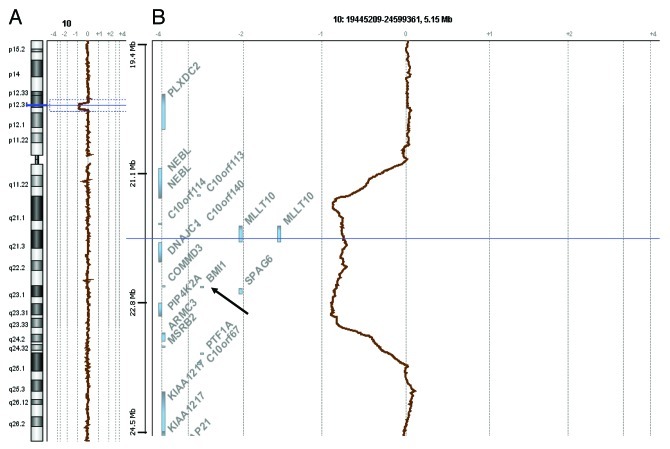
Recent articles in the Journal of Experimental Medicine and in Cell Cycle by the laboratory of A. Iwama established a role for polycomb gene BMI1 in leukemogenesis using a mouse model.1,2 Polycomb proteins regulate gene expression through the control of chromatin structure and repressive histone marks. They play major roles in development and stem cell biology. They distribute into two complexes called polycomb repressive complexes 1 and 2 (PRC1 and PRC2). PRC1 contains several proteins, including BMI1. PRC1 monoubiquitylates lysine 119 of histone H2A via the ubiquitin ligases RING1 and RNF2.3 PRC2 contains EED, EZH2 and SUZ12 proteins. PRC2 is a histone methyltransferase that trimethylates lysine 27 of histone H3, resulting in the H3K27me3 mark, which specifies transcriptional repression. Recent studies have shown that components of PRC2, especially EZH2, are targeted by inactivating mutations in human myeloid malignancies.4-7 PRC2 genes are mutated in acute stages, such as acute myeloid leukemia (AML), and chronic stages, such as myelodysplastic syndromes (MDS) or myeloproliferative neoplasms (MPN). The latter comprise polycythemia vera (PV), essential thrombocythemia (ET) and primary myelofibrosis (PMF). EZH2 is mutated in 13% of PMF cases and in 12% of cases with MDS/MPN features.5 Actually, through these direct inactivating mutations in its components or through alterations in PRC2-interacting proteins such as ASXL1,8 PRC2 appears as a key tumor suppressor complex in myeloid diseases. No such results have been reported yet for PRC1 genes in human samples. In contrast, because the loss of Bmi1 gene induces a defect in the self-renewal capacity of hematopoietic stem cells9 it was thought that PRC1 and PRC2 played opposite roles in leukemogenesis. Oguro and colleagues1 have just demonstrated that in an Ink4a/Arf-l- mouse the loss of the Bmi1 gene generated a disease similar to PMF. Mice repopulated with Bmi1-/-Ink4a-Arf−/− hematopoietic cells developed a lethal disease with clinical features observed in human PMF. This exciting result suggests that alterations of PRC1 complex components could, according to the genetic context (an abrogation of p16 and p19 tumor-suppressor control in the mouse model of Oguro and colleagues), play a role in the development of myeloid diseases similar to that of PRC2. PRC1 and PRC2 silencing functions are coordinated, and the two complexes may exert their leukemogenic effect through the deregulation of the same loci. Indeed, Oguro and colleagues identified oncogenic loci repressed by PRC1, such as Hmga2, and further demonstrated that Hmga2 promoter is also under the control of EZH2.1,2 A consequence of these results in human medicine is that alterations or deregulation of PRC1 components and controlled loci may substantially increase the proportion of hematopoietic diseases with defects in polycomb network genes. To determine whether BMI1 may be altered in human MPNs, we studied the genome of 35 PMF cases by using array-comparative genomic hybridization (aCGH) on high-density oligonucleotide microarrays (Hu-244A, Agilent Technologies), as previously described.10 We also searched for mutations in BMI1 in 77 MPN cases comprising 4 PV, 20 ET and 36 myelofibrosis (MF) (25 PMF, 8 post-ET MF, 3 post-PV MF), 3 MPN/MDS, 3 MPN-unclassifiable, 3 blast phase post-ET, 5 blast phase post-ET MF, 3 blast phase PMF using Sanger DNA sequencing of BMI1 coding exons 2 to 10. We did not find any mutations in BMI1. We found a heterozygous deletion of chromosome region 10p12.31 encompassing the BMI1 locus and seven other genes (Fig. 1) in case HD-1095, a 64-year-old man diagnosed with PMF. The HD-1095 sample had a JAK2V617F mutation (50–60% allele burden), was not mutated in ASXL1, DNMT3A, IDH1, IDH2 and TET2, and displayed other small chromosomal deletions that had led to the loss of SOCS2, FBXL18 and MYB, but not of CDKN2A/INK4A (not shown). To our knowledge, this is the first example of BMI1 locus alteration in human cancer. BMI1 deletion could lead to haploinsufficiency; unfortunately, RNA was not available for this sample. We studied BMI1 mRNA expression in 25 PMF profiled by Affymetrix microarrays. BMI1 expression in PMF was similar to that in normal blood (not shown), suggesting that the BMI1 locus is not inactivated by hypermethylation as a general rule. In this series studied by aCGH we did not find alterations of the HMGA2 locus, but we had previously reported one case of HMGA2 3′UTR breakage and mRNA overexpression in an MPN case.11 Finally, to determine whether BMI1 alteration could be found in other myeloid diseases, we searched for a deletion of the locus in 253 non-PMF samples (73 MPNs, 53 chronic myelomonocytic leukemias, 63 MDSs, and 64 AMLs) by using aCGH but found no other deleted case. Our results show that, although structural alterations of BMI1 and downstream effectors are rare, they do occur in human MPN samples. This supports both the findings of Oguro and colleagues in the mouse and the potential role of PRC1 in leukemogenesis. Whether other mechanisms of downregulation of BMI1 or alterations in other PRC1 components can be found in hematopoietic diseases remains to be demonstrated.
Figure 1. aCGH profile of chromosome 10 in one PMF case showing loss of the BMI1 region. (A) Chromosome 10 ideogram and aCGH profile of HD-1095 case. (B) zoom on the 10p12.31 band showing the deletion that spans chr10:21,274,386–23,428,386, according to hg18 UCSC (http:/genome.ucsc.edu), and includes the BMI1 gene (arrow).
Acknowledgments
This work was supported by Inserm, Institut Paoli-Calmettes and grants from Association pour la Recherche contre le Cancer and Association Laurette Fugain.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21114
References
- 1.Oguro H, et al. J Exp Med. 2012;209:445–54. doi: 10.1084/jem.20111709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mochizuki-Kashio M, et al. Cell Cycle. 2012;11:2043–4. doi: 10.4161/cc.20533. [DOI] [PubMed] [Google Scholar]
- 3.Bracken AP, et al. Nat Rev Cancer. 2009;9:773–84. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 4.Brecqueville M, et al. Blood Cancer J. 2011;1:e33. doi: 10.1038/bcj.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ernst T, et al. Nat Genet. 2010;42:722–6. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 6.Nikoloski G, et al. Nat Genet. 2010;42:665–7. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 7.Score J, et al. Blood. 2012;119:1208–13. doi: 10.1182/blood-2011-07-367243. [DOI] [PubMed] [Google Scholar]
- 8.Abdel-Wahab O, et al. ASH December 2011; Abstract 405; ASXL1 Mutations Promote Myeloid Transformation Through Inhibition of PRC2-Mediated Gene Repression. [DOI] [PMC free article] [PubMed]
- 9.Oguro H, et al. J Exp Med. 2006;203:2247–53. doi: 10.1084/jem.20052477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gelsi-Boyer V, et al. BMC Cancer. 2008;8:299. doi: 10.1186/1471-2407-8-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Etienne A, et al. Cancer Genet Cytogenet. 2007;176:80–8. doi: 10.1016/j.cancergencyto.2007.03.009. [DOI] [PubMed] [Google Scholar]