Abstract
Resistance to anti-neoplastic agents is the major cause of therapy failure, leading to disease recurrence and metastasis. E2F1 is a strong inducer of apoptosis in response to DNA damage through its capacity to activate p53/p73 death pathways. Recent evidence, however, showed that E2F1, which is aberrantly expressed in advanced malignant melanomas together with antagonistic p73 family members, drives cancer progression. Investigating mechanisms responsible for dysregulated E2F1 losing its apoptotic function, we searched for genomic signatures in primary and late clinical tumor stages to allow the prediction of downstream effectors associated with apoptosis resistance and survival of aggressive melanoma cells. We identified miR-205 as specific target of p73 and found that upon genotoxic stress, its expression is sufficiently abrogated by endogenous DNp73. Significantly, metastatic cells can be rescued from drug resistance by selective knockdown of DNp73 or overexpression of miR-205 in p73-depleted cells, leading to increased apoptosis and the reduction of tumor growth in vivo. Our data delineate an autoregulatory circuit, involving high levels of E2F1 and DNp73 to downregulate miR-205, which, in turn, controls E2F1 accumulation. Finally, drug resistance associated to this genetic signature is mediated by removing the inhibitory effect of miR-205 on the expression of Bcl-2 and the ATP-binding cassette transporters A2 (ABCA2) and A5 (ABCA5) related to multi-drug resistance and malignant progression. These results define the E2F1-p73/DNp73-miR-205 axis as a crucial mechanism for chemoresistance and, thus, as a target for metastasis prevention.
Keywords: ABC transporters, DNp73, E2F1, cancer, chemoresistance, melanoma mouse model, miR-205, p73
Introduction
Malignant melanoma is the most aggressive form of skin cancer highly resistant to conventional chemotherapy.1 Various genes are differentially regulated between benign nevi and melanomas, but abundant expression of the E2F1 transcription factor has been primarily observed in advanced lesions and tumor metastases correlating with cancer aggressiveness, therapy failure and poor patient survival prognosis.2,3 E2F1 is a unique member of the E2F family of proteins as it is involved in cell cycle progression and apoptosis induction in response to DNA damage. Recently, we have shown that deregulated E2F1 acts as driving force in melanoma progression and promotes tumor invasion and metastasis independently from its other cellular activities.4 This aggressive behavior of the transcription factor in malignant cells is partially mediated through the induction of the epidermal growth factor receptor (EGFR) pathway.4 However, it is well-established that E2F1 stimulates the expression of the tumor suppressor p73 and its N-terminally truncated isoforms (named DNp73) via direct transactivation of the TP73 gene.5,6 While full-length p73 inhibits cancer development by inducing cell cycle arrest and apoptosis through its ability to bind p53 DNA target sites, DNp73 acts as antagonist of wild-type p53 family members by either directly interfering with DNA binding or forming inactive heteromeric complexes with transcriptionally active p73.7 Strikingly, in melanoma metastases, DNp73 isoforms are strongly upregulated in conjunction with full-length p73 compared with primary tumors,3 whereas downregulation or mutation of wild-type p73 is not noted in cutaneous melanoma. We have previously shown that DNp73 exhibits anti-apoptotic activity in human melanoma cells and demonstrated that specific suppression of individual isoforms enhances the sensitivity toward cytotoxic drugs.8 As overexpression of E2F1 significantly correlates with DNp73 levels in advanced stages of cancer and E2F1 promotes malignant progression instead of inducing apoptosis, we considered the E2F1-DNp73 axis as a potentially widespread mechanism of drug resistance in human skin cancer, albeit specific targets are unheard-of.
Emerging evidence suggests that endogenous noncoding microRNAs (miRs) play an important role in tumorigenesis and progression.9 MiRs negatively regulate gene expression at the posttranscriptional level through an RNA interference pathway by binding to 3′-untranslated regions (UTR) of mRNAs of target genes or by directly binding to the mRNA.10 Based on the targets and their functions, miRs are considered as oncogenes or tumor suppressors, and, as such, are relevant for cancer development. Thus, recent data suggest that miR signatures have a potential as clinically relevant biomarkers of prognosis in metastatic melanoma with the ability to identify high-risk patients most likely to benefit from adjuvant therapy and/or elevated surveillance.11 Many factors are responsible for abnormal miR expression in cancer, including promoter methylation, mutations, chromosomal aberrations, defects in miRNA biogenesis and changes in the expression of transcription factors.12 Studies indicate that p53 is involved in transcriptional regulation of tumor suppressing miRs and is also able to influence the maturation process during miRNA biogenesis.13 As opposed to p53, only little is known about p73 regulation of miRs. Whereas p53’s tumor suppressor activity is often abolished in melanomas after acquiring apoptotic defects during cancer progression,14 p73 is, as described above, dysregulated in advanced metastatic tumors.
Given the importance of p73 in E2F1-induced apoptosis and suppression of survival, as well as the miRs’ share in cancer aggressiveness, we searched for p73-regulated miRs. We identified miR-205 as a specific target of p73/DNp73 and investigated its regulation in metastatic skin cancer. We provide evidence that miR-205 mediates the DNA damage response of p73, whereas its repression by rather low DNp73 levels is crucial for E2F1 to induce chemoresistance. Moreover, the established miR-205/E2F1 feedback circuit provides a plausible explanation for the aberrant expression of E2F1 in highly chemotherapy-resistant metastatic melanomas.
Results
MiR-205 is regulated by p73 but not p53
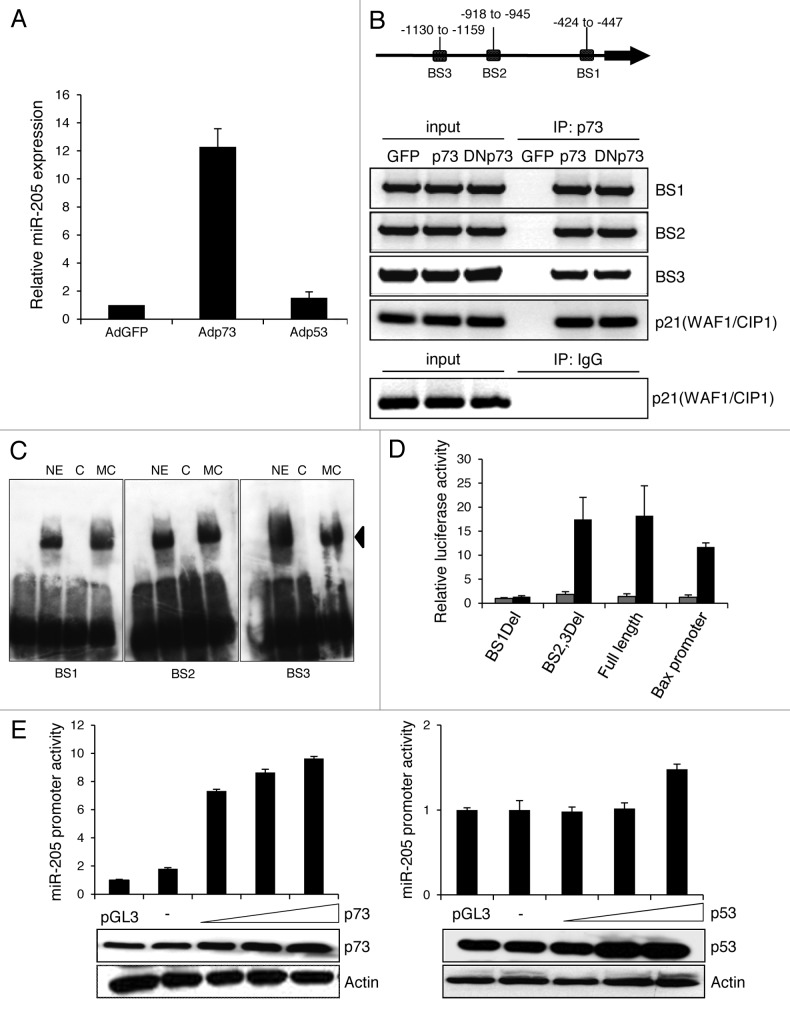
To identify microRNAs that are regulated by p73 in malignant melanoma cells, TaqMan microarrays were performed in non-metastatic SK-Mel-29 melanoma cells infected with adenoviral vector expressing p73, p53 or GFP as control. After p73 but not p53 or GFP overexpression, miR-205 was upregulated 10-fold (data not shown). The validity of these results was further confirmed by measuring miR-205 expression by TaqMan qRT-PCR (Fig. 1A). Encouraged after observing the strong induction of miR-205 expression through p73, we next focused on understanding the mechanism involved in p73-dependent regulation of miR-205. p73 might activate miR transcription via its N-terminal transactivation domain known to stimulate a wide variety of genes. We conducted an in silico screen for p73-responsive elements on the miR-205 promoter selecting 5 kb upstream of the gene. Assuming identical binding sites for p73 and p53, we applied the p53MH algorithm previously used to identify novel p53 responsive genes to search for corresponding elements in the miR-205 promoter. We found three conserved putative p73 motifs and named those BS1, BS2 and BS3 starting proximal from the miR-205 transcription start site (Fig. 1B, upper panel) and verified these using chromatin immunoprecipitation assay on SK-Mel-29 (Fig. 1B, lower panel). DNA-protein complexes were immunoprecipitated with antibodies against p73 or IgG. PCR analysis demonstrated that p73 binds to all three binding sites as good as DNp73, which lacks the transactivation domain, in a manner comparable with its binding to the p21(WAF1/CIP1) consensus sequence used as positive control. p73 binding was also confirmed in gel mobility shift assays using oligonucleotides corresponding to BS1-BS3. One predominant nuclear complex for each site was completely abolished by 50-fold molar excess of unlabled oligos but not by the probe in which the p73 site was mutated, indicating that every binding site of the miR-205 promoter directly interacts with p73 (Fig. 1C). No binding was seen in the absence of nuclear extract. To identify the promoter region mediating p73 responsiveness, a panel of luciferase reporter constructs containing the full-length and deleted fragments were tested after cotransfecting SK-Mel-29 cells with p73. As shown in Figure 1D, the main loss of promoter activity was observed with the BS1Del mutant, suggesting that the p73 binding site at position -427 to -445 is important for transactivation of miR-205. Promoter upregulation in response to ectopic p73 occurred in a dose-dependent manner reaching a 10-fold increase of luciferase activity compared with endogenous levels (Fig. 1E, left), while equal amounts of p53 had no significant effect (Fig. 1E, right).
Figure 1. p73 induces miR-205 expression. (A) Expression of miR-205 in SK-Mel-29 cells infected with Adp53, Adp73 or AdGFP (set as 1) was determined by TaqMan qRT-PCR. (B) Scheme of the miR-205 promoter (top). Three putative p73 binding sites (BS) are indicated as BS1, BS2 and BS3 relative to the pre-miR sequence. In vivo binding of p73 to miR-205 promoter regions was detected by ChIP using anti-p73 (ER-15) or control IgG (bottom). PCR primers for the p21(WAF1/CIP1) promoter were used as positive control. Input lane, 10% of total chromatin. (C) Biotin-labeled miR-205 p73-binding sites were incubated with nuclear extract and a 50-fold molar excess of cold double-stranded self-competitor or an equimolar amount of mutant site competitor. The arrow indicates a specific band competed by cognate competitor. Lane 1, without nuclear extract; NE, nuclear extract; C, competitor; MC, mutant competitor. (D) Luciferase assay of SK-Mel-29 cotransfected with 1 µg miR-205 promoter, BS1Del and BS2,3Del deletion constructs and 0.5 µg of p73 expression plasmid (black bars) or pcDNA (gray bars). The p73-responsive human Bax promoter construct was used as positive control. Luciferase activities are expressed as the fold activation relative to cotransfection of the promoter construct with empty vector following normalization to total protein concentrations in the cell extract at 36 h after transfection. Error bars indicate the mean ± SD. E, Luciferase activities measured after cotransfection of the miR-205 promoter with increasing amounts (0.25, 0.50, 1 µg) of p73 (left) and p53 (right) expression plasmid or 1 µg pcDNA that served as control. Results are shown as fold increase compared with pGL3basic which was set as 1. Expression levels of p73, p53 and actin in different samples are indicated by western blot.
DNp73 interferes with miR-205 expression in melanoma cells
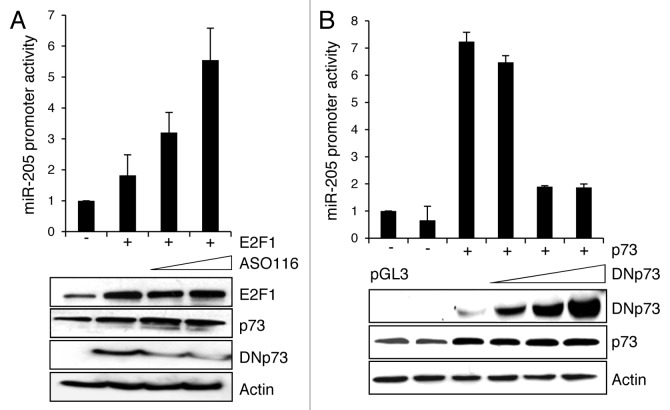
p73 is positively regulated by the transcription factor E2F1,15 and E2F1 upregulation has been noticed in metastatic melanoma samples.3 Since E2F1 can likewise induce antagonistic DNp73 isoforms, its effect on miR-205 promoter activation was studied in the presence or absence of the DNp73-specific inhibitor ASO116.8 As shown by reporter assays in SK-Mel-29 cells, E2F1 overexpression has basically no influence on the miR-205 promoter, unless endogenous DNp73 is specifically knocked down (Fig. 2A). The increase in luciferase activity induced by E2F1, which itself does not bind to the miR-205 promoter (data not shown), is directly correlated with DNp73 knockdown, with strongest activation at the highest dose of ASO116. We further clarified the role of DNp73 on miR-205 expression in promoter assays in combination with full-length p73. In a dose-dependent manner, p73-driven activity of the miR-205 promoter decreased in response to DNp73 (Fig. 2B). This indicates that inhibitory DNp73 is responsible for miR-205 depletion in melanoma cells.
Figure 2. Inhibition of miR-205 stimulation by DNp73. Luciferase assay of SK-Mel-29 cells co-transfected with p73-responsive miR-205 reporter construct and (A) 1 µg E2F1 expression plasmid in the presence of 0.1 and 0.5 µM of ASO116 or (B) 1 µg pcDNAp73 with 0.1, 0.5 and 1 µg of DNp73 construct. pcDNA served as control. Promoter activity of the control was normalized to 1. Standard deviation of the mean is indicated by error bars. Protein expression including DNp73 knockdown was validated by immunoblot using actin for equal loading.
MiR-205 and p73/DNp73 expression inversely correlates in melanocytes, melanoma cells and skin cancer tissues
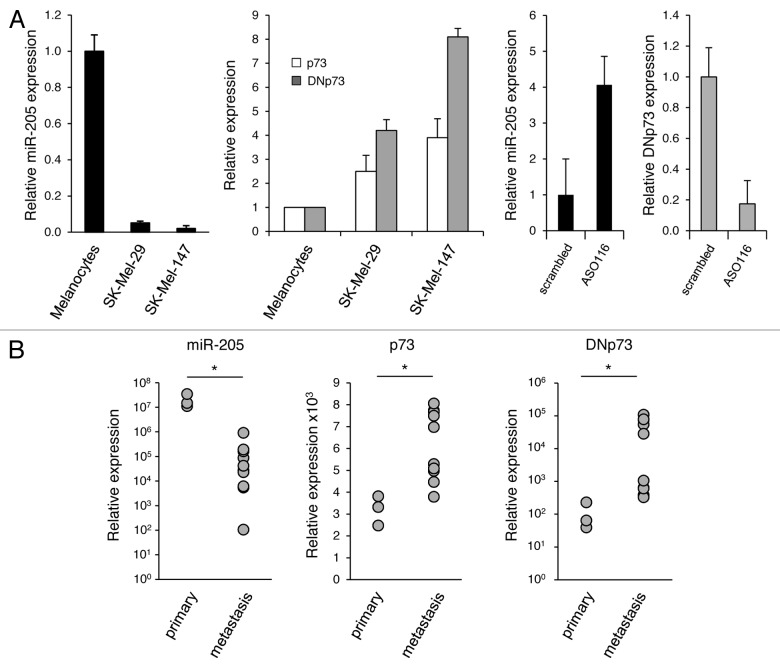
We previously reported that p73ΔExon2/3, which is coexpressed with p73, is the most abundant DNp73 species in melanoma metastases and patient-derived metastatic tumor cell lines.3 Based on the above findings, we analyzed the levels of p73, DNp73 and miR-205 expression in melanocytes vs. non- or highly metastatic melanoma cell lines,4 and primary skin tumors vs. metastases. While melanocytes showed the highest abundance of miR-205, its expression was severely downregulated in malignant cells with the strongest reduction in aggressive SK-Mel-147 (Fig. 3A, left). This inversely correlates with p73/DNp73 mRNA levels, which are highest in SK-Mel-147 cells (Fig. 3A, center). Consistently, specific silencing of endogenous DNp73 in these cells caused a 3-fold upregulation of miR-205 (Fig. 3A, right). In accordance with the cell culture data, we detected a significant loss of miR-205 expression in melanoma metastases from patients in comparison to primary melanoma, whereas p73/DNp73 transcripts are consistently higher in the metastases group (Fig. 3B). The inverse correlation between high DNp73 and low miR-205 expression in metastatic skin tumors corresponds with the finding that amino-truncated p73 isoforms lacking the transactivation domain can inhibit wild-type p73 and implies the direct competition of p73-mediated stimulation of miR-205 to contribute to pathogenesis of human aggressive melanoma.
Figure 3. Downregulation of miR-205 in metastatic melanoma tissues and cell lines correlates with high levels of p73/DNp73. qRT-PCR analysis of endogenous miR-205, p73 and DNp73 expression levels in (A) melanocytes, non-metastatic SK-Mel-29 or metastatic SK-Mel-147 w/o ASO116 treatment and (B) in a cohort of primary vs. metastatic melanoma tissues (*, miR-205, p < 0.001; p73, p = 0.001; DNp73, p = 0.026).
MiR-205 mediates the DNA damage response of p73
The sensitivity of cancer cells to genotoxic drugs is linked to p73 activation,16 whereas high DNp73 levels are associated with adverse clinico-pathological characteristics and failure of chemotherapy.17,18 We investigated the regulation of this miRNA by p73/DNp73 during cisplatin treatment of melanoma cells. In SK-Mel-147, expression of miR-205 significantly increased upon drug treatment (Fig. 4A), but not when expression of p73 is abrogated through shp73 prior to chemotherapy, indicating that upregulation of miR-205 upon DNA damage is p73-dependent. FACS analysis of cells treated with cisplatin and/or Adp73 revealed a considerable reduction of p73-mediated apoptosis after knockdown of miR-205 by antagomiR-205. Therefore, the p73 response to chemotherapy does in part rely on miR-205 induction (Fig. 4B). Melanoma cells showing diminished apoptosis after p73 knockdown retained their sensitivity to cisplatin when miR-205 was overexpressed (Fig. 4C, left). Equally, DNp73-mediated repression of drug-induced cell death could be resolved via enforced expression of miR-205 or specific depletion of inhibitory DNp73 by ASO116 (Fig. 4C, center), allowing the conclusion that in advanced melanoma cells, DNp73 causes loss of chemosensitivity by downregulating miR-205 expression.
Figure 4. MiR-205 mediates the DNA damage response of p73. (A) Relative expression of miR-205 in cisplatin treated SK-Mel-147 cells with and without p73 knockdown compared with undamaged cells that express shRNA-control (set as 1). (B and C) The percentage of apoptotic SK-Mel-147 after cisplatin treatment was measured by FACS analysis following overexpression of p73 and antagomiR-205 (B), shp73 and miR-205 (C, left) or ASO116, DNp73 and miR-205 (C, center). Lentiviral expression of miR-205 was confirmed by qPCR compared with miR-control transduced cells (C, right). Bars represent the mean ± SD of three independent experiments. Statistical significance was determined by Student’s t-test. P-values of less than 0.05 were deemed statistically significant. (D) Tumors established in nude mice by subcutaneous injection of 5 x 106 SK-Mel-147 cells stably expressing miR-205 or miR-control and infected ex vivo with Adshp73 were treated with cisplatin (arrows) and monitored for tumor growth. Each group, n = 5. Graphs represent the tumor volume at indicated time points after tumor cell injection. AdshGFP-infected cells were used as control. Student’s t-test (two-sided) indicates statistically significant differences between the groups 6 weeks post-injection. (shp73 vs. shp73-miR-205, p = 0.008); (shp73-miRNA-control vs. shp73-miR-205, p = 0.004); (shp73 vs. shGFP, p = 0.016).
In addition, we analyzed the effect of miR-205 regulation by antagonistic p73/DNp73 on chemosensitization of tumors growing in vivo. SK-Mel-147 cells stably expressing miR-control or miR-205 were infected with AdshGFP or Adshp73 and subcutaneously injected into nude mice. Upon tumor growth, mice were treated with cisplatin. As shown in Figure 4D, drug treatment affected tumor growth in all experimental groups. Whereas growth inhibition in response to chemotherapy was less pronounced in p73-knockdown cells and those with miR-control, p73-depleted tumors expressing miR-205 markedly decreased in size over time, indicating that responsiveness to apoptotic stimuli can be rescued through miR-205. A substantial tumor growth reduction was also observed for the AdshGFP group, but to a lesser extent than in the shp73 plus miR-205 group.
MiR-205 repression by DNp73 provides a mechanism for E2F1 to confer chemoresistance
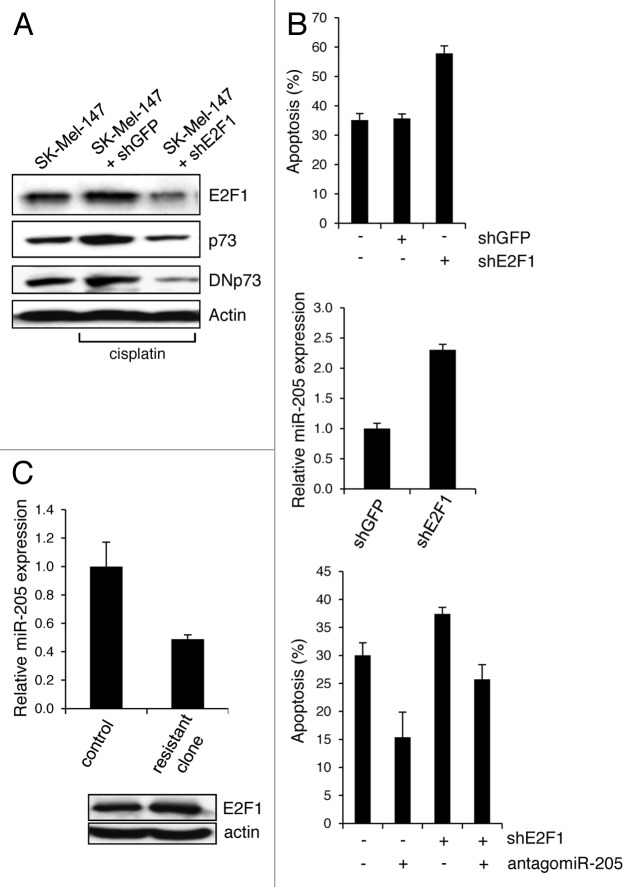
Next, we investigated whether stimulation of the DNp73-miR-205 axis contributes to the emergence of an E2F1-induced drug-insensitive cancer phenotype. Therefore, endogenously high E2F1 was selectively knocked down in chemoresistant SK-Mel-147 cells, which resulted in a decrease of p73 and even stronger of DNp73 (Fig. 5A) with a concomitant increase in the amount of apoptotic cells following cisplatin treatment (Fig. 5B, top). While this is accompanied by significantly enhanced miR-205 expression (Fig. 5B, central), inhibition of miR-205 in these cells reduces the apoptotic effect (Fig. 5B, bottom). In addition, drug-resistant SK-Mel-147 clones were selected upon increased levels of cisplatin treatment from 1 µM to 50 µM. As shown in Figure 5C, chemoresistant cells express high levels of E2F1 in combination with low levels of miR-205, indicating that E2F1-mediated chemoresistance of melanoma cells is dependent on the suppression of miR-205.
Figure 5. E2F1 induces chemoresistance via DNp73-mediated reduction of miR-205. (A) Protein levels of endogenous E2F1, p73 and DNp73 in cisplatin-treated SK-Mel-147 cells expressing shE2F1 or control shGFP compared with untreated cells. Actin was used for equal loading. (B) The percentage of apoptotic cells shown in (A) following knockdown of E2F1 and/or miR-205 in response to cisplatin was measured by FACS analysis (top and bottom panel). shE2F1-dependent changes in miR-205 expression are indicated (central panel). Statistical significance was determined by Student’ s t-test (p < 0.05). (C) Relative miR-205 expression in cisplatin-resistant SK-Mel-147 clones compared with parental cells set as 1 (right). Bar graphs indicate the mean ± SD of three independent experiments. E2F1 levels were analyzed by western blot (bottom).
MiR-205 targets Bcl-2 and ABCA2/5 transporters
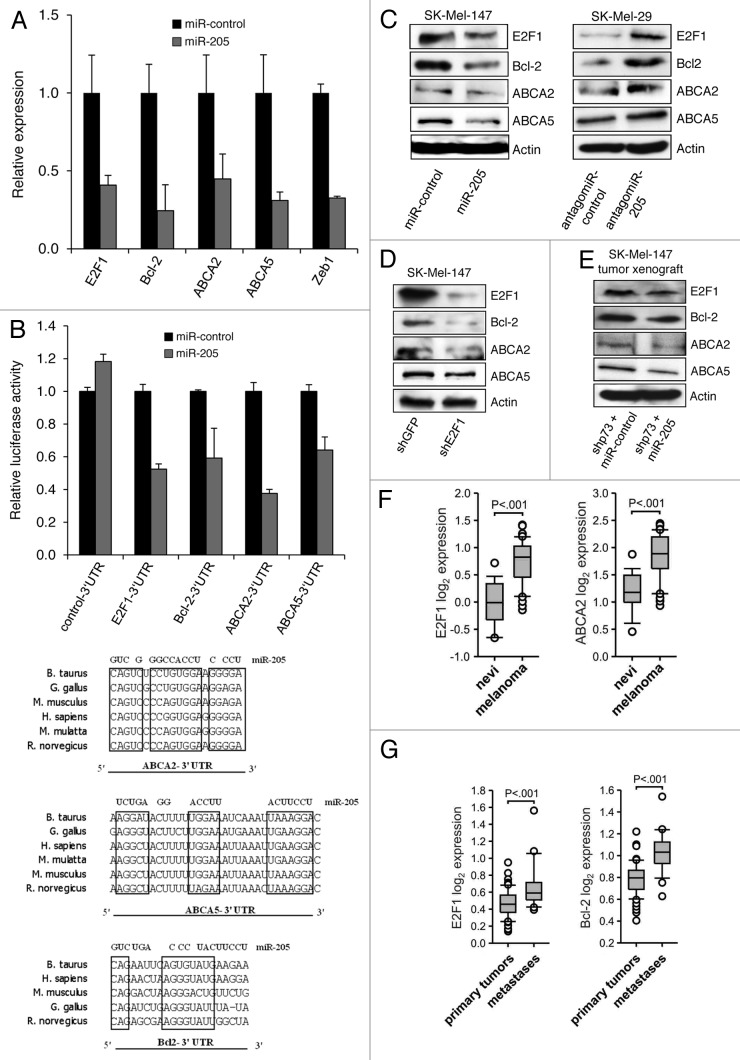
To identify molecular mechanisms by which the E2F1-DNp73-miR-205 axis causes resistance, we performed a bioinformatic analysis using several computational algorithms, including TargetScan and PicTar, to search for target genes of miR-205. The predicted list of potential targets was narrowed down by Gene Ontology and/or STRING 9.0 and subsequently subjected to literature-based analysis. We found that the 3′UTR of the Bcl-2 gene, a negative regulator of p53/p73-dependent apoptotic genes20 and the ATP-binding cassette transporters 2 (ABCA2) and 5 (ABCA5) associated with drug resistance21 have predicted miRNA-responsive elements containing regions that matched the seed sequences of miR-205. To verify that they are direct targets of miR-205, gene expression was analyzed in metastatic SK-Mel-147 cells resistant to chemotherapy after ectopic expression of miR-205 (Fig. 6A). MiRNA overexpression led to significantly decreased transcript levels of all three endogenous genes compared with cells expressing scrambled miRNA (miR-control) similar to ZEB1, a known target of miR-205. In addition, upregulation of miR-205 in SK-Mel-147 cells with very high E2F1 levels induced a strong reduction of E2F1 mRNA expression. Next, Bcl-2, ABCA2, -5 and E2F1 3′UTRs containing miR-205-responsive elements were cloned into the pEZX-MT01 luciferase reporter vector. Cotransfection of these constructs with miR-205 expression plasmid decreased the luciferase activity compared with the scrambled-miR (Fig. 6B). A significant reduction of Bcl-2, ABCA2 and -5 was also observed on the protein level in stable miR-205 expressing SK-Mel-147 (Fig. 6C), while their levels rose after knockdown of miR-205 in SK-Mel-29 cells (Fig. 6C, right). As loss of E2F1 leads to decreased expression of these miR-205 targets (Fig. 6D), our findings clearly suggest that the increased sensitivity for apoptosis after knockdown of E2F1 shown in Figure 5B (upper panel) depends on the downregulation of these genes. This is also evident from SK-Mel-147-based xenograft tumors shown in Figure 4D that considerably decreased in size upon overexpression of miR-205 in conjunction with reduced levels of Bcl-2, ABCA2 and ABCA5 after chemotherapy (Fig. 6E). Due to these results, we can conclude that the E2F1-DNp73-miR-205 axis is active in cancer cells progressing to metastases by targeting molecules involved in drug resistance.
Figure 6. Bcl-2 and the ATP-binding cassette transporters ABCA2 and ABCA5 are direct targets of miR-205. (A) qRT-PCR analysis of ABCA2, ABCA5, Bcl-2, E2F1 and ZEB1 in SK-Mel-147 infected with lentivirus expressing miR-205 (gray bars). Values are calculated after normalization with mean expression levels in cells overexpressing miR-control (set as 1; black bars). (B, upper panel) Reporter contructs containing predicted miR-205 binding sites in the Bcl-2, ABCA2 and ABCA5 3′UTR were assayed. E2F1 3′UTR was used as positive control. The miR-205 seed sequence is conserved in six different species (lower panel). (C) The expression levels of E2F1 and miR-205 target proteins in stable miR-205 or miR-control SK-Mel-147 cells (left) as well as in SK-Mel-29 cells at 48 h after knockdown of miR-205 compared with cells treated with antagomiR-control (right). Actin was used for equal loading. (D-E) E2F1, Bcl-2, ABCA2 and ABCA5 protein levels at 48 h after knockdown of E2F1 in SK-Mel-147 cells (D) or in primary tumors growing 6 weeks after cell injection in the xenograft model shown in Figure 4D. (E) Using actin as loading control. (F and G) Bioinformatic analysis of published microarray data showed a correlation of E2F1 and ABCA2 or Bcl-2 transcript levels in malignant melanomas (n = 24) vs. benign nevi (n = 18)23 (D) and prostate cancer metastases (n = 24) vs. primary tumors (n = 64)24 (E). In each graph, the solid lines within the boxes represent the median value and boxes show the 25th to 75th percentile range of mRNA levels. Bars represent 95% confidence intervals, with circles representing outliers. All statistical tests were one-sided.
However, considering that the miR-205 target Bcl-2 has been shown to be a direct target of E2F1,22 we examined the possibility whether, in addition to the effects of the E2F1-DNp73-miR-205 axis on these genes, ABCA2 and ABCA5 are also directly regulated by the transcription factor. We observed an increased ABCA5, but not ABCA2, mRNA expression in the presence of cycloheximide after E2F1 activation in SK-Mel-29 cells, suggesting that for this gene also a direct mechanism is involved (Fig. S1). In support of our experimental data, bioinformatic analysis of two published microarray studies23,24 using Oncomine database revealed that high E2F1 expression in melanoma tissues compared with skin nevi significantly correlates with increased ABCA2 levels in tumor vs. normal tissue (Fig. 6F). Similar data were obtained from prostate cancer samples, where high E2F1 and Bcl-2 levels are found in metastases as opposed to primary tumors (Fig. 6G). Apart from skin cancer, these clinical data define E2F1 and miR-205 as crucial regulators of genes associated with chemoresistance in other aggressive tumors, underscoring their possible role in malignant progression.
Discussion
Increasing evidence supports the view that non-coding microRNAs play an important role in cancer regulation and, consequently, modulation of their activity could be a new treatment option. Owing to previous reports, expression of miR-205 is confined to normal epithelial cells, whereas it is reduced or completely eliminated in matching tumor specimens.25-27 Initially described to be associated with the absence of vascular invasion in breast cancer,28 miR-205 was shown to suppress cell growth and invasion of this tumor.29 According to its tumor suppressor function, miR-205 is able to inhibit proliferation mediated by the HER receptor family, thereby improving the response to specific targeted therapies.30 Supported by a previous study,26 we have shown that miR-205 is significantly downregulated in cutaneous melanoma metastases compared with primary lesions as well as in highly metastatic melanoma cell lines. This confirms its role as important inhibitor of tumor progression via its ability to regulate the EMT process by targeting the E-cadherin transcriptional repressor ZEB1.31 Among factors mediating downregulation of miRs in cancer cells are promoter methylation, chromosomal aberrations and mutations in transcription factors regulating them. Searching for microRNAs influenced in skin cancer cells by p53 family members, we found that miR-205 expression is specifically upregulated by direct binding of p73 to its promoter.
MiR-205 suppresses melanoma cell proliferation and induces senescence by inhibiting E2F1.26 The E2F1 transcription factor expression is aberrantly high in advanced melanomas2,3 and was lately identified as driving force of invasion and metastasis in malignant skin tumors.4 In accordance with the high E2F1 abundance in metastatic cancers, only low levels of miR-205 were detectable in metastatic melanoma cells. E2F1 is responsible for the increased expression of both p73 and its antagonistic DNp73 isoforms, as evident from significantly higher levels of these factors in advanced tumors that directly correlate with E2F1 abundance.3 Given that DNp73 can transdominantly inhibit p73 targets, we investigated its ability to interfere with p73-mediated miR-205 expression and found a pronounced downregulation of this microRNA after enforced DNp73 expression, which explains the low miR-205 levels in the presence of high E2F1 activity.
Accordingly, since ectopic DNp73 expression in melanoma cells blocks their chemosensitivity, upregulation of miR-205 could rescue apoptosis defects acquired by endogenously high DNp73. This supports that DNp73-mediated repression of miR-205 is one mechanism by which drug resistance is achieved. As E2F1 itself stimulates DNp73 expression, it appears obvious that high levels of E2F1 can induce chemoresistance via the DNp73-miR-205 axis. Consequently, our data clearly show that knockdown of E2F1 leads to DNp73 downregulation with a concomitant rise of miR-205. Ultimately, this results in enhanced sensitization to genotoxic drugs of otherwise resistant melanoma cells. Vice versa, selected chemoresistant cell clones expressed high levels of E2F1 in conjunction with reduced levels of miR-205, indicating that the loss of responsiveness to apoptotic stimuli is due to an E2F1-promoted inhibition of miR-205. The aggressiveness of these resistant clones presumably raises with ongoing drug treatment via increased E2F1-DNp73-miR-205 signaling and eventually elevated E2F1 levels due to its release from negative regulation by miR-205.26 This is in line with clinical surveys demonstrating a frequent upregulation of E2F1 in high-grade tumors with chemoresistance and infaust prognosis.32 Beyond that, the relevance of high E2F1 levels for the induction of apoptosis resistance is substantiated by reports on engineered chemoresistant cervical, prostate and breast cancer cell lines with considerably higher levels of E2F1 compared with sensitive parental cells.33,34
We recently reported that E2F1-induced melanoma progression is mediated through increased expression and activation of EGFR, resulting in enhanced AKT phosphorylation.4 Interestingly, miR-205 overexpression reduces phospho-AKT and raises apoptosis sensitivity, which can be restored through E2F1 upregulation.26 Taking into account that miR-205 suppresses tumors by attenuating E2F1 expression in melanoma cells,26 our study proposes an autoregulatory circuit where E2F1 expression is controlled by p73/DNp73-dependent regulation of miR-205. In this regulatory loop, p73 activity is responsible for the decline of E2F1 by inducing miR-205 expression, while DNp73 protein via its inhibitory function on miR-205 results in E2F1 accumulation.
Our results define miR-205 as negative regulator of anti-apoptotic Bcl-2 and, importantly, ABCA2 and ABCA5 genes of the ATP-binding cassette transporter superfamily responsible for exclusion of conventional cytotoxic antitumor agents and molecularly targeted drugs from compound-treated tumor cells, resulting in the development of multidrug resistance in human cancer.21 Our data revealed a considerable reduction of these novel targets after overexpression of miR-205 in metastatic melanoma cells to levels comparable with the miR-205 target ZEB131 and established tumor xenografts, while their expression increased when miR-205 was knocked down in less aggressive tumor cells. Although definite functions of ABCA2 and ABCA5 in advanced skin cancer remain vague, another study in malignant mesotheliomas showed that blocking ERK1/2, which are critical for multi-drug resistance and survival, leads to the inhibition of both ABC proteins,35 underscoring that their upregulation after miR-205 depletion is critical for E2F1/DNp73-induced drug resistance. Furthermore, augmented ABCA2 levels in chemoresistant prostate and ovarian cancer cells were linked to enhanced estramustine efflux.36 Reflecting the proposed link between E2F1-induced chemoresistance and metastasis,37 it is striking that both miR-205 targets, ABCA2 and the related ABCA5 lysosomal ABC transporter, have been also associated with metastatic progression.35,38 As downregulation of miR-205 seems to be critical in tumor progression,31 further studies may elucidate whether the E2F1-DNp73-miR-205-ABCA2/ABCA5 axis represents a novel mechanism by which E2F1 drives metastatic disease.
Our data explain the rationale for downregulation of miR-205 in aggressive skin cancer prone to E2F1 and DNp73 and strongly support the relevance of this regulatory axis for chemoresistance. Moreover, the negative E2F1-miR-205-E2F1 feedback loop provides an intriguing explanation for high E2F1 levels in late stage cancer. This renders the DNp73-miR-205 axis a novel target for antimetastatic therapy.
Materials and Methods
Cell culture, adenoviral infection and antisense oligonucleotide transfection
Cell lines were cultured in DMEM with sodium pyruvate, 2 mM L-glutamine, 100 µg/ml penicillin, 100 U/ml streptomycin and 1.25 µg/ml amphotericin B. Melanocytes were purchased from Sigma and maintained according to supplier’s instructions. The adenoviral (Ad) vectors AdshGFP, Adshp73, AdGFP and Adp73 have been described previously.39 Adenovirus infection was performed at a multiplicity of infection (MOI) that allowed 100% transduction of target cells. Oligonucleotide (ON) LNA modifications were conducted by BioTeZ GmbH. The ASO sequence used was ASO116 5′-GATTGAACTGGGCCTGCAGC-3′.8 Transfections were performed with Lipofectamine 2000 (Invitrogen).
RNA and miR extraction
Samples from patients with primary (n = 3) and metastatic melanoma (n = 10) were obtained under a protocol approving the patients informed consent. RNA was extracted using the RNeasy Mini Kit (Qiagen), and miR extraction was performed with the mirVana™ miRNA Isolation Kit (Applied Biosystems).
Western blotting
Cells were lysed in RIPA buffer, and total protein concentration was quantified by Bradford assay (Bio-Rad). Equal amounts of protein were separated by SDS-PAGE and transferred to nitrocellulose membranes (Amersham Biosciences). Samples were probed with antibodies against p73 (ER-15) and E2F1 (KH-95) from BD Biosciences, Bcl-2 (Calbiochem), ABCA2 (Abcam), ABCA5 (Acris) and Actin (Sigma).
Cloning of promoter and 3′UTRs constructs and luciferase reporter assay
A small genomic DNA fragment containing miR-205 and a scramble miR sequence were amplified by PCR and cloned into PmeI and MluI restriction sites of the lentiviral vector pWPXL. VSV-G envelope pseudotyped lentiviral vector particles were produced by cotransfection of HEK293T cells using the plasmids pMD2.G and psPAX2 (Addgene) as described.40 Stable miR-205 or miR-control SK-Mel-147 cells were generated using these lentiviruses. Luciferase reporter gene constructs containing the miR-205 promoter sequence pGL3.miR-205 or the deletion mutant pGL3.miR-205-BS1Del (generated from the full-length promoter by TthIIII and BstEII restriction) and pGL3.miR-205-BS2,3Del were amplified by PCR from SK-Mel-29 genomic DNA and cloned into the pGL3-basic vector (Promega) using KpnI and XhoI restriction sites. Expression plasmids for wild-type p73, p73Δex2,3 (DNp73), E2F1 and the control Bax reporter construct have been described.39 Cells were transfected using Effectene (Qiagen). Luciferase activity was measured 36 h after transfection using the Luciferase Reporter Assay System (Promega) and normalized to total protein concentration in the cell extract. pEZX-MT01 vector expressing the E2F1 3′UTR and the control vectors were purchased from GeneCopoeia. The 3′UTRs of the ABCA2, ABCA5 and Bcl-2 mRNAs containing the sequences complementary to miR-205 seed sequence were inserted downstream of the luciferase reporter pEZX-MT01 vector into the AsiSI and XhoI sites. SK-Mel-147 cells were transfected using Turbofect tranfection reagent and the reporter activity was assayed 48 h post-transfection. Primers are shown in Supplementary Materials, Table S1.
Chromatin immunoprecipitation (ChIP)
ChIP assay was performed essentially as described.4 Protein-DNA complexes were immunoprecipitated using anti-p73 (BD Biosciences) or control IgG. Primers are shown in Supplementary Materials, Table S1.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed with nuclear extracts from SK-Mel-29 cells using biotin end-labeled (BrightStar BioDetect Kit, Ambion) double-stranded oligonucleotides corresponding to the BS1 (-424 to -447), BS2 (-918 to -945) and BS3 containing element (-1130 to -1159) in the miR-205 promoter as described.41 For oligonucleotide sequences see Table S1 in Supplementary Materials.
TaqMan array and qRT-PCR
MicroRNA array was performed using TaqMan Array Human MicroRNA Panel v2.0 with 7900HT Fast Real-Time PCR System (Applied Biosystems). Specific microRNA primers were used for the detection of hsa-miR-205 and hsa-miRNA-RNU6B purchased from Applied Biosystems. Relative miR expression was calculated by the comparative Ct method using RNU6B for normalization.
For qRT-PCR, RNA was reverse transcribed with Omniscript RT Kit (Qiagen). cDNA samples were mixed with iQTM SYBR® Green Supermix and analyzed on iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad). Relative gene expression was calculated by the comparative Ct method using B2M for normalization.
Apoptosis assay
For apoptosis quantification, cells were harvested after indicated times after infection and cisplatin treatment, fixed in 70% ethanol and stained for DNA content with propidium iodide. Analysis was performed in a FACSCalibur flow cytometer (BD Biosciences) using CellQuest software.
Animal studies
Athymic BALB/c-nu/nu mice of 6 to 8 weeks old (Charles River) were subcutaneously injected into the right flank with 5 x 106 SK-Mel-147 tumor cells stably expressing miR-control or miR-205 and infected with AdshGFP or Adshp73. Mice with tumors received a single intratumoral injection of cisplatin (6 mg/kg) followed by three intraperitoneal injections at day 25, 28, 31 and 34. Mice were monitored for tumor regression over 6 weeks. All animal experiments were performed according to the Institutional Animal Care and Use Committee.
Statistical analysis
Statistical significance was calculated by paired Student’s t-test. All statistical tests employed in this study were two-sided
Supplementary Material
Acknowledgments
The authors thank Anja Stoll and Ilona Klampfuß for technical assistance, Manfred Kunz for providing patient tumor samples and Ottmar Herchenröder for critical reading of the manuscript. This work is supported by funding provided by the Rostock University Medical Center for the project Systems Medicine of Cancer Invasion and Metastasis.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author’s Contributions
V.A., B.M.P. designed research; V.A., S.E., B.S.K. performed research; D.E., US, M.S. analyzed data; and B.M.P., V.A. wrote the paper.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21476
References
- 1.Soengas MS, Lowe SW. Apoptosis and melanoma chemoresistance. Oncogene. 2003;22:3138–51. doi: 10.1038/sj.onc.1206454. [DOI] [PubMed] [Google Scholar]
- 2.Halaban R, Cheng E, Smicun Y, Germino J. Deregulated E2F transcriptional activity in autonomously growing melanoma cells. J Exp Med. 2000;191:1005–16. doi: 10.1084/jem.191.6.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuve S, Wagner SN, Schittek B, Pützer BM. Alterations of DeltaTA-p73 splice transcripts during melanoma development and progression. Int J Cancer. 2004;108:162–6. doi: 10.1002/ijc.11552. [DOI] [PubMed] [Google Scholar]
- 4.Alla V, Engelmann D, Niemetz A, Pahnke J, Schmidt A, Kunz M, et al. E2F1 in melanoma progression and metastasis. J Natl Cancer Inst. 2010;102:127–33. doi: 10.1093/jnci/djp458. [DOI] [PubMed] [Google Scholar]
- 5.Pützer BM, Tuve S, Tannapfel A, Stiewe T. Increased DeltaN-p73 expression in tumors by upregulation of the E2F1-regulated, TA-promoter-derived DeltaN’-p73 transcript. Cell Death Differ. 2003;10:612–4. doi: 10.1038/sj.cdd.4401205. [DOI] [PubMed] [Google Scholar]
- 6.Seelan RS, Irwin M, van der Stoop P, Qian C, Kaelin WG, Jr., Liu W. The human p73 promoter: characterization and identification of functional E2F binding sites. Neoplasia. 2002;4:195–203. doi: 10.1038/sj.neo.7900237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stiewe T, Theseling CC, Pützer BM. Transactivation-deficient Delta TA-p73 inhibits p53 by direct competition for DNA binding: implications for tumorigenesis. J Biol Chem. 2002;277:14177–85. doi: 10.1074/jbc.M200480200. [DOI] [PubMed] [Google Scholar]
- 8.Emmrich S, Wang W, John K, Li W, Pützer BM. Antisense gapmers selectively suppress individual oncogenic p73 splice isoforms and inhibit tumor growth in vivo. Mol Cancer. 2009;8:61. doi: 10.1186/1476-4598-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mueller DW, Bosserhoff AK. Role of miRNAs in the progression of malignant melanoma. Br J Cancer. 2009;101:551–6. doi: 10.1038/sj.bjc.6605204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–4. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 11.Segura MF, Belitskaya-Lévy I, Rose AE, Zakrzewski J, Gaziel A, Hanniford D, et al. Melanoma MicroRNA signature predicts post-recurrence survival. Clin Cancer Res. 2010;16:1577–86. doi: 10.1158/1078-0432.CCR-09-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, et al. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–83. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–33. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 14.Soengas MS, Capodieci P, Polsky D, Mora J, Esteller M, Opitz-Araya X, et al. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature. 2001;409:207–11. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
- 15.Stiewe T, Pützer BM. Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat Genet. 2000;26:464–9. doi: 10.1038/82617. [DOI] [PubMed] [Google Scholar]
- 16.Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin WG., Jr. Chemosensitivity linked to p73 function. Cancer Cell. 2003;3:403–10. doi: 10.1016/S1535-6108(03)00078-3. [DOI] [PubMed] [Google Scholar]
- 17.Müller M, Schilling T, Sayan AE, Kairat A, Lorenz K, Schulze-Bergkamen H, et al. TAp73/DeltaNp73 influences apoptotic response, chemosensitivity and prognosis in hepatocellular carcinoma. Cell Death Differ. 2005;12:1564–77. doi: 10.1038/sj.cdd.4401774. [DOI] [PubMed] [Google Scholar]
- 18.Stiewe T, Zimmermann S, Frilling A, Esche H, Pützer BM. Transactivation-deficient DeltaTA-p73 acts as an oncogene. Cancer Res. 2002;62:3598–602. [PubMed] [Google Scholar]
- 19.Deyoung MP, Ellisen LW. p63 and p73 in human cancer: defining the network. Oncogene. 2007;26:5169–83. doi: 10.1038/sj.onc.1210337. [DOI] [PubMed] [Google Scholar]
- 20.Jiang M, Milner J. Bcl-2 constitutively suppresses p53-dependent apoptosis in colorectal cancer cells. Genes Dev. 2003;17:832–7. doi: 10.1101/gad.252603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gatti L, Cossa G, Beretta GL, Zaffaroni N, Perego P. Novel insights into targeting ATP-binding cassette transporters for antitumor therapy. Curr Med Chem. 2011;18:4237–49. doi: 10.2174/092986711797189682. [DOI] [PubMed] [Google Scholar]
- 22.Gomez-Manzano C, Mitlianga P, Fueyo J, Lee H-Y, Hu M, Spurgers KB, et al. Transfer of E2F-1 to human glioma cells results in transcriptional up-regulation of Bcl-2. Cancer Res. 2001;61:6693–7. [PubMed] [Google Scholar]
- 23.Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, et al. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res. 2005;11:7234–42. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- 24.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–9. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 25.Bhatnagar N, Li X, Padi SK, Zhang Q, Tang MS, Guo B. Downregulation of miR-205 and miR-31 confers resistance to chemotherapy-induced apoptosis in prostate cancer cells. Cell Death Dis. 2010;1:e105. doi: 10.1038/cddis.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dar AA, Majid S, de Semir D, Nosrati M, Bezrookove V, Kashani-Sabet M. miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J Biol Chem. 2011;286:16606–14. doi: 10.1074/jbc.M111.227611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–20. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 28.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 29.Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009;19:439–48. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iorio MV, Casalini P, Piovan C, Di Leva G, Merlo A, Triulzi T, et al. microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 2009;69:2195–200. doi: 10.1158/0008-5472.CAN-08-2920. [DOI] [PubMed] [Google Scholar]
- 31.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 32.Engelmann D, Pützer BM. Translating DNA damage into cancer cell death-A roadmap for E2F1 apoptotic signalling and opportunities for new drug combinations to overcome chemoresistance. Drug Resist Updat. 2010;13:119–31. doi: 10.1016/j.drup.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 33.Hirano G, Izumi H, Kidani A, Yasuniwa Y, Han B, Kusaba H, et al. Enhanced expression of PCAF endows apoptosis resistance in cisplatin-resistant cells. Mol Cancer Res. 2010;8:864–72. doi: 10.1158/1541-7786.MCR-09-0458. [DOI] [PubMed] [Google Scholar]
- 34.Millour J, de Olano N, Horimoto Y, Monteiro LJ, Langer JK, Aligue R, et al. ATM and p53 regulate FOXM1 expression via E2F in breast cancer epirubicin treatment and resistance. Mol Cancer Ther. 2011;10:1046–58. doi: 10.1158/1535-7163.MCT-11-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shukla A, Hillegass JM, MacPherson MB, Beuschel SL, Vacek PM, Pass HI, et al. Blocking of ERK1 and ERK2 sensitizes human mesothelioma cells to doxorubicin. Mol Cancer. 2010;9:314. doi: 10.1186/1476-4598-9-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laing NM, Belinsky MG, Kruh GD, Bell DW, Boyd JT, Barone L, et al. Amplification of the ATP-binding cassette 2 transporter gene is functionally linked with enhanced efflux of estramustine in ovarian carcinoma cells. Cancer Res. 1998;58:1332–7. [PubMed] [Google Scholar]
- 37.Engelmann D, Pützer BM. The dark side of E2F1: in transit beyond apoptosis. Cancer Res. 2012;72:571–5. doi: 10.1158/0008-5472.CAN-11-2575. [DOI] [PubMed] [Google Scholar]
- 38.Mack JT, Helke KL, Normand G, Green C, Townsend DM, Tew KD. ABCA2 transporter deficiency reduces incidence of TRAMP prostate tumor metastasis and cellular chemotactic migration. Cancer Lett. 2011;300:154–61. doi: 10.1016/j.canlet.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.John K, Alla V, Meier C, Pützer BM. GRAMD4 mimics p53 and mediates the apoptotic function of p73 at mitochondria. Cell Death Differ. 2011;18:874–86. doi: 10.1038/cdd.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salmon P, Trono D. Production and titration of lentiviral vectors. Curr Protoc Hum Genet 2007; Chapter 12:Unit 12.10. [DOI] [PubMed] [Google Scholar]
- 41.Engelmann D, Knoll S, Ewerth D, Steder M, Stoll A, Pützer BM. Functional interplay between E2F1 and chemotherapeutic drugs defines immediate E2F1 target genes crucial for cancer cell death. Cell Mol Life Sci. 2010;67:931–48. doi: 10.1007/s00018-009-0222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.