Abstract
Comment on: Formentini L, et al. Mol Cell 2012; 45:731-42.
Keywords: ATPase inhibitory factor 1, H+-ATP synthase, cancer, glycolysis, mitochondria, oxidative phosphorylation, reactive oxygen species, retrograde signaling
Mitochondria of normal eukaryotic cells synthesize most ATP requirements needed to support cellular activity. They also participate in Ca2+ and reactive oxygen species (ROS) signaling and in the execution of cell death. The structure and molecular composition of mitochondria vary largely among the different cellular types of mammals.1 The final mitochondrial phenotype results from gene expression programs that are regulated at both the transcriptional2 and post-transcriptional3 levels.
A key component of mitochondria in energy conservation, ROS signaling and the execution of cell death is the H+-ATP synthase, a reversible engine of oxidative phosphorylation that catalyzes the synthesis of ATP using as driving force the proton gradient generated by the respiratory chain4 (Fig. 1A). Its catalytic subunit (β-F1-ATPase, dark blue in Fig. 1) forms part of the soluble F1-ATPase domain (Fig. 1A). β-F1-ATPase is significantly diminished in cancer and provides a bioenergetic signature of disease progression and of the response to chemotherapy.5 In carcinomas of the lung, colon and breast, the downregulation of β-F1-ATPase is also accompanied by an increased expression of the ATPase inhibitory factor 1 (IF1),6 a physiological inhibitor of the H+-ATP synthase.7
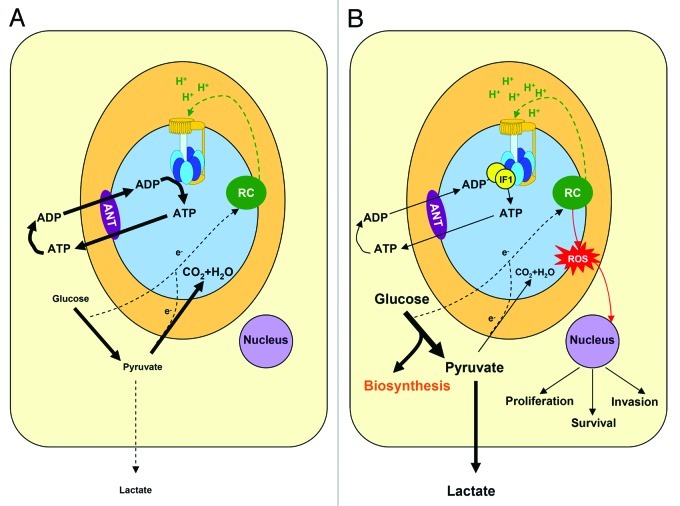
Figure 1. IF1 regulates energy metabolism in cancer cells. A, In normal aerobic cells the oxidation of glucose to CO2 and H2O is the source of electrons (e-) that feeds the respiratory chain (RC) for the generation of the proton gradient (H+, green). In oxidative phosphorylation, the H+-ATP synthase (F0, yellow rotor and F1, blue membrane protruding head) uses the proton gradient for the synthesis of ATP. The H+-ATP synthase supplies most of the ATP needed to sustain cellular activity. In the presence of oxygen the production of lactate (aerobic glycolysis) is low. B, The overexpression of IF1 in cancer cells inhibits the H+-ATP synthase and limits the flux of ATP being synthesized in oxidative phosphorylation. Consequently, aerobic glycolysis is stimulated and glucose derived carbon skeletons are diverted for biosynthetic purposes. In cancer cells, the oxidation of pyruvate in mitochondria is restrained. Inhibition of the H+-ATP synthase promotes an increase in the mitochondrial membrane potential (H+, green) and the subsequent production of superoxide radical (ROS, red). Mitochondrial ROS signal to the nucleus of the cell features of the cancer phenotype such as the promotion of proliferation, invasion and survival. The oxidation of glutamine in cancer cells is not taken into consideration for simplicity of the schematic. ANT, adenine nucleotide translocase.
IF1 is a highly conserved protein encoded in the nuclear ATPIF1 gene. Alternative splicing generates three different isoforms of IF1. The expression of IF1 in different normal human tissues has been shown to vary largely,6 from very high levels in the heart, to intermediate expression in the liver and negligible levels in breast, colon and lung. In contrast, mitochondria of prevalent human carcinomas have an overwhelming content of IF1.6
The physiological function of IF1 in normal hypoxic cells is to inhibit the hydrolase activity of the H+-ATP synthase,7 i.e., its reverse functioning when mitochondrial matrix pH drops below neutrality. In this situation, residues 48 to 56 stabilize the formation of a coiled-coil region between two IF1 molecules to generate the active IF1 inhibitory dimmer. The structure of the inhibited F1-ATPase complex with bound IF1 in the presence of ATP has been solved.8 Residues 1–13 in IF1 stabilize the binding of the inhibitor to the αβ-interface in the F1-ATPase domain, blocking rotary catalysis of the H+-ATP synthase.8
More recently, we have described that overexpression of IF1 in cells with negligible content of the protein results in the inhibition of the ATP synthetic activity of the H+-ATP synthase and the switch to an increased aerobic glycolysis6,9 (Fig. 1B). On the contrary, silencing of IF1 enhances the H+-ATP synthase activity and reduces aerobic glycolysis.6,9 These findings strongly support that IF1 is a master regulator of energy metabolism playing a crucial role in mediating the metabolic switch experienced by cancer cells in order to favor the diversion of carbon skeletons for biosynthetic processes (Fig. 1B).
How is IF1 upregulated in cancer? The answer to this question is presently unknown. One can speculate that hypoxia, activating mutations in cancer genes or other epigenetic events of the tumor microenvironment might directly control the expression of IF1. In any case, it is likely that different cell type-specific programs of gene regulation will mediate IF1 accretion in different carcinomas.
An additional question that deserves further investigation is why the high overexpression of IF1 in some normal human tissues is not blocking by mass-action ratio the synthase activity of the H+-ATP synthase. One can speculate that a mechanism of post-translational modification exists in addition to the well-known pH-regulated binding of IF1 to F1-ATPase.7 Such a mechanism should override the inhibition promoted by the very high content of IF1 in these tissues. In this situation, it is likely that the carcinomas that overexpress IF1 either lack this sort of regulation and/or the mechanism results inactivated by oncogenesis.
Moreover, and in addition to the role of IF1 in rewiring energy metabolism (Fig. 1B), the overexpression of IF1 in human cancer cells also triggers a retrograde signal to the nucleus to establish the appropriate adaptive cellular program needed for tumor development9 (Fig. 1B). Indeed, IF1-mediated inhibition of the H+-ATP synthase results in mitochondrial hyperpolarization (Fig. 1B) and the subsequent production of superoxide radical9 (ROS in Fig. 1B). Remarkably, it has been demonstrated that the ROS-mediated response in colon cancer cells signals to the nucleus an NFκB-dependent adaptation that includes enhanced proliferation, invasion and cell survival.9 Conversely, permanent IF1-knockdown in HeLa cells triggers an increase in cell death response when exposed to ischemia or high ROS.10 Overall, IF1 is the mitochondrial factor that contributes to the acquisition of other hallmarks of the cancer phenotype. Due to the heterogeneity of mitochondria in mammals, we anticipate that the development of conditional and tissue-specific IF1 transgenic mouse models will contribute to unveil the specific mechanism of action of IF1 in different cellular types. Experiments in this regard are already in their way.
Finally, we stress that new insights into the mechanisms that regulate the expression and/or activity of IF1 will open-up a promising venue for the development of a targeted treatment of cancer metabolism, short-cutting in this way the winding trail that we are experiencing to successfully translate basic cancer knowledge into the clinics.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21387
References
- 1.Mootha VK, et al. Cell. 2003;115:629–40. doi: 10.1016/S0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- 2.Scarpulla RC. Physiol Rev. 2008;88:611–38. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 3.Willers IM, et al. Biochim Biophys Acta 2011; 1807:543-51;. [DOI] [PubMed]
- 4.Boyer PD. Annu Rev Biochem. 1997;66:717–49. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 5.Cuezva JM, et al. Biochim Biophys Acta 2009; 1792:1145-58;. [DOI] [PubMed]
- 6.Sánchez-Cenizo L, et al. J Biol Chem. 2010;285:25308–13. doi: 10.1074/jbc.M110.146480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gledhill JR, et al. Proc Natl Acad Sci USA. 2007;104:15671–6. doi: 10.1073/pnas.0707326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabezón E, et al. Nat Struct Biol. 2003;10:744–50. doi: 10.1038/nsb966. [DOI] [PubMed] [Google Scholar]
- 9.Formentini L, et al. Mol Cell. 2012;45:731–42. doi: 10.1016/j.molcel.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 10.Fujikawa M, et al. J Biol Chem. 2012;287:18781–7. doi: 10.1074/jbc.M112.345793. [DOI] [PMC free article] [PubMed] [Google Scholar]