Abstract
In most non-photosynthetic eukaryotes it has been demonstrated a conserved signal transduction pathway, namely TOR-S6K, that coordinates growth and cell proliferation. This pathway targets the translational apparatus to induce selective translation of ribosomal mRNAs as well as stimulate the cell cycle transition through the G1/S phase. Thus, by activation of this pathway through environmental signals, nutrients, stress, or specific growth factors, such as insulin or insulin-like growth factors (IGF), this pathway allows organisms to regulate growth and cell division. In plants, evidence has shown that TOR protein has been highly conserved through evolution, being involved in growth and cell proliferation control as well. Particularly in maize, a peptide named ZmIGF has been found in actively growing tissues. It targets the maize TOR pathway at the same extent as insulin and, by doing so it induces growth, as well as ribosomal proteins and DNA synthesis. Thus, higher metazoans and plants seem to conserve similar biochemical paths to regulate cell growth through equivalent targets that conduce to activation of the TOR-S6K pathway. Recent research shows evidence that supports this proposal by uncovering the ZmIGF receptor in maize, providing further means for analyzing the role of the conserved TOR signaling pathway in this plant.
Keywords: TOR pathway, ZmIGF maize growth factor, cell growth and proliferation, ribosome biogenesis, translational control
Signal Transduction by TOR Pathway
A mechanism that modulates growth and proliferation through signal transduction has been reported for both, animals1,2 and plants.3,4 It is referred as the TOR pathway and it is well characterized in no photosynthetic eucaryotes. Several inputs such as nutrients, stress, cell energy level and growth factors stimulates or represses this signaling network,5-7 where the serine/threonine protein kinase TOR plays a central role. Insulin and insulin-like growth factors (IGFs) are effectors of this growth regulatory pathway,8 which selectively activates the translational machinery regulating protein synthesis, ribosome biogenesis and ultimately cell growth.9-11 In plants, an increasing number of reports describing several components of the TOR pathway, such as TOR, RAPTOR and S6K1 and S6K2 kinases5,12,13 and other proteins involved in regulate TOR activity, such as FKBP1214 or the translationally controlled tumor protein (TCTP)15 are found across the literature (Fig. 1). Particularly in maize, orthologs of TOR and S6K kinases have been described and characterized.13,16 However, there are few reports regarding the plant growth factor that activates this pathway. Our research group has, recently found and isolated a small peptide from actively growing maize tissues, namely the maize insulin-like growth factor (ZmIGF),17 that has proved to perform this role. Indeed, ZmIGF as well as insulin, have shown to stimulate growth through the TOR signal transduction pathway13 as well as to increment DNA and protein synthesis in maize seedlings and callus.4,18 Further, this factor stimulates cell mitotic transition from G1 to S phase on geminating seeds (unpublished data). Moreover ZmIGF as well as insulin induce phosphorylation of the TOR and S6K kinases, main components of this pathway (Fig. 1), and accelerate the synthesis of specific proteins, mainly ribosomal proteins.
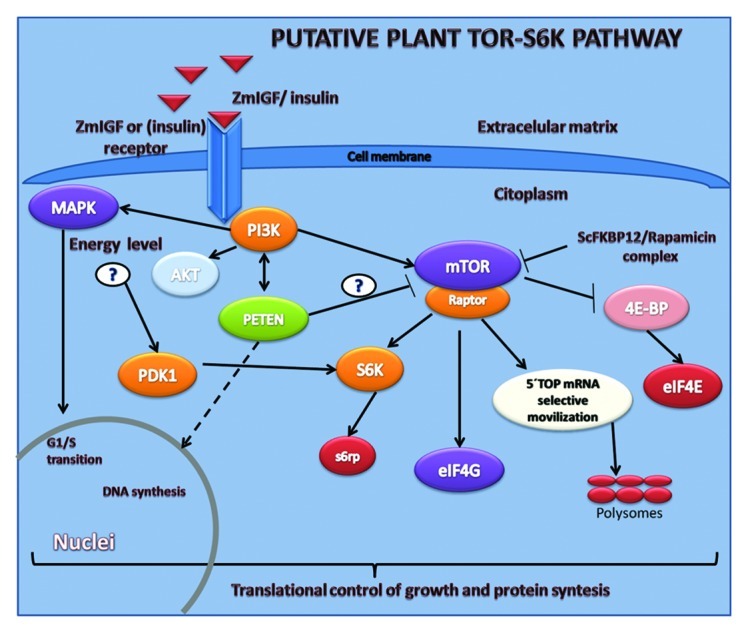
Figure 1. Putative plant PI3K/TOR-S6K pathway. Pathway activation by plant IGFs or insulin turns on a signaling cascade of reactions through the phosphorylation of kinases, mainly TOR and S6K, ultimately targeting ribosome synthesis. Arrowheads denote activation and truncated lines inhibition, interrogation marks denote no confirmed function (Modified from Sotelo et al.).18
These data are consistent with the large selective movilization of 5′ top mRNA into polysomes achieved for selective translation in maize embryos activated by either effector during germination.19 Lately, purification of this factor to homogeneity showed that, ZmIGF is a 5.7 kDa peptide, with similar functional structure as insulin, demonstrated by its specific recognition by insulin antibodies20 and by circular dichroism data.17 Recently, unpublished data from our research group have demonstrated that the transcriptome patterns of stimulated maize calli, either by ZmIGF or insulin, showed discrete changes as compared with their control non-stimulated tissue. However, these changes were identical regardless the effectors tested. These data suggested the presence of a unique receptor at the cell membrane level that recognizes, both, ZmIGF as well as insulin, for activation of this signaling pathway. Competition experiments performed between ZmIGF and insulin at the maize protoplast membranes, showed that the ZmIGF receptor is the initial target of the pathway, since both, ZmIGF or insulin, compete for the membrane receptor site, within the maize calli cells before internalizing the signal (in preparation).
In summary, the maize growth factor ZmIGF or insulin, targets TOR pathway in plants, as well as in animals, suggesting high conservation for the effector and the receptor structures. Taken together all these data indicate that an ancient peptide such as insulin, or its analog ZmIGF, regulates growth and cell division by targeting animal as well as plant membrane receptors, suggesting high conservation of the TOR pathway in photosynthetic as well as non-photosynthetic eukaryotes from yeast through mammals and plants.
Acknowledgments
Work supported by CONACYT grant 101327.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/19993
References
- 1.Fingar DC, Richardson CJ, Tee AR, Cheatham L, Tsou C, Blenis J. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol Cell Biol. 2004;24:200–16. doi: 10.1128/MCB.24.1.200-216.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shahbazian D, Roux PP, Mieulet V, Cohen MS, Raught B, Taunton J, et al. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J. 2006;25:2781–91. doi: 10.1038/sj.emboj.7601166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robaglia C, Menand B, Lei Y, Sormani R, Nicolaï M, Gery C, et al. Plant growth: the translational connection. Biochem Soc Trans. 2004;32:581–4. doi: 10.1042/BST0320581. [DOI] [PubMed] [Google Scholar]
- 4.Dinkova TD, Reyes de la Cruz H, García-Flores C, Aguilar R, Jiménez-García LF, Sánchez de Jiménez E. Dissecting the TOR-S6K signal transduction pathway in maize seedlings: relevance on cell growth regulation. Physiol Plant. 2007;130:1–10. doi: 10.1111/j.1399-3054.2007.00862.x. [DOI] [Google Scholar]
- 5.Mahfouz MM, Kim S, Delauney AJ, Verma DP. Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell. 2006;18:477–90. doi: 10.1105/tpc.105.035931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao X-H, Majithia A, Huang X, Kimmel AR. Growth control via TOR kinase signaling, an intracellular sensor of amino acid and energy availability, with crosstalk potential to proline metabolism. Amino Acids. 2008;35:761–70. doi: 10.1007/s00726-008-0100-3. [DOI] [PubMed] [Google Scholar]
- 7.Petersen J. TOR signalling regulates mitotic commitment through stress-activated MAPK and Polo kinase in response to nutrient stress. Biochem Soc Trans. 2009;37:273–7. doi: 10.1042/BST0370273. [DOI] [PubMed] [Google Scholar]
- 8.Giannakou ME, Partridge L. Role of insulin-like signalling in Drosophila lifespan. Trends Biochem Sci. 2007;32:180–8. doi: 10.1016/j.tibs.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Vanhaesebroeck B, Alessi DR. The PI3K-PDK1 connection: more than just a road to PKB. Biochem J. 2000;346:561–76. doi: 10.1042/0264-6021:3460561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, et al. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005;19:2199–211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolaï M, et al. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 2007;8:864–70. doi: 10.1038/sj.embor.7401043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, et al. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci U S A. 2002;99:6422–7. doi: 10.1073/pnas.092141899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reyes de la Cruz H, Aguilar R, Sanchez de Jimenez E. Functional characterization of maize ribosomal S6 protein kinase (ZmS6K), a plants ortholog of methazoan p70 (S6K) Biochem. 2004;43:533–9. doi: 10.1021/bi035222z. [DOI] [PubMed] [Google Scholar]
- 14.Sormani R, Yao L, Menand B, Ennar N, Lecampion C, Meyer C, et al. Saccharomyces cerevisiae FKBP12 binds Arabidopsis thaliana TOR and its expression in plants leads to rapamycin susceptibility. BMC Plant Biol. 2007;7:26–33. doi: 10.1186/1471-2229-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berkowitz O, Jost R, Pollmann S, Masle J. Characterization of TCTP, the translationally controlled tumor protein, from Arabidopsis thaliana. Plant Cell. 2008;20:3430–47. doi: 10.1105/tpc.108.061010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agredano-Moreno LT, Reyes de la Cruz H, Martínez-Castilla LP, Sánchez de Jiménez E. Distinctive expression and functional regulation of the maize (Zea mays L.) TOR kinase ortholog. Mol Biosyst. 2007;3:794–802. doi: 10.1039/b705803a. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez CD, Rodriguez A, Aguilar CR, Sanchez de Jiménez E. Biochemical characterization of a maize novel peptide hormone. Protein Pept Lett. 2011;18:84–91. doi: 10.2174/092986611794328636. [DOI] [PubMed] [Google Scholar]
- 18.Sotelo R, Garrocho-Villegas V, Aguilar CR, Calderon ME, Sanchez de Jimenez E. Coordination of cell growth and cell division in maize (Zea mays L.) relevance of the conserved TOR signal transduction pathway. In Vitro Cell Dev Biol Plant. 2010;46:578–86. doi: 10.1007/s11627-010-9293-8. [DOI] [Google Scholar]
- 19.Jiménez-López S, Mancera-Martínez E, Donayre-Torres A, Rangel C, Uribe L, March S, et al. Expression profile of maize (Zea mays L.) embryonic axes during germination: translational regulation of ribosomal protein mRNAs. Plant Cell Physiol. 2011;52:1719–33. doi: 10.1093/pcp/pcr114. [DOI] [PubMed] [Google Scholar]
- 20.García Flores C, Aguilar R, Reyes de la Cruz H, Albores M, Sánchez de Jiménez E. A maize insulin-like growth factor signals to a transduction pathway that regulates protein synthesis in maize. Biochem J. 2001;358:95–100. doi: 10.1042/0264-6021:3580095. [DOI] [PMC free article] [PubMed] [Google Scholar]


