Abstract
One of the most fundamental questions in the control of gene expression is how epigenetic patterns of DNA methylation and histone modifications are established. Our recent studies demonstrate that histone deacetylase HDA6 integrates DNA methylation and histone modifications in gene silencing by interacting with DNA methyltransferase MET1 and histone demethylase FLD, suggesting that regulatory crosstalk between histone modifications and DNA methylation could be mediated by the interaction of various epigenetic modification proteins.
Keywords: DNA Methylation, FLD, HDA6, MET1, histone deacetylation, histone demethylation
DNA cytosine methylation and histone modifications including acetylation, methylation, phosphorylation as well as ubiquitination are important epigenetic mechanisms that have profound roles in gene regulation and development processes.1-3 The functional consequences of histone modifications can be direct, causing structural changes to chromatin, or indirect, acting through the recruitment of effector proteins. All histone modifications are removable, which may therefore provide a flexible way for regulation of gene expression.
DNA cytosine methylation of genes is often associated with transcriptional gene silencing. In Arabidopsis, cytosine methylation occurs at CG, CHG and CHH sites.4 The Arabidopsis thaliana genome encodes three classes of DNA cytosine methyltransferases. The function of DRM2 is to control de novo methylation.5 MET1, the ortholog of the mammalian DNMT1, maintains CG methylation; whereas CMT3 is plant specific methyltransferase, which functions to control the maintenance of CHG methylation.6
Histone acetylation levels are determined by the action of histone acetyltransferases and histone deacetylases (HDACs). Plant HDACs can be grouped into three major families, RPD3/HDA1, SIR2 and HD2.7,8 It was reported that gene silencing could be relieved by treatment with either HDAC inhibitors or an inhibitor of DNA methylation.9-11 In addition, inhibiting cytosine methylation induces histone acetylation, whereas inhibiting histone deacetylation causes the loss of cytosine methylation.12 These findings suggest that histone deacetylation and DNA methylation may act coordinately in gene silencing. In mammals, HDACs can be recruited by high DNA methylation levels, via association with methyl-DNA binding domain-containing proteins or via direct recruitment by the DNA methyltransferase DNMT1,13,14 suggesting a tight interplay between histone deacetylation and DNA methylation.
HDA6 interacts with MET1 Mediating Crosstalk of Hitone Deacetylation and DNA Methylation
HDA6 was first identified to be involved in transgene silencing through an auxin-responsive element mutant screening in Arabidopsis.15 HDA6 mutant alleles axe1–1 to axe1–5 displayed increased expression of the auxin-responsive reporter genes in the absence of auxin treatment, suggesting a role of HDA6 in gene silencing.15 Furthermore, HDA6 was also identified as an essential component in RNA-directed DNA methylation (RdDM).16,17 The HDA6 mutant allele rts1 exhibits a reactivation of RdDM-silenced promoters and results in reduced cytosine methylation in symmetric sequence contexts, highlighting a function for HDA6 in methylation maintenance.16,17 Recently, To et al. (2011) reported that HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1.18 In addition, HDA6 and MET1 co-target to the heterochromatin sites and maintain heterochromatin silencing.18
More recently, we show that a subset of transposons was transcriptionally reactivated in hda6 mutants, associated with elevated histone H3 and H4 acetylation as well as increased H3K4Me3 and H3K4Me2.19 Decreased DNA methylation of the transposons was also detected in hda6 mutants, suggesting that HDA6 silences the transposons by regulating histone acetylation and methylation as well as the DNA methylation status. Similarly, transcripts of some of these transposons were also increased in the met1 mutant, with decreased DNA methylation. Furthermore, H4 acetylation, H3K4Me3, H3K4Me2, and H3K36Me2 were enriched at the co-regulated transposons in the met1 and hda6met1 mutants. Protein-protein interaction analysis indicated that HDA6 physically interacts with MET1 in vitro and in vivo. These results suggest that HDA6 and MET1 interact directly and act together to silence transposons by modulating DNA methylation, histone acetylation, and histone methylation status.19
HDA6 interacts with Histone Demethylase FLD
Histone methylation is dynamically regulated by histone methylases and demethylases. There are two types of demethylases with distinct mechanisms, amine oxidation by lysine-specific demethylase1 (LSD1) and hydroxylation by Jumonji C (JmjC) domain-containing proteins, removing methyl groups from methylated lysine residues. The mammalian histone demethylase, LSD1, is an integral component of histone deacetylase corepressor complexes in which HDACs and LSD1 may cooperate to remove activating acetyl and methyl histone modifications.20,21 HDAC inhibitors can diminish histone demethylation activity, whereas the abrogation of LSD1 activity by mutations can decrease the deacetylation activity,21 suggesting that the enzymatic activities of HDACs and LSD1 are closely linked. The Arabidopsis homolog of LSD1, FLD, was found to promoter flowering in Arabidopsis by repressing FLC expression.22 In addition, two Arabidopsis FLD homologs, LDL1 and LDL2, act in partial redundancy with FLD to promote flowering by repress FLC expression.23 Our recent study indicates that HDA6 is physically associated with FLD in Arabidopsis,24 suggesting that histone deacetylases and demethylases may cross-talk and mutually affect each other’s activities to coordinate regulate gene expression in plant development and in response to environmental changes.
Conclusions and Perspectives
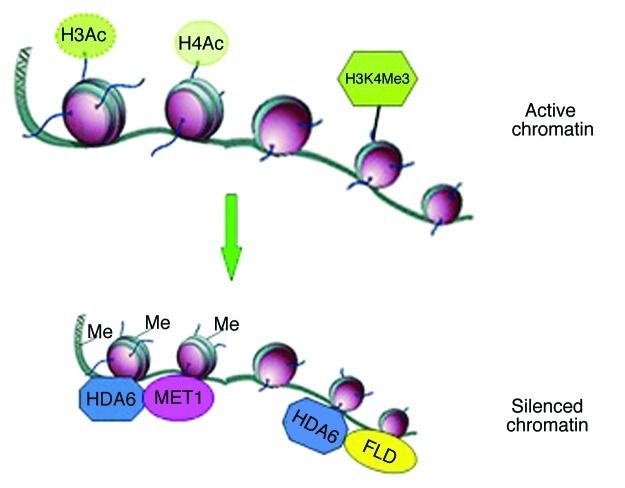
Our studies demonstrate that HDA6 integrates DNA methylation and histone modifications in gene silencing by interacting with MET1 and FLD (Fig. 1). More recently, OTU6 was found to directly interact with the histone lysine demethylase LDL1/KDM1C in planta,25 suggesting that regulatory crosstalk between histone demethylation and deubiquitination through the direct interaction between LDL1/KDM1C and OTU6. Taken together, data described above indicate that the crosstalk among different epigenetic modifications could be mediated by the interaction of various histone modification proteins and complexes.
Figure 1. Gene silencing mediated by HDA6. The gene activation markers, H3 acetylation, H4 acetylation and H3K4 tri-methylation are associated with active chromatin. HDA6 can mediate gene silencing through interacting with MET1 and FLD, resulting in DNA methylation, histone deacetylation and histone demethylation.
Acknowledgments
This study was funded by grants from the National Science Council of Taiwan (98–2628-B-002–016-MY3 and 99–2321-B-002–027-MY3) and National Taiwan University (10R80917–5).
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/19994
References
- 1.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 2.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–16. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 3.Fukuda H, Sano N, Muto S, Horikoshi M. Simple histone acetylation plays a complex role in the regulation of gene expression. Brief Funct Genomic Proteomic. 2006;5:190–208. doi: 10.1093/bfgp/ell032. [DOI] [PubMed] [Google Scholar]
- 4.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–20. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao X, Jacobsen SE. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol. 2002;12:1138–44. doi: 10.1016/S0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- 6.Chan SWL, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–60. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- 7.Pandey R, Müller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, et al. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002;30:5036–55. doi: 10.1093/nar/gkf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alinsug MV, Yu C-W, Wu K. Phylogenetic analysis, subcellular localization, and expression patterns of RPD3/HDA1 family histone deacetylases in plants. BMC Plant Biol. 2009;9:37. doi: 10.1186/1471-2229-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen ZJ, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 1997;11:2124–36. doi: 10.1101/gad.11.16.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pikaart MJ, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12:2852–62. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selker EU. Trichostatin A causes selective loss of DNA methylation in Neurospora. Proc Natl Acad Sci U S A. 1998;95:9430–5. doi: 10.1073/pnas.95.16.9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, Neves N, et al. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell. 2004;13:599–609. doi: 10.1016/S1097-2765(04)00064-4. [DOI] [PubMed] [Google Scholar]
- 13.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–9. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 14.Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 15.Murfett J, Wang XJ, Hagen G, Guilfoyle TJ. Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell. 2001;13:1047–61. doi: 10.1105/tpc.13.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aufsatz W, Mette MF, van der Winden J, Matzke M, Matzke AJ. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 2002;21:6832–41. doi: 10.1093/emboj/cdf663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aufsatz W, Stoiber T, Rakic B, Naumann K. Arabidopsis histone deacetylase 6: a green link to RNA silencing. Oncogene. 2007;26:5477–88. doi: 10.1038/sj.onc.1210615. [DOI] [PubMed] [Google Scholar]
- 18.To TK, Kim JM, Matsui A, Kurihara Y, Morosawa T, Ishida J, et al. Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet. 2011;7:e1002055. doi: 10.1371/journal.pgen.1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, Yu CW, Duan J, Luo M, Wang K, Tian G, et al. HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol. 2012;158:119–29. doi: 10.1104/pp.111.184275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–64. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 21.Lee MG, Wynder C, Bochar DA, Hakimi MA, Cooch N, Shiekhattar R. Functional interplay between histone demethylase and deacetylase enzymes. Mol Cell Biol. 2006;26:6395–402. doi: 10.1128/MCB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He Y, Michaels SD, Amasino RM. Regulation of flowering time by histone acetylation in Arabidopsis. Science. 2003;302:1751–4. doi: 10.1126/science.1091109. [DOI] [PubMed] [Google Scholar]
- 23.Jiang D, Yang W, He Y, Amasino RM. Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell. 2007;19:2975–87. doi: 10.1105/tpc.107.052373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu CW, Liu X, Luo M, Chen C, Lin X, Tian G, et al. HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol. 2011;156:173–84. doi: 10.1104/pp.111.174417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krichevsky A, Zaltsman A, Lacroix B, Citovsky V. Involvement of KDM1C histone demethylase-OTLD1 otubain-like histone deubiquitinase complexes in plant gene repression. Proc Natl Acad Sci U S A. 2011;108:11157–62. doi: 10.1073/pnas.1014030108. [DOI] [PMC free article] [PubMed] [Google Scholar]