Abstract
Comment on: Cho EC, et al. EMBO J 2012; 31:1785-97.
Keywords: E2F-1, PRMT5, growth, methylation
The transcription factor E2F-1 is an important regulator of cell fate, which is able to promote cell cycle progression and DNA repair, but also drive p53-dependent and -independent apoptosis.1 Accordingly, E2F-1 has been reported to have both tumor-suppressive and oncogenic properties.2 For example, E2F-1−/− mice suffer from an increased susceptibility to tumor formation, which is consistent with a role for E2F-1 as a tumor suppressor.3 In contrast, loss of E2F-1 reduces tumorigenesis and extends the lifespan of RB1+/− mice, arguing that E2F-1 also contributes to tumor progression.4 These facts clearly show the importance of a tight control of E2F-1 in order to avoid tumorigenesis. However, the mechanisms that control the opposing outcomes of E2F-1 activity remain largely unknown. Our recent studies have shown that arginine methylation of E2F-1 regulates its biological activity and raises the possibility that this event might influence tumorigenesis.5
Over the last decade, arginine methylation has been shown to play an important role in the functional regulation of a diverse number of proteins. It is implicated in many processes, such as chromatin regulation, transcriptional control, DNA repair and RNA processing.6 Interestingly, recent research highlights an important role for the arginine methyltransferase PRMT5 in growth-promoting and pro-survival pathways.7 For example, hypermethylation of histones H3R8 and H4R3 by PRMT5 triggers the transcriptional silencing of the tumor suppressor gene nonmetastatic 23.8 In addition, phosphorylation of MEP50 by the cyclinD1/CDK4 complex stimulates MEP50/PRMT5 activity, which triggers H4R3 hypermethylation, DNA rereplication and transformation.9 Further, methylation of the tumor suppressor PDCD4 by PRMT5 is important for enhanced tumor growth in a xenograft tumor model.10 Previously, we identified the tumor suppressor p53 as a PRMT5 target.11 DNA damage-induced p53 arginine methylation impacts on the biochemical properties of p53 and the cellular outcome of the p53 response, as it favors cell cycle arrest over apoptosis.11
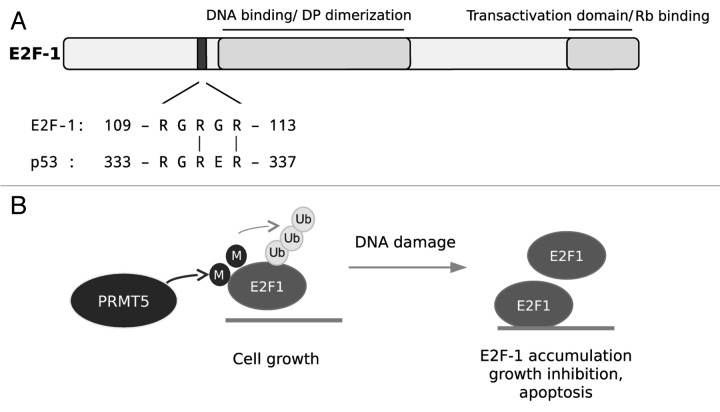
Interestingly, we identified a short sequence motif in E2F-1, namely RGRGR, which is similar to the RGRER motif in the p53 sequence targeted by PRMT5 arginine methylation (Fig. 1). Due to the similarity of the sequence motifs and the roles of PRMT5 and E2F-1 in growth control, we considered that E2F-1 might be a substrate for PRMT5. Indeed, we found that PRMT5 symmetrically dimethylates E2F-1 in vitro and in tumor cells. Depletion of PRMT5 leads to enhanced E2F-1 protein levels, a reduction in E2F-1 polyubiquitination and increased expression of several E2F-1 target genes, which correlated with increased DNA binding activity of E2F-1.
Figure 1. Model of E2F-1 regulation by PRMT5: (A) Sequence motifs targeted by PRMT5. (B) PRMT5-dependent E2F-1 methylation promotes E2F-1 polyubiquitination and negatively regulates the DNA binding activity of E2F-1. PRMT5-mediated E2F-1 methylation is important for PRMT5-dependent cell growth and cell survival. Upon DNA damage, E2F-1 methylation disappears, providing a mechanism that might contribute to E2F-1 stabilization, growth inhibition and apoptosis induction.
The molecular mechanisms by which PRMT5 influences E2F-1 protein stability are currently not known. The methylation mark might act as a docking site for proteins that alter the biochemical properties of E2F-1 or for proteins that displace binding partners, which normally act to protect E2F-1 from proteasomal degradation. Further, there might be crosstalk with other posttranslational modifications, which finally favor E2F-1 polyubiquitination. Interestingly, the levels of E2F-1 arginine methylation are reduced upon DNA damage, a condition under which E2F-1 is known to be stabilized and promote DNA repair or apoptosis.1 This suggests, that a loss of E2F-1 arginine methylation might be crucial for DNA damage-induced E2F-1 stabilization and function.
As PRMT5 influences E2F-1’s biochemical properties, it was important to assess the role of PRMT5 in E2F-1-dependent growth control. Depletion of PRMT5 led to decreased cell growth and enhanced apoptosis. Remarkably, co-depletion of E2F-1 rescued these effects, arguing that PRMT5 exerts its growth-promoting and cell-survival effects at least in part through negative regulation of E2F-1. In support of this finding, the expression of the pro-apoptotic E2F-1 target p73 was found to be crucial for apoptosis upon PRMT5 depletion. The exact mechanism by which PRMT5 influences cell growth and survival needs further investigation. For example, we do not know whether PRMT5 activity changes the promoter binding specificity of E2F-1, or if PRMT5 modulates the E2F-1-dependent transcriptome.
Together, our recent results argue that the negative regulation imposed on E2F-1 by PRMT5 is important for PRMT5′s growth-promoting activity. Interestingly, analysis of E2F-1 and PRMT5 expression in tumor biopsies suggests that in a subgroup of colorectal cancer, high levels of PRMT5 coincided with low levels of E2F-1, and this profile occurs in a group of tumors with poor clinical outcome. This raises the possibility that arginine methylation contributes to tumorigenesis by influencing the E2F pathway.
Our recent study supports the idea that PRMT5 might be a valuable target for cancer therapies,7 and that its expression in tumor biopsies in combination with the expression of other proteins, such as E2F-1, might serve as a prognostic marker. In addition, the fact that PRMT5 regulates both E2F-1 and p53 in a DNA damage-dependent manner, and that it favors cell survival in both cases, hints toward a new level of crosstalk between the E2F-1 and p53 pathway. However, further research is necessary for a comprehensive understanding of the PRMT5 methylation-dependent crosstalk between signaling pathways in both normal and cancer cells.
Acknowledgments
Supported by Cancer Research UK, the Medical Research Council, Leukaemia Research Fund and EU.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21467
References
- 1.Polager S, et al. Trends Cell Biol. 2008;18:528–35. doi: 10.1016/j.tcb.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Engelmann D, et al. Cancer Res. 2012;72:571–5. doi: 10.1158/0008-5472.CAN-11-2575. [DOI] [PubMed] [Google Scholar]
- 3.Yamasaki L, et al. Cell. 1996;85:537–48. doi: 10.1016/S0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 4.Yamasaki L, et al. Nat Genet. 1998;18:360–4. doi: 10.1038/ng0498-360. [DOI] [PubMed] [Google Scholar]
- 5.Cho EC, et al. EMBO J. 2012;31:1785–97. doi: 10.1038/emboj.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bedford MT, et al. Mol Cell. 2005;18:263–72. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Karkhanis V, et al. Trends Biochem Sci. 2011;36:633–41. doi: 10.1016/j.tibs.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pal S, et al. Mol Cell Biol. 2004;24:9630–45. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aggarwal P, et al. Cancer Cell. 2010;18:329–40. doi: 10.1016/j.ccr.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Powers MA, et al. Cancer Res. 2011;71:5579–87. doi: 10.1158/0008-5472.CAN-11-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jansson M, et al. Nat Cell Biol. 2008;10:1431–9. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]