrase (int). The S9 template DNA was first digested with TasI (Fermentas), and self-ligated using T4 DNA ligase (New England Biolabs). The PCR product was cloned into the pGEMT-easy TA vector (Promega), yielding pS9att3. Alignments of the inserts with available C. perfringens genomes enabled precise identification of the attachment site locus. Primers inv8 (corresponding to to phage sequence) and S9attBP-3 (homologous to C. perfringens ATCC 13124 sequence facing the integration site) were used to confirm the prophage location. For confirmation of prophage presence or absence in the identified locus, PCR with S9attBP-3 and S9attBP1-1 was performed on genomic DNA of the ΦS9 lysogen, and of a ΦS9-cured strain (S9ΔΦS9) (Fig. 4).
A similar strategy was used to identify the attB and attP of phage ΦS63. After digestion of S63 DNA with MboI (Fermentas) and self-ligation, inverse PCR was performed using primers orf23_fw, orf23_rev, both homologous to downstream sequence of the putative ΦS63 integrase gene. Using alignments with sequence obtained by inverse PCR with primer pair int_fw and int_rev corresponding to the phage integrase, the transition point from phage to host DNA was identified. Results were confirmed by sequencing of PCR products generated with primer combinations S63_att23 + orf23_fw and S63_att24 + int_fw, as well as sigK_upstr and sigK_dstr on C. perfringens lysogen DNA (Fig. 4).
Bioinformatic analyses
CLC Genomics Workbench Version 5.1 (CLC, Aarhus, Denmark) was used for analysis of nucleotide (nt) and amino acid (aa) sequences. The BLAST algorithms44 were used for similarity searches in the non-redundant protein and nucleotide sequence databases available through the NCBI website (http://www.ncbi.nlm.nih.gov). HHPred (http://toolkit.tuebingen.mpg.de/hhpred) was used for additional homology and structure predictions. The integrated ClustalW algorithm of CLC Genomics Workbench was used for multiple sequence alignments and comparisons.45 InterProScan (http://www.ebi.ac.uk/InterProScan/) was used to identify conserved domains in translated Orfs, and TmHMM protein analysis software (version 2.0) was used to predict transmembrane domains.46 Putative tRNAs genes were identified using tRNAScan SE.47
Nucleotide sequence accession numbers
The DNA sequences reported here appear in GenBank under accession number AY082069 (ΦS9), JQ660954 (ΦS63) and JQ660953 (partial sequence of sigK gene of strain S63).
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to D. Mahony, Dalhousie University, Halifax, Canada for the gift of C. perfringens strains S9 and S63.
Supplemental Material
Supplemental materials may be found here:
www.landesbioscience.com/journals/bacteriophage/article/21363
Footnotes
Previously published online: www.landesbioscience.com/journals/bacteriophage/article/21363
References
- 1.Petit L, Gibert M, Popoff MR. Clostridium perfringens: toxinotype and genotype. Trends Microbiol. 1999;7:104–10. doi: 10.1016/S0966-842X(98)01430-9. [DOI] [PubMed] [Google Scholar]
- 2.Briggs DC, Naylor CE, Smedley JG, 3rd, Lukoyanova N, Robertson S, Moss DS, et al. Structure of the food-poisoning Clostridium perfringens enterotoxin reveals similarity to the aerolysin-like pore-forming toxins. J Mol Biol. 2011;413:138–49. doi: 10.1016/j.jmb.2011.07.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitadokoro K, Nishimura K, Kamitani S, Fukui-Miyazaki A, Toshima H, Abe H, et al. Crystal structure of Clostridium perfringens enterotoxin displays features of beta-pore-forming toxins. J Biol Chem. 2011;286:19549–55. doi: 10.1074/jbc.M111.228478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amimoto K, Noro T, Oishi E, Shimizu M. A novel toxin homologous to large clostridial cytotoxins found in culture supernatant of Clostridium perfringens type C. Microbiology. 2007;153:1198–206. doi: 10.1099/mic.0.2006/002287-0. [DOI] [PubMed] [Google Scholar]
- 5.Paredes-Sabja D, Sarker N, Sarker MR. Clostridium perfringens tpeL is expressed during sporulation. Microb Pathog. 2011;51:384–8. doi: 10.1016/j.micpath.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Myers GS, Rasko DA, Cheung JK, Ravel J, Seshadri R, DeBoy RT, et al. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 2006;16:1031–40. doi: 10.1101/gr.5238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T, et al. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc Natl Acad Sci U S A. 2002;99:996–1001. doi: 10.1073/pnas.022493799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bannam TL, Teng WL, Bulach D, Lyras D, Rood JI. Functional identification of conjugation and replication regions of the tetracycline resistance plasmid pCW3 from Clostridium perfringens. J Bacteriol. 2006;188:4942–51. doi: 10.1128/JB.00298-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyamoto K, Fisher DJ, Li J, Sayeed S, Akimoto S, McClane BA. Complete sequencing and diversity analysis of the enterotoxin-encoding plasmids in Clostridium perfringens type A non-food-borne human gastrointestinal disease isolates. J Bacteriol. 2006;188:1585–98. doi: 10.1128/JB.188.4.1585-1598.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beres SB, Sylva GL, Barbian KD, Lei B, Hoff JS, Mammarella ND, et al. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc Natl Acad Sci U S A. 2002;99:10078–83. doi: 10.1073/pnas.152298499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 12.Brüssow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zimmer M, Scherer S, Loessner MJ. Genomic analysis of Clostridium perfringens bacteriophage phi3626, which integrates into guaA and possibly affects sporulation. J Bacteriol. 2002;184:4359–68. doi: 10.1128/JB.184.16.4359-4368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oakley BB, Talundzic E, Morales CA, Hiett KL, Siragusa GR, Volozhantsev NV, et al. Comparative genomics of four closely related Clostridium perfringens bacteriophages reveals variable evolution among core genes with therapeutic potential. BMC Genomics. 2011;12:282. doi: 10.1186/1471-2164-12-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seal BS, Fouts DE, Simmons M, Garrish JK, Kuntz RL, Woolsey R, et al. Clostridium perfringens bacteriophages ΦCP39O and ΦCP26F: genomic organization and proteomic analysis of the virions. Arch Virol. 2011;156:25–35. doi: 10.1007/s00705-010-0812-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volozhantsev NV, Verevkin VV, Bannov VA, Krasilnikova VM, Myakinina VP, Zhilenkov EL, et al. The genome sequence and proteome of bacteriophage ΦCPV1 virulent for Clostridium perfringens. Virus Res. 2011;155:433–9. doi: 10.1016/j.virusres.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 17.Volozhantsev NV, Oakley BB, Morales CA, Verevkin VV, Bannov VA, Krasilnikova VM, et al. Molecular Characterization of Podoviral Bacteriophages Virulent for Clostridium perfringens and Their Comparison with Members of the Picovirinae. PLoS One. 2012;7:e38283. doi: 10.1371/journal.pone.0038283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller RW, Skinner EJ, Sulakvelidze A, Mathis GF, Hofacre CL. Bacteriophage therapy for control of necrotic enteritis of broiler chickens experimentally infected with Clostridium perfringens. Avian Dis. 2010;54:33–40. doi: 10.1637/8953-060509-Reg.1. [DOI] [PubMed] [Google Scholar]
- 19.Morales CA, Oakley BB, Garrish JK, Siragusa GR, Ard MB, Seal BS. Complete genome sequence of the podoviral bacteriophage ΦCP24R, which is virulent for Clostridium perfringens. Arch Virol. 2012;157:769–72. doi: 10.1007/s00705-011-1218-2. [DOI] [PubMed] [Google Scholar]
- 20.Stewart AW, Johnson MG. Increased numbers of heat-resistnat spores produced by two strains of Clostridium perfringens bearing temperate phage s9. J Gen Microbiol. 1977;103:45–50. doi: 10.1099/00221287-103-1-45. [DOI] [PubMed] [Google Scholar]
- 21.Ackermann HW. Tailed bacteriophages: the order caudovirales. Adv Virus Res. 1998;51:135–201. doi: 10.1016/S0065-3527(08)60785-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loessner MJ, Inman RB, Lauer P, Calendar R. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol Microbiol. 2000;35:324–40. doi: 10.1046/j.1365-2958.2000.01720.x. [DOI] [PubMed] [Google Scholar]
- 23.Klumpp J, Dorscht J, Lurz R, Bielmann R, Wieland M, Zimmer M, et al. The terminally redundant, nonpermuted genome of Listeria bacteriophage A511: a model for the SPO1-like myoviruses of gram-positive bacteria. J Bacteriol. 2008;190:5753–65. doi: 10.1128/JB.00461-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dorscht J, Klumpp J, Bielmann R, Schmelcher M, Born Y, Zimmer M, et al. Comparative genome analysis of Listeria bacteriophages reveals extensive mosaicism, programmed translational frameshifting, and a novel prophage insertion site. J Bacteriol. 2009;191:7206–15. doi: 10.1128/JB.01041-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kilcher S, Loessner MJ, Klumpp J. Brochothrix thermosphacta bacteriophages feature heterogeneous and highly mosaic genomes and utilize unique prophage insertion sites. J Bacteriol. 2010;192:5441–53. doi: 10.1128/JB.00709-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casjens SR, Gilcrease EB, Winn-Stapley DA, Schicklmaier P, Schmieger H, Pedulla ML, et al. The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. J Bacteriol. 2005;187:1091–104. doi: 10.1128/JB.187.3.1091-1104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goh S, Ong PF, Song KP, Riley TV, Chang BJ. The complete genome sequence of Clostridium difficile phage phiC2 and comparisons to phiCD119 and inducible prophages of CD630. Microbiology. 2007;153:676–85. doi: 10.1099/mic.0.2006/002436-0. [DOI] [PubMed] [Google Scholar]
- 28.Govind R, Fralick JA, Rolfe RD. Genomic organization and molecular characterization of Clostridium difficile bacteriophage PhiCD119. J Bacteriol. 2006;188:2568–77. doi: 10.1128/JB.188.7.2568-2577.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakaguchi Y, Hayashi T, Kurokawa K, Nakayama K, Oshima K, Fujinaga Y, et al. The genome sequence of Clostridium botulinum type C neurotoxin-converting phage and the molecular mechanisms of unstable lysogeny. Proc Natl Acad Sci U S A. 2005;102:17472–7. doi: 10.1073/pnas.0505503102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paredes CJ, Alsaker KV, Papoutsakis ET. A comparative genomic view of clostridial sporulation and physiology. Nat Rev Microbiol. 2005;3:969–78. doi: 10.1038/nrmicro1288. [DOI] [PubMed] [Google Scholar]
- 31.Piggot PJ, Hilbert DW. Sporulation of Bacillus subtilis. Curr Opin Microbiol. 2004;7:579–86. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Haraldsen JD, Sonenshein AL. Efficient sporulation in Clostridium difficile requires disruption of the sigmaK gene. Mol Microbiol. 2003;48:811–21. doi: 10.1046/j.1365-2958.2003.03471.x. [DOI] [PubMed] [Google Scholar]
- 33.Kunkel B, Losick R, Stragier P. The Bacillus subtilis gene for the development transcription factor sigma K is generated by excision of a dispensable DNA element containing a sporulation recombinase gene. Genes Dev. 1990;4:525–35. doi: 10.1101/gad.4.4.525. [DOI] [PubMed] [Google Scholar]
- 34.Stragier P, Kunkel B, Kroos L, Losick R. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science. 1989;243:507–12. doi: 10.1126/science.2536191. [DOI] [PubMed] [Google Scholar]
- 35.Sonenshein AL, Haraldsen JD, Dupuy B. RNA Polymerase and Alternative σ Factors. In: Dürre P, ed. Handbook on Clostridia. Boca Raton: CRC Press, 2005. [Google Scholar]
- 36.Kimura T, Amaya Y, Kobayashi K, Ogasawara N, Sato T. Repression of sigK intervening (skin) element gene expression by the CI-like protein SknR and effect of SknR depletion on growth of Bacillus subtilis cells. J Bacteriol. 2010;192:6209–16. doi: 10.1128/JB.00625-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu S, Cutting S, Kroos L. Sporulation protein SpoIVFB from Bacillus subtilis enhances processing of the sigma factor precursor Pro-sigma K in the absence of other sporulation gene products. J Bacteriol. 1995;177:1082–5. doi: 10.1128/jb.177.4.1082-1085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahony DE, Kalz GG. A temperate bacteriophage of Clostridium perfringens. Can J Microbiol. 1968;14:1085–93. doi: 10.1139/m68-183. [DOI] [PubMed] [Google Scholar]
- 39.Adams MH. Methods of study of bacterial viruses. Bacteriophages. New York: Interscience publishers, Inc., 1959:443-57. [Google Scholar]
- 40.Yamamoto KR, Alberts BM, Benzinger R, Lawhorne L, Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970;40:734–44. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]
- 41.Steven AC, Trus BL, Maizel JV, Unser M, Parry DA, Wall JS, et al. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J Mol Biol. 1988;200:351–65. doi: 10.1016/0022-2836(88)90246-X. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Russell DW. Molecular Cloning—A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press, 2001. [Google Scholar]
- 43.Ochman H, Gerber AS, Hartl DL. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–3. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 46.Durbin R, Eddy S, Krogh A. G. M. Biological sequence analysis: probabilistic models of proteins and nucleic acids. Cambridge, UK: Cambridge University Press, 1998. [Google Scholar]
- 47.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–64. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.