Abstract
Many asymmetrically dividing cells segregate the poles of the mitotic spindle non-randomly between their two daughters. In budding yeast, the protein Kar9 localizes almost exclusively to the astral microtubules emanating from the old spindle pole body (SPB) and promotes its movement toward the bud. Thereby, Kar9 orients the spindle relative to the division axis. Here, we show that beyond perturbing Kar9 distribution, activation of the spindle assembly checkpoint (SAC) randomizes SPB inheritance. Inactivation of the B-type cyclin Clb5 led to a SAC-dependent defect in Kar9 orientation and SPB segregation. Furthermore, unlike the Clb4-dependent pathway, the Clb5- and SAC-dependent pathways functioned genetically upstream of the mitotic exit network (MEN) in SPB specification and Kar9-dependent SPB inheritance. Together, our study indicates that Clb5 functions in spindle assembly and that the SAC controls the specification and inheritance of yeast SPBs through inhibition of the MEN.
Keywords: Clb5, Kar9 asymmetry, SPB inheritance, SPB specification, mitotic exit network, spindle assembly checkpoint
Introduction
As in many stem cells of higher eukaryotes, the mitotic spindle of budding yeast is non-randomly positioned and oriented with respect to the cleavage-site. This leads to non-random segregation of the spindle pole bodies (SPBs, the yeast equivalent of the centrosome) between mother and bud. Concomitantly, migration of the spindle to the bud neck and its alignment along the mother-bud axis ensure proper segregation of sister chromatids between the mother and the bud (reviewed in refs. 1–3). Two functionally overlapping pathways enforce the proper positioning of the spindle. First, the adenomatous polyposis coli-related protein Kar9 mediates the migration of the metaphase spindle to the bud neck and its alignment with the mother-bud axis together with the yeast EB1 protein Bim14-6 and the type V myosin Myo2.7 Second, the dynein-dependent pathway takes over at anaphase entry and promotes the elongation of the spindle along the mother-bud axis.8-10 In contrast to dynein, the Kar9-pathway mediates not only the alignment of the spindle with the division axis, but also ensures its orientation along this axis by directing the old spindle pole body toward the bud. Thereby, it promotes its inheritance by the daughter.11,12 Deciphering how the Kar9 pathway controls the position of the spindle in the cell, therefore, provides an excellent model for understanding the mechanisms of spindle rotation and centrosome inheritance during asymmetric cell divisions.
Through its interactions with Bim1 and Myo2, Kar9 links astral microtubules to and mediates their movement along actin cables.13,14 Importantly, Kar9 preferentially accumulates on the astral microtubules emanating from the old spindle pole body (SPB) to promote pulling of the spindle toward the future cleavage site, its alignment along the mother-bud axis and the orientation of the old SPB toward the bud.11,13,15 Therefore, the asymmetry of Kar9 distribution is crucial for the alignment of the spindle, while the orientation of this asymmetry toward the old SPB drives the non-randomness of SPB inheritance. However, how this asymmetry is achieved and regulated is not fully understood.
Previous studies have identified several players in the establishment of spindle asymmetry. First, Kar9 is phosphorylated by Cdk1, which acts in complex with the cyclin Clb413,16,12. Preventing phosphorylation leads to an increase of Kar9 levels on the astral microtubules associated with the new pole, and, hence, the spindle becomes more symmetrical. As a consequence, both SPBs have the tendency to orient toward the bud, and the spindle fails to stably align with the division axis. In addition to Clb4, the cyclin Clb5 has also been implicated in Kar9 asymmetry.17,18 Clb5 and Kar9 interact in a two-hybrid assay, and Kar9 was identified as an in vitro substrate of Cdk1/Clb5.19 However, Clb5 was not required for phosphorylation of Kar9 in vivo.13 Thus, how exactly Clb5 promotes Kar9 asymmetry is not fully understood.
Second, the mitotic exit network (MEN) and the SPB protein Nud1 promote the orientation of Kar9 asymmetry toward the old SPB.15 This regulation involves the direct phosphorylation of Kar9 by the redundant MEN kinases Dbf2 and Dbf20. In contrast to Cdk1/Clb4, MEN and Nud1 activity is correlated with SPB age and stabilizes Kar9 asymmetry specifically toward the old SPB. This bias for the old SPB facilitates proper SPB inheritance, and, accordingly, MEN and Nud1 mutant cells segregate SPBs more randomly.
Third, the spindle assembly checkpoint (SAC) negatively regulates Kar9 asymmetry.20 The central function of the SAC is to prevent anaphase as long as all kinetochores are not correctly attached to the spindle (reviewed in ref. 21). Accordingly, many kinetochore mutants arrest at the SAC. In these mutant cells, Kar9 localizes more symmetrically to both asters in a SAC-dependent manner.20 However, how the SAC controls Kar9 asymmetry is unknown. While MEN/Nud1 and Cdk1/Clb4 are extra-nuclear and directly target Kar9, the kinetochores and the components of the SAC reside inside the dividing nucleus, away from Kar9 (reviewed in ref. 22). How the SAC communicates with Kar9 is currently unknown. To gain insight into this question, we investigated how the SAC controls Kar9 localization on the metaphase spindle.
Results
The SAC functions independently of Cdk1/Clb4 in the control of Kar9 asymmetry
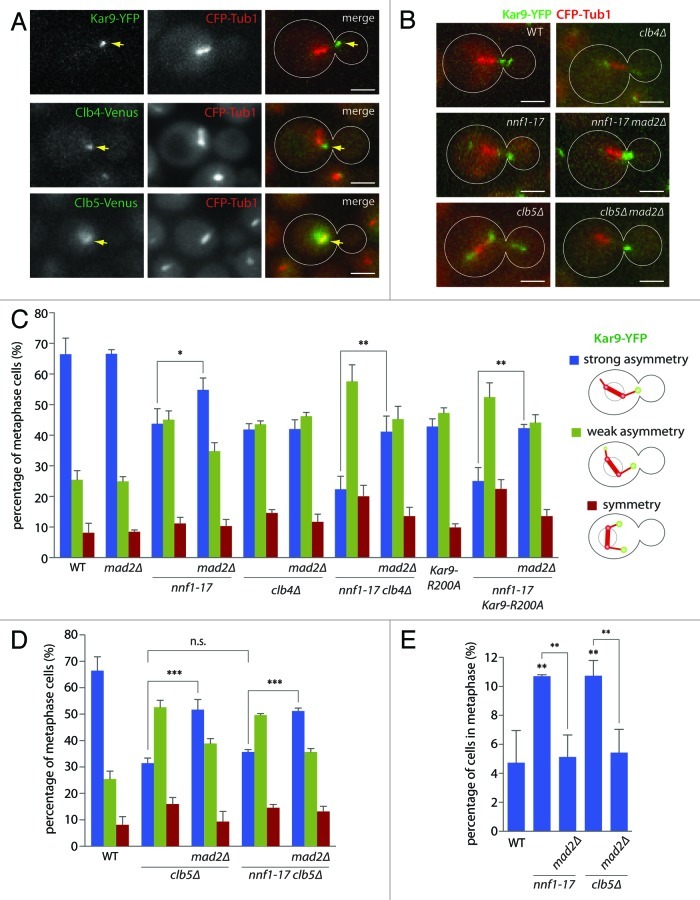
As reported,20 activation of the SAC by the kinetochore mutant nnf1–17 causes symmetric localization of Kar9 on metaphase spindles (Fig. 1B and C). This phenotype was measured as in ref 15: Cells are sorted in one of the following three classes: (1) “strong asymmetry” for cells carrying Kar9 exclusively on one side of the spindle throughout a 100s movie, (2) “symmetry,” when roughly equal amounts of Kar9 appeared on both sides of the spindle over the course of the movie, (3) “weak asymmetry” for intermediate cases. Shifting nnf1–17 mutant cells to the restrictive temperature led to a decrease in strongly asymmetric cells (43.8 ± 4.9% compared with 66.5 ± 5.3% in WT, p < 0.001). As described,20 the effect of the nnf1–17 mutation on Kar9 localization was partially suppressed by inactivating the SAC, using the mad2∆ mutation (54.8 ± 3.9% of strongly asymmetric nnf1–17 mad2∆ mutant cells, p < 0.05 compared with nnf1–17). The mad2∆ mutation on its own left Kar9 asymmetry unaffected (66.6 ± 1.4% of strong asymmetry). Together, these data confirm that activation of the SAC in kinetochore mutant cells causes a Kar9 asymmetry defect.
Figure 1. The SAC controls Kar9 asymmetry independently of Cdk1/Clb4, but in a Cdk1/Clb5-dependent manner. (A) Representative images of metaphase cells expressing Kar9-YFP, Clb4-Venus or Clb5-Venus together with CFP-Tub1. Scale bar is 2 µm. (B) Representative images of metaphase cells expressing Kar9-YFP in various mutant backgrounds. (C and D) Quantification of Kar9 asymmetry in nnf1–17 mutant background in combination with mutations affecting Cdk1/Clb4-dependent phosphorylation of Kar9 (C) or Cdk1/Clb5 activity (D). Cells coexpressed Kar9-YFP and CFP-Tub1 and were either MAD2 or mad2∆. Quantification as described in the text, cells were shifted to 37°C for 50 min before imaging. Stars indicate p-values obtained from one-way ANOVA comparing “strong asymmetry” (3 clones with total n > 115). Scale bar is 2 µm. (E) Quantification of metaphase arrest in nnf1–17 and clb5∆ cells in either MAD2 or mad2∆ background. Percentage of metaphase cells over the total number of cells was determined. Stars indicate p-values obtained from one-way ANOVA comparing to WT or as indicated (three clones with total n > 1100).
To better understand how the SAC impacts on Kar9 asymmetry, we tested whether it acted through one of the known pathways. To this end, we combined the nnf1–17 allele with mutations affecting the known regulators of Kar9. The cyclin-dependent kinase Cdk1 phosphorylates Kar9 and thereby promotes the asymmetric accumulation of Kar9 on the metaphase spindle. In this process, Cdk1 acts in complex with the cyclin Clb4, whereas Clb5 was not required for in vivo phosphorylation.13 As previously reported, both Kar9 and Clb4 localize to astral microtubules in metaphase cells,13,17 (Fig. 1A, top and middle panel), whereas Clb5 remains nuclear (Fig. 1A, bottom panel), until it disappears at the metaphase to anaphase transition.23 Interestingly, the Clb5-GFP signal was enriched along the metaphase spindle, consistent with reference 24. Despite their different localizations, both Clb4 and Clb5 have been implicated in the control of Kar9 asymmetry.12 Thus, we first tested whether one of these cyclins acted in the SAC-dependent pathway for Kar9 regulation.
First, we assessed whether the effect of the clb4∆ mutation was mediated by the SAC. Deleting MAD2 in clb4∆ mutant cells did not suppress the symmetric distribution of Kar9 (41.9 ± 1.9% vs. 42.0 ± 3.0% of strong asymmetry for clb4∆ and clb4∆ mad2∆ mutant cells, respectively, Fig. 1C), indicating that SAC did not mediate the effects of the clb4∆ mutation in Kar9 asymmetry. In addition, combination of the nnf1–17 and clb4∆ mutations caused additive effects on Kar9 distribution: only 22.4 ± 4.2% of the nnf1–17 clb4∆ mutant cells displayed strong Kar9 asymmetry, which was significant less than in the nnf1–17 and clb4∆ single mutant strains (43.8 ± 4.9% and 41.9 ± 1.9%, respectively, p < 0.001). Similarly, introducing a version of Kar9 that cannot be phosphorylated on Ser197 by Cdk1 (Kar9-R200A)15 caused additive effects with the nnf1–17 mutation alone (25.1 ± 4.4% strong asymmetry for nnf1–17 Kar9-R200A mutant cells). Finally, the inactivation of MAD2 in the nnf1–17 clb4∆ and the nnf1–17 Kar9-R200A double mutant cells partially restored Kar9 asymmetry (p < 0.01 between the double and the triple mutants), bringing it back to the level of symmetry observed in clb4∆ and Kar9-R200A single mutant cells. Thus, these data establish that Cdk1/Clb4 and SAC control Kar9 asymmetry independently of each other.
Disruption of the cyclin CLB5 caused a strong symmetry phenotype (31.5 ± 1.9% of cells showing strong asymmetry, p < 0.001 compared with WT, Fig. 1D). However, in contrast to the phenotype of the clb4∆ mutant cells, the effect of the clb5∆ mutation was significantly suppressed upon deletion of MAD2 (51.7 ± 3.8% of strongly asymmetric cells, p < 0.001). Thus, the effect of the clb5∆ mutation occurs at least partially through activation of the SAC. In agreement with this notion, introducing the nnf1–17 allele in clb5∆ mutant cells did not further decrease Kar9 asymmetry (36.7 ± 0.5% of strong asymmetry), and deletion of MAD2 largely restored asymmetry in this double mutant (51.2 ± 1.1% of strong asymmetry). Thus, our data suggest that Cdk1/Clb5 function is required to satisfy the SAC, and that this is at least part of the mechanism through which it contributes to the establishment of Kar9 asymmetry.
Since these data suggested that the clb5∆ mutation caused SAC activation, we tested this possibility further, and analyzed whether this mutation caused a SAC-dependent arrest prior to anaphase. Thus, we determined the percentage of nnf1–17 and clb5∆ mutant cells that accumulate in metaphase in an asynchronous population (Fig. 1E). Both mutant strains, showed a significant increase in metaphase cells compared with wild type cells (10.7 ± 0.1% and 10.7 ± 1.1 for nnf1–17 and clb5∆, respectively, compared with 4.7 ± 2.2% in WT, p < 0.01). Furthermore, in both cases this phenotype depended on Mad2 (5.1 ± 1.5% and 5.4 ± 1.6% for nnf1–17 mad2∆ and clb5∆ mad2∆, respectively, p < 0.01 compared with the single mutant cells). Thus, function of both Nnf1 and Clb5 is required for SAC satisfaction.
The SAC acts upstream of the MEN in metaphase
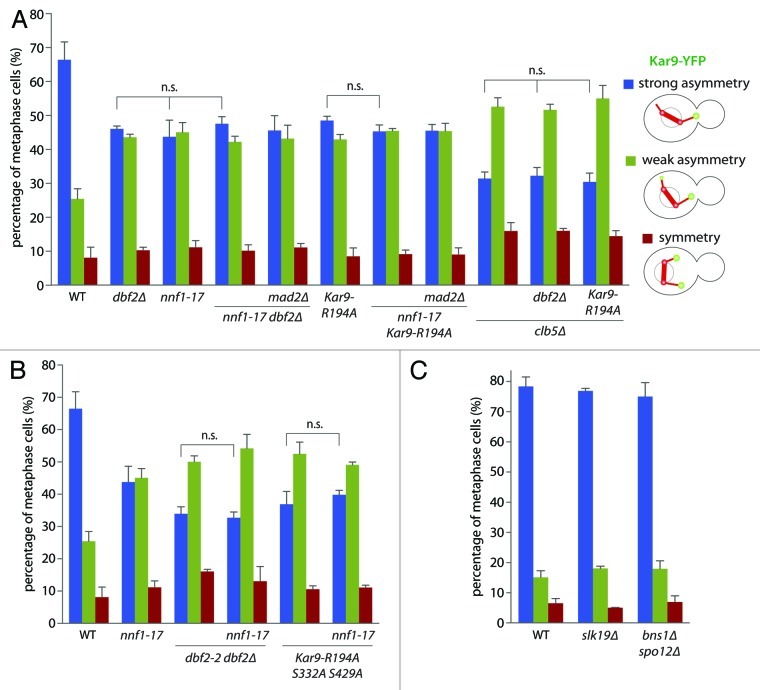
Next, we asked how the effects of SAC activation and MEN disruption relate to each other in the context of Kar9 regulation (Fig. 2A and B). Thus, we combined the nnf1–17 allele with the dbf2∆ mutation, which disrupts the MEN kinase Dbf2, the dbf2–2 dbf2∆ double mutation, which disrupts both Dbf kinases, and with a mutant form of Kar9 that cannot be phosphorylated by Dbf2 and Dbf20 on Ser197 (KAR9-R194A). In all three cases, no additive effects were observed upon adding the nnf1–17 mutation (47.6 ± 2.1% and 45.4 ± 1.9% strong asymmetry in nnf1–17 dbf2∆ and nnf1–17 KAR9-R194A respectively vs. 43.8 ± 4.9% in nnf1–17). The same observation was made in an nnf1–17 Kar9-R194A S332A S429A mutant strain, where all Dbf consensus-sites are mutated. These data indicate that SAC activation and MEN inactivation affect Kar9 asymmetry through the same mechanism, and hence that they may function in the same genetic pathway.
Figure 2. The SAC inhibits the MEN during metaphase. (A) and (B) Cells expressing the nnf1–17 allele in combination with mutations affecting Dbf2-dependent phosphorylation of Kar9. Cells coexpressed Kar9-YFP and CFP-Tub1 and were either MAD2 or mad2∆, as well as CLB5 or clb5∆. All quantifications were performed as in Figure 1C, cells were shifted to 37°C for 50 min before imaging. Stars indicate p-values obtained from one-way ANOVA comparing “strong asymmetry” (three clones with total n > 130). (C) Analysis of Kar9 asymmetry in FEAR mutant cells at 22°C as in (A).
To determine whether SAC acts upstream or downstream of the MEN, we next investigated the effect of the mad2∆ mutation in this context (Fig. 2A). If the SAC acts downstream of the MEN, the mad2∆ mutation would restore Kar9 asymmetry in the nnf1–17 dbf2∆ and nnf1–17 KAR9-R194A mutant cells. However, this was not the case (45.6 ± 4.4% of strong asymmetry in nnf1–17 dbf2∆ mad2∆ and 45.5 ± 1.9% in nnf1–17 KAR9-R194A mad2∆ cells). Together with the fact that deleting MAD2 did not restore Kar9 asymmetry in MEN mutants,15 we conclude that in metaphase, SAC controls Kar9 distribution by acting upstream of MEN, which is inhibited in response to kinetochore defects. Importantly, Kar9 asymmetry was less affected in nnf1–17 mutant cells compared with cells carrying mutations that fully disrupt MEN activity.15 Thus, we conclude that either the SAC only partially inhibits MEN activity, or the nnf1–17 allele does not fully activate the SAC.
Since both MEN and Clb5 functioned in the same pathway as the SAC, we next introduced the clb5∆ mutation in cells carrying either dbf2∆ or KAR9-R194A and assessed Kar9 asymmetry in these cells (Fig. 2A). No phenotypic enhancement was observed in the dbf2∆ clb5∆ and KAR9-R194A clb5∆ double mutant compared with the clb5∆ single mutant cells (32.3 ± 2.4% and 30.5 ± 2.6% strong asymmetry in clb5∆ dbf2∆ and clb5∆ Kar9-R194A, respectively, vs. 31.5 ± 1.9%). Thus, together our data indicate that Clb5 acts in the control of Kar9 asymmetry by promoting SAC satisfaction, thereby releasing the MEN from SAC inhibition.
One likely candidate for mediating the inhibitory effect of the SAC on the MEN is the Cdc14 early anaphase release network (FEAR, reviewed in ref. 25). The FEAR consists of the yeast separase Esp1 and the downstream acting factors Slk19 and Spo12/Bns1 and promotes cell cycle progression at the metaphase-to-anaphase transition. Since Esp1 is subject to SAC-dependent inhibition, we wondered whether mutations of FEAR components displayed the same phenotypes as SAC-activating mutations. However, neither slk19∆ nor bns1∆ spo12∆ mutant cells displayed any defects in Kar9 asymmetry (76.9 ± 0.9% and 75.1 ± 4.6%, respectively, compared with 78.4 ± 3.2% in WT, Fig. 2C). Thus, the SAC-dependent inhibition of the FEAR does not seem to play a role in the control of Kar9 distribution.
Inhibition of the MEN by the SAC perturbs SPB segregation
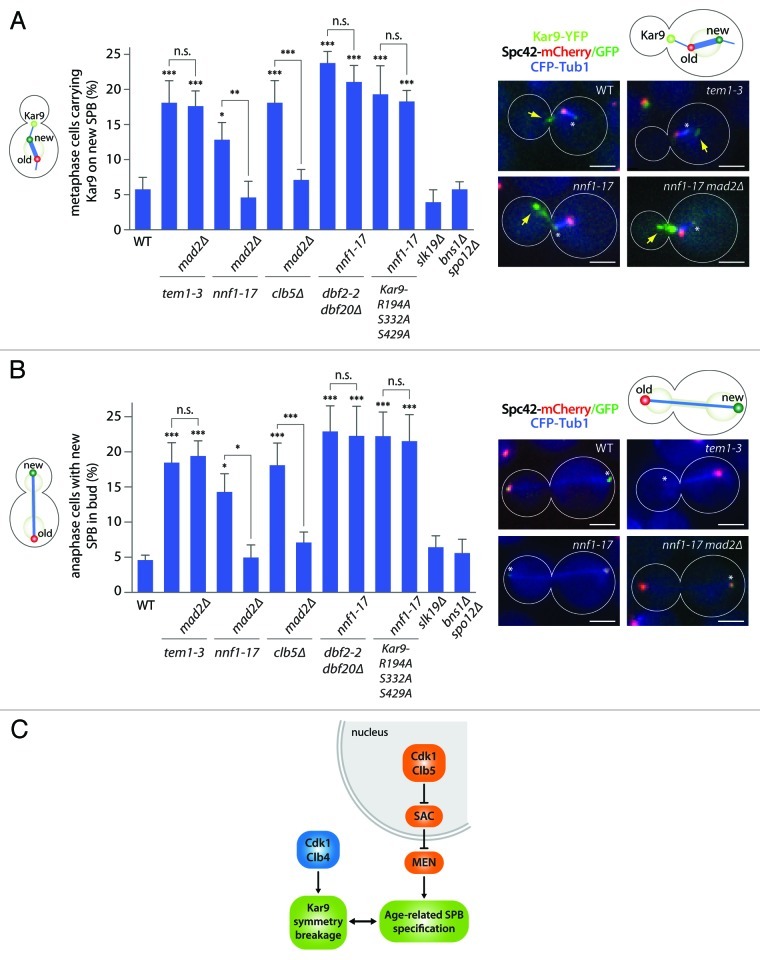
As shown previously, the MEN controls SPB specification and inheritance through the regulation of Kar9 localization.15 Cells expressing mutant forms of MEN proteins orient Kar9 distribution more randomly on the metaphase spindle with respect to SPB age, i.e., many cells carry more Kar9 on astral microtubules of the new SPB than on those of the old SPB. Since our results implicate Clb5 and the SAC in the control of MEN activity, we wondered whether SAC activation also interfered with the specification of SPBs. Therefore, we took advantage of a recombinase-based, tag-exchange reporter15 to determine Kar9 distribution on astral microtubules associated with the old and new SPBs upon SAC activation. In metaphase cells, 12.8 ± 2.5% of the nnf1–17 mutant cells showed more Kar9 on astral microtubules emanating from the new SPB, compared with 5.8 ± 1.7% in wild type cells (p < 0.05), while the tem1–3 mutant cells showed a slightly stronger defect (18.1 ± 3.1%, Fig. 3A). Unlike in tem1–3 cells, the phenotype of the nnf1–17 mutant allele was suppressed by the mad2∆ mutation (4.6 ± 2.3% for nnf1–17 mad2∆ mutant cells, p < 0.01 compared with nnf1–17). Similarly, the clb5∆ mutation caused mis-orientation of Kar9 asymmetry (18.1 ± 3.2%), and this phenotype depended on Mad2 (7.1 ± 1.5% for clb5∆ mad2∆ vs. 17.6 ± 2.2% for tem1–3 mad2∆ mutant cells). Therefore, the nnf1–17 and clb5∆ mutations cause SAC-dependent defects in SPB specification. In agreement with the idea that SAC affects Kar9 distribution through the MEN, adding the nnf1–17 mutation to the dbf2–2 dbf20∆ or the Kar9-R194A S332A S429A mutant cells did not further randomize Kar9 orientation (21.1 ± 2.3% and 18.3 ± 1.6% for nnf1–17 dbf2–2 dbf20∆ and nnf1–17 Kar9-R194A S332A S429A mutant cells compared with 23.8 ± 1.6% and 19.3 ± 4.1% in the respective NNF1 strains). Again, mutating the FEAR components Slk19 and Bns1/Spo12 did not affect Kar9 orientation (Fig. 2C). Together, we conclude that activation of the SAC in response to the nnf1–17 or clb5∆ mutations inhibits the MEN in metaphase and leads to SPB specification defects. This response was independent of FEAR function.
Figure 3. SAC activation perturbs faithful segregation of the old SPB to the bud. (A) Metaphase cells expressing nnf1–17, tem1–3 or deletions of the cyclins CLB4 and CLB5 in combination with either MAD2 or mad2∆. In addition, cells expressing dbf2–2 dbf20∆ or Kar9-R194A S332A S429A in either NNF1 or nnf1–17 background as well as FEAR mutant cells were analyzed. Cells coexpressed a switchable mCherry/GFP-tag allowing distinction of old and new SPB, as well as Kar9-YFP and CFP-Tub1. Stars indicate p-values obtained from one-way ANOVA comparing to WT or as indicated (three clones with total n > 125). Asterisks mark the new SPB, yellow arrows indicate Kar9 fluorescence. Scale bar is 2 µm. (B) Same strains and conditions as for (A), but anaphase cells were imaged. Three clones with total n > 87 cells. (C) Schematic drawing of the pathways controlling Kar9 asymmetry and SPB specification in metaphase.
Next, we tested the consequence of the randomized orientation of Kar9 asymmetry on SPB inheritance by determining the fate of the SPBs at anaphase (Fig. 3B). Compared with wild type, where very few daughter cells inherited the new SPB (4.6 ± 0.7% of anaphase cells), 14.3 ± 2.6% of the nnf1–17 and 19.0 ± 2.9% of the clb5∆ mutant cells displayed this phenotype (p < 0.01 and p < 0.001, respectively, compared with WT). In both strains, this phenotype was suppressed by the mad2∆ mutation: 5.0 ± 1.8% and 5.7 ± 2.2% of anaphase cells segregated the new SPB to the bud in nnf1–17 mad2∆ and clb5∆ mad2∆ double mutant cells (p < 0.01 and p < 0.001, respectively, compared with nnf1–17 and clb5∆ alone). For comparison, 18.5 ± 2.8% of tem1–3 mis-segregated SPBs, and this effect did not depend on Mad2 (19.4 ± 2.2% of tem1–3 mad2∆ mutant cells). In addition, expressing nnf1–17 in dbf2–2 dbf20∆ or Kar9-R194A S332A S429A mutant cells did not further perturb SPB inheritance (22.3 ± 4.2% and 21.5 ± 3.8% for nnf1–17 dbf2–2 dbf20∆ and nnf1–17 Kar9-R194A S332A S429A mutant cells compared with 22.9 ± 3.7% and 22.2 ± 3.4% in the respective NNF1 strains). Inhibition of the FEAR had no effect on SPB segregation. Thus, our data establish that Clb5 and the SAC act upstream of the MEN to control Kar9 distribution and proper SPB inheritance.
Discussion
Curiously, SAC activation affects the distribution of the spindle positioning factor Kar9,20 indicating that the active SAC induces a signal, which, in addition to preventing anaphase onset, reaches into the cytoplasm. Our data establish that the SAC acts in parallel to the Cdk1/Clb4 complex, which promotes Kar9 asymmetry by directly phosphorylating it, and through inhibition of the MEN. How the SAC inhibits the MEN pathway remains to be discovered.
Together, these observations confirm the idea that MEN controls the age-related specification of the SPBs in metaphase cells, while Cdk1/Clb4 acts independently of SPB age in the breakage of Kar9 symmetry.15 In metaphase cells carrying the kinetochore mutant allele nnf1–17, Kar9 asymmetry was more frequently mis-oriented, and Kar9 accumulated with a lower preference for the old SPB. This phenotype required Mad2 function and was perfectly mimicked by mutations preventing Kar9 phosphorylation by the MEN. Kar9 mis-orientation caused also the mis-segregation of the new SPB to the bud. Thus, together with the finding that the MEN mediates SAC effects, our data provide new evidence for MEN acting on SPB-specification during metaphase. In contrast, cells lacking Clb4 do not display any such SPB age-related phenotypes.15
Next to its direct role in complex with the cyclin Clb4, Cdk1 also acts together with Clb5 in the control of Kar9 asymmetry.17,18 While Clb4 is required for in vivo phosphorylation of Kar9, it remained unclear how Clb5 acts in this process. Consistent with its nuclear localization, our data suggest that Clb5 function is required for the satisfaction of the SAC and therefore regulates Kar9 distribution mostly indirectly (Fig. 1D). Accordingly, clb5∆ mutant cells displayed mis-oriented Kar9 asymmetry with respect to SPB age and randomized SPB inheritance, and both these effects were Mad2-dependent (Fig. 3A and B). To our knowledge, this is the first evidence that Clb5 contributes to SAC satisfaction in yeast. Several mechanisms may account for this observation. First of all, the delay in DNA replication caused by the clb5∆ mutation26 may cause a delay in the duplication of the centromere region and, hence, in kinetochore duplication and bipolar attachment. However, it is also tempting to speculate that Clb5 might directly contribute either to the proper regulation of kinetochore function or in the control of SAC silencing. Clb5 enrichment in the spindle region supports these two last possibilities, and testing them will be an interesting topic for future studies.
Deletion of CLB5 led to a defect that was more pronounced than the expression of the kinetochore mutant allele nnf1–17, and comparable to the effect of tem1–3 (Fig. 1D).15 This suggests that the clb5∆, but not the nnf1–17, mutation leads to full activation of the SAC and to complete inhibition of the MEN. However, while disruption of Mad2 in cells carrying the clb5∆ mutation fully suppressed the defect in Kar9 orientation and SPB inheritance (Fig. 3A and B), it did not completely restore the asymmetry of Kar9 (Fig. 1D). Furthermore, the enrichment for metaphase cells was equally strong for nnf1–17 as for clb5∆ mutant cells (Fig. 1E). This suggests that the clb5∆ mutation also has a SAC-independent effect on Kar9 asymmetry. This additional role of Clb5 promotes Kar9 asymmetry irrespective of SPB age, since the specific enrichment of Kar9 on the old SPB and SPB inheritance were completely restored by the mad2∆ mutation. As suggested by others, this role could involve direct targeting of Kar9 by the Cdk1/Clb5 complex16,12 in addition to its function in SAC satisfaction.
Our study describes a pathway, by which the SAC controls the distribution of Kar9 on the metaphase spindle (Fig. 3C). We identify the MEN as the primary target of the SAC in the regulation of Kar9 asymmetry. The relevance of this mechanism might be to delay cell cycle progression upon kinetochore-microtubule attachment defects not only at the metaphase-to-anaphase transition, but also by inhibiting mitotic exit. Alternatively, it is also tempting to speculate that MEN inhibition by the SAC coordinates the dynamics of both astral microtubules and the spindle with each other. Kar9 redistribution could lead to enhanced spindle movements,13 which may, in turn, facilitate the retrieval of detached and lost chromosomes by the spindle. In summary, the observation that the behavior of astral components is tightly coordinated with events happening inside the spindle was not anticipated, and suggests that the spindle functions much more as an integrated entity than previously thought.
Finally, our study suggests that SAC satisfaction might not be the limiting step to allow the metaphase-to-anaphase transition in budding yeast. Indeed, in wild type cells Kar9 asymmetry is established early in metaphase and maintained relatively robustly throughout the duration of metaphase, suggesting that SAC is already satisfied during all that time. The fact that anaphase is triggered long after establishment of Kar9 asymmetry indicates that in budding yeast, other events, such as the completion of DNA replication, might determine the timing of anaphase onset more than SAC satisfaction. Thus, our data provide also useful insights and tools for future studies about the timing of SAC satisfaction and about what actually determines the timing of the metaphase-to-anaphase transition in yeast.
Materials and Methods
Strains and growth conditions
Media and genetic methods as described.27 Yeast strains are listed in Table S1, all strains derived from S288C or were backcrossed 3×. Fluorescently labeled proteins were tagged at their endogenous loci.28 CFP-Tub1 was inserted at the URA3 locus and gdp:Cre-EBD78 at the LEU2 locus. The switchable mCherry/GFP-cassette was constructed as described.15 Kar9 alleles were constructed by site-directed mutagenesis (pfu-Turbo, Stratagene) on a pRS314 plasmid, amplified by PCR, and after integration verified by PCR and sequencing. Deletions were performed as described.28 Fluorophore switch was induced with estradiol 4 h before imaging.
Fluorescence microscopy
100s time-lapse microscopy was performed using an Olympus BX51 microscope, TILLVision software (TILLPhotonics). For the localization of GFP-, YFP- and CFP-labeled proteins, Z-stacks of five layers (step size 0.3 µm), and maximum projections were used. Fluorescence microscopy was performed with a monochromator PolychromIV as light source and a CCD camera (Imago, TillPhotonics). Images were analyzed with ImageJ.
Supplementary Material
Acknowledgments
We thank P. Meraldi for critical discussions, G. Euskirchen for strains and reagents and the Light Microscopy Centre of the ETH Zurich for technical support. M.H., J.L. and Y.B. were supported by ETHZ and grants from the Swiss National Science Foundation and the European Research Council to Y.B.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21504
References
- 1.Kusch J, Liakopoulos D, Barral Y. Spindle asymmetry: a compass for the cell. Trends Cell Biol. 2003;13:562–9. doi: 10.1016/j.tcb.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–74. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- 3.Barral Y, Liakopoulos D. Role of spindle asymmetry in cellular dynamics. Int Rev Cell Mol Biol. 2009;278:149–213. doi: 10.1016/S1937-6448(09)78004-9. [DOI] [PubMed] [Google Scholar]
- 4.Miller RK, Cheng SC, Rose MD. Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol Biol Cell. 2000;11:2949–59. doi: 10.1091/mbc.11.9.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee L, Tirnauer JS, Li J, Schuyler SC, Liu JY, Pellman D. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science. 2000;287:2260–2. doi: 10.1126/science.287.5461.2260. [DOI] [PubMed] [Google Scholar]
- 6.Korinek WS, Copeland MJ, Chaudhuri A, Chant J. Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science. 2000;287:2257–9. doi: 10.1126/science.287.5461.2257. [DOI] [PubMed] [Google Scholar]
- 7.Yin H, Pruyne D, Huffaker TC, Bretscher A. Myosin V orientates the mitotic spindle in yeast. Nature. 2000;406:1013–5. doi: 10.1038/35023024. [DOI] [PubMed] [Google Scholar]
- 8.Carminati JL, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–41. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol. 2000;149:863–74. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grava S, Schaerer F, Faty M, Philippsen P, Barral Y. Asymmetric recruitment of dynein to spindle poles and microtubules promotes proper spindle orientation in yeast. Dev Cell. 2006;10:425–39. doi: 10.1016/j.devcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 11.Pereira G, Tanaka TU, Nasmyth K, Schiebel E. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 2001;20:6359–70. doi: 10.1093/emboj/20.22.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore JK, D’Silva S, Miller RK. The CLIP-170 homologue Bik1p promotes the phosphorylation and asymmetric localization of Kar9p. Mol Biol Cell. 2006;17:178–91. doi: 10.1091/mbc.E05-06-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liakopoulos D, Kusch J, Grava S, Vogel J, Barral Y. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell. 2003;112:561–74. doi: 10.1016/S0092-8674(03)00119-3. [DOI] [PubMed] [Google Scholar]
- 14.Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, et al. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–4. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- 15.Hotz M, Leisner C, Chen D, Manatschal C, Wegleiter T, Ouellet J, et al. Spindle pole bodies exploit the mitotic exit network in metaphase to drive their age-dependent segregation. Cell. 2012;148:958–72. doi: 10.1016/j.cell.2012.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maekawa H, Usui T, Knop M, Schiebel E. Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J. 2003;22:438–49. doi: 10.1093/emboj/cdg063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maekawa H, Schiebel E. Cdk1-Clb4 controls the interaction of astral microtubule plus ends with subdomains of the daughter cell cortex. Genes Dev. 2004;18:1709–24. doi: 10.1101/gad.298704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore JK, Miller RK. The cyclin-dependent kinase Cdc28p regulates multiple aspects of Kar9p function in yeast. Mol Biol Cell. 2007;18:1187–202. doi: 10.1091/mbc.E06-04-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–8. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- 20.Leisner C, Kammerer D, Denoth A, Britschi M, Barral Y, Liakopoulos D. Regulation of mitotic spindle asymmetry by SUMO and the spindle-assembly checkpoint in yeast. Curr Biol. 2008;18:1249–55. doi: 10.1016/j.cub.2008.07.091. [DOI] [PubMed] [Google Scholar]
- 21.Lew DJ, Burke DJ. The spindle assembly and spindle position checkpoints. Annu Rev Genet. 2003;37:251–82. doi: 10.1146/annurev.genet.37.042203.120656. [DOI] [PubMed] [Google Scholar]
- 22.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–93. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 23.Shirayama M, Tóth A, Gálová M, Nasmyth K. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature. 1999;402:203–7. doi: 10.1038/46080. [DOI] [PubMed] [Google Scholar]
- 24.Huisman SM, Smeets MF, Segal M. Phosphorylation of Spc110p by Cdc28p-Clb5p kinase contributes to correct spindle morphogenesis in S. cerevisiae. J Cell Sci. 2007;120:435–46. doi: 10.1242/jcs.03342. [DOI] [PubMed] [Google Scholar]
- 25.Rock JM, Amon A. The FEAR network. Curr Biol. 2009;19:R1063–8. doi: 10.1016/j.cub.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwob E, Böhm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–44. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 27.Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology (New York: Academic Press). 1991. [Google Scholar]
- 28.Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, et al. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15(10B):963–72. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.