Abstract
Cln1 and Cln2 are very similar but not identical cyclins. In this work, we tried to describe the molecular basis of the functional distinction between Cln1 and Cln2. We constructed chimeric cyclins containing different fragments of Cln1 and Cln2 and performed several functional analysis that make it possible to distinguish between Cln1 or Cln2. We identified that region between amino acids 225 and 299 of Cln2 is not only necessary but also sufficient to confer Cln2 specific functionality compared with Cln1. We also studied Cln1 and Cln2 subcellular localization identifying additional differences between them. Both cyclins are distributed between the nucleus and the cytoplasm, but Cln1 shows stronger nuclear accumulation. Nuclear import of both cyclins is mediated by the classical nuclear import pathway and by sequences in the N-terminal end of the proteins. For Cln2, but not for Cln1, a nuclear export mechanism mediated by karyopherin Msn5 has been identified. Strikingly, Cln2 export depends on a Msn5-dependent NES between amino acids 225 and 299. In fact, the introduction of this region confers to Cln1 an export mechanism dependent on Msn5; importantly, this causes the gain of Cln2-specific cytosolic functions and the impairment of nuclear function. In short, a region from Cln2 controlling an Msn5-dependent nuclear export mechanism confers a specific functionality to Cln2 compared with Cln1.
Keywords: Cln1, Cln2, Msn5, cell cycle, cyclins
Introduction
In S.cerevisiae, a single cyclin-dependent kinase (CDK), Cdc28, is associated with nine different cyclins to drive progression through the cell cycle: G1/S cyclins Cln1–3, S-phase cyclins Clb5–6 and G2/M cyclins Clb1–4. Cyclins basically play two major functions: activating the CDK and conferring a specific functionality to the CDK by a combination of factors, including the periodic expression and degradation of cyclins, interactions with specific substrates and inhibitors and the control of the subcellular location of the kinase.
Different cyclins play different roles in cell cycle regulation (although extended functional redundancy exists between some of them). This functional distinction could be due to quantitative or intrinsic qualitative causes. Quantitative differences are given basically by a difference in the time of the expression and/or degradation. For instance, the functional differences between Clb5 and Clb6 in the regulation of DNA replication are simply due to the quantitative differences caused by a distinct degradation mechanism.1 In other cases, the functional differences between cyclins also reflect qualitative differences. For instance, when expressed under the control of the CLB5 promoter, the mitotic Clb2 cyclin is able to control the S phase but not as efficiently as Clb5 does. This is due to intrinsic differences between these cyclins in the recognition of S phase substrates2-4 and inhibition by the Swe1 kinase.5,6
Functional differences have also been described for the G1 cyclins. The Cln1, Cln2 and Cln3 cyclins are responsible for the regulation of Start, a major control point at the G1/S transition, when yeast cells decide whether or not to initiate a new round of cell division.7 Execution of Start consists in the activation of a transcriptional program, which implies the coordinated expression of a large number of genes involved in budding, spindle pole body duplication and DNA replication, the events that are activated at this initial cell cycle step.8,9 Cln1, Cln2 and Cln3 play different roles in cell cycle initiation. Cln3 is involved in the initial activation of the transcriptional program. As a result of this transcriptional wave, kinases Cln1-Cdc28 and Cln2-Cdc28 accumulate and act to maintain the transcriptional program active and to trigger Start events. This functional distinction is primarily due to differences in the time of the expression of the cyclins: Cln3 is the only cyclin present in G1, whereas Cln1 and Cln2 are expressed as part of the Start transcriptional wave. However, intrinsic differences between these cyclins also exist, because Cln2 and Cln3 differ even when both are expressed under the control of the CLN3 promoter.10 One important factor that determines the function of a cyclin-Cdc28 kinase is its localization, and this is highlighted for Cln2 and Cln3, because the localization pattern contributes to Cln2 and Cln3′s ability to affect different processes. The Cln3-Cdc28 complex is associated with the endoplasmic reticulum membrane until late G1, when it enters the nucleus to activate transcription.11 The introduction of a nuclear export signal (NES) into Cln3 renders the Cln3-Cdc28 complex unable to trigger cell cycle initiation. As regards Cln2, it is present in both the nucleus and the cytoplasm. The forced location of the cyclin has allowed us to distinguish between functions that require a nuclear or a cytoplasmic localization of Cln2-Cdc28. Thus, nuclear Cln2 might be needed in processes like DNA synthesis or SPB replication, whereas the role in budding involves a cytosolic location of the cyclin. However, some Cln2 functions, like the rescuing of the lethality of a swi4 swi6 mutant strain (Swi4 and Swi6 are components of the SBF transcription factor of the Start transcriptional program), could imply the existence of shuttling between the nucleus and the cytosol.12-14
Cln1 and Cln2 are the most similar cyclins in yeast. They show a 57% sequence identity (72% in the N-terminal region). Cln1 and Cln2 and their associated kinase activity are strongly periodic, with a peak during the G1/S transition.15,16 The levels of both proteins are controlled by the same molecular mechanisms: the CLN1 and CLN2 genes are expressed periodically during the G1/S transition by the transcription factor SBF,17-19 and the Cln1 and Cln2 proteins are degraded via the SCFGrr1 ubiquitin-ligase.20-22 Compared with single mutants, the cln1 cln2 double mutant shows severe growth defects,23,24 indicating a functional overlap between these two cyclins. In fact, the extensive work performed on these cyclins has revealed that they take part in many common functions, such as induction of growth polarization and budding,25-27 SPB duplication,28 degradation of CDK-inhibitors Far1 and Sic1,29-32 inactivation of the APC ubiquitin-ligase complex,33,34 repression of pheromone-induced transcription or stimulation of pseudohyphal growth.35 However, in addition to the numerous studies that highlight the similarity between Cln1 and Cln2, several functional differences between them have also been described. Thus, Cln1 is more important than Cln2 in adapting cell size to new carbon sources36 and pseudohyphal development,37,38 but these functional distinctions seem to be caused by quantitative differences in gene expression. On the other hand, meiosis is blocked more efficiently in the presence of Cln2 than in Cln1,39 and Cln2 plays the primary role in the morphogenetic processes leading to budding during the G1/S transition.40 More recently, cyclin-specific docking motifs that bind preferentially to Cln2 have been described in signaling proteins, such as Ste5 and Ste20.41 In this work, we attempt to describe new molecular mechanisms contributing to the functional distinction between Cln1 and Cln2.
Results
The region between amino acids 225 and 299 of Cln2 is necessary to confer specific functionality
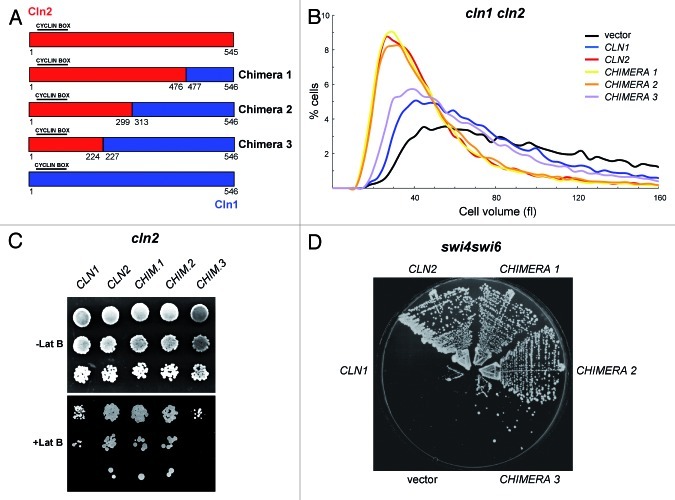
In a first attempt to characterize the mechanism underlining the functional distinction between the Cln1 and Cln2 cyclins, we searched for elements in their protein sequence that are responsible for such specificity. The approach consisted in the construction of chimeric proteins designed to substitute the equivalent regions between the cyclins based on sequence alignment (Fig. S1). Initially, three chimeras were constructed by exchanging increasing regions from the C-terminal end of Cln2 for the corresponding fragments of Cln1 (Fig. 1A). All the constructed chimeras retained cyclin activity, as deduced from the recovery of the severe growth defect of a cln1 cln2 mutant strain (Fig. S2). Next, we tested whether the proteins presented Cln1- or Cln2-specific functionality by means of three different experimental approaches. First, cell size was analyzed in cultures of the cln1 cln2 mutant cells expressing the different chimeras. The cln1 cln2 mutant cells were larger than normal. The introduction of Cln1, Cln2 or any of the three chimeras led to a reduction in cell size, thus confirming the functionality of the chimeric proteins, but, more importantly, differences in the effect of the different cyclins were detected (Fig. 1B). It was known that the cln2 mutant, but not the cln1 mutant, showed increased cell size (Fig. S3A).40 In agreement with this, introduction of Cln2 allowed the cln1 cln2 mutant cells to recover a normal size similar to that of the wild-type cells, whereas the cln1 cln2 cells expressing Cln1 were still significantly larger. As regards the chimeric cyclins, the expression of chimeras 1 and 2 caused an identical cell size reduction to that caused by Cln2, suggesting that there were no functional differences between these chimeras and Cln2. However, the effect observed with chimera 3 was milder than that observed with Cln2 and resembled more the effect obtained with Cln1. Hence the specific function of Cln2 related to cell size control was impaired in chimera 3. Similar results were observed when cyclins were introduced into a cln2 mutant strain (Fig. S3B).
Figure 1. Functional analisys of chimeric cyclins 1–3. (A) Diagram of chimeric cyclins 1, 2 and 3. All cyclins are HA-epitope tagged at the C terminus. Expression is driven by the promoter corresponding to the cyclin at the N-terminal end. (B) Cell size distribution in exponentially growing cultures of the cln1 cln2 mutant strain (JCY847) transformed with an empty vector or a centromeric plasmid containing the CLN1, CLN2, CHIMERA1, CHIMERA2 or CHIMERA3 gene. (C) 10-fold serial dilutions from exponentially growing cultures of the cln2 mutant strain (JCY846) transformed with an empty vector or a centromeric plasmid containing the CLN1, CLN2, CHIMERA1, CHIMERA2 or CHIMERA3 gene were spotted onto YPD medium and YPD medium supplemented with 25 μM latrunculin B and incubated at 28° for 3 d. (D) Cells of the swi4ts swi6 mutant strain (K2003) transformed with an empty vector or a centromeric plasmid containing the CLN1, CLN2, CHIMERA1, CHIMERA2 or CHIMERA3 gene were streaked onto YPD plates and incubated at 35° for 3 d.
A second approach to test Cln2-specific functionality consisted in analyzing the sensitivity to latrunculin B of the cln2 mutant strain transformed with the plasmids containing the cyclins genes. The cln2 mutant, compared with the cln1 mutant, presents hypersensitivity to the perturbation of the actin cytoskeleton by latrunculin B.40 Therefore, the expression of Cln2, but not of Cln1, allowed the cln2 mutant cells to overcome hypersensitivity to latrunculin B (Fig. 1C). The expression of those chimeras that function as well as Cln2 was expected to allow the cln2 mutant cells to properly grow in the presence of the drug. This was the case for chimeras 1 and 2. However, similarly to that observed for Cln1, the cells containing chimera 3 still manifested greater sensitivity to the drug than the cells expressing Cln2 or chimera 1 or 2. These results reinforce the idea that Cln2-specific functionality is affected in chimera 3.
Finally, the ability of the chimeric cyclins to suppress the lethality of the Start mutant swi4ts swi6 was studied. This strain was unable to grow at 35°. This lethality was suppressed by a centromeric plasmid bearing the CLN2, the CHIMERA1 or the CHIMERA2 gene, but not by a plasmid bearing the CLN1 or the CHIMERA3 gene (Fig. 1D). In short, the analysis of cyclin functionality by three different approaches was consistent and allowed us to state that the chimeras 1 and 2 cyclins displayed a specific functionality characteristic of Cln2, whereas the chimera 3 cyclin had lost this specific function, at least partially. Taking into account how the chimeras had been constructed, we can conclude that the region between amino acids 225 and 299 of the Cln2 cyclin is necessary for its specific function.
The region between amino acids 225 and 299 of Cln2 is sufficient to confer Cln2-specific functionality to Cln1
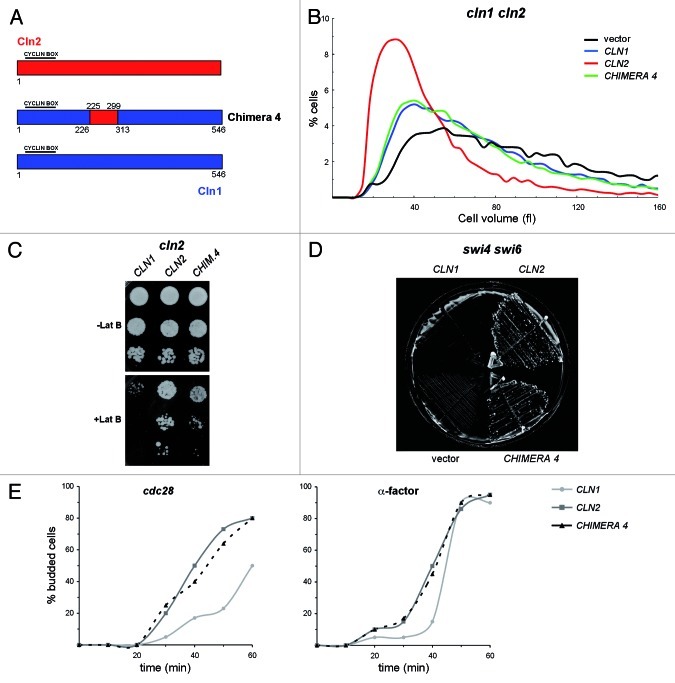
In order to investigate whether the introduction of the 225–299 region of Cln2 into Cln1 would be sufficient to confer Cln2-specific functionality to the cyclin, the chimera 4 was constructed (Fig. 2A). Whether chimera 4 behaved as Cln2 or Cln1 was tested by the same functionality assays described above. The cln2 and cln1 cln2 cells transformed with chimera 4 have a larger than normal cell size, very similar to that observed with Cln1 (Fig. 2B; Fig. S3C). Thus, chimera 4 was unable to properly control cell size. Nevertheless, the introduction of chimera 4 into the cln2 mutant cells improved resistance to latrunculin B (Fig. 2C). Moreover, chimera 4 perfectly suppressed the lethality of the swi4ts swi6 mutant at the restrictive temperature (Fig. 2D). These results demonstrate that region 225–299 of Cln2 was indeed sufficient to confer Cln2-specific functions to Cln1. The fact that the chimera functionality type differed according to the assay probably reflects that the functional differences between Cln2 and Cln1 involve diverse mechanisms, and that the 225–299 region of Cln2 is responsible for most of, but not all, the Cln2-specific functions.
Figure 2. Functional analisys of chimeric cyclin 4. (A) Diagram of chimeric cyclin 4. All cyclins are HA-epitope tagged at the C terminus. Expression is driven by the promoter corresponding to the cyclin at the N-terminal end. (B) Cell size distribution in exponentially growing cultures of the cln1 cln2 mutant strain (JCY847) transformed with an empty vector or a centromeric plasmid containing the CLN1, CLN2 or CHIMERA4 gene. (C) 10-fold serial dilutions from exponentially growing cultures of the cln2 mutant strain (JCY846) transformed with an empty vector or a centromeric plasmid containing the CLN1, CLN2 or CHIMERA4 gene were spotted onto YPD medium and YPD medium supplemented with 25 μM latrunculin B and incubated at 28° for 3 d. (D) Cells of the swi4ts swi6 mutant strain (K2003) transformed with an empty vector or a centromeric plasmid containing the CLN1, CLN2 or CHIMERA4 gene were streaked onto YPD plates and incubated at 37° for 3 d. (E) Exponentially growing cultures of the cdc28ts cln1 cln2 (JCY1048) and the cln1 cln2 (JCY847) mutant strains transformed with a centromeric plasmid containing the CLN1, CLN2 or CHIMERA4 gene were arrested at the G1 phase by incubation at 37° or in the presence of 5 μg/mL α-factor for 3 h, respectively. The budding index was determined at the indicated time after release from cell cycle arrest.
To further investigate the capability of the 225–299 fragment of Cln2 to confer a specific functionality, an additional functional assay was performed. Cln2 is the major regulator of budding, whereas Cln1 is only relevant when Cln2 is not present; for this reason, inactivation of Cln2, but not of Cln1, results in a specific delay in budding initiation.40 Budding kinetics was analyzed in the cln1 cln2 mutant transformed with CLN1, CLN2 or CHIMERA 4 plasmids after the release from a G1 arrest induced by either α-factor or a cdc28 mutation. The results revealed that, as expected, the cln1 cln2 cells transformed with CLN1 showed a delay in bud emergence in comparison to the cells transformed with CLN2. Interestingly, chimera 4 abolished the budding delay, since those cells expressing chimera 4 behaved like those expressing Cln2 (Fig. 2E). Thus, the 225–299 region mediates the specific function of Cln2 in budding.
In conclusion, our results demonstrate that the region of Cln2 between amino acids 225 and 229 is not only necessary, but is also sufficient to confer this cyclin specific functions. The next aim was to determine the mechanism by which this occurs.
The specific functionality of cyclins is not caused by differences in protein level
We observed in western blot analysis that the detected protein level of Cln2 was higher than that of Cln1. It was possible that the Cln2-specific functionality might be related to quantitative differences in the protein level. However, the analysis of the cellular content of the chimeric cyclins ruled out this possibility: chimeras 1 and 2, which are functionally equivalent to Cln2, were markedly less abundant than Cln2 (Fig. S4). Moreover, chimera 4 gained Cln2-specific functions, but its cellular content was roughly identical to that of Cln1. Therefore, there was no correlation between protein level and functionality.
Karyopherins involved in the nuclear-cytoplasmic transport of the Cln1 and Cln2 cyclins
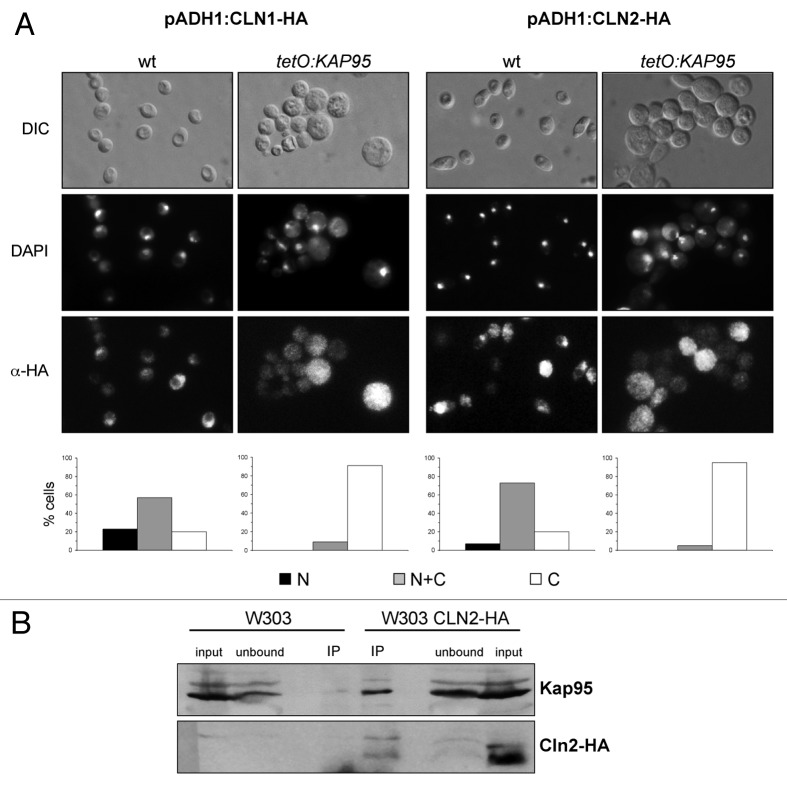
Previous studies by our group have reported a difference between the subcellular localization of Cln1 and Cln2: although both cyclins were located in the nucleus and the cytoplasm, Cln1 presented a stronger nuclear accumulation in relation to Cln2.40 More recently, single cell analysis has revealed subtle differences between Cln1 and Cln2 localization, with the latter being partially relocated to the cytoplasm during the budding period.42 In the functional distinction between Cln1 and Cln2 context, we decided to characterize the spatial regulation of both cyclins in detail. The first aim was to identify the karyopherins (the carrier proteins responsible for the active transport of proteins between the nucleus and the cytoplasm) involved in their transport. A systematic analysis of the localization of Cln1 and Cln2 was performed in all the β-karyopherin mutants. The nuclear signals of Cln1 and Cln2 were detected in all strains, except for the tetO7: KAP95 and tetO7: CSE1 strains incubated in the presence of doxycycline in order to shut off the tetO7 promoter (Fig. 3A and results not shown). Kap95 and Cse1 are involved, together with α-importin Kap60, in the classical nuclear import pathway.43,44 Our result indicates, therefore, a role of the classical nuclear import pathway in the regulation of the Cln1 and Cln2 cyclins.
Figure 3. Analysis of the regulation of the subcellular localization of Cln1 and Cln2 by karyopherin Kap95. (A) Cells from exponentially growing cultures of the wild type (W303–1a) and the tetO7:KAP95 (JCY970) strains transformed with a plasmid expressing a HA-tagged version of the Cln1 or Cln2 protein under the control of the ADH1 promoter, were incubated in the presence of 5 µg/ml doxycycline for 8 h and assayed by indirect immunofluorescence. The DIC image, the DAPI staining of DNA and the HA indirect-fluorescence signals are shown. Graphs represent protein localization derived from more than 100 cells from at least three independent cultures (N, fluorescence signal only in the nucleus; N+C, fluorescence signal in nucleus and cytoplasm; C, fluorescence signal in cytoplasm). (B) Cln2 was immunoprecipitated with anti-HA antibody from crude extracts of cells expressing a HA-tagged Cln2 (JCY1357) and the control strain (W303–1a). The presence of Kap95 and HA-tagged Cln2 in the input, unbound and immunoprecipitated fractions was determined by western analysis with a specific anti-Kap95 or anti-HA antibody.
The physical interaction between Cln2 and the Kap95 karyopherin was tested by a co-immunoprecipitation assay. Cln2-HA, expressed at the endogenous level, was immunoprecipitated, and the presence of Kap95 in the purified fraction was analyzed. As seen in Figure 3B, Kap95 specifically co-immunoprecipitated with Cln2-HA. This result demonstrates that Cln2 physically interacts with Kap95 in vivo, which reinforces the conclusion that the classical import pathway is responsible for the nuclear import of the Cln2 cyclin.
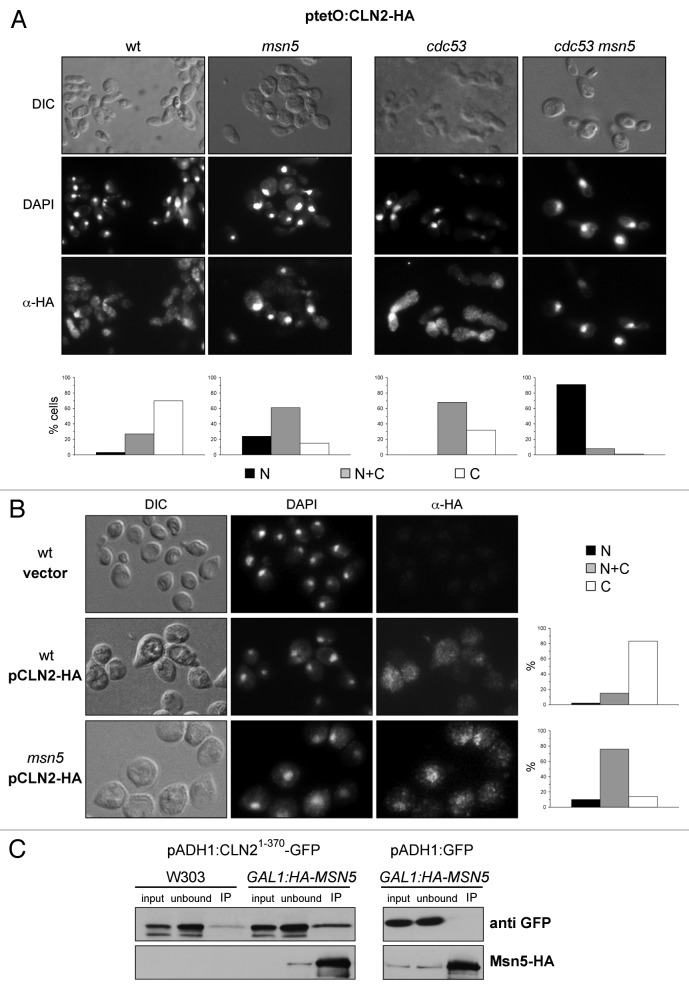
A second result obtained when analyzing the localization of Cln1 and Cln2 in the karyopherin mutants was the increased nuclear accumulation of Cln2 in the case of the msn5 mutant strain (Fig. 4A). To better detect Cln2 in the immunofluorescence assay, subcellular localization was also analyzed in a mutant strain in the SCF degradation pathway, cdc53–1, which renders high levels of Cln2. Notably, inactivation of Msn5 in the cdc53–1 mutant cells led to a dramatic nuclear accumulation of Cln2 (Fig. 4A). No changes in Cln1 localization were observed in the absence of Msn5 (Fig. S5). The previous results were obtained under Cln2 overexpression conditions. We also analyzed the role of Msn5 in Cln2 localization under endogenous conditions. Cultures were synchronized, and Cln2 localization was investigated in G1/S cells (the cell cycle period when Cln2 is expressed). While a nuclear signal was recognizable only in 17% of the wild-type cells, a nuclear signal was detected in approximately 86% of the msn5 mutant cells (Fig. 4B). Our results support that Msn5 is also involved in the nuclear export of Cln2, but that it is not involved in the control of Cln1 subcellular localization.
Figure 4. Analysis of the regulation of the subcellular localization of Cln2 by karyopherin Msn5. (A) Cells from exponentially growing cultures of the wild type (W303–1a), msn5 (JCY1366), cdc53 (MTY740) and cdc53 msn5 (JCY684) strains transformed with a plasmid expressing a HA-tagged version of the Cln2 protein under the control of the tetO2 promoter, were assayed by indirect immunofluorescence as described in Figure 4. (B) Cells from exponentially growing cultures of the wild type (W303–1a) and the msn5 (JCY1366) strains transformed with an empty vector or a centromeric plasmid expressing a HA-tagged version of the Cln2 protein at endogenous level, were assayed by indirect immunofluorescence. (C) Left panel: Msn5 was immunoprecipitated with an anti-HA antibody from crude extracts of cells of the GAL1:HA-MSN5 (JCY313) and the control (W303–1a) strains transformed with a plasmid expressing a GFP tagged Cln21–370 protein. The presence of Mns5 and Cln21–370 in the input, unbound and immunoprecipitated fractions was determined by western analysis with an anti-HA or anti-GFP antibody respectively. Right panel: the immunoprecipitation assay was repeated with the GAL1:HA-MSN5 (JCY313) strain transformed with a control plasmid expressing the GFP protein.
Finally, the physical interaction between a stable truncated Cln2 protein (Cln21–370) and Msn5 was investigated. As seen in Figure 4C, Cln21–370 was specifically purified in Msn5 immunoprecipitates. This result demonstrates that Cln2 physically interacts with Msn5 in vivo, which further supports that Msn5 is the exportin of Cln2.
Identification of the region involved in the nuclear export of Cln2 cyclin
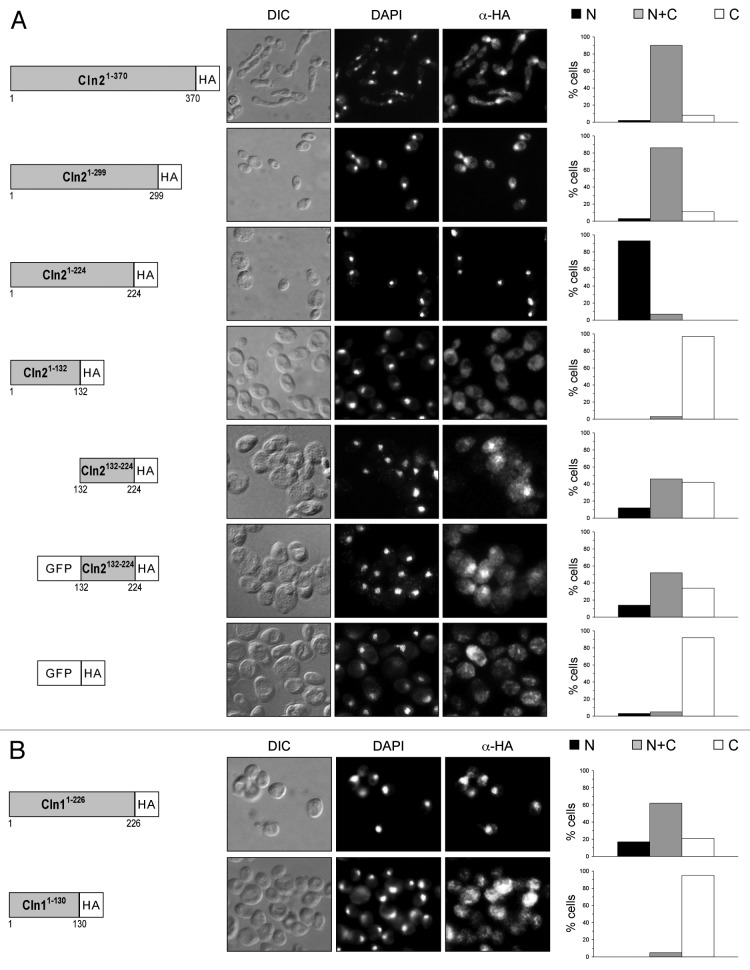
Once the karyopherins responsible for the control of Cln1 and Cln2 localization were identified, the next step was to characterize the nuclear localization signals (NLS) and the nuclear export signals (NES) that mediate this transport. The strategy followed involved the expression of truncated versions of the cyclin tagged with the HA epitope (Fig. 5A). The analysis of the localization of Cln21–370 and Cln21–299 revealed that both truncated proteins were distributed between both the nucleus and the cytosol. On the contrary, only a strong nuclear signal was detected for the truncated version of Cln21–224. This suggested that the region of Cln2 between amino acids 225 and 299 might contain a nuclear export signal.
Figure 5. Analysis of the subcellular localization of truncated versions of cyclins. (A) Cells from exponentially growing cultures of the wild type strain expressing HA-tagged Cln21–370 (JCY1128), Cln21–299 (JCY1385), Cln21–224 (JCY1125), Cln21–132 (JCY1389), Cln2132–224 (JCY1392), GFP-Cln2132–224 (JCY1393) or GFP (MCY220) under the control of the GAL1 promoter, and (B) cells from exponentially growing cultures of the wild type strain expressing HA-tagged Cln11–226 (JCY1517) or Cln11–130 (JCY1518) under the control of the GAL1 promoter, were assayed by indirect immunofluorescence as described in Figure 4.
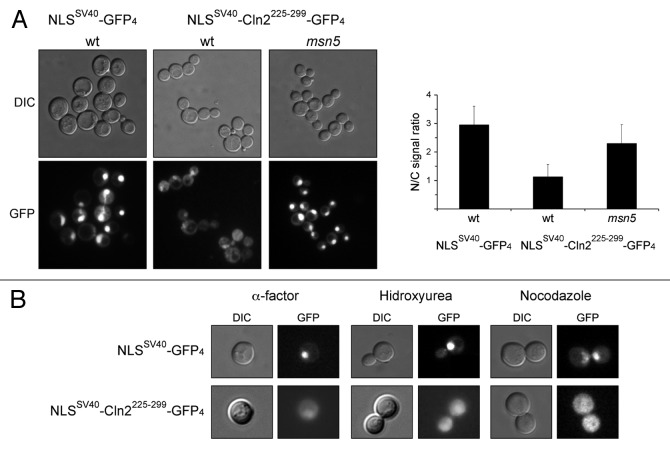
To definitively verify this hypothesis, the Cln2225–299 fragment was added to a nuclear protein composed of four copies of GFP fused to the NLS of SV40 (NLSSV40 -GFP4), and the ability of this Cln2 fragment to drive the nuclear export of this chimeric protein was tested (Fig. 6). Only a strong nuclear signal was detected in the case of the control NLSSV40-GFP4. However, cytosolic signal was also observed in the case of the NLSSV40-Cln2225–299-GFP4 protein. In fact, the nuclear/cytosolic signal ratio notably decreased when the Cln2225–299 region was introduced into the NLSSV40-GFP4 protein. This result is consistent with this region mediating the nuclear export of the protein. Interestingly, inactivation of Msn5 raised the nuclear/cytosolic signal ratio, indicating that Msn5 is required for the nuclear export of the NLSSV40-Cln2225–299-GFP4 protein. In conclusion, our results support that the 225–299 region of Cln2 functions as a NES that depends on Msn5 exportin.
Figure 6. Characterization of a Cln2 fragment with an Msn5-dependent NES activity. (A) Exponentially growing cells of the wild type (W303–1a) and msn5 (JCY1366) strains transformed with plasmids pNLSSV40-GFP4 or pNLSSV40-CLN2225–299-GFP4 were analyzed by fluorescence microscopy. GFP signal and DIC images are shown. Graph represents the nuclear/cytosolic intensity ratio average derived from at least three cultures and two independent clones of pNLSSV40-CLN2225–299-GFP4 plasmid (approximately 50 cells were analyzed in each culture) for each strain. (B) Exponentially growing cells of the wild type (W303–1a) strain transformed with plasmids pNLSSV40-GFP4 or pNLSSV40-CLN2225–299-GFP4 were synchronized in G1, S or G2/M phase by incubation in the presence of 5 μg/mL α-factor, 200mM HU or 10 μg/mL nocodazole for 3 h, respectively, and analyzed by fluorescence microscopy.
One question that arises when considering the subcellular localization of cell cycle regulators is whether this spatial control is cell cycle-regulated. As seen in Figure 5A, no differences were found in the localization of Cln21–299 and Cln21–370 between cells in the different cell cycle stages. In agreement with this, the NLSSV40-Cln2225–299-GFP4 protein was present in the cytosol of those cells arrested in the G1, S or G2/M phase (Fig. 6B). These observations indicate that the nuclear export mediated by Cln2225–299 is not cell cycle-regulated.
Identification of the region involved in the nuclear import of the Cln2 cyclin
We followed the same strategy described above to identify the NLS of Cln2. A nuclear signal was detected for the truncated Cln21–224 protein, indicating that this region contains sufficient information to direct this protein into the nucleus (Fig. 5A). However, no nuclear signal was detected in the case of the Cln21–132 variant. This suggests that the region between amino acids 132 and 224 is required for Cln2 import. In fact, a nuclear signal was observed in the assays of Cln2132–224, suggesting that this fragment contains sufficient information to direct its import into the nucleus. Caution with this result is necessary, because this truncated protein is below the diffusion limit of the nuclear pore complex, thus the result could be due to the passive diffusion of the protein. However, the fact that nuclear signals were absent in the case of Cln21–132, a protein whose estimated molecular weight is also below the diffusion limit, and that nuclear signals were observed in the case of Cln2132–224-GFP, but not for GFP, supports that the region between amino acids 132 and 224 of Cln2 contains information to direct the nuclear accumulation of a protein.
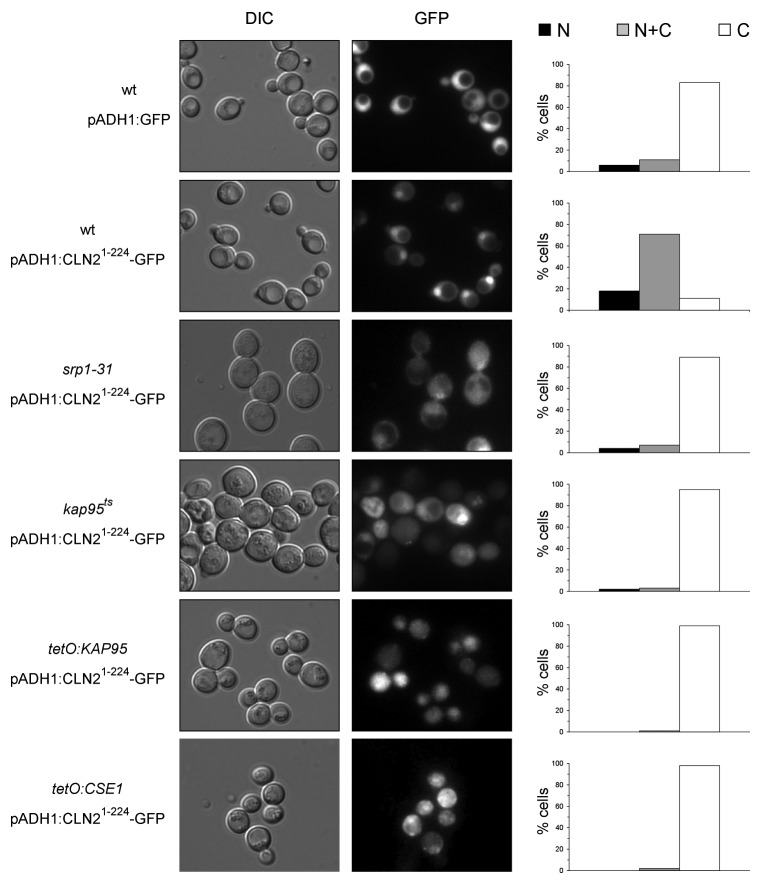
The results described above indicate that Cln2 is imported into the nucleus via the classical nuclear import pathway. Because of that, the localization of the Cln21–224-GFP in mutant strains in this pathway was investigated. The results indicate that, unlike what was observed in the wild-type cells, Cln21–224-GFP was never detected in the nucleus of the mutant cells in Kap95, Kap60/Srp1 or Cse1 karyopherins (Fig. 7). This indicates that nuclear import directed by fragment 1–224 of Cln2 is dependent on a functional classical nuclear import pathway. The classical NLS sequences recognized by this pathway are well characterized, consisting of one or two short clusters of basic amino acids.44,45 Surprisingly, no resembling sequences were present in the Cln21–224 fragment, thus raising the possibility that an adaptor protein could mediate the recognition of Cln2 by Kap95-Kap60. One possible candidate was the Cln2-Cdc28 inhibitor Far1, since the nuclear import of Clb5 cyclin depends on binding to its CDK inhibitor Sic1.46 However, the nuclear localization of Cln21–224 was not altered in the far1 mutant strain (Fig. S6).
Figure 7. Characterization of a Cln2 fragment with classical NLS activity. Exponentially growing cells of the wild type (W303–1a), srp1ts, kap95ts (SWY1313), tetO7:KAP95 (JCY970) and tetO7:CSE1 (JCY972) strains transformed with plasmids pADH1:GFP or pADH1: CLN21–224-GFP were analyzed by fluorescence microscopy. GFP signal and DIC images are shown. Graphs show protein localization as described in Figure 4.
Identification of the region involved in the nuclear import of the Cln1 cyclin
To identify the sequences in Cln1 involved in the spatial regulation of the cyclin, truncated versions of the Cln1 cyclin were obtained, and their subcellular localization was analyzed. Cln11–226 was detected in both the cytoplasm and the nucleus (Fig. 5B). However, the nuclear signal was lost in the case of Cln11–130-HA. These results are similar to those described above for Cln2, and they strongly suggest the existence of a region responsible for the Cln1 nuclear import between amino acids 1 and 226, as occurs for Cln2.
Involvement of spatial regulation mechanisms in the functional distinction between Cln1 and Cln2
In previous sections, we describe the existence of differences in the function and subcellular localization between Cln1 and Cln2. Remarkably, the fragment 225–229 of Cln2, which confers a specific function to Cln2, coincides with an Msn5-dependent NES, an export mechanism that is specific of Cln2. The question arises as to whether this Msn5-mediated nuclear export mechanism could be related to the functional differences observed between Cln1 and Cln2.
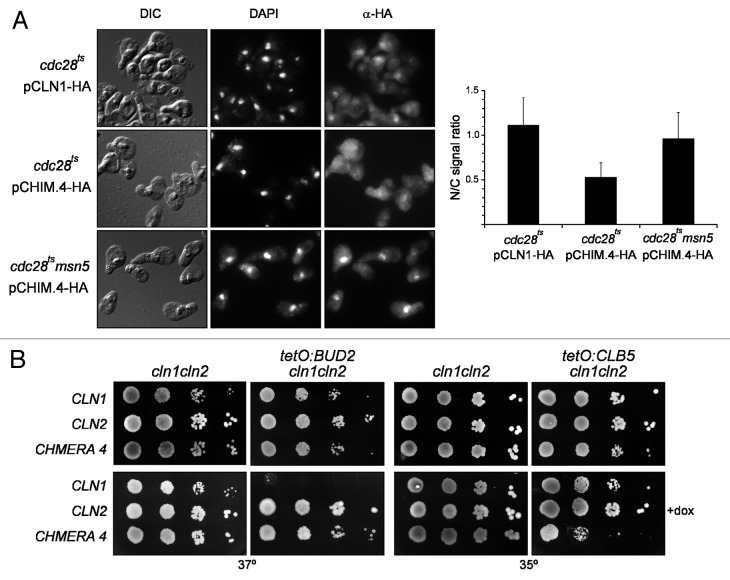
We first investigated whether the 225–229 region of Cln2 was capable of altering the spatial regulation of Cln1. We observed that, whereas Cln1 was located in the cytoplasm and the nucleus, the nuclear signal was absent or very weak in the case of chimera 4 (Fig. 8A). This is consistent with the idea that chimera 4 contains an NES, which results in a mostly cytosolic localization. Next we tested whether the localization of chimera 4 was regulated by Msn5. As shown in Figure 8A, the nuclear signal was clearly detected in the msn5 mutant cells expressing chimera 4. All these results are consistent with the presence of an NES recognized by Msn5 in chimera 4. Thus, chimera 4 is subject to a specific Cln2 nuclear export mechanism that is absent in Cln1.
Figure 8. Analysis of the subcellular localization of Chimera 4 protein. (A) Cells from exponentially growing cultures of the cdc28 and the cdc28 msn5 (JCY1474) strains transformed with a centromeric plasmid expressing a HA-tagged version of the Cln1 or Chimera 4 protein at endogenous level were assayed by indirect immunofluorescence. Graph represents the nuclear/cytosolic intensity ratio average derived from at least three independent cultures. (B) 10-fold serial dilutions from exponentially growing cultures of the cln1 cln2 (JCY847), tetO7:BUD2 cln1 cln2 (JCY1538) and tetO7:CLB5 cln1 cln2 (JCY1534) transformed with a centromeric plasmid containing the CLN1, CLN2 or CHIMERA4 gene were spotted onto YPD or YPD medium supplemented with 5 μg/mL doxycyclin and incubated at the indicated temperature for 3 d.
We describe above how chimera 4 was able to suppress the lethality of the swi4ts swi6 mutant, a function that has been associated with the existence of Cln2 shuttling between the cytosol and the nucleus,12 and the growth defect in the presence of latrunculine B and the budding delay of the cln2 mutant, aspects that are expected to involve a cytosolic function of the cyclin. In contrast, chimera 4 cannot properly control the cell size, a function that has been associated with a nuclear localization of Cln2.13 These results suggest that the introduction of the Cln2 NES confers to Cln1 the Cln2 specific cytosolic functions. To further support this idea, new cytosolic and nuclear functions were analyzed. In particular, the suppression of the growth defects of the bud2 or clb5 mutant strains were used to test for cytosolic or nuclear functions of the cyclin, respectively.12,13 We introduced the tetO7: BUD2 or the tetO7: CLB5 genes into the cln1 cln2 strain, and we transformed those cells with plasmids expressing Cln1, Cln2 or chimera 4. First, it must be highlighted that Cln2 and chimera 4, but not Cln1, suppressed the growth defect of the bud2 mutant (Fig. 8B). This result is consistent with previous results of resistance to latrunculine B or budding kinetics, indicating that Cln2 and chimera 4 perform a cytosolic function that Cln1 is unable to perform, or at least not as efficiently. On the other hand, the clb5 mutant cells containing chimera 4 grew significantly worse than those containing Cln1, indicating that chimera 4 performs a nuclear function less efficiently than Cln1. Thus, the introduction of the NES containing Cln2 fragment 225–299 into Cln1 leads to the gain of cytosolic functions and to the impairment of nuclear functions.
In short, all our observations support a model in which the presence of a nuclear export mechanism in Cln2 confers this cyclin a differential functionality if compared with Cln1.
Discussion
In S. cerevisiae, a single CDK, Cdc28, associates with nine different cyclins in order to govern progression through the cell cycle. Therefore, it is the cyclin subunit of the complex that confers functional specificity to the different cyclin-Cdc28 kinases. Previous works by our group have revealed a functional distinction between Cln1 and Cln2 in the G1/S transition control.40 In this manuscript, we attempt to characterize the mechanism that explains this difference.
Cln1 and Cln2 are expressed at the same cell cycle time, which rules out a functional distinction based on a difference in the time of their expression. Another explanation might lie in the difference in protein levels between the two cyclins. Nevertheless, this hypothesis can also be ruled out, because chimera 1 and chimera 2, whose levels are lower than Cln2, present the Cln2-specific functionality. One particularly remarkable case is chimera 4, since it is basically a Cln1 cyclin but a small fragment from Cln2; although its protein level is similar to Cln1, it shows rather a functionality similar to Cln2. Hence, the specific function of Cln2 does not seem to be related to a higher level of cyclin either. So, quantitative differences do not lead to the functional distinction between Cln1 and Cln2; rather, functional specialization must reflect qualitative intrinsic differences.
It might be expected that a region in Cln2 is responsible for its specific functionality toward Cln1. In fact, a small region between amino acids 225 and 299 of Cln2 is necessary and sufficient to give its specific functionality to the cyclin. Intrinsic functional specificity can be determined, at least in part, by the differences in the subcellular localization that targets the CDK to specific locations. It is interesting to note that the 225–299 region of Cln2 contains an export signal activity depending on exportin Msn5. This correlation strongly suggests that the existence of an export mechanism for Cln2 is the basis of its specific functionality compared with Cln1. This conclusion is supported by two fundamental facts. First, the introduction of the 225–299 region of Cln2 into Cln1 (chimera 4) results in a more prominent cytosolic localization in an Msn5-dependent way. Second, the resulting cyclin is able to gain new functions that are specific for Cln2 but only those associated with a cytosolic localization. Sequence comparison between Cln1 and Cln2 revealed a high degree of sequence identity in this region except for a fragment flanked by short stretches of Ser or Thr that is present only in Cln1 (Fig. S1); this suggests that the presence of this fragment in Cln1 could disrupt a Msn5-dependent nuclear export mechanism.
The proposed model is consistent with the differences in location observed between Cln1 and Cln2. Cln2 shows a more prominent cytoplasmic localization than Cln1, which probably reflects the existence of a nuclear export mechanism that might explain why Cln2 performs better than Cln1 functions related to budding. Proteomic approaches have identified proteins interacting with Cln2 and substrates of Cln2-Cdc28, such as Cdc24, Boi1 or Rga2, which are involved in budding.47,48 These proteins are located on the cell surface in order to trigger polarization during budding, and their phosphorylation by Cln2-Cdc28 can justify the need for Cln2 in the cytosol during budding. Cln1-Cdc28 might prove less efficient in performing these phosphorylations, probably because its more prominent nuclear localization. The introduction of a nuclear export activity into Cln1 may unbalance Cln1 distribution toward an increase in the cytosolic fraction of the protein, thus allowing the Cln1 cyclin to perform functions that it normally carries out inefficiently.
On the other hand, it cannot be ruled out that the 225–299 region of Cln2 might confer a specific functionality through a mechanism independent of NES activity. The intrinsic functional specificity between cyclins could be determined by differences in the recognition of specific targets. Very recently, it was described that some Cln targets involved in the response to α mating factor like Ste5 and Ste20 contain cyclin docking domains that are better recognized by Cln2 than by Cln1.41 It is possible that the 225–299 fragment of Cln2, but not the equivalent region from Cln1, might directly contribute to the recognition of the specific CDK substrates involved in morphogenesis and in the process of budding independently of, or in addition to, its effect in cyclin localization.
Spatial regulation adds a new dimension to protein function control. In the case of cell cycle control, evidence has accumulated emphasizing the importance of controlling the subcellular localization of some cell cycle regulators. This work characterizes the spatial regulation of the Cln1 and Cln2 cyclins. They had been previously described to be located in the nucleus and the cytoplasm; nonetheless, the karyopherins and signals involved in their transport had not been analyzed. The results presented herein have allowed us to describe Msn5 as the exportin of Cln2, but not of Cln1, and that the classical nuclear import pathway mediates the nuclear import of both cyclins. In the case of Cln3, the other G1 cyclin, its import into the nucleus depends on a canonical classical NLS,14 so it is conceivable that Cln3 might also be imported by the classical import pathway. Thus, the classical import pathway is responsible for the entrance of the three G1 cyclins to the nucleus. In addition, the nuclear import of the complete transcriptional machinery of the Start transition is also mediated by this pathway, which explains why the classical import pathway is essential for the execution of Start at the initiation of a new round of division.49
The classical import pathway transports those proteins carrying the classical nuclear localization signal. These signals consist of either a short cluster of basic amino acids (monopartite) or two short clusters of basic amino acids separated by 10–12 residues (bipartite). Surprisingly, no resembling sequence can be found in Cln1 or Cln2. This resembles the case of mammalian cyclin E, the functional homolog of Cln1 and Cln2. It has been described that nuclear targeting information is contained entirely within cyclin E, which binds to and can be imported into nuclei by the importin α-β heterodimer; however, no obvious classical NLS can be identified in the cyclin E sequence.50 Different hypothetical scenarios to account for this apparent contradiction can be proposed. It is possible that a still unidentified new signal recognized by the classical import pathway may exist in Cln1, Cln2 or cyclin E. Kap95 in yeast and importin β in mammals contribute to the import of some proteins by recognizing an import signal other than the classical NLS.51 This is a specific function of Kap95, independent of its role in the classical import pathway. Hence, this does not apply to Cln1 and Cln2, since our results indicate that Kap60 and Cse1 are involved in the transport of Cln1 and Cln2. On the other hand, the nuclear import of a multiprotein complex could depend on the reconstitution of an NLS by residues from different subunits of the complex. Perhaps a classical NLS recognized by Kap95-Kap60 may appear after binding of the cyclin to the CDK. However, this is not likely the case for Cln2, because, despite the Cln2-Cdc28 complex being imported more efficiently than the cyclin, Cln2 is still able to enter the nucleus on its own.14 Alternatively, recognition of Cln1 and Cln2 by Kap60-Kap95 may involve additional bridging proteins containing a classical NLS. A good candidate was the Cln1,2-Cdc28 inhibitor Far1. The nuclear import of the yeast S-phase Clb5 requires interaction with CKI Sic1. Similarly, cyclin D1-CDK4 in mammalian cells might use the classical NLS of CKI p21 for its nuclear import. Nevertheless, Cln1 and Cln2 locate in the nucleus, even in the absence of Far1. Thus, bridging proteins other than CKI could be involved in the nuclear import of Cln1 and Cln2.
In spite of the similarity in the nuclear import regulation of Cln1 and Cln2 in relation to the region of the protein with the targeting information and the karyopherins involved in their transport, it must be noted that differences in the nuclear import can exist. Cln21–224 shows an exclusively nuclear localization, whereas the equivalent Cln11–226 truncated protein is distributed between the nucleus and the cytoplasm. Although another explanation can be considered, it is possible that this observation reflects a different strength in the NLS signals present in both cyclins.
Previous works have connected Msn5 to cell cycle control. Msn5 is the karyopherin responsible for the nuclear export of cell cycle transcription factors Swi6, Whi5 and Swi552-54 or of other cell cycle regulators like the CKI inhibitor Far1 or the APC ubiquitin ligase activator Cdh1.55,56 Our results reinforce the involvement of karyopherin Msn5 in cell cycle regulation, especially in the Start transition. The minimal NES identified in some of the Msn5 cargoes often involves long protein regions of approximately 100 amino acids.53,55,57,58 This is also the case for Cln2. The nuclear export mediated by Msn5 often requires the phosphorylation of the cargo proteins in critical residues.53,57-59 Interestingly, it has been suggested that Cln2 phosphorylation by Cdc28 may regulate its localization, since the mutant version of Cln2 at Cdc28 phosphorylation sites shows a nuclear accumulation of the cyclin.14 This could suggest that nuclear export is enhanced upon the phosphorylation of Cln2. This would help relocate the Cln2-Cdc28 complex, once activated, out of the nucleus, when it may be required in the cytosol for the budding process.
In conclusion, we propose that a spatial regulation mechanism, the existence of a nuclear export activity, contributes to the specific functionality of the Cln2-Cdc28 complex if compared with Cln1-Cdc28. This example is one of many others that have, in recent years, illustrated the relevance of spatial regulation in cell cycle control.
Materials and Methods
Yeast strains and growth conditions
The yeast strains used in this study are shown in Table S1. The cln1, cln2 and msn5 mutant strains were obtained by integrating a DNA fragment amplified from the pFA6a series plasmids60 or from strains containing the msn5Δ3::HIS3 or cln2::LEU2 cassette. Strains expressing from the GAL1 promoter the Cln2 or Cln1 protein or truncated versions of them C-terminal tagged with HA, were constructed in a two-step process by integrating at the appropriate position DNA fragments amplified from the pFA6a series plasmids. The substitution of the KAP95, CSE1, CLB5 and BUD2 promoters by the tetO7 promoter was obtained by integrating a DNA fragment amplified from plasmid pCM225 (a gift from Dr. E. Herrero). To repress the tetO7 promoter, doxycicline was added to a concentration of 5 μg/mL.
Plasmids
Centromeric plasmids pCLN1 and pCLN2, containing HA-tagged versions of Cln1 and Cln2, respectively, and plasmids ptetO:CLN1 and ptetO:CLN2 overexpressing the CLN1 and CLN2 genes under the control of the tetO2 promoter, were a gift from Dr. M. Aldea. The plasmids containing the chimeric cyclin genes were constructed in a two-step process by cloning the appropriate DNA fragments obtained by PCR amplification with oligonucleotide bearing restriction sites. For chimeras 1, 2 and 3, a XbaI-HindIII fragment amplified from pCLN1 (from nucleotide 1428, 936 or 831 to 1638, respectively) fused to a sequence coding for three copies of the HA epitope was first cloned in Xba-HindIII-cleaved YCplac33; this plasmid was then digested with KpnI-XbaI and a KpnI-XbaI DNA fragment amplified from pCLN2 (from nucleotide -604 to 1427, 897 or 792, respectively) was subsequently cloned. For chimera 4, a SphI-HindIII fragment containing a region of the CHIMERA2 gene from nucleotide 673 was first cloned in SphI-HindIII-cleaved YCplac33; this plasmid was then digested with KpnI-SphI and a KpnI-SphI DNA fragment amplified from pCLN1 (from nucleotide -333 to 678) was subsequently cloned. At least two independent clones from each cyclin construct were used in the functional analysis. All clones were checked by sequencing around the ligation points, and one clone in each case was completely sequenced along the coding region.
pADH1:GFP plasmid was constructed in a three-step process. First, ADH1 promoter (-1473, +3) amplified from genomic DNA with oligos containing the EcoRI and KpnI restriction sites was cloned in EcoRI-KpnI digested YCplac33. Next, the GFP(S65T) coding region without start and stop codons, amplified from pFA6a-GFP(S65T) using oligos containing a BamHI or a XbaI restriction site, was cloned in frame by BamHI-XbaI digestion. Finally, the ADH1 terminator including the stop codon (+772, +1011), amplified from pFA6a-GFP(S65T) with oligos containing a SalI or PstI site, was introduced by SalI-PstI digestion. pADH1:Cln21–224-GFP, and pADH1:Cln21–370-GFP plasmids were obtained cloning in frame in pADH1:GFP by KpnI-BamHI digestion the appropriate CLN2 coding fragment amplified from pCLN2 with oligos containing the indicated restriction sites. pADH1:CLN1-HA and pADH1:CLN2-HA were obtained removing the GFP coding region from pADH1-GFP by KpnI-SalI digestion and introducing the CLN1 or CLN2 coding fragments fused to three copies of the HA epitope amplified from pCLN1 or pCLN2 with oligos containing a KpnI or SalI sites.
pNLSSV40-Cln2225–299-GFP4 plasmid was obtained by cloning the appropiate fragment amplified from pCLN2 with a forward oligo containing a KpnI restriction site and the NLSSV40 coding sequence and a reverse oligo containing a BamHI site in KpnI-BamHI digested pNLSSV40-GFP4.53
Fluorescence microscopy
HA-tagged proteins were detected by indirect immunofluorescence basically as described.61 Samples were incubated overnight with the primary antibody anti-HA 3F10 (Roche) at 4°C and with the secondary antibody Alexa 546-labeled goat anti-rat antibody (Molecular Probes) for 1 h at 4°C. An additional incubation with a third antibody Alexa 546-labeled donkey anti-goat antibody (Molecular Probes) for 1 h at 4°C is included in the detection of cyclins at endogenous level. GFP tagged proteins were analyzed in living cells grown on selective medium. Samples were analyzed in an Axioskop 2 Fluorescence Microscope (Zeiss Inc.), and pictures were taken using an AxioCam MRm (Zeiss Inc.). Where indicated, localization of proteins was monitored by visual inspection. At least 100 cells from three independent experiments were scored as cells with fluorescence signal only in the nuclei (N), in the nuclei and the cytoplasm (N+C) or only in the cytoplasm (C). Where indicated, a quantitative estimation of the relative intensity of the nuclear and cytosolic signal was achieved by determining the average pixel intensity in the nuclear and the cytosolic region of the cell and calculating the (N-C)/C value in at least 100 cells from three independent experiments using Axiovission v4.7 software (Zeiss. Inc.).
Miscellaneous
Cell size analysis was performed as described.40 Immunoprecipitation and western blot analysis were performed as previously described.54 Antibodies used were anti-HA 3F10 or 12C5A (Roche), anti-Kap95 (Santa Cruz), anti-GFP (Roche) and anti-tubulin (Serotec Ltd.).
Supplementary Material
Acknowledgments
We are very grateful to Dr. J.R. Pringle, Dr. D. M. Tyers, Dr. J. Thorner, Dr. G. Fink, Dr. E. Herrero and Dr. M. Aldea for kindly provide materials. This work was supported by Grants BFU2007–67929-C02–02 and BFU2010–20927 from the Spanish Governement (co-financed by ERDF from European Union) and ACOMP/2012/219 from Generalitat Valenciana. I.Q. was recipient of a Predoctoral Fellowship from the Spanish Governement.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21505
References
- 1.Jackson LP, Reed SI, Haase SB. Distinct mechanisms control the stability of the related S-phase cyclins Clb5 and Clb6. Mol Cell Biol. 2006;26:2456–66. doi: 10.1128/MCB.26.6.2456-2466.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cross FR, Yuste-Rojas M, Gray S, Jacobson MD. Specialization and targeting of B-type cyclins. Mol Cell. 1999;4:11–9. doi: 10.1016/S1097-2765(00)80183-5. [DOI] [PubMed] [Google Scholar]
- 3.Donaldson AD. The yeast mitotic cyclin Clb2 cannot substitute for S phase cyclins in replication origin firing. EMBO Rep. 2000;1:507–12. doi: 10.1093/embo-reports/kvd108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–8. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- 5.Hu F, Aparicio OM. Swe1 regulation and transcriptional control restrict the activity of mitotic cyclins toward replication proteins in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2005;102:8910–5. doi: 10.1073/pnas.0406987102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keaton MA, Bardes ESG, Marquitz AR, Freel CD, Zyla TR, Rudolph J, et al. Differential susceptibility of yeast S and M phase CDK complexes to inhibitory tyrosine phosphorylation. Curr Biol. 2007;17:1181–9. doi: 10.1016/j.cub.2007.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mendenhall MD, Hodge AE. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1191–243. doi: 10.1128/mmbr.62.4.1191-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. doi: 10.1016/S1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 9.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–97. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine K, Huang K, Cross FR. Saccharomyces cerevisiae G1 cyclins differ in their intrinsic functional specificities. Mol Cell Biol. 1996;16:6794–803. doi: 10.1128/mcb.16.12.6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vergés E, Colomina N, Garí E, Gallego C, Aldea M. Cyclin Cln3 is retained at the ER and released by the J chaperone Ydj1 in late G1 to trigger cell cycle entry. Mol Cell. 2007;26:649–62. doi: 10.1016/j.molcel.2007.04.023. [DOI] [PubMed] [Google Scholar]
- 12.Edgington NP, Futcher B. Relationship between the function and the location of G1 cyclins in S. cerevisiae. J Cell Sci. 2001;114:4599–611. doi: 10.1242/jcs.114.24.4599. [DOI] [PubMed] [Google Scholar]
- 13.Miller ME, Cross FR. Distinct subcellular localization patterns contribute to functional specificity of the Cln2 and Cln3 cyclins of Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:542–55. doi: 10.1128/MCB.20.2.542-555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller ME, Cross FR. Mechanisms controlling subcellular localization of the G(1) cyclins Cln2p and Cln3p in budding yeast. Mol Cell Biol. 2001;21:6292–311. doi: 10.1128/MCB.21.18.6292-6311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–84. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wittenberg C, Sugimoto K, Reed SI. G1-specific cyclins of S. cerevisiae: cell cycle periodicity, regulation by mating pheromone, and association with the p34CDC28 protein kinase. Cell. 1990;62:225–37. doi: 10.1016/0092-8674(90)90361-H. [DOI] [PubMed] [Google Scholar]
- 17.Bean JM, Siggia ED, Cross FR. High functional overlap between MluI cell-cycle box binding factor and Swi4/6 cell-cycle box binding factor in the G1/S transcriptional program in Saccharomyces cerevisiae. Genetics. 2005;171:49–61. doi: 10.1534/genetics.105.044560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasmyth K, Dirick L. The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell. 1991;66:995–1013. doi: 10.1016/0092-8674(91)90444-4. [DOI] [PubMed] [Google Scholar]
- 19.Ogas J, Andrews BJ, Herskowitz I. Transcriptional activation of CLN1, CLN2, and a putative new G1 cyclin (HCS26) by SWI4, a positive regulator of G1-specific transcription. Cell. 1991;66:1015–26. doi: 10.1016/0092-8674(91)90445-5. [DOI] [PubMed] [Google Scholar]
- 20.Barral Y, Jentsch S, Mann C. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- 21.Schweitzer K, Cocklin R, Garrett L, Desai F, Goebl M. The ubiquitin ligase SCFGrr1 is necessary for pheromone sensitivity in Saccharomyces cerevisiae. Yeast. 2005;22:553–64. doi: 10.1002/yea.1234. [DOI] [PubMed] [Google Scholar]
- 22.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–19. doi: 10.1016/S0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 23.Dirick L, Böhm T, Nasmyth K. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 1995;14:4803–13. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stuart D, Wittenberg C. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 1995;9:2780–94. doi: 10.1101/gad.9.22.2780. [DOI] [PubMed] [Google Scholar]
- 25.Benton BK, Tinkelenberg AH, Jean D, Plump SD, Cross FR. Genetic analysis of Cln/Cdc28 regulation of cell morphogenesis in budding yeast. EMBO J. 1993;12:5267–75. doi: 10.1002/j.1460-2075.1993.tb06222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cvrcková F, Nasmyth K. Yeast G1 cyclins CLN1 and CLN2 and a GAP-like protein have a role in bud formation. EMBO J. 1993;12:5277–86. doi: 10.1002/j.1460-2075.1993.tb06223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lew DJ, Reed SI. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–20. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haase SB, Winey M, Reed SI. Multi-step control of spindle pole body duplication by cyclin-dependent kinase. Nat Cell Biol. 2001;3:38–42. doi: 10.1038/35050543. [DOI] [PubMed] [Google Scholar]
- 29.Henchoz S, Chi Y, Catarin B, Herskowitz I, Deshaies RJ, Peter M. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 1997;11:3046–60. doi: 10.1101/gad.11.22.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peter M, Gartner A, Horecka J, Ammerer G, Herskowitz I. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell. 1993;73:747–60. doi: 10.1016/0092-8674(93)90254-N. [DOI] [PubMed] [Google Scholar]
- 31.Schneider BL, Yang QH, Futcher AB. Linkage of replication to start by the Cdk inhibitor Sic1. Science. 1996;272:560–2. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]
- 32.Schwob E, Böhm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–44. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 33.Amon A, Tyers M, Futcher B, Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. doi: 10.1016/0092-8674(93)90722-3. [DOI] [PubMed] [Google Scholar]
- 34.Huang JN, Park I, Ellingson E, Littlepage LE, Pellman D. Activity of the APC(Cdh1) form of the anaphase-promoting complex persists until S phase and prevents the premature expression of Cdc20p. J Cell Biol. 2001;154:85–94. doi: 10.1083/jcb.200102007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahn SH, Tobe BT, Fitz Gerald JN, Anderson SL, Acurio A, Kron SJ. Enhanced cell polarity in mutants of the budding yeast cyclin-dependent kinase Cdc28p. Mol Biol Cell. 2001;12:3589–600. doi: 10.1091/mbc.12.11.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flick K, Chapman-Shimshoni D, Stuart D, Guaderrama M, Wittenberg C. Regulation of cell size by glucose is exerted via repression of the CLN1 promoter. Mol Cell Biol. 1998;18:2492–501. doi: 10.1128/mcb.18.5.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loeb JD, Kerentseva TA, Pan T, Sepulveda-Becerra M, Liu H. Saccharomyces cerevisiae G1 cyclins are differentially involved in invasive and pseudohyphal growth independent of the filamentation mitogen-activated protein kinase pathway. Genetics. 1999;153:1535–46. doi: 10.1093/genetics/153.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madhani HD, Galitski T, Lander ES, Fink GR. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc Natl Acad Sci USA. 1999;96:12530–5. doi: 10.1073/pnas.96.22.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Purnapatre K, Piccirillo S, Schneider BL, Honigberg SM. The CLN3/SWI6/CLN2 pathway and SNF1 act sequentially to regulate meiotic initiation in Saccharomyces cerevisiae. Genes to Cells. Devoted to Molecular & Cellular Mechanisms. 2002;7:675–91. doi: 10.1046/j.1365-2443.2002.00551.x. [DOI] [PubMed] [Google Scholar]
- 40.Queralt E, Igual JC. Functional distinction between Cln1p and Cln2p cyclins in the control of the Saccharomyces cerevisiae mitotic cycle. Genetics. 2004;168:129–40. doi: 10.1534/genetics.104.029587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhaduri S, Pryciak PM. Cyclin-specific docking motifs promote phosphorylation of yeast signaling proteins by G1/S Cdk complexes. Curr Biol. 2011;21:1615–23. doi: 10.1016/j.cub.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ball DA, Marchand J, Poulet M, Baumann WT, Chen KC, Tyson JJ, et al. Oscillatory dynamics of cell cycle proteins in single yeast cells analyzed by imaging cytometry. PLoS ONE. 2011;6:e26272. doi: 10.1371/journal.pone.0026272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hahn S, Maurer P, Caesar S, Schlenstedt G. Classical NLS proteins from Saccharomyces cerevisiae. J Mol Biol. 2008;379:678–94. doi: 10.1016/j.jmb.2008.04.038. [DOI] [PubMed] [Google Scholar]
- 44.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–71. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 46.Rossi RL, Zinzalla V, Mastriani A, Vanoni M, Alberghina L. Subcellular localization of the cyclin dependent kinase inhibitor Sic1 is modulated by the carbon source in budding yeast. Cell Cycle. 2005;4:1798–807. doi: 10.4161/cc.4.12.2189. [DOI] [PubMed] [Google Scholar]
- 47.Archambault V, Chang EJ, Drapkin BJ, Cross FR, Chait BT, Rout MP. Targeted proteomic study of the cyclin-Cdk module. Mol Cell. 2004;14:699–711. doi: 10.1016/j.molcel.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 48.McCusker D, Denison C, Anderson S, Egelhofer TA, Yates JR, 3rd, Gygi SP, et al. Cdk1 coordinates cell-surface growth with the cell cycle. Nat Cell Biol. 2007;9:506–15. doi: 10.1038/ncb1568. [DOI] [PubMed] [Google Scholar]
- 49.Taberner FJ, Igual JC. Yeast karyopherin Kap95 is required for cell cycle progression at Start. BMC Cell Biol. 2010;11:47. doi: 10.1186/1471-2121-11-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J, Kornbluth S. All aboard the cyclin train: subcellular trafficking of cyclins and their CDK partners. Trends Cell Biol. 1999;9:207–10. doi: 10.1016/S0962-8924(99)01577-9. [DOI] [PubMed] [Google Scholar]
- 51.Fries T, Betz C, Sohn K, Caesar S, Schlenstedt G, Bailer SM. A novel conserved nuclear localization signal is recognized by a group of yeast importins. J Biol Chem. 2007;282:19292–301. doi: 10.1074/jbc.M700217200. [DOI] [PubMed] [Google Scholar]
- 52.Queralt E, Igual JC. Cell cycle activation of the Swi6p transcription factor is linked to nucleocytoplasmic shuttling. Mol Cell Biol. 2003;23:3126–40. doi: 10.1128/MCB.23.9.3126-3140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taberner FJ, Quilis I, Igual JC. Spatial regulation of the start repressor Whi5. Cell Cycle. 2009;8:3010–8. doi: 10.4161/cc.8.18.9621. [DOI] [PubMed] [Google Scholar]
- 54.Taberner FJ, Quilis I, Sendra J, Bañó MC, Igual JC. Regulation of cell cycle transcription factor Swi5 by karyopherin Msn5. Biochim Biophys Acta. 2012;1823:959–70. doi: 10.1016/j.bbamcr.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 55.Blondel M, Alepuz PM, Huang LS, Shaham S, Ammerer G, Peter M. Nuclear export of Far1p in response to pheromones requires the export receptor Msn5p/Ste21p. Genes Dev. 1999;13:2284–300. doi: 10.1101/gad.13.17.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jaquenoud M, van Drogen F, Peter M. Cell cycle-dependent nuclear export of Cdh1p may contribute to the inactivation of APC/C(Cdh1) EMBO J. 2002;21:6515–26. doi: 10.1093/emboj/cdf634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boustany LM, Cyert MS. Calcineurin-dependent regulation of Crz1p nuclear export requires Msn5p and a conserved calcineurin docking site. Genes Dev. 2002;16:608–19. doi: 10.1101/gad.967602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeVit MJ, Johnston M. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr Biol. 1999;9:1231–41. doi: 10.1016/S0960-9822(99)80503-X. [DOI] [PubMed] [Google Scholar]
- 59.Kaffman A, Rank NM, O’Neill EM, Huang LS, O’Shea EK. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature. 1998;396:482–6. doi: 10.1038/24898. [DOI] [PubMed] [Google Scholar]
- 60.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–61. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 61.Juanes MA, Martínez-Garay CA, Igual JC, Bañó MC. Targeting and membrane insertion into the endoplasmic reticulum membrane of Saccharomyces cerevisiae essential protein Rot1. FEMS Yeast Res. 2010;10:639–47. doi: 10.1111/j.1567-1364.2010.00653.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.