Abstract
The let-7 family contains 12 members, which share identical seed regions, suggesting that they may target the same mRNAs. It is essential to develop a means that can regulate the functions of all members. Using a DNA synthesis technique, we have generated an anti-let-7 sponge aiming to modulate the function of all members. We found that products of the anti-let-7 construct could bind and inactivate all members of the let-7 family, producing decoy and decay effects. To test the role of the anti-let-7 sponge, we stably expressed the anti-let-7 construct in two types of cells, the breast carcinoma cells MT-1 and the oldest and most commonly used human cervical cancer cell line, HeLa cells. We found that expression of anti-let-7 increased cell survival, invasion and adhesion, which corroborate with known functions of let-7 family members. We further identified a novel target site across all species of the let-7 family in hyaluronan synthase 2 (HAS2). HAS2 overexpression produced similar effects as the anti-let-7 sponge. Silencing HAS2 expression by siRNAs produced opposite effects to anti-let-7 on cell survival and invasion. The ability of anti-let-7 to regulate multiple members of the let-7 family allows us to observe their multiple functions using a single reagent. This approach can be applied to other family members with conserved sequences.
Keywords: let-7, anti-sense, cell invasion, microRNA, siRNA
Introduction
MiRNAs are non-coding RNAs that are made up of 18–25 nucleotides and are able to modulate gene expression, primarily negativity by either inducing the degradation or repressing translation of the target mRNAs.1 MiRNAs are highly conserved across different species, although they can be expressed in a tissue specific manner and perform essential functions in regulating diverse cellular processes, including cell proliferation,2,3 cell differentiation,4 senescence,5 apoptosis,6 cell division,7 migration,8 morphogenesis,9 tissue development,10 tumor growth,11 angiogenesis12,13 and metastasis.14 There are over 1,000 miRNAs that have been sequenced and reported, and it is estimated that one third of genes are regulated by miRNAs, as one miRNA can regulate the expression of many genes.15 miRNAs bind through imperfect base pairing to the 3' untranslated region (3'UTR) of target mRNAs. The binding specificity and efficiency is believed to be determined by a 6–7 nucleotide sequence near the 5′ region of miRNAs.15 This sequence is called the “seed sequence”16 and is the initial binding site of the miRNA to the 3'UTR of the target mRNA.17 The degree of complementation of the seed sequence of miRNAs to the target mRNA’s 3'UTR also determines whether the mRNA is degraded or translation of protein is repressed.17,18
The let-7 family members have been extensively studied and are classified as tumor suppressor miRNAs, because they have a lower expression in a variety of tumors compared with the normal tissues.19-21 In lung cancer, let-7 greatly reduces growth in different lung cancer cell lines and in xenografts in lung cancer mouse models.22 In breast cancer, let-7 was drastically reduced in tumor-initiating cells compared with non-tumor-initiating cells, and its expression level increased with differentiation.20 Furthermore, when let-7 was overexpressed in breast tumor-initiating cells, there was a reduction in cell proliferation, mammosphere formation, tumor growth and metastasis.20 Additionally, the inhibitory effect of let-7 was detected when the antisense of let-7 oligonucleotides increased the self renewal ability of non-tumor-initiating cells.20 It’s function as a tumor suppressor is further strengthened by the observations that it can downregulate several oncogenes.23-25
Different let-7 family members are involved in the suppression of cancer and are frequently lost in cancer tissues. Individually, let-7a represses gastric and colon cancer and suppresses breast cancer cell migration and invasion.26,27 Let-7b suppresses metastasis in melanoma.28 Let-7c can suppress metastasis in colorectal cancer, because it represses expression of PBX3,29 which is also a target of Let-7d.30 Let-7e targets cyclin D1 and plays a role in breast cancer cell cycle progression.31 Let-7f can target MYH9 and inhibits invasion and metastasis of gastric cancer.32 The levels of let-7g are much lower in metastatic breast and hepatocellular carcinomas compared with normal cells,33,34 because it can target the 3'UTRs of Bcl-xL, collagen type 1 α 2 and c-Myc, resulting in an increase in cell apoptosis and decrease in migration and proliferation.34-36
The fact that the seed regions of all let-7 members are identical suggests that all members of the family could target the same mRNAs and play similar functions. Thus, overexpression of one member or inhibition of the function of one member using an antisense approach may not produce a potent effect on phenotypic functions. It would be ideal to greatly increase the levels of all members to study their function, but this is impossible at present, because there is no such a system available. We developed an expression construct producing a transcript that can bind and inhibit the function of all members of the family. This approach may be used to study the functions of other miRNA families. It may also be a useful means in the development of reagents for intervention of miRNA functions.
Results and Discussion
Generation of an antisense sponge construct that decreases endogenous let-7 expression
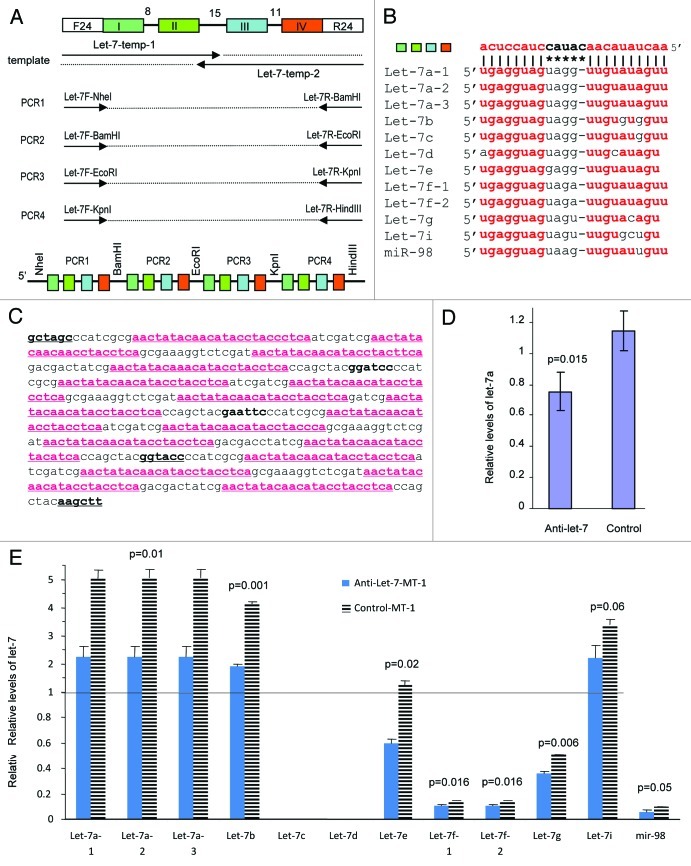
A miRNA sponge was initially developed by Ebert and colleagues to inhibit miRNA function.37 To study the function of endogenous let-7 family members, we generated a construct anti-let-7 aiming to function as a sponge to decoy and decay all members of the let-7 family. The construct was generated by linking four fragments of PCR products (Fig. 1A). Each PCR fragment contained four binding sites for let-7, resulting in 16 binding sites present in the insert. Due to the high degrees of conservation among the let-7 members, the construct can produce transcripts that are able to bind to the different let-7 family members (Fig. 1B). After molecular cloning, the DNAs were sequenced to confirm their identity (Fig. 1C). Since the transcript is highly complementary to all 12 members of the let-7 family, it is conceivable that the transcript can serve as a decoy for the mature miRNAs of the family. It is known that miRNAs with a central loop can bind better to the target.38 Similarly, we expect that the transcript of the anti-let-7 sponge can bind to all members of the let-7 family with high affinity, serving as a decoy in arresting the functions of all members.
Figure 1. Development of a sponge that can decay levels of let-7 family. (A) Strategy to generate the anti-let-7 construct is shown. It contains 16 motifs that can bind to the different members of the let-7 family. Each PCR product contains four different antisense sequences that can bind to the let-7 family members. It was generated by ligating together four different fragments to have multiple copies of the binding regions. (B) Alignment of one of the anti-let-7 sequences with the 12 members of the let-7 family members. Complete matching of the seed regions with the anti-let-7 sequence and high degrees of matching between the tail regions are shown. (C) Sequence of the anti-let-7 insert. The four different antisense constructs are underlined, which are capable of binding to the 12 members of let-7 family. (D) The efficiency of the anti-let-7 construct in HeLa cells was tested. The construct was able to decrease the levels of the mature miRNAs of the let-7 family members as seen through real-time PCR. Error bars, SD (n = 3). (E) The efficiency of the anti-let-7 construct was tested in MT-1 cells. The construct was able to downregulate mature miRNAs of the let-7 family member from basal levels that was found in the control cells as seen through real-time PCR.
To test the effects of the anti-let-7 construct, we stably expressed it in human breast cancer cell line MT-1 and cervical cancer cell line HeLa. The effect of the anti-let-7 plasmid on let-7 levels was analyzed using real-time PCR. We analyzed let-7a in HeLa cells and observed that transfection of the anti-let-7 construct decreased let-7a levels (Fig. 1D). We then analyzed all members of the let-7 family in MT-1 cells, and found that ten of the 12 members could be detected. Transfection with the anti-let-7 construct resulted in decreased levels of the detectable members of let-7 as compared with the cells transfected with the control vector (Fig. 1E). Let-7a appeared to be the predominant member. The decrease in levels of let-7 members in the cells transfected with the anti-let-7 construct indicated that the product of anti-let-7 could decay let-7 members. It has been extensively reported that miRNA can decay the levels of the targets.39-41 The mechanism by which the binding transcripts decay miRNA is less clear. Nevertheless, we have found that expression of versican 3′UTR causes a decrease in miRNA levels.42
Expression of anti-let-7 enhances cell adhesion, cell invasion and survival
The effects of the anti-let-7 sponge on cell activities were examined in MT-1 cells and HeLa cells. MT-1 cells transfected with the anti-let-7 construct displayed greater adhesive capacity than cells transfected with the control vector (Fig. 2A, left). The former was able to spread on the plates in serum-free medium, while the mock cells could only adhere to the plates (Fig. 2A, right). This suggests that the anti-let-7-transfected cells might have expressed higher levels of adhesion molecules than the control cells. The anti-let-7 cells also showed greater activity of adhesion than the control cells (Fig. 2B).
Figure 2. Expression of anti-let-7 increased cell adhesion and invasion. (A) MT-1 cells transfected with the anti-let-7 construct were cultured in tissue culture plates for cell adhesion assay. The anti-let-7 cells had an increased amount of cells attach to the plate compared with the control cells. Attached cells were counted for statistical analysis. Typical photographs of the cells are shown. The MT-1 cells adhered well to the plates and with each other. Error bars, SD (n = 6). (B) The adhesion assay was also performed in HeLa cells stably transfected with the anti-let-7 construct. The anti-let-7 cells had an increased amount of cells attached to the plates compared with the control cells. Error bars, SD (n = 6). (C) Cell invasion assays were performed in the MT-1 cells transfected with the control vector or the anti-let-7 construct. The invaded cells were stained and counted for statistical analysis. The anti-let-7 cells displayed greater activity in invading through the Matrigel coated trans-well membranes compared with the control cells. Typical photographs of invasion are shown (right). Error bars, SD (n = 6). (D) Cell invasion assays were also performed in the HeLa cells transfected with the control vector or the anti-let-7 construct. Expression of the anti-let-7 promoted cell invasion. Error bars, SD (n = 6).
In cell invasion experiments, we observed that the anti-let-7-transfected MT-1 cells had a faster rate of invasion through Matrigel-coated transwell membranes compared with the control cells (Fig. 2C). Similarly, the anti-let-7-transfected HeLa cells also showed greater activity of invasion than the control cells (Fig. 2D). It has been reported that downregulation of let-7c and let-7g could result in increased metastasis and distorted ECM structures.33,43 Since all members of the let-7 family share an identical seed region, they may target similar mRNAs and play similar roles in mediating cell activities. However, in different cell types and different environments, expression levels of the members may be different: some members may be dominant others in one cell types, but it could be vice versa in a different cell type.
We further examined whether or not expression of the anti-let-7 construct affected cell survival, and found that the anti-let-7 MT-1 cells survived significantly longer in nutrition depletion than the control cells (Fig. 3A). The effect of anti-let-7 on cell survival was even stronger in HeLa cells (Fig. 3B). Cancer cell survival is crucial to tumor development, invasion and metastasis. The cells that can survive in stress conditions are believed to be cancer stem-like cells. It is possible that these are the cells that survived in the serum-free medium.
Figure 3. Expression of anti-let-7 enhanced cell survival. (A) Cell survival was tested in the anti-let-7 and control MT-1 cells. Cells were grown in serum-free conditions for 12 d. The anti-let-7 cells had an increased amount of surviving cells compared with the control cells. Error bars, SD (n = 3). (B) HeLa cells transfected with the anti-let-7 and control plasmids were cultured in serum-free medium for 14 d. The anti-let-7 cells displayed increased rated of survival compared with the control cells. Error bars, SD (n = 3). (C) HeLa cells transfected with the anti-let-7 and control plasmids were cultured in 5% serum-containing medium for proliferation assay. Transfection with anti-let-7 increased cell proliferation. Error bars, SD (n = 3). (D) The cells were also subjected to cell cycle analysis. Decreased number of the anti-let-7 cells were detected in the G1 phase as compared with the control cells. (E) The experiments were repeated three times, and the results were subjected to statistical analysis.
We then used HeLa cell to examine the effect of the anti-let-7 construct on cell proliferation, and found that the anti-let-7 cells grew significantly faster than the control cells (Fig. 3C). Cell cycle analysis indicated that fewer anti-let-7 cells were detected in the G1 phase, while more cells were detected in the G2 and S phases than the control cells (Fig. 3D). The experiments were repeated three times for statistical analysis. It showed that there was a significant difference in each phase of the cell cycle (Fig. 3E). This correlates with the current understanding that let-7 is a tumor suppressor miRNA that induces apoptosis44 and plays roles in drug resistance of cancer cells.2
Targeting of HAS2 by let-7 is disrupted by anti-let-7 molecule
The observed roles in cell survival, adhesion and invasion are due to anti-let-7 decreasing endogenous let-7 family members that are expressed in MT-1 and HeLa cells. There have been a number of targets of let-7 family identified. In our studies, we were interested in a target that may mediate the phenotypes described above. A well-known function of let-7 family is their inhibitory effects on tumor growth. Thus, the reported targets can be surveyed to explain the phenotypes in cell survival and invasion, since both phenotypes are tightly linked to tumor development. However, the effect of cell adhesion on tumor growth is more complicated; it can enhance or inhibit tumor development. The target in the cell lines involved in our studies may be associated with adhesion molecules.
Using different software available, we analyzed potential targets of let-7. We were interested in targets that not only play a role in tumor cell survival and invasion, but are also involved in cell adhesion. This is because our results showed that expression of the anti-let-7 construct promoted cell survival, invasion and adhesion. One software (http://pictar.mdc-berlin.de/cgi-bin/PicTar_vertebrate.cgi?action=Search%20for%20targets%20of%20a%20miRNA&name2=hsa-let-7a) predicts that hyaluronan synthase 2 (HAS2) is a potential target of let-7. HAS2 is an inner plasma membrane-bound molecule that is responsible for producing hyaluronic acid, a well-studied cell adhesion molecule that is involved in the formation of the ECM and functions in wound healing and tumor growth and metastasis. It has been known that hyaluronan synthase 2 plays important roles in tumor growth and progression.45,46 The HAS2 3'UTR contains a binding site for let-7 (Fig. 4A).
Figure 4. Targeting analysis of anti-let-7 on HAS2. (A) HAS2 3'UTR (nucleotides 2467–2488) was found to be a potential target of let-7. The binding interaction between let-7a and HAS is shown in the diagram. (B) Upper: cell lysate was prepared from the MT-1 cells transfected with anti-let-7 or the control vector. Upregulation of HAS2 was seen in the anti-let-7 transfected cells compared with the control. β-actin levels served as a loading control. The intensities of each protein band were scanned with a densitometer and the relative intensities are shown below each band normalized with the β-actin bands. Lower: cell lysate from HeLa cells transfected with the construct was also analyzed on western blot probed with anti-c-Myc and anti-cyclin D1 antibodies. Expression of both c-Myc and cyclin D1 was upregulated in the anti-let-7-transfected cells. (C) Upper: alignment of the let-7 targeting HAS2 3'UTR located at nucleotides 2467–2488 across Bos tauru (NM_174079.2), Callithrix jacchus (XM_002759268.1), Equus caballus (NM_001081801.1), Gallus gallus (NM_204806.1), Homo sapiens (NM_005328), Macaca mulatta (XM_001098841.2), Mus musculus (NM_008216.3), Nomascus leucogenys (XM_003256164.1), Pan troglodytes (XM_528222.3), Rattus norvegicus (NM_013153.1), Silurana tropicalis (NM_001127085.1) and Xenopus laevis (NM_001090368.1). The seed regions for let-7-HAS2 interactions are in the box. Lower: conservation of the sequences is shown across all species. (D) Left: a luciferase construct was generated with the 3′ UTR sequence of HAS2, harboring the target site of let-7, producing Luc-HAS2. Mutations were also generated on the potential target sequence (underlined), resulting in a mutant construct Luc-HAS2-mut. Middle: MT-1 cells were co-transfected with let-7a and a luciferase reporter construct. The luciferase reporter vector was used as a control. Luciferase activity assays indicted that the let-7a repressed the luciferase activities of Luc-HAS2, which were abolished with the mutant construct, when the potential let-7 binding site was mutated. Asterisks indicate significant differences. ** p < 0.01. Error bars, SD (n = 3). Right: the luciferase assays were also performed with HeLa cells and similar results were obtained.
Western blot analysis showed that the levels of HAS2 were higher in the anti-let-7 MT-1 and HeLa cells than in the control cells (Fig. 4B, upper). This is because the anti-let-7 transcript could bind and inhibit the functions of endogenous let-7 miRNAs. As a consequence, HAS2, which is usually involved in binding to let-7 and repressed by the miRNAs, is free for translation reaching higher levels in the transfected cells. It has been reported that let-7 can target cyclin D1 and c-Myc.47,48 To confirm that the sponge construct functioned properly, we analyzed the levels of these proteins and detected upregulation of c-Myc and cyclin D1 (Fig. 4B, lower). These results showed that the sponge construct could suppress the function of let-7 members.
Sequence analysis of the reported HAS2 nucleotide sequences in different organisms indicated that the target sites are highly conserved across different species (Fig. 4C). This result also suggests that the target site is of evolutional importance in the regulation of protein expression. In order to further confirm the targeting of HAS2 by let-7, luciferase assays were performed. In the pMir-luciferase construct, the 3′UTR of HAS2 was inserted into the luciferase reporter vector, producing a luciferase construct Luc-HAS2 (Fig. 4D, left). The detail sequences of the luciferase constructs are provided (Fig. S1). In addition, a mutant construct was generated, in which the let-7 target sites were mutated producing the mutant construct Luc-HAS2-mut. When these plasmids were transfected along with the let-7 oligonucleotide into MT-1 cells, there was significant downregulation of luciferase activities seen in the cells transfected with let-7 oligonucleotide as compared with the cells transfected with the control oligo (Fig. 4D, middle). However, when the mutant Luc-HAS2-mut and Let-7 were transfected together, there was a reverse effect in luciferase activity. Similar results were obtained when HeLa cells were transfected with these construct (Fig. 4D, right). This result confirmed the direct targeting of HAS2 by let-7. A diagram summarized the targeting of HAS2 by let-7, which is modulated by the anti-let-7 transcript is provide (Fig. 5).
Figure 5. A diagram showing how the anti-let-7 sponge works. Overexpression of the sponge construct would bind extensively the endogenous let-7 family members. As a consequence, the potential target (e.g., hyaluronan synthase 2 or HAS2) is free for translation.
siRNAs against HAS2 affect cell survival and invasion
Four different siRNA sequences were designed to target HAS2 (Fig. 6A). Cells transfected with all four siRNAs were found to produce lower levels of HAS2 protein than cells transfected with the control oligo (Fig. 6B). The functional changes previously observed in MT-1 and HeLa cells, when the cells were transfected with the anti-let-7 construct, were reversed with the transfection of these siRNAs (Fig. 6C). There was a significant reduction in cell survival in the siRNA-transfected cells compared with the control oligonucleotide-transfected cells. Similarly, in cell invasion assays, there was a significant reduction in cell invasion in the siRNA-transfected cells compared with the control (Fig. 6D; Fig S2).
HAS2 overexpression mimics the function of anti-let-7
To corroborate the effects of the anti-let-7, the construct of the HAS2 coding region was overexpressed for functional tests. We confirmed the increased expression of HAS2 in MT-1 and HeLa cells compared with the cells transfected with a control vector by western blot analysis (Fig. 7A). The cells were subjected to survival assays. We found that expression of HAS2 increased survival of both MT-1 and HeLa cells (Fig. 7B). In cell invasion assay, we found that expression of HAS2 promoted invasion as compared with the control (Fig. 7C). These results indicated that HAS2, along with the molecules previously identified as being targeted by let-7 family members, is an important molecule in mediating cell survival, cell migration and adhesion of both MT-1 and HeLa cells.
Figure 6. Silencing of HAS2 expression. (A) Four siRNA sequences were generated to downregulate HAS2 expression. (B) MT-1 cells were transiently transfected with HAS2 siRNA constructs and a control vector. Expression of endogenous HAS2 was silenced by siRNAs when compared with the control which was confirmed by western blot analysis. β-actin was detected on the same blot as a loading control. The intensities of each protein band was scanned with a densitometer and the relative intensities are shown below each band normalized with the β-actin bands. (C) MT-1 cells and HeLa cells were transfected with the siRNA constructs followed by cell survival assays. Silencing of HAS2 decreased cell survival. Error bars, SD (n = 3). (D) MT-1 cells and HeLa cells were transfected with the siRNA constructs followed by cell invasion assays. Silencing of HAS2 decreased activities of invasion. Error bars, SD (n = 6).
In summary, we have developed an approach to inhibit the functions of all members of let-7 family, one of the most extensively studied miRNA families. Although this family has been significantly studied, there has been no molecule that has been reported to possess the ability to regulate the functions of all members in the family. Our experiments demonstrated that ectopic expression of the anti-let-7 construct increases cell survival, invasion and adhesion. This is a novel approach to investigate the role of the entire let-7 family. Additionally, we have identified a novel target for let-7. Molecular analysis indicated that expression of HAS2 was repressed by let-7. The targeting of HAS2 by let-7 was confirmed by western blot, luciferase and siRNA assays. Since each family member of let-7 can potentially target many mRNAs, it is possible that other mRNAs are involved and can contribute to the observed phenotypes. Our assay of expressing a single antisense molecule to study multiple members of the let-7 miRNA family is useful in studying the functions of an entire miRNA family. Using this approach, it is possible to uncover the combinatorial roles of different miRNAs in the let-7 family, which can be extended to study other miRNA families. Our assay system can also function as a model approach to miRNA families and their gene regulation for the development of gene therapy targeting many family members.
Methods
Construct generation
To generate a fragment of cDNA containing four binding sites for members of the let-7 family, we designed and synthesized two large oligos to serve as primers. They were Let7-temp-1 (5 cgcgtgccgg gctagc ccatcgcg aactatacaacatacctacctca atcgatcg aactatacaacatacctacctca gcgaaaggtctcgat aac) and Let7-temp-2 (5' accgcggtac ggatcc gtagctgg tgaggtag gtatg ttgtatagtt cgatagtcgtc tgaggtag gtatg ttgtatagtt atcgagacctttcgc). Both primers were mixed in a PCR. They annealed with each other and extended to produce a small fragment of cDNA. This cDNA was used as a template in four PCRs: PCR-1 using primers Let7-F-NheI (5' cgcgtgccgg gctagc ccatcgcg) and Let7-R-BamHI (5' accgcggtac ggatcc gtagctgg); PCR-2 using primers Let7-F-BamHI (5' cgcgtgccgg ggatcc ccatcgcg) and Let7-R-EcoRI (5' accgcggtac gaattc gtagctgg); PCR-3 using primers Let7-F-EcoRI (5' cgcgtgccgg gaattc ccatcgcg) and Let7-R-KpnI (5' accgcggtac ggtacc gtagctgg); PCR-4 using primers Let7-F-KpnI (5' cgcgtgccgg ggtacc ccatcgcg) and Let7-R-HindIII (5' accgcggtac aagctt gtagctgg). All PCR products were cloned, and the inserts were recovered with the restriction enzymes indicated in the primers. All fragments were finally ligated into the NheI- and HindIII-opened pEGFP-N1 plasmid producing an anti-let-7 expression construct.
A luciferase reporter vector (pMir-Report; Ambion) was used to generate the luciferase constructs. A fragment of the 3'UTR of HAS2 was cloned using two primers, HAS2-N3'SacI (5' ccc ggg gag ctc tgatct tccatgtttt gacgtttg) and HAS2-C3'MluI (5' ggg ccc acg cgt ggc tag tgaggta tata gggc), by PCR. The PCR products were then digested with SacI and MluI, and the fragment was inserted into a SacI- and MluI-digested pMir-Report Luciferase plasmid to obtain a luciferase construct Luc-HAS2. A mutant construct was generated with two primers HAS2-N3'SacI and HAS2-C3'MluI-Mut (5' ggg ccc acg cgt ggc tag actccat tata gggc aaaa cac ttt c) using similar approach.
To serve as a negative control, a non-related sequence was amplified from the coding sequence of the chicken versican G3 domain using two primers, chver10051SpeI and chver10350SacI, as described previously.49 It is expected that there is no endogenous miRNA bind to this fragment as it is in the coding region. The PCR product was then inserted into a SpeI- and SacI-digested pMir-Report Luciferase plasmid.
RT-PCR
Total RNAs were extracted from cell cultures with mirVana miRNA Isolation Kit following the instructions of the manufacturer (Ambion). The RNAs were subjected to reverse transcription. Real-time PCR was performed as previously described.50 For mature microRNA analysis, total RNA was extracted from ~1 × 106 cells using mirVana™ miRNA Isolation Kit (Applied Biosystems), followed by cDNA synthesis using 2 µg RNA. Successive PCR was performed by QuantiMir RT Kit (System bioscience) using 1 µl cDNA as template. Two primers let-7PCRN (5'ccgtcagatccgctagcccatcgc) and let-7PCRC (5' ctg cag aat tcg aag ctt gta gct) were used for the detection of the Let-7 family members. The primers used for the detection of HAS2 mRNA were HAS2-N3'SacI and HAS2-C3'MluI. The primers used as real-time PCR controls were human-U6RNAf (5' gtgct cgcttcggca gcacatatac) and human-U6RNAr (5' aa aaa tat gg aa cgc ttc acga atttg).
Cell survival assay
Cells were seeded at the density of 1 × 105 cells/well in 6-well plates with 2 ml culture medium containing 10% FBS. After 24 h, cells were washed twice with PBS and then incubated in serum-free medium. After 10 and 18 d, cells were harvested and the viable cells were counted after Trypan Blue staining.
Invasion assay
Cell transwell membrane inserts were placed in 24-well plate and coated with 100 µl of Matrigel that had been diluted (1:10 fold dilution). 5 × 104 cells were placed in serum-free media inside the inserts, while media supplemented with 10% FBS was placed at the bottom of the inserts and in the wells of the plates. Cells were incubated at 37°C and were allowed to invade and migrate through the Matrigel and the membrane pores in the inserts. After 48 h (and 72 h for siRNA experiments), the upper Matrigel layer and cells were removed, while the cells on the surface of the lower side of the membrane were fixed and stained. Cells that migrated onto the lower surface were counted from representative areas for quantification.
Cell adhesion
HeLa cells were plated at a density of 1 × 105 cells per well in a 6-well plate in serum-free DMEM and assayed after 4 h. MT-1 cells were plated at a density of 1 × 105 cells per well in a 6-well plate in serum-free RPMI and assayed after overnight incubation. The unattached cells were removed. The cells that adhered to the plates were counted for quantification.
Luciferase assay
U343 cells were cultured on 24-well tissue culture plates at a density of 3 × 104 cells per well in DMEM containing 10% FBS. The cultures were maintained at 37°C for 24 h, followed by co-transfection with the luciferase reporter constructs, corresponding miRNA mimics and Renilla luciferase construct using Lipofectamine 2000. A dual-luciferase reporter system developed by Promega (E1960) was used to analyze luciferase activities. After gene transfection and expression, the cells were lysed using 100 μl of passive lysis buffer per well on a shaker for 30 min, and the lysates were centrifuged for supernatant collection. Twenty μl of the supernatants was then mixed with 100 μl of LAR II, and then firefly luciferase activities were measured using a luminometer. For the internal control, 100 μl of Stop-and-Go reagent was added to the samples. Renilla luciferase activities were then measured in the same tube. Luciferase activities between different treatments were compared after normalization with Renilla luciferase activities. The let-7a oligonucleotides were used at a concentration of 100 nM.
Western blot
Protein expression was analyzed by western blot. Briefly, cells were seeded onto a 6-well plate at 2 × 105 cells per well overnight. They were then transfected with 1 μg of anti-let-7 or the control vector to create stable cell lines. In addition, the anti-let-7 cells were transfected with siRNA against HAS2 or control oligonucleotide. The control was a scrambled RNA sequence provided in the kit. Proteins were extracted 48 h after transfection by lysing in 60 μl lysis buffer containing protease inhibitors (150 mM NaCl, 25 mM TRIS-HCl, pH 8.0, 0.5 M EDTA, 20% Triton X-100, 8 M Urea and 1× protease inhibitor cocktail). Proteins separated on SDS-PAGE were transferred to nitrocellulose membranes followed by incubating with antibodies against HAS2 (Santa Cruz, SC34067) at a dilution of 1:1000 at 4°C overnight. Secondary goat antibody was used at 1:5000 dilution to incubate with the membranes at room temperature for 2 h. After detection of the protein bands, the blots were stripped and re-probed with mouse monoclonal antibody against β-actin (Sigma, A5316) to confirm equal sample loading. After secondary antibody incubation, the blot was washed and detected using ECL kit (Millipore).
Statistical analysis
The results (mean values ± SD) of all the experiments were subjected to statistical analysis by t-test. The level of significance was set at p < 0.05 and p < 0.01.(Fig. 7)
Figure 7. Ectopic expression of HAS2 increased cell survival and invasion. (A) MT-1 cells were transfected with the HAS2 expression construct. Cell lysate prepared from the HAS2- and vector-transfected cells were subjected to western blot analysis probed with anti-HAS2 antibody confirming increased expression of HAS2. (B) The anti-let-7 MT-1 cells were transfected with the HAS2 expression construct followed by cell survival assay. Introduction of HAS2 into the anti-let-7 cells enhanced cell survival. Error bars, SD (n = 3). (C) The anti-let-7 MT-1 cells were transfected with the HAS2 construct followed by cell invasion assay. Introduction of HAS2 into the anti-let-7 cells enhanced cell invasion. Error bars, SD (n = 6).
Supplementary Material
Acknowledgments
This work was supported by grants from Canadian Institutes of Health Research (MOP-102635, MOP-111171) to B.B.Y. who is the recipient of a Career Investigator Award (CI 7418) from the Heart and Stroke Foundation of Ontario. Z.J.R. was supported by an Ontario Graduate Scholarship.
Glossary
Abbreviations:
- DMEM
Dulbecco’s modified Eagle’s medium
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- siRNA
small interfering RNA
- miRNA
microRNA
- 3'UTR
3' untranslated region
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21503
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Viticchiè G, Lena AM, Latina A, Formosa A, Gregersen LH, Lund AH, et al. MiR-203 controls proliferation, migration and invasive potential of prostate cancer cell lines. Cell Cycle. 2011;10:1121–31. doi: 10.4161/cc.10.7.15180. [DOI] [PubMed] [Google Scholar]
- 3.Shatseva T, Lee DY, Deng Z, Yang BB. MicroRNA miR-199a-3p regulates cell proliferation and survival by targeting caveolin-2. J Cell Sci. 2011;124:2826–36. doi: 10.1242/jcs.077529. [DOI] [PubMed] [Google Scholar]
- 4.Kawasaki H, Taira K. Retraction: Hes1 is a target of microRNA-23 during retinoic-acid-induced neuronal differentiation of NT2 cells. Nature. 2003;426:100. doi: 10.1038/nature02141. [DOI] [PubMed] [Google Scholar]
- 5.Marasa BS, Srikantan S, Martindale JL, Kim MM, Lee EK, Gorospe M, et al. MicroRNA profiling in human diploid fibroblasts uncovers miR-519 role in replicative senescence. Aging (Albany NY) 2010;2:333–43. doi: 10.18632/aging.100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 8.Shan SW, Lee DY, Deng Z, Shatseva T, Jeyapalan Z, Du WW, et al. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat Cell Biol. 2009;11:1031–8. doi: 10.1038/ncb1917. [DOI] [PubMed] [Google Scholar]
- 9.Wang CH, Lee DY, Deng Z, Jeyapalan Z, Lee SC, Kahai S, et al. MicroRNA miR-328 regulates zonation morphogenesis by targeting CD44 expression. PLoS ONE. 2008;3:e2420. doi: 10.1371/journal.pone.0002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–20. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 11.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang L, Deng Z, Shatseva T, Yang J, Peng C, Du WW, et al. MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-β8. Oncogene. 2011;30:806–21. doi: 10.1038/onc.2010.465. [DOI] [PubMed] [Google Scholar]
- 13.Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA. 2007;104:20350–5. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–10. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 15.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 16.Latronico MV, Catalucci D, Condorelli G. Emerging role of microRNAs in cardiovascular biology. Circ Res. 2007;101:1225–36. doi: 10.1161/CIRCRESAHA.107.163147. [DOI] [PubMed] [Google Scholar]
- 17.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 18.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–4. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 19.Cairo S, Wang Y, de Reyniès A, Duroure K, Dahan J, Redon MJ, et al. Stem cell-like micro-RNA signature driven by Myc in aggressive liver cancer. Proc Natl Acad Sci USA. 2010;107:20471–6. doi: 10.1073/pnas.1009009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 21.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–64. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 23.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 24.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–70. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 25.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–9. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim SJ, Shin JY, Lee KD, Bae YK, Sung KW, Nam SJ, et al. MicroRNA let-7a suppresses breast cancer cell migration and invasion through downregulation of C-C chemokine receptor type 7. Breast Cancer Res. 2012;14:R14. doi: 10.1186/bcr3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang F, Zhang P, Ma Y, Yang J, Moyer MP, Shi C, et al. NIRF is frequently upregulated in colorectal cancer and its oncogenicity can be suppressed by let-7a microRNA. Cancer Lett. 2012;314:223–31. doi: 10.1016/j.canlet.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 28.Fu TY, Chang CC, Lin CT, Lai CH, Peng SY, Ko YJ, et al. Let-7b-mediated suppression of basigin expression and metastasis in mouse melanoma cells. Exp Cell Res. 2011;317:445–51. doi: 10.1016/j.yexcr.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 29.Pereira F, Barbáchano A, Singh PK, Campbell MJ, Muñoz A, Larriba MJ. Vitamin D has wide regulatory effects on histone demethylase genes. Cell Cycle. 2012;11:1081–9. doi: 10.4161/cc.11.6.19508. [DOI] [PubMed] [Google Scholar]
- 30.Ramberg H, Alshbib A, Berge V, Svindland A, Taskén KA. Regulation of PBX3 expression by androgen and Let-7d in prostate cancer. Mol Cancer. 2011;10:50. doi: 10.1186/1476-4598-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitra D, Das PM, Huynh FC, Jones FE. Jumonji/ARID1 B (JARID1B) protein promotes breast tumor cell cycle progression through epigenetic repression of microRNA let-7e. J Biol Chem. 2011;286:40531–5. doi: 10.1074/jbc.M111.304865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang S, He L, Zhao X, Miao Y, Gu Y, Guo C, et al. MicroRNA let-7f inhibits tumor invasion and metastasis by targeting MYH9 in human gastric cancer. PLoS ONE. 2011;6:e18409. doi: 10.1371/journal.pone.0018409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qian P, Zuo Z, Wu Z, Meng X, Li G, Wu Z, et al. Pivotal role of reduced let-7g expression in breast cancer invasion and metastasis. Cancer Res. 2011;71:6463–74. doi: 10.1158/0008-5472.CAN-11-1322. [DOI] [PubMed] [Google Scholar]
- 34.Shimizu S, Takehara T, Hikita H, Kodama T, Miyagi T, Hosui A, et al. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J Hepatol. 2010;52:698–704. doi: 10.1016/j.jhep.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 35.Ji J, Zhao L, Budhu A, Forgues M, Jia HL, Qin LX, et al. Let-7g targets collagen type I alpha2 and inhibits cell migration in hepatocellular carcinoma. J Hepatol. 2010;52:690–7. doi: 10.1016/j.jhep.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lan FF, Wang H, Chen YC, Chan CY, Ng SS, Li K, et al. Hsa-let-7g inhibits proliferation of hepatocellular carcinoma cells by downregulation of c-Myc and upregulation of p16(INK4A) Int J Cancer. 2011;128:319–31. doi: 10.1002/ijc.25336. [DOI] [PubMed] [Google Scholar]
- 37.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–6. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye W, Lv Q, Wong CK, Hu S, Fu C, Hua Z, et al. The effect of central loops in miRNA:MRE duplexes on the efficiency of miRNA-mediated gene regulation. PLoS ONE. 2008;3:e1719. doi: 10.1371/journal.pone.0001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Behm-Ansmant I, Rehwinkel J, Izaurralde E. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb Symp Quant Biol. 2006;71:523–30. doi: 10.1101/sqb.2006.71.013. [DOI] [PubMed] [Google Scholar]
- 40.Vohradsky J, Panek J, Vomastek T. Numerical modelling of microRNA-mediated mRNA decay identifies novel mechanism of microRNA controlled mRNA downregulation. Nucleic Acids Res. 2010;38:4579–85. doi: 10.1093/nar/gkq220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choe J, Cho H, Chi SG, Kim YK. Ago2/miRISC-mediated inhibition of CBP80/20-dependent translation and thereby abrogation of nonsense-mediated mRNA decay require the cap-associating activity of Ago2. FEBS Lett. 2011;585:2682–7. doi: 10.1016/j.febslet.2011.07.047. [DOI] [PubMed] [Google Scholar]
- 42.Lee DY, Jeyapalan Z, Fang L, Yang J, Zhang Y, Yee AY, et al. Expression of versican 3′-untranslated region modulates endogenous microRNA functions. PLoS ONE. 2010;5:e13599. doi: 10.1371/journal.pone.0013599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han HB, Gu J, Zuo HJ, Chen ZG, Zhao W, Li M, et al. Let-7c functions as a metastasis suppressor by targeting MMP11 and PBX3 in colorectal cancer. J Pathol. 2012;226:544–55. doi: 10.1002/path.3014. [DOI] [PubMed] [Google Scholar]
- 44.Wang S, Tang Y, Cui H, Zhao X, Luo X, Pan W, et al. Let-7/miR-98 regulate Fas and Fas-mediated apoptosis. Genes Immun. 2011;12:149–54. doi: 10.1038/gene.2010.53. [DOI] [PubMed] [Google Scholar]
- 45.Kosaki R, Watanabe K, Yamaguchi Y. Overproduction of hyaluronan by expression of the hyaluronan synthase Has2 enhances anchorage-independent growth and tumorigenicity. Cancer Res. 1999;59:1141–5. [PubMed] [Google Scholar]
- 46.Okuda H, Kobayashi A, Xia B, Watabe M, Pai SK, Hirota S, et al. Hyaluronan synthase HAS2 promotes tumor progression in bone by stimulating the interaction of breast cancer stem-like cells with macrophages and stromal cells. Cancer Res. 2012;72:537–47. doi: 10.1158/0008-5472.CAN-11-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao C, Sun G, Li S, Lang MF, Yang S, Li W, et al. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci USA. 2010;107:1876–81. doi: 10.1073/pnas.0908750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–8. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee DY, Shatseva T, Jeyapalan Z, Du WW, Deng Z, Yang BB. A 3′-untranslated region (3’UTR) induces organ adhesion by regulating miR-199a* functions. PLoS ONE. 2009;4:e4527. doi: 10.1371/journal.pone.0004527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.LaPierre DP, Lee DY, Li SZ, Xie YZ, Zhong L, Sheng W, et al. The ability of versican to simultaneously cause apoptotic resistance and sensitivity. Cancer Res. 2007;67:4742–50. doi: 10.1158/0008-5472.CAN-06-3610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.