Abstract
The nematode C. elegans responds to infection by the fungus Drechmeria coniospora with a rapid increase in the expression of antimicrobial peptide genes. To investigate further the molecular basis of this innate immune response, we took a two-dimensional difference in-gel electrophoresis (2D-DIGE) approach to characterize the changes in host protein that accompany infection. We identified a total of 68 proteins from differentially represented spots and their corresponding genes. Through class testing, we identified functional categories that were enriched in our proteomic data set. One of these was “protein processing in endoplasmic reticulum,” pointing to a potential link between innate immunity and endoplasmic reticulum function. This class included HSP-3, a chaperone of the BiP/GRP78 family known to act coordinately in the endoplasmic reticulum with its paralog HSP-4 to regulate the unfolded protein response (UPR). Other studies have shown that infection of C. elegans can provoke a UPR. We observed, however, that in adult C. elegans infection with D. coniospora did not induce a UPR, and conversely, triggering a UPR did not lead to an increase in expression of the well-characterized antimicrobial peptide gene nlp-29. On the other hand, we demonstrated a specific role for hsp-3 in the regulation of nlp-29 after infection that is not shared with hsp-4. Epistasis analysis allowed us to place hsp-3 genetically between the Tribbles-like kinase gene nipi-3 and the protein kinase C delta gene tpa-1. The precise function of hsp-3 has yet to be determined, but these results uncover a hitherto unsuspected link between a BiP/GRP78 family protein and innate immune signaling.
Keywords: Drechmeria coniospora, MAPK, gene regulation, proteomics, signal transduction
Introduction
The nematophagous fungus Drechmeria coniospora infects various species of nematodes. Its spores adhere to the surface of a worm, germinate and perforate the cuticle. The worm’s body is then totally invaded by the fungus, rapidly causing death (reviewed by Engelmann and Pujol1). When D. coniospora infects Caenorhabditis elegans this triggers the expression of a large number of genes including those encoding antimicrobial peptides (AMPs) of the NLP family.2-4 The induction of several nlp genes is dependent upon a protein kinase C delta (PKC∂)/p38 MAPK pathway that can be activated in the epidermis either by infection or by sterile wounding.5 In both cases, signaling passes via TPA-1, a PKC∂ that acts upstream of TIR-1, the nematode ortholog of SARM, and a MAPK cassette constituted of a MAP3K (NSY-1), MAP2K (SEK-1) and the p38 MAPK PMK-1. This then acts upstream of the STAT-like transcription factor STA-2 to regulate nlp gene expression.6 The elements that contribute to signaling upstream of TPA-1 have only been partially characterized. Wounding and infection require G-protein signaling upstream of TPA-1, while infection specifically involves the Tribbles-like kinase NIPI-3.5,7
Part of the innate defenses against intestinal pathogens and toxins are also mediated by a p38 MAPK cascade that shares many but not all of the elements that act in the epidermis;8-14 reviewed by Partridge et al.15 and Coleman and Mylonakis.16 Intestinal infection or exposure to bacterial toxins can also induce an unfolded protein response (UPR); this too is linked to the p38 pathway.17,18
The UPR in C. elegans is divided into constitutive and inducible pathways, the former being essential during development.19,20 Part of the UPR involves activation of the endoribonuclease IRE-1 that leads to the production of an alternatively spliced isoform of the mRNA of the transcription factor XBP-1.21 Compounds such as thapsigargin, dithiothreitol and tunicamycin that perturb endoplasmic reticulum (ER) homeostasis trigger a UPR and lead to IRE-1 activation. The subsequent production of the specific form of XBP-1 then leads to the expression throughout the organism of a large number of genes, many involved in metabolism, or the secretory pathway, including chaperones.19-21 Feeding worms the bacterial pore-forming toxin Cry5B also activates IRE-1 and upregulates chaperone expression specifically in the intestine. This requires the p38 MAPK signaling cassette.17 The Gram-negative bacterial pathogen Pseudomonas aeruginosa also induces IRE-1-mediated splicing of xbp-1 mRNA in larvae and consequent chaperone gene expression, in a p38 MAPK-dependent manner.18 As the UPR-deficient xbp-1 mutants arrest as larvae when cultured on P. aeruginosa,18 in this case, it was suggested that the ER cannot cope with the combined developmental and defense demands placed upon it (reviewed in Ewbank and Pujol22).
In addition to the UPR-mediated changes in protein maturation, turnover and trafficking, the innate immune response may also affect the activity, post-translational modification and subcellular localization of signal transduction proteins. These can be analyzed at a global level through proteomic approaches. Indeed, there have already been a number of informative studies addressing the changes in the proteome that accompany infection of C. elegans by several different bacterial pathogens.23-25
To extend our characterization of the response of C. elegans to D. coniospora we have now compared the proteomes of infected and control worms using two-dimensional difference gel electrophoresis (2D-DIGE). This fluorescence-based method allows two different protein samples tagged with two distinct fluorescent dyes to be run on the same gel, thereby improving comparative quantitation. We decided to focus on a single time point, early in the infection, with the hope of detecting changes in proteins involved in signal transduction, rather than finding proteins altered by the pathophysiological consequences of infection. We found that few changes were detected in whole extracts, but after fractionation we detected changes in many proteins. For one of these candidates, a C. elegans BiP/GRP78 homolog, we defined a novel role in the regulation of AMP gene expression.
Results
Protein fractionation reveals changes in the proteome induced by fungal infection
The infection of C. elegans by D. coniospora induces significant changes in gene expression within a matter of hours26 (and unpublished data). We used a standard 2D-DIGE approach to identify alterations at the protein level in C. elegans after 5 h of infection with D. coniospora. As with any 2D gel approach, with DIGE it is appropriate to refer to changes in representation, rather than stating that a protein is more or less abundant, unless all protein spots are identified and quantified. With whole animal extracts (FT), when we used a narrow-range pH gradient for isoelectric focusing, although 890 protein spots were detected, we observed no differences between extracts of infected and control worms. With a broad-range pH gradient that allowed 1,478 spots to be resolved, just three differentially represented proteins were detected. Only one of these was present in sufficient quantities to allow identification; it corresponded to the galectin LEC-6 (see Materials and Methods for access to data). We therefore adopted a more laborious approach, separating the extracts into four fractions (F1, F2, F3 and FNS), and performing DIGE as above for each one. The fractionation allowed many more spots to be detected (9,246 in total), and revealed differences in intensity for 67 and 103 spots in the narrow- and broad-range pH gradient gels, respectively. All these spots were excised and analyzed by mass-spectrometry, leading to an identification of a protein from 98 spots (Tables 1 and 2). This clearly illustrates the interest of combining DIGE with a prior protein fractionation approach.
Table 1. Protein identification from narrow pH (4–7) gel.
| Fractions | F1 | F2 | F3 | FNS | FT | Total |
|---|---|---|---|---|---|---|
| Number of detected spots |
867 |
933 |
883 |
1,016 |
890 |
4,589 |
| Number of differentially represented spots |
12 |
14 |
13 |
28 |
0 |
67 |
| Number of identified spots | 4 | 6 | 7 | 26 | 0 | 43 |
Table 2. Protein identification from broad pH (3–10) gel.
| Fraction | F1 | F2 | F3 | FNS | FT | Total |
|---|---|---|---|---|---|---|
| Number of detected spots |
1,588 |
1,238 |
1,083 |
1,638 |
1,478 |
7,025 |
| Number of differentially represented spots |
35 |
13 |
7 |
45 |
3 |
103 |
| Number of identified spots | 20 | 1 | 0 | 33 | 1 | 55 |
Classification of differentially represented proteins
In some cases, the same protein was identified from more than one spot, either within the same fraction on the same gel, or from different fractions and/or gels. As a consequence, the 98 characterized spots corresponded to 67 individual proteins that were differentially-represented between infected and control worms (Table 3). We used WormMart27 (WS220) to match each of the 68 identified proteins (LEC-6 and the 67 others) with its corresponding C. elegans gene (Table 3). They fall into many different structural and functional classes (Table S1). We therefore performed two complementary bioinformatic analyses to find common themes. We first used the KEGG database28 to determine whether there was an over-representation of higher-level systemic functions within the list of 68 genes. The most populated categories (Table 3) were “implicated in a metabolic pathway” (13 genes), and “protein processing in endoplasmic reticulum” (8 genes). We then used EASE,29 with our extensive in-house annotations culled from the C. elegans literature and referenced to WS220.4 There were 24 functional classes identified as significantly enriched (p < 0.001, Fisher exact test; see Materials and Methods). Among these classes, 6 were related to the response of C. elegans to infection, with a further 10 linked to aging and stress-resistance, including to the insulin/DAF-2 pathway (Tables 4and S2). Given the intimate connection between stress-resistance and susceptibility to infection, a part of the observed protein changes could thus be directly or indirectly associated with an innate immune response. The EASE analysis also revealed a potential connection with protein processing in the endoplasmic reticulum, as had been seen with KEGG.
Table 3. List of identified proteins from spots with different intensities.
| Spot N° Range pH 4–7 |
Spot N° Range pH 3–10 |
Wormpep name | Worm base ID | Gene name |
|---|---|---|---|---|
| |
F1-740 |
ZK455.1 |
WBGene00000040 |
aco-1* |
| F2-401; FNS-17 |
FNS-677 |
C34E10.6 |
WBGene00000229 |
atp-2* |
| FNS-489 |
FNS-1706 |
F47B10.1 |
WBGene00009812 |
F47B10.1* |
| |
FNS-1926 |
K10B3.7 |
WBGene00001685 |
gpd-3* |
| |
FNS-1965 |
F33H1.2 |
WBGene00001686 |
gpd-4* |
| |
FNS-1717 |
H28O16.1 |
WBGene00010419 |
H28O16.1* |
| FNS-228 |
|
Y45G12B.1 |
WBGene00021562 |
nuo-5* |
| F3-516 |
|
K12G11.3 |
WBGene00010790 |
sodh-1* |
| FNS-455 |
|
Y49A3A.2 |
WBGene00013025 |
vha-13* |
| F3-516 |
|
Y39G8B.1 |
WBGene00012722 |
Y39G8B.1* |
| F2-821 |
F2-1119 |
Y69A2AR.18 |
WBGene00022089 |
Y69A2AR.18* |
| FNS-401 |
FNS-1522 |
ZK829.4 |
WBGene00014095 |
ZK829.4* |
| FNS-198 |
|
F40F9.6 |
WBGene00009583 |
aagr-3*† |
| FNS-463 |
|
Y38A10A.5 |
WBGene00000802 |
crt-1† |
| FNS-315 |
|
C15H9.6 |
WBGene00002007 |
hsp-3† |
| FNS-411 |
|
C07A12.4 |
WBGene00003963 |
pdi-2† |
| |
F1-776 |
Y113G7A.3 |
WBGene00004754 |
sec-23† |
| FNS-239 |
|
T05E11.3 |
WBGene00011480 |
T05E11.3† |
| F3-142 |
|
T14G8.3 |
WBGene00011771 |
T14G8.3† |
| FNS-157 |
|
T24H7.2 |
WBGene00020781 |
T24H7.2† |
| F1-463 |
F1-1375; F1-1389; FNS-739; FNS-1617 |
M03F4.2 |
WBGene00000066 |
act-4 |
| F1-460 |
|
T25C8.2 |
WBGene00000067 |
act-5 |
| |
FNS-1450 |
B0334.3 |
WBGene00007143 |
B0334.3 |
| F2-100 |
|
C08H9.2 |
WBGene00007463 |
C08H9.2 |
| |
F1-1246; FNS1717 |
C44B7.10 |
WBGene00016630 |
C44B7.10 |
| |
F1-1234 |
C07H6.5 |
WBGene00000479 |
cgh-1 |
| |
FNS-2317 |
T03E6.7 |
WBGene00000776 |
cpl-1 |
| |
FNS-2295 |
F58G1.4 |
WBGene00010266 |
dct-18 |
| F3-526; F3-531 |
FNS-1996 |
C18A11.7 |
WBGene00001000 |
dim-1 |
| FNS-684 |
|
F54H12.6 |
WBGene00018846 |
eef-1B.1 |
| FNS-684 |
|
Y41E3.10 |
WBGene00012768 |
eef-1B.2 |
| FNS-209 |
FNS-996; FNS-1002; FNS-1018; FNS-1019; FNS-1055 |
F25H5.4 |
WBGene00001167 |
eef-2 |
| |
|
F09B12.3 |
WBGene00008607 |
F09B12.3 |
| |
|
F57F4.4 |
WBGene00019017 |
F57F4.4 |
| FNS-285; FNS-286; FNS-287; FNS-288 |
|
T21G5.3 |
WBGene00001598 |
glh-1 |
| FNS-489 |
|
C26D10.2 |
WBGene00001840 |
hel-1 |
| FNS-390 |
|
Y22D7AL.5 |
WBGene00002025 |
hsp-60 |
| |
F1-1012 |
F10C1.2 |
WBGene00002053 |
ifb-1 |
| FNS-401 |
|
M6.1 |
WBGene00002056 |
ifc-2 |
| |
FNS-1051 |
C43C3.1 |
WBGene00002067 |
ifp-1 |
| FNS-503 |
|
F57B9.6 |
WBGene00002083 |
inf-1 |
| |
F1-553; F1-579 |
K08H10.2 |
WBGene00010695 |
K08H10.2 |
| |
FNS-1284 |
Y71H2AM.19 |
WBGene00002244 |
laf-1 |
| |
F1-533; F1-579 |
K08H10.1 |
WBGene00002263 |
lea-1 |
| |
FT-1877 |
Y55B1AR.1 |
WBGene00002269 |
lec-6 |
| |
F1–1012 |
DY3.2 |
WBGene00003052 |
lmn-1 |
| |
FNS-1051 |
Y48C3A.7 |
WBGene00003119 |
mac-1 |
| F2-821 |
|
Y69A2AR.30 |
WBGene00003161 |
mdf-2 |
| F3-731 |
|
C36E6.3 |
WBGene00003369 |
mlc-1 |
| |
F1-362; F1-367 |
R07G3.3 |
WBGene00019940 |
npp-21 |
| FNS-134 |
FNS-739 |
F54F2.1 |
WBGene00003929 |
pat-2 |
| |
F2-1199 |
R05G6.7 |
WBGene00019900 |
R05G6.7 |
| F2-564; F3-516 |
|
F25H2.10 |
WBGene00004408 |
rla-0 |
| F2-584 |
|
B0041.4 |
WBGene00004415 |
rpl-4 |
| |
F1-741 |
T22F3.3 |
WBGene00020696 |
T22F3.3 |
| F2-456; FNS-463 |
F1-1115; FNS1669 |
K01G5.7 |
WBGene00006536 |
tbb-1 |
| F2-401; FNS-455 |
|
C36E8.5 |
WBGene00006537 |
tbb-2 |
| |
FNS-1567 |
Y71H2AM.23 |
WBGene00007000 |
tufm-1 |
| FNS-784 |
|
F40G9.3 |
WBGene00006715 |
ubc-20 |
| F1-114; F3-97 |
F1-463; FNS-373 |
F11C3.3 |
WBGene00006789 |
unc-54 |
| |
F1-1234; F1-1246 |
F08B6.4 |
WBGene00006819 |
unc-87 |
| |
F1-908 |
Y54E10A.9 |
WBGene00006888 |
vbh-1 |
| |
FNS-534 |
K09F5.2 |
WBGene00006925 |
vit-1 |
| |
FNS-501; FNS-517; FNS-518 |
C42D8.2 |
WBGene00006926 |
vit-2 |
| |
FNS-518; FNS-532 |
F59D8.1 |
WBGene00006927 |
vit-3 |
| |
FNS-531; FNS-534 |
F59D8.2 |
WBGene00006928 |
vit-4 |
| |
FNS-1126; FNS-1129 |
K07H8.6 |
WBGene00006930 |
vit-6 |
| FNS-831 | Y48A6B.3 | WBGene00012964 | Y48A6B.3 |
* Proteins implicated in a metabolic pathway. †Protein processing in endoplasmic reticulum.
Table 4. Functional classification by EASE of differentially represented proteins.
| Gene category | Infection | Stress | List hits | Population hits | Probability |
|---|---|---|---|---|---|
| Down ≥ 2x daf-2 (D6); Halaschek-Wiener 2005 |
|
X |
25 |
234 |
7.6E-31 |
| Proteome changes S. aureus; Bogaerts 2010 |
X |
|
17 |
109 |
7.2E-24 |
| Differentially expressed proteins in crt-1;cnx-1 vs N2 at 20°C; Lee 2006 |
|
|
7 |
13 |
9.0E-15 |
| Protein expression; Kim 2001 |
|
|
14 |
446 |
4.7E-10 |
| Proteome changes Aeromonas h.; Bogaerts 2010 |
X |
|
7 |
64 |
2.8E-09 |
| Down ≥ 2x dauer; Halaschek-Wiener 2005 |
|
X |
6 |
36 |
2.9E-09 |
| Differentially expressed proteins in crt-1;cnx-1 vs N2 at 25°C; Lee 2006 |
|
|
4 |
12 |
7.2E-08 |
| Up > 1.75x in M. luteus vs. Pseudomonas sp; Coolon 2009 |
X |
|
6 |
69 |
1.7E-07 |
| Glycoproteins GaL6 binding; Kaji 2007 |
|
|
9 |
287 |
8.0E-07 |
| Heat shock; Kim 2001 |
|
X |
4 |
25 |
1.8E-06 |
| Cell structural, muscle; Kim 2001 |
|
|
9 |
332 |
2.6E-06 |
| Down after organophosphorus pesticide chlorpyrifos + diazinon; Vinuela 2010 |
|
X |
5 |
65 |
3.5E-06 |
| Regulated down_daf-2 mutant and RNAi,Class2,-IGF1; Murphy 2003 |
|
X |
7 |
222 |
1.4E-05 |
| Down > 1.75x in Pseudomonas spp vs E. coli; Coolon 2009 |
X |
|
4 |
44 |
1.8E-05 |
| Down after organophosphorus pesticide diazinon; Vinuela 2010 |
|
X |
5 |
121 |
7.2E-05 |
| Regulated down_Bt toxin, Cry5B; Huffman 2004 |
|
X |
8 |
442 |
0.00017 |
| Differentially expressed proteins in crt-1 vs N2 at 20°C; Lee 2006 |
|
|
2 |
7 |
0.00026 |
| Regulated down_Cadmium; Huffman 2004 |
|
X |
7 |
388 |
0.00046 |
| Energy generation; Kim 2001 |
|
|
4 |
104 |
0.00052 |
| Up > 1.75x in B. megaterium vs. Pseudomonas sp; Coolon 2009 |
X |
|
3 |
45 |
0.00055 |
| Down ≥ 2x oxidative stress; Park 2009 |
|
X |
2 |
13 |
0.00095 |
| DNA synthesis; Kim 2001 |
|
|
7 |
440 |
0.00096 |
| Up ≥ 2x by PA14 8h; Troemel 2006 |
X |
|
5 |
233 |
0.00146 |
| Overlap Between oxidative stress and aging—downregulated genes by oxidative stress; Park 2009 | X | 3 | 66 | 0.00169 |
Fungal infection in adults does not provoke the UPR
The proteins linked to protein processing in the endoplasmic reticulum included the calreticulin CRT-1, the protein disulphide isomerase PDI-2 and HSP-3. HSP-3 and the closely related HSP-4 represent the worm’s BiP/GRP78 homologs. All these proteins function in the ER to ensure the correct folding of nascent polypeptides and are important components of the UPR. Given the reported link between the UPR and resistance to bacterial toxins and infection,17,18,22,30,31 we decided to investigate whether the UPR is involved in the host response to D. coniospora infection.
A direct measure of the activation of the UPR is provided by the detection of a specific UPR-associated alternatively spliced isoform of the transcription factor XBP-1. In contrast to the splicing of xbp-1 observed when young adult worms were treated with the UPR-inducing drug tunicamycin, the alternatively spliced isoform of xbp-1 was not detected following D. coniospora infection (Fig. 1A). Another indicator of the UPR is an increased expression of hsp-3 and hsp-4. In C. elegans, the UPR is often monitored in vivo using a phsp-4::GFP transgene reporter, which has a lower constitutive expression and higher level of induction during a UPR than phsp-3::GFP.32 In contrast to tunicamycin-treated worms, there was neither induction of an phsp-4::GFP transgene reporter after infection (Fig. 1B), nor increase of the hsp-4 transcript as measured by qRT-PCR (data not shown). This is consistent with previous genome-wide transcriptome studies that found that the expression of hsp-4 (and hsp-3) was not significantly altered following infection with D. coniospora.3,4 When worms carrying a pnlp-29::GFP transgene reporter were exposed to tunicamycin, strong GFP expression was observed in young larvae. This is consistent with a previous microarray study that reported the induction of a number of epidermal AMP genes, including nlp-29, in L2 larvae treated with tunicamycin.20 A marked increase in reporter gene expression was also seen in young larvae carrying a pnlp-30::GFP reporter transgene. On the other hand, no induction of either of these reporters was seen in L4 or adult worms (Fig. 1B and data not shown). Similar results were obtained using the UPR-inducing agents dithiothreitol and thapsigargin (data not shown).
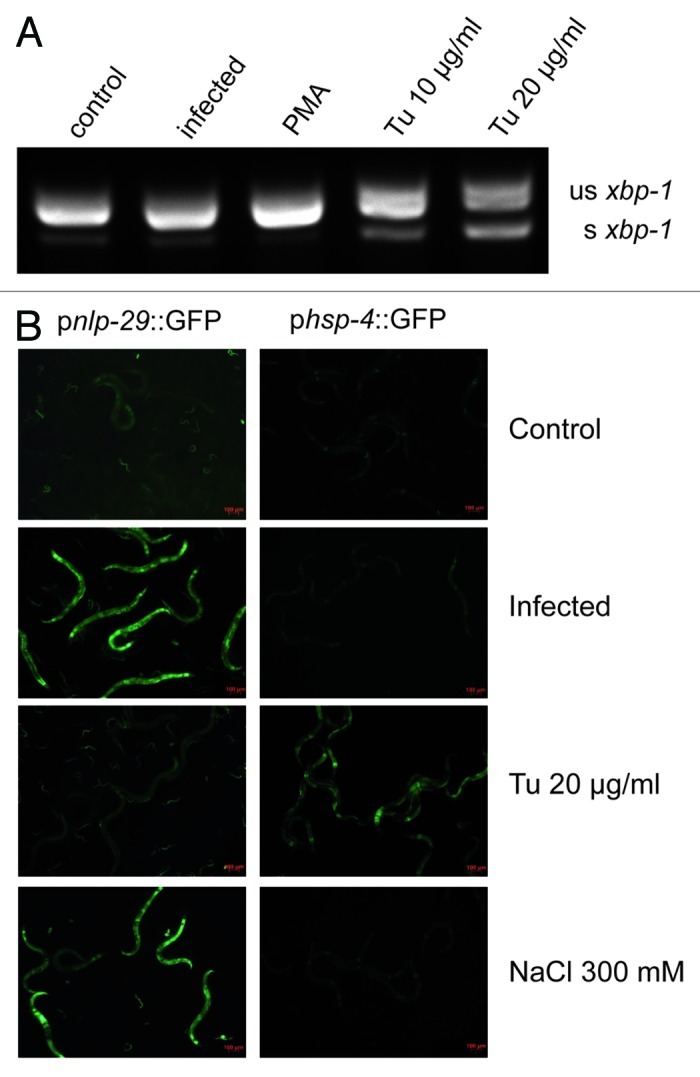
Figure 1. Fungal infection of adult worms does not induce a UPR. (A) RT-PCR analysis of xbp-1 splicing. Under standard culture conditions (control), a 220 bp amplicon from an unspliced (us) xbp-1 transcript is detected, together with very low levels of a 197 bp amplicon from a spliced (s) transcript. The abundance of this smaller band does not increase after infection with D. coniospora (infection) or PMA treatment (PMA), but is clearly increased upon UPR-induction with tunicamycin (Tu). (B) The green fluorescence in transgenic worms carrying a pnlp-29::GFP (strain IG274; left column) or a phsp-4::GFP (IG1320; right column) reporter was observed after infection, exposure to tunicamycin, or high salt. While infection and osmotic stress induced high level of pnlp-29::GFP expression, tunicamycin induced phsp-4::GFP.
We also tested whether direct activation of effector genes in the epidermis would trigger a UPR. PMA activates the PKCδ TPA-1 that controls multiple AMP genes, including nlp-29.7 It provokes very high levels of AMP gene expression within 4 h (unpublished results). Treating worms with PMA for 5 h did not lead to splicing of xbp-1 nor to induction of hsp-4 or the phsp-4::GFP transgene reporter (Fig. 1A, data not shown). The expression of many epidermal genes, including some AMPs, is strongly upregulated by osmotic stress.3,33 Although exposure to high salt did induce a pnlp-29::GFP transgene reporter as expected, it did not cause a measurable increase in phsp-4::GFP expression (Fig. 1B). Thus, neither fungal infection nor the strong induction of gene expression by PMA or salt provokes a UPR, and conversely a UPR does not trigger the expression of anti-fungal immune effectors in adult C. elegans.
hsp-3 regulates nlp-29 AMP gene expression
While the results described above suggested that the UPR did not play a direct role in the antifungal innate immune response, the representation of a number of ER-resident proteins is modulated by infection. This led us to assay directly the role of the corresponding genes in the regulation of nlp-29 by RNAi. While several of the tested genes had an effect (results not shown), hsp-3 stood out for its strong effect, essentially totally blocking the induction of pnlp-29::GFP normally observed upon infection in adult worms (Fig. 2A). A similar abrogation of reporter gene expression was seen in an atf-6 mutant, but not in a pek-1 mutant background (Fig. S1).
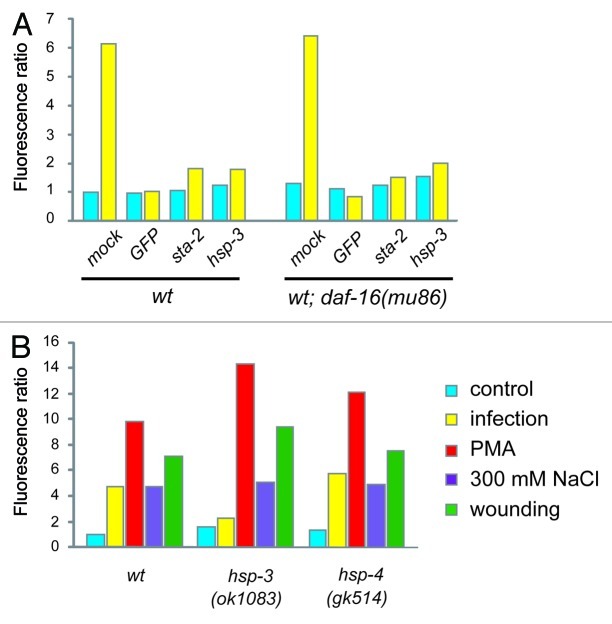
Figure 2. A specific role for hsp-3 in the regulation of nlp-29. (A) Quantification of the effect of control (K04G11.3), GFP, hsp-3 and sta-2 RNAi on pnlp-29::GFP expression in a wild-type or daf-16(mu86) mutant background. For reasons given elsewhere,5 in this and the subsequent graphs, error bars are not shown. Data are representative of three independent experiments. (B) Quantification of pnlp-29::GFP expression in hsp-3(ok1083) and hsp-4(gk514) mutant backgrounds following different treatments. In all cases, quantification was with the COPAS Biosort. The normalized average ratio of green to red fluorescence is shown. The analysis was restricted to worms with a TOF above 450. The number of worms analyzed here and in subsequent figures is given in the Supplemental Material.
The mRNA sequence of the second BiP/GRP78 gene in C. elegans, hsp-4, is highly similar to that of hsp-3 (1472/1873 nucleotides identical, including several contiguous stretches of more than 21 nucleotides) and would thus be predicted to be targeted by the hsp-3 RNAi construct. At the same time, there is a reciprocal control of hsp-3 and hsp-4, such that a decrease in hsp-3 expression normally leads to an increase in the level of hsp-4, and vice versa.34 As RNAi with hsp-4 did block pnlp-29::GFP induction upon infection in adult worms (data not shown), we sought to discriminate between the two genes using available null mutants. We observed a strong reduction in pnlp-29::GFP expression only in an hsp-3 mutant, not in an hsp-4 mutant background (Fig. 2B). Attempts to establish a hsp-3;hsp-4 strain were confounded by the fact that homozygous double mutants were sterile. When we inactivated hsp-3 by RNAi in the hsp-4 mutant background, the adult worms were sterile, and the induction of pnlp-29::GFP expression upon infection was blocked (data not shown).
In C. elegans, fertility and pathogen resistance are interlinked, via the FOXO transcription factor DAF-16,35,36 which also plays a role in the UPR.37 We therefore assayed the effect of hsp-3 RNAi on pnlp-29::GFP expression in a daf-16 mutant background. Loss of daf-16 had no effect on the abrogation of pnlp-29::GFP expression provoked by hsp-3 RNAi, or by RNAi with the STAT-like transcription factor sta-2, previously characterized for its role in nlp-29 expression6 (Fig. 2A), indicating that the effect of hsp-3, and of sta-2, is independent of daf-16.
We then determined the specificity of the effect of hsp-3 on reporter gene expression. In clear contrast to the near-complete block of pnlp-29::GFP expression after infection, in an hsp-3 mutant the induction of the reporter gene was at least as strong as in the wild-type background when triggered by PMA, salt or wounding. In the hsp-4 mutant, however, no effect was seen under any of the experimental conditions (Fig. 2B). These results underline the specific role hsp-3 plays in regulating pnlp-29::GFP only after infection, and place hsp-3 genetically upstream of, or parallel to, the PKCδ TPA-1.
hsp-3 acts downstream of nipi-3 to regulate nlp-29 AMP gene expression
The only previously known component of the innate immune signaling pathways that regulates nlp-29 expression specifically upon infection is the Tribbles-like kinase nipi-3. Overexpression of nipi-3 leads to an induction of pnlp-29::GFP.5 This induction was blocked in the hsp-3 mutant background, placing hsp-3 genetically downstream of nipi-3 (Fig. 3A). Consistent with this result, hsp-3 did not block the increased expression of pnlp-29::GFP provoked by an activated form of GPA-12 that triggers TPA-1 independently of NIPI-37 (Fig. 3B). Together these results indicate that hsp-3 acts between nipi-3 and tpa-1 to control the expression of nlp-29 upon fungal infection (Fig. 4).
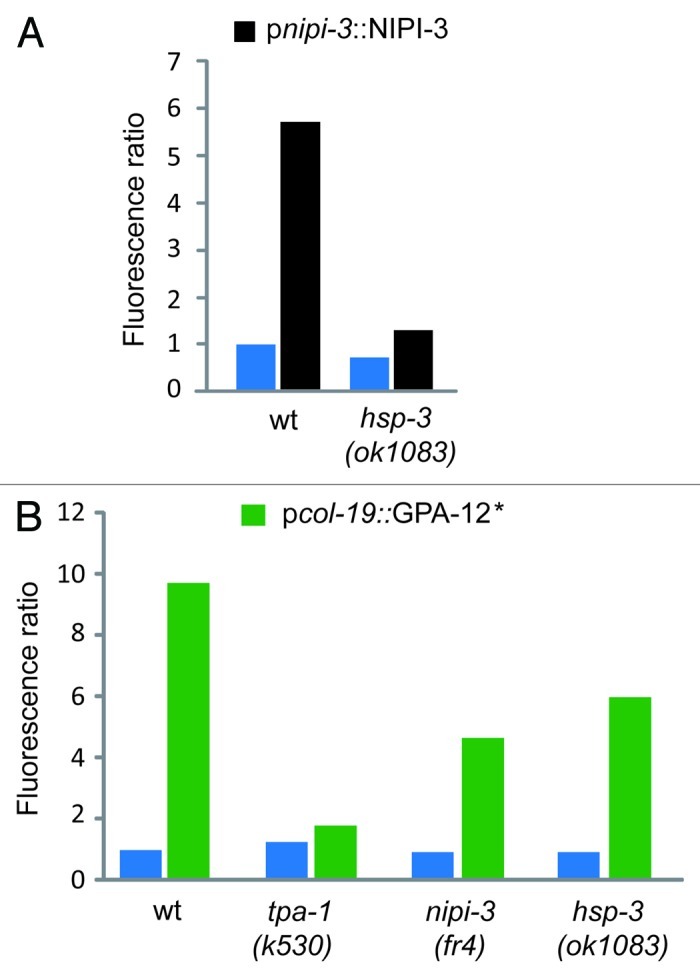
Figure 3.hsp-3 acts genetically downstream of nipi-3 but not of gpa-12. (A) pnlp-29::GFP reporter expression was quantified in wt and hsp-3(ok1083) mutant worms with (black bars) or without (blue bars) copies of a transgene containing nipi-3 under the control of its own promoter. (B) Quantification of pnlp-29::GFP reporter expression in wt, tpa-1(k530), nipi-3(fr4) and hsp-3(ok1083) mutant worms with (green bars) or without (blue bars) copies of a transgene containing a gain-of-function (*) allele of gpa-12 under the control of the epidermis-specific col-19 promoter. Both pnipi-3::NIPI-3 and pcol-19::GPA-12* transgenes provoke a robust nlp-29 upregulation in the absence of infection in adult worms. Quantification was with the COPAS Biosort. The normalized average ratio of green fluorescence to time of flight (TOF) is shown. The analysis was restricted to worms with a TOF between 450 and 650.
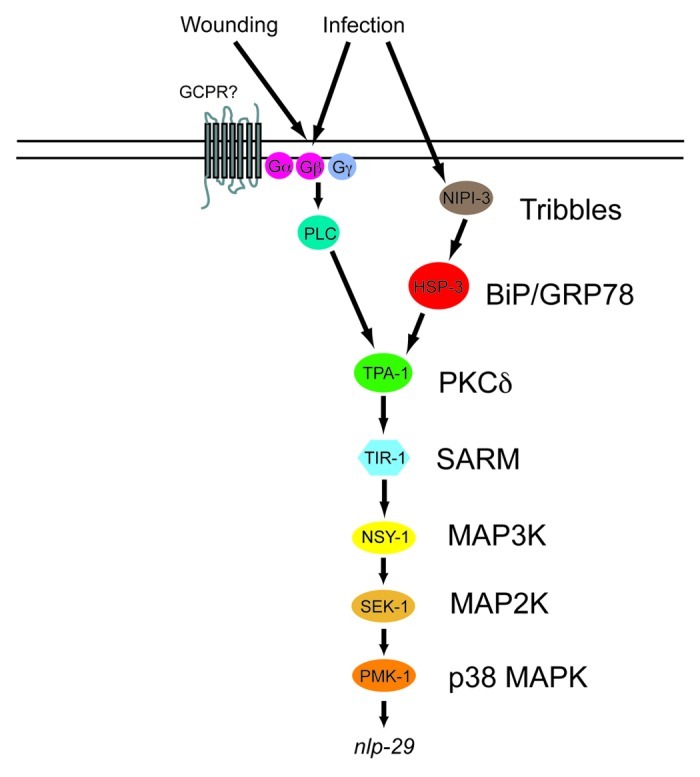
Figure 4. Model of the control of nlp-29 expression. Signals perceived upon D. coniospora infection and injury are transduced by a PKCδ - p38 MAPK pathway to regulate the expression of nlp-29. HSP-3 functions between NIPI-3 and the PKCδ TPA-1. Many other known regulatory elements, including the OSM-11/WNK-1/GCK-3 pathway48 and the recently described pseudokinase NIPI-449 have been omitted for the sake of clarity.
Discussion
Much of our previous characterization of the innate immune response of C. elegans to D. coniospora has been focused on the host transcriptional changes that accompany infection. In Drosophila, the expression of many components of immune signaling pathways are themselves highly regulated upon infection.38,39 In contrast, none of 18 genes known to influence nlp-29 expression, including the p38 MAPK cascade components nsy-1, sek-1 and pmk-1, show a marked change in their expression level after infection.4 In a previous DIGE-based pilot study, we identified RACK-1 as a factor involved in the regulation of anti-fungal defenses.7 In an attempt to identify additional candidates, we extended the approach and undertook a comprehensive proteomic study of the changes that accompany fungal infection.
A number of other comparative gel-based proteomic studies have been performed using C. elegans40,41 including two looking at protein changes upon bacterial infection of the intestine.23,24 It is striking that certain proteins, such as ACT-4, SODH-1, VHA-13 and PDI-2, appear in almost every published list. This may reflect an intrinsic bias in the approach, since the measured expression level4 for the genes corresponding to the proteins that we identified as differentially represented was very significantly higher than that of genes in general, (71.7% > 5 dcpm vs. 6.6% for all transcripts; p < 0.001 binomial test). It may also result from the fact that all these analyses used whole-animal extracts, potentially masking tissue-specific biologically relevant variations in protein abundance, and underlines the interest for developing efficient and simple methods to allow protein extraction from a specific C. elegans tissue.
If a particular spot on a gel increases or decreases in intensity, one cannot always infer that the total level of the corresponding protein was changed. For example, post-translational modifications may render a protein more difficult to extract, so that spot intensity does not reflect protein abundance. Similarly, post-translational modifications may also lead to an alteration of the sub-cellular localization of a protein, which may cause a protein to be found in different extraction fractions, and thereby affect spot intensity. As many proteins give rise to multiple spots, generally because of post-translational modifications, only if all the spots for a given protein were identified and quantified from each fraction would one be able to quantitate protein abundance. The development of alternative methods, such as metabolic labeling coupled to mass spectrometry holds considerable promise for profiling changes during pathogenic challenge in C. elegans.25 In the meantime, caution needs to be exercised when analyzing gel-based proteomic results. For this reason, until we have further functional evidence for a role in innate immunity for the various candidate proteins we identified, any discussion of a putative role would be premature.
The exception is HSP-3, which clearly has a specific function in regulating AMP gene expression. The hsp-3 gene is expressed at a high-level and is unchanged by infection,4 so the total level of HSP-3 may not change upon infection. We have not established the change in HSP-3 (e.g., degradation, phosphorylation, etc.) that leads to a change in intensity of the corresponding spot. It is noteworthy that in an atf-6 mutant background there is a marked reduction of pnlp-29::GFP expression after infection. This may reflect a role for ATF-6 in regulating HSP-3 levels.
During development, hsp-3 has an unambiguous role in the UPR.19,42 The data presented here indicate that the immune function of hsp-3 is independent of its function in the UPR. There is, however, evidence for a link between the UPR and AMP gene regulation in larvae. As mentioned above, a number of genes, including cnc-4, fipr-26, nlp-28 and nlp-29 are induced in L2 larvae after treatment with tunicamycin, apparently independently of xbp-1.20 But although tunicamycin does provoke upregulation of pnlp-29::GFP in larvae, D. coniospora infection does not induce phsp-4::GFP expression either in larvae or adults. Further, this UPR-induced expression of pnlp-29::GFP is independent of the p38 MAPK pathway, as it is observed in pmk-1 mutant background, as well as in a tpa-1 and nipi-3 mutant backgrounds (data not shown), and overall, there is only a minimal overlap between the genes upregulated by tunicamycin and D. coniospora infection.3,20 So the relationship between anti-fungal innate immunity and the UPR is not straightforward.
It is interesting, nonetheless, to speculate on how HSP-3 might exert its influence on AMP expression. There are several plausible models that are based on the idea that although genetically hsp-3 is positioned between nipi-3 and tpa-1, it seems unlikely that it plays a direct role in signal transduction. HSP-3 might be needed to ensure the correct intracellular localization of NIPI-3, itself, or of a protein that acts downstream of NIPI-3 and upstream of TPA-1. It may therefore be worthwhile to look at NIPI-3 localization in wild-type and hsp-3 mutant worms. The presence of two almost identical BiP/GRP78 proteins in the nematode is intriguing, as mammals, for example, only have one. It is conceivable that HSP-3 has a UPR-independent function outside the ER. Interestingly, one of the areas of sequence divergence between the two proteins is at the C-terminus; where HSP-3 has the ER retention signal KDEL sequence, HSP-4 has HDEL.
There are many examples of heat shock proteins playing a more or less direct role in innate immune responses.43 For example, they can function as endogenous danger signals to indicate cell stress and tissue damage to the immune system. As another example, the conserved SGT1/HSP90 complex binds NLR proteins, and modulates innate immune signaling in plants and animals,44 although it should be noted that there are no obvious NLR proteins in C. elegans. A study of the intracellular localization of HSP-3, and of the consequences of artificially expressing it in the cytoplasm could be merited. It is interesting to note that in an hsp-3 mutant there is some residual induction of the nlp-29 reporter gene, but this is fully abolished if the mutants are subject to RNAi against hsp-4 (C.C., unpublished observations). On the other hand, we have shown that loss of hsp-4 function alone has no effect on nlp-29 reporter gene expression. This suggests that hsp-4 can partially compensate for the absence of hsp-3.
It has been shown that during C. elegans development, the activation of a p38 MAPK pathway that follows intestinal infection with the P. aeruginosa strain PA14 causes a UPR.18 This is believed to be a consequence of the increased expression of innate immune effectors that overload the protein folding machinery in the ER. The results we have presented here show that infection of adult C. elegans by D. coniospora and the resultant induction of a large number of defense proteins in the epidermis does not provoke a UPR. This might reflect a relatively low constitutive activity of the secretory pathway in the epidermis, and therefore a buffering capacity in adult animals to cope with the consequences of infection. It will be interesting to dissect further the complex interplay between developmental, physiological (e.g., production of digestive enzymes in the intestine) and induced processes that put stress on the ER, both in C. elegans and other organisms. Additional study is also required to understand fully the UPR-independent role of BiP/GRP78 in innate immunity in C. elegans and to determine whether it might play any such role in other organisms.
Materials and Methods
Strains and culture condition
Worms were grown and maintained on nematode growth medium (NGM) and cultured with the E. coli strain OP50, as described.45 The hsp-3(ok1083), hsp-4(gk514), daf-16(mu86), tpa-1(k530), atf-6(ok551), pek-1(ok275) mutants were obtained from the Caenorhabditis Genetics Center (CGC). The strain SJ17 (xbp-1(zs12) III; zsIs4[phsp-4::GFP] V)21 was the kind gift of Dr Eric Chevet.
Reporter gene constructs and transgenic lines
IG274 (wt; frIs7[pnlp-29::GFP, pcol-12::DsRed] IV) is described elsewhere.5 IG981 [hsp-3(ok1083) X; frIs7 IV], IG982 [hsp-4(gk514) II; frIs7 IV] and IG1161 [daf-16(mu86) I; frIs7 IV], IG983 [atf-6(ok551) II; frIs7 IV], IG1424 [pek-1(ok275) II; frIs7 IV], were obtained by crossing the mutants hsp-3(ok1083), hsp-4(gk514), daf-16(mu86), atf-6(ok551) and pek-1(ok275) with IG274. The strain IG1320 (wt; zsIs4[phsp-4::GFP] V) was obtained by backcrossing the strain SJ17 with N2. The strain IG1363 (wt; frEx486[(pcol-19::GPA-12*), pNP21(pBunc-53::GFP)]) was the kind gift of Dr Nathalie Pujol. The strains IG1361 [tpa-1(k530) frIs7 IV; frEx486], IG1364 [hsp-3(ok1083) X; frIs7 IV; frEx486] and IG1365 [nipi-3(fr4) X; frIs7 IV; frEx486] were obtained by crossing respectively tpa-1(k530), hsp-3(ok1083) and nipi-3(fr4) with IG1363.
Splicing of xbp-1
N2 worms were grown and maintained on NGM plates with OP50. When they reached the young adult stage, worms were infected with D. coniospora by transferring them to NGM/OP50 plates previously spread with a dense suspension of spores. These had been freshly harvested in M9 buffer. Otherwise uninfected young adult worms were tranferred onto NGM/OP50 plates containing 10 µg/mL or 20 µg/mL tunicamycin (Sigma) or 1 µg/mL PMA (Sigma). After 5 h worms were harvested and RNA extracted with Trizol as described.46 Reverse transcription used High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and PCR analyses were performed as described.17 Samples were normalized by Q-PCR as described26 with eft-217 as an internal control.
RNAi
All RNAi feeding experiments were performed essentially as described,47 using clones from the Ahringer library. All RNAi clones were sequence verified before use. The experiments were performed with worms cultured on OP50 until the L2 stage.
Infection, wounding, osmotic stress, PMA stress
Infection, and wounding were performed as described.5 For exposure to osmotic stress, PMA and tunicamycin, compounds were added to NGM plates to a final concentration of 300 mM for NaCl, 1 µg/mL for PMA and 10 or 20 µg/mL for tunicamycin. Worms were grown and maintained on NGM plates with OP50. When they reached the young adult stage, worms were transferred onto the appropriate modified NGM/OP50 plates. Similar conditions were used to assay the induction of GFP expression in the strains IG274 and IG1320 (shown in Fig. 1B), except images were taken after only 5 h.
Biosorter
The quantification of fluorescent reporter gene (GFP) expression was performed with the COPAS Biosort (Union Biometrica), essentially as described.26 Generally, animals were analyzed for length (time of flight), optical density (extinction), green fluorescence, and red fluorescence (if appropriate).
Protein extraction
A synchronized population of L4 IG274 worms was infected with D. coniospora. After 5 h, when there was a clear induction of GFP, indicative of the innate immune response to a productive infection, worms were harvested by washing plates with M9 buffer. Worms were pelleted by decantation, washed twice in the M9 buffer, then twice with PBS buffer. Proteins were extracted from a pellet of 500 µL of worms either by sonication in 8 M urea, 2 M thiourea, 4% (w/v) CHAPS, pH 8.5, containing a phosphatase inhibitor cocktail (Roche), or using the 2D fractionation kit (Amersham) according to the manufacturer’s instructions.
Protein fractionation, labeling, gel electrophoresis and identification
Full experimental details are publically available at http://miapegeldb.expasy.org/experiment/118. The comprehensive set of analytical data from this study is available at the World-2DPAGE database http://world-2dpage.expasy.org/repository/0042.
Supplementary Material
Acknowledgments
We thank J. Belougne for worm sorting. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). We thank E. Chevet and N. Pujol for providing strains, and S. Granjeaud, P. Pierre and the members of the Ewbank lab for discussion and critical comments. This work was funded by institutional grants from INSERM and the CNRS.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/virulence/article/20384
References
- 1.Engelmann I, Pujol N. Innate immunity in C. elegans. Adv Exp Med Biol. 2010;708:105–21. doi: 10.1007/978-1-4419-8059-5_6. [DOI] [PubMed] [Google Scholar]
- 2.Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, Kohara Y, et al. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol. 2004;5:488–94. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- 3.Pujol N, Zugasti O, Wong D, Couillault C, Kurz CL, Schulenburg H, et al. Anti-fungal innate immunity in C. elegans is enhanced by evolutionary diversification of antimicrobial peptides. PLoS Pathog. 2008;4:e1000105. doi: 10.1371/journal.ppat.1000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engelmann I, Griffon A, Tichit L, Montañana-Sanchis F, Wang G, Reinke V, et al. A comprehensive analysis of gene expression changes provoked by bacterial and fungal infection in C. elegans. PLoS One. 2011;6:e19055. doi: 10.1371/journal.pone.0019055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pujol N, Cypowyj S, Ziegler K, Millet A, Astrain A, Goncharov A, et al. Distinct innate immune responses to infection and wounding in the C. elegans epidermis. Curr Biol. 2008;18:481–9. doi: 10.1016/j.cub.2008.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dierking K, Polanowska J, Omi S, Engelmann I, Gut M, Lembo F, et al. Unusual regulation of a STAT protein by an SLC6 family transporter in C. elegans epidermal innate immunity. Cell Host Microbe. 2011;9:425–35. doi: 10.1016/j.chom.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 7.Ziegler K, Kurz CL, Cypowyj S, Couillault C, Pophillat M, Pujol N, et al. Antifungal innate immunity in C. elegans: PKCdelta links G protein signaling and a conserved p38 MAPK cascade. Cell Host Microbe. 2009;5:341–52. doi: 10.1016/j.chom.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–6. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- 9.Kurz CL, Shapira M, Chen K, Baillie DL, Tan MW. Caenorhabditis elegans pgp-5 is involved in resistance to bacterial infection and heavy metal and its regulation requires TIR-1 and a p38 map kinase cascade. Biochem Biophys Res Commun. 2007;363:438–43. doi: 10.1016/j.bbrc.2007.08.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liberati NT, Fitzgerald KA, Kim DH, Feinbaum R, Golenbock DT, Ausubel FM. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc Natl Acad Sci U S A. 2004;101:6593–8. doi: 10.1073/pnas.0308625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huffman DL, Abrami L, Sasik R, Corbeil J, van der Goot FG, Aroian RV. Mitogen-activated protein kinase pathways defend against bacterial pore-forming toxins. Proc Natl Acad Sci U S A. 2004;101:10995–1000. doi: 10.1073/pnas.0404073101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren M, Feng H, Fu Y, Land M, Rubin CS. Protein kinase D is an essential regulator of C. elegans innate immunity. Immunity. 2009;30:521–32. doi: 10.1016/j.immuni.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shivers RP, Pagano DJ, Kooistra T, Richardson CE, Reddy KC, Whitney JK, et al. Phosphorylation of the conserved transcription factor ATF-7 by PMK-1 p38 MAPK regulates innate immunity in Caenorhabditis elegans. PLoS Genet. 2010;6:e1000892. doi: 10.1371/journal.pgen.1000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Partridge FA, Gravato-Nobre MJ, Hodgkin J. Signal transduction pathways that function in both development and innate immunity. Dev Dyn. 2010;239:1330–6. doi: 10.1002/dvdy.22232. [DOI] [PubMed] [Google Scholar]
- 16.Coleman JJ, Mylonakis E. The tangled web of signaling in innate immunity. Cell Host Microbe. 2009;5:313–5. doi: 10.1016/j.chom.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bischof LJ, Kao CY, Los FC, Gonzalez MR, Shen Z, Briggs SP, et al. Activation of the unfolded protein response is required for defenses against bacterial pore-forming toxin in vivo. PLoS Pathog. 2008;4:e1000176. doi: 10.1371/journal.ppat.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richardson CE, Kooistra T, Kim DH. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature. 2010;463:1092–5. doi: 10.1038/nature08762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon A, et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/S0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- 20.Shen X, Ellis RE, Sakaki K, Kaufman RJ. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet. 2005;1:e37. doi: 10.1371/journal.pgen.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–6. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 22.Ewbank JJ, Pujol N. Cellular homeostasis: coping with ER overload during an immune response. Curr Biol. 2010;20:R452–5. doi: 10.1016/j.cub.2010.03.023. [DOI] [PubMed] [Google Scholar]
- 23.Bogaerts A, Beets I, Temmerman L, Schoofs L, Verleyen P. Proteome changes of Caenorhabditis elegans upon a Staphylococcus aureus infection. Biol Direct. 2010;5:11. doi: 10.1186/1745-6150-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bogaerts A, Temmerman L, Boerjan B, Husson SJ, Schoofs L, Verleyen P. A differential proteomics study of Caenorhabditis elegans infected with Aeromonas hydrophila. Dev Comp Immunol. 2010;34:690–8. doi: 10.1016/j.dci.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Simonsen KT, Møller-Jensen J, Kristensen AR, Andersen JS, Riddle DL, Kallipolitis BH. Quantitative proteomics identifies ferritin in the innate immune response of C. elegans. Virulence. 2011;2:120–30. doi: 10.4161/viru.2.2.15270. [DOI] [PubMed] [Google Scholar]
- 26.Zugasti O, Ewbank JJ. Neuroimmune regulation of antimicrobial peptide expression by a noncanonical TGF-beta signaling pathway in Caenorhabditis elegans epidermis. Nat Immunol. 2009;10:249–56. doi: 10.1038/ni.1700. [DOI] [PubMed] [Google Scholar]
- 27.Schwarz EM, Antoshechkin I, Bastiani C, Bieri T, Blasiar D, Canaran P, et al. WormBase: better software, richer content. Nucleic Acids Res. 2006;34(Database issue):D475–8. doi: 10.1093/nar/gkj061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38(Database issue):D355–60. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hosack DA, Dennis G, Jr., Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haskins KA, Russell JF, Gaddis N, Dressman HK, Aballay A. Unfolded protein response genes regulated by CED-1 are required for Caenorhabditis elegans innate immunity. Dev Cell. 2008;15:87–97. doi: 10.1016/j.devcel.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun J, Singh V, Kajino-Sakamoto R, Aballay A. Neuronal GPCR controls innate immunity by regulating noncanonical unfolded protein response genes. Science. 2011;332:729–32. doi: 10.1126/science.1203411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caruso ME, Jenna S, Bouchecareilh M, Baillie DL, Boismenu D, Halawani D, et al. GTPase-mediated regulation of the unfolded protein response in Caenorhabditis elegans is dependent on the AAA+ ATPase CDC-48. Mol Cell Biol. 2008;28:4261–74. doi: 10.1128/MCB.02252-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohlfing AK, Miteva Y, Hannenhalli S, Lamitina T. Genetic and physiological activation of osmosensitive gene expression mimics transcriptional signatures of pathogen infection in C. elegans. PLoS One. 2010;5:e9010. doi: 10.1371/journal.pone.0009010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapulkin WJ, Hiester BG, Link CD. Compensatory regulation among ER chaperones in C. elegans. FEBS Lett. 2005;579:3063–8. doi: 10.1016/j.febslet.2005.04.062. [DOI] [PubMed] [Google Scholar]
- 35.TeKippe M, Aballay A. C. elegans germline-deficient mutants respond to pathogen infection using shared and distinct mechanisms. PLoS One. 2010;5:e11777. doi: 10.1371/journal.pone.0011777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyata S, Begun J, Troemel ER, Ausubel FM. DAF-16-dependent suppression of immunity during reproduction in Caenorhabditis elegans. Genetics. 2008;178:903–18. doi: 10.1534/genetics.107.083923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henis-Korenblit S, Zhang P, Hansen M, McCormick M, Lee SJ, Cary M, et al. Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc Natl Acad Sci U S A. 2010;107:9730–5. doi: 10.1073/pnas.1002575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci U S A. 2001;98:12590–5. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002;21:2568–79. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee W, Kim KR, Singaravelu G, Park BJ, Kim DH, Ahnn J, et al. Alternative chaperone machinery may compensate for calreticulin/calnexin deficiency in Caenorhabditis elegans. Proteomics. 2006;6:1329–39. doi: 10.1002/pmic.200500320. [DOI] [PubMed] [Google Scholar]
- 41.Jeong PY, Na K, Jeong MJ, Chitwood D, Shim YH, Paik YK. Proteomic analysis of Caenorhabditis elegans. Methods Mol Biol. 2009;519:145–69. doi: 10.1007/978-1-59745-281-6_10. [DOI] [PubMed] [Google Scholar]
- 42.Urano F, Calfon M, Yoneda T, Yun C, Kiraly M, Clark SG, et al. A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. J Cell Biol. 2002;158:639–46. doi: 10.1083/jcb.200203086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osterloh A, Breloer M. Heat shock proteins: linking danger and pathogen recognition. Med Microbiol Immunol. 2008;197:1–8. doi: 10.1007/s00430-007-0055-0. [DOI] [PubMed] [Google Scholar]
- 44.Kadota Y, Shirasu K, Guerois R. NLR sensors meet at the SGT1-HSP90 crossroad. Trends Biochem Sci. 2010;35:199–207. doi: 10.1016/j.tibs.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 45.Stiernagle T. Maintenance of C. elegans WormBook. http://www.wormbook.org: The C. elegans Research Community ed, 2006:1551-8507.
- 46.Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S, Kohara Y, et al. Inducible antibacterial defense system in C. elegans. Curr Biol. 2002;12:1209–14. doi: 10.1016/S0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- 47.Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–12. doi: 10.1016/S0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 48.Lee KZ, Kniazeva M, Han M, Pujol N, Ewbank JJ. The fatty acid synthase fasn-1 acts upstream of WNK and Ste20/GCK-VI kinases to modulate antimicrobial peptide expression in C. elegans epidermis. Virulence. 2010;1:113–22. doi: 10.4161/viru.1.3.10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Labed SA, Omi S, Gut M, Ewbank JJ, Pujol N. The Pseudokinase NIPI-4 Is a Novel Regulator of Antimicrobial Peptide Gene Expression. PLoS One. 2012;7:e33887. doi: 10.1371/journal.pone.0033887. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.