Abstract
Occult hepatitis B virus infection is characterized by the absence of detectable hepatitis B surface antigen (HBsAg) in the serum, despite detectable HBV DNA. Investigations of the mechanisms underlying the development of occult HBV infection are lacking in the current literature, although viral mutations in the surface region, resulting in decreased HBsAg expression or secretion, represent one potential mechanism. Wild-type HBsAg expression vectors were constructed from genotype-matched chronic HBV sequences. Site-directed mutagenesis was then utilized to introduce three genotype A mutations – M103I, K122R, and G145A – associated with occult HBV infection in vivo, alone and in combination, into the wild-type HBsAg vectors. Transfection of Huh7 and HepG2 cell lines was performed, and cell culture supernatants and cell lysates were collected over 7 days to assess the effects of these mutations on extracellular and intracellular HBsAg levels. The G145A mutation resulted in significantly decreased extracellular and intracellular HBsAg expression in vitro. The most pronounced reduction in HBsAg expression was observed when all 3 mutations were present. The mutations evaluated in vitro in the current study resulted in decreased HBsAg expression and potentially increased hepatic retention and/or decreased hepatic secretion of synthesized HBsAg, which could explain the lack of HBsAg detection that is characteristic of occult HBV infection in vivo.
Keywords: G145A, HBV/HIV co-infection, HBsAg mutants, hepatitis B surface antigen (HBsAg), occult hepatitis B virus
Introduction
Greater than 2 billion people have been exposed to hepatitis B virus (HBV) worldwide, and more than 350 million individuals are currently living with chronic HBV infection (CHBV)(1). While C-HBV infection is diagnosed by detection of serum hepatitis B surface antigen (HBsAg)(2), occult HBV infection (O-HBV) is characterized by a lack of detectable HBsAg in the serum despite low-level HBV replication(3). O-HBV infection was once defined by antibodies against HBV core (anti-HBc) alone(4), although cases have now been reported in which HBV DNA is the only detectable marker of O-HBV infection(5). The clinical relevance of O-HBV has not been thoroughly explored and remains controversial(6); however, several studies have suggested a potential association of O-HBV infection with increased incidence of hepatocellular carcinoma (HCC) and severity of liver fibrosis(7-11). In addition, O-HBV is transmissible via blood transfusion in humans and primates, as well as solid organ transplantation(12-15), from which the recipient may develop either C-HBV or O-HBV.
Globally, studies have reported rates of O-HBV infection from 0% – 89.5%, depending on the population, laboratory methods, and inclusion/exclusion criteria(16,17). In the US, O-HBV prevalence ranges from 0% – 45% with higher rates in HIV and/or HCV co-infected patients(18,19). Few studies have included virologic comparisons between C-HBV and O-HBV patients from the same cohort, and functional analysis of mutations associated with O-HBV infection is rarely performed. Potential mechanisms resulting in O-HBV infection include 1) complexes of HBsAg with antibodies against surface (anti-HBs) that circulate in the serum but are not readily detected by standard HBsAg ELISAs, 2) interference of HBV replication by other viruses, 3) altered host immunologic responses, and/or 4) mutations within the HBV genome that change HBsAg expression(20). We previously identified mutations associated with O-HBV infection by comparing viral sequences from O-HBV infections and genotype-matched C-HBV infections from the same cohort(21). Three O-HBV mutations – M103I, K122R, and G145A – from a genotype A infection were significant by signature pattern analysis. M103I had been previously identified in 2 O-HBV infections, although these patients harbored genotype C or D(22). K122R – which indicates a sub-serotype change from d to y – has also been identified in one genotype A O-HBV infection(22). Finally, G145A was reported in one O-HBV infection of unknown genotype(23). Since these small HBsAg mutations have not been well characterized, the current study evaluated their effects on HBsAg synthesis and secretion in vitro.
Materials and Methods
Mutations Associated with O-HBV Infection
O-HBV-infected patients were identified by evaluating patient sera for markers of HBV infection, including real-time PCR for HBV DNA and ELISAs for serologic markers (HBsAg, anti-HBs, and anti-HBc)(5). Mutations associated with O-HBV were identified in the HBV Pre-Surface, Surface, and Polymerase regions as described previously(21). Three mutations within the small HBsAg – M103I, K122R, and G145A – from an individual with a genotype A infection, were evaluated in the current study, since genotype A represents the most common HBV genotype in the US(24). The most common amino acids at these positions among the representative GenBank reference sequences are listed in Table 1.
Table 1. Most Common Amino Acids at Positions of Interest by Genotype.
Five GenBank references for each HBV genotype (A-H), used previously to identify mutations associated with O-HBV infection(21), were screened to identify the most common amino acid present at positions 103, 122, and 145 of the small HBsAg compared to the mutations associated with OHBV being examined.
| Position | Mutation | A | B | C | D | E | F | G | H |
|---|---|---|---|---|---|---|---|---|---|
| 103 | I | M* | M | M | M | M* | M | M | M |
| 122 | R | K* | K | K* | R | R | K | K | K |
| 145 | A | G | G | G | G | G | G | G | G |
indicates that the amino acid position was variable for the given genotype.
HBsAg Expression Vector Construction
The O-HBV-infected patient was matched to 2 C-HBV-infected, HIV-positive individuals from the same cohort, based on HBV genotype, age <5 years apart, gender, ethnicity, and HCV serostatus. HBV viral load for the O-HBV patient was 7.61×105 IU/mL, while the C-HBV patients were 1.48×107 and 9.74×102 IU/mL. First round PCR amplification was performed as previously described(25) with the Picomaxx High Fidelity PCR System (Agilent, Santa Clara, CA). Second round primers – sHBs-EcoRI-F (5' – CGA ATT CGG GAC CCT GTG ACG AAC) and sHBs-KpnI-R (5' – GGG GTA CCC ATC TCT TTG TTT TGT TAG) – designed to amplify the small S region (721bp, nt138-859, according to X97848), contained EcoRI and KpnI sites to facilitate cloning into pCMV-HA (Clontech, Mountain View, CA), producing 5'hemagglutinin (HA)-tagged small HBsAg proteins. Site-directed mutagenesis introduced O-HBV-associated mutations – separately or in combination – into each C-HBV expression vector (Table 2) using the QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent).
Table 2. Genotype A HBsAg Expression Vectors.
Wild-type partial S ORF sequences were derived from 2 genotype A C-HBV infections and ligated into the pCMV-HA vector. The wild-type vectors served as positive controls and were the backbones into which mutations associated with O-HBV infection were introduced by site-directed mutagenesis. A total of 17 vectors were utilized in these experiments as listed above.
| Genotype A HBsAg Expression Vectors | |
|---|---|
| Wild-Type 1 | Wild-Type 2 |
| Wild-Type 1 + M103I mutation | Wild-Type 2 + M103I mutation |
| Wild-Type 1 + K122R mutation | Wild-Type 2 + K122R mutation |
| Wild-Type 1 + G145A mutation | Wild-Type 2 + G145A mutation |
| Wild-Type 1 + M103I + K122R mutations | Wild-Type 2 + M103I + K122R mutations |
| Wild-Type 1 + M103I + G145A mutations | Wild-Type 2 + M103I + G145A mutations |
| Wild-Type 1 + K122R + G145A mutations | Wild-Type 2 + K122R + G145A mutations |
| Wild-Type 1 + M103I + K122R + G145A mutations | Wild-Type 2 + M103I + K122R + G145A mutations |
| pCMV-HA alone (negative control) | |
Transfection and HBsAg Quantitation
Huh7 and HepG2 cells were maintained in DMEM (10% FBS, 1% penicillin/streptomycin) and RPMI-1640 (4% FBS, 4mM L-glutamine), respectively. Huh7 cells were seeded at 5×104 cells/well in 24-well plates, while HepG2 cells were seeded at 1×105 cells/well. 24 hours later, cells were transfected in duplicate with 2ug DNA/well with 3uL FuGENE6 (Roche, Indianapolis, IN). Media were changed after 24 hours, and collected every 48 hours thereafter. Supernatants collected on days 3 and 5 were pooled for the day 5 sample, while supernatants collected on days 3, 5, and 7 were pooled for the day 7 sample, and clarified by centrifugation at 10,000rpm for 5 minutes. Cells were washed three times with PBS and lysed with 200uL lysis buffer – Tris-HCl (50mmol/L, pH 7.8), NaCl (150mmol/L), EDTA (1mmol/L), 0.5% Nonidet P-40 (NP-40), and 1/100th volume of protease inhibitors (Complete Mini Protease Inhibitor Cocktail, Roche). Plates were frozen, thawed, and lysates were clarified by centrifugation at 13,000g for 3 minutes. Cell viability post-transfection was determined by trypan blue staining on day 7. Cells were transfected with 2ug wild-type vector 1, G145A, M103I+K122R+G145A, or pCMV-HA. After 7 days, cells were washed, trypsinized, and resuspended in fresh media before staining with trypan blue and counting. Mean number of cells were determined for each construct and log-transformed for comparison (Supplemental Figure 1A). Additionally, cells were seeded to determine transfection efficiency. 50ng of plasmid p0Z-17, encoding for β–galactosidase, was co-transfected along with 2ug vector, while pCMV-HA alone served as a negative control. On day 7 post-transfection, cells were washed twice with PBS, and fixed for 5 minutes with 2% formaldehyde. Cells were incubated with 5mM phosphate ferricyanide, 5mM phosphate ferrocyanide, 2mM MgCl2, and 1mg/mL X-gal in PBS, for 2 hours at 37°C. Five 25X fields of view were counted per well to determine the number of blue cells/well. The average was calculated and compared between constructs (Supplemental Figure 1B).
HBsAg was quantitated by ELISA (Biochain, Hayward, CA) with recombinant HBsAg (adr subtype) standards from 500ng/mL-0.25ng/mL with a lower limit of detection of 0.5ng/mL. The OD450 was measured and compared to a standard curve from 0.25ng/mL-10ng/mL. Samples with OD450 >10ng/mL were diluted and re-measured. Total nanograms of HBsAg were corrected for the proper dilution and lysateHBsAg:supernatantHBsAg ratios were calculated for each construct.
Statistical Analysis
For each cell type, compartment, wild-type group, and time point, data were analyzed by a one-factor, repeated-measures ANOVA. Type of mutant (with wild-type as the reference level) was the repeated factor, and comparisons of each mutant condition to wild-type were performed using Dunnett's procedure for comparing multiple treatments to a common control condition. The maximum family-wise type I error rate for Dunnett's procedure was set at α=0.05.
Results
Wild-type HBsAg Expression
Supernatant and lysate HBsAg levels from transfected Huh7 and HepG2 cells were assessed for wild-type (i.e., C-HBV) expression vectors on days 3, 5, and 7 post-transfection. Overall, HBsAg expression was higher in Huh7 cells (Figures 1A-B compared to 1C-D). No significant differences were observed between wells with regards to viable cell number (Supplemental Figure 1A) or transfection efficiency (Supplemental Figure 1B). Wild-type supernatant HBsAg levels in Huh7 cells increased between days 3 and 5 post-transfection and decreased by day 7 (Figure 1A). Lysate HBsAg levels in Huh7 cells showed a similar trend (Figure 1B). These trends were also observed in HepG2 cells (Figures 1C-D) and did not vary when a second wild-type expression vector was utilized (Supplemental Figure 2). However, overall HBsAg levels were higher in the supernatant, while lysate HBsAg levels were lower, indicating increased HBsAg secretion with the second C-HBV backbone, presumably due to the 2 amino acid mutations (G164E and S212F) that differentiate the C-HBV sequences.
Figure 1.
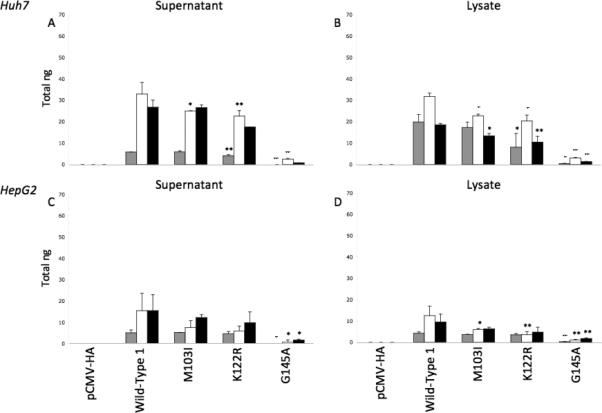
Total ng of HBsAg produced by transfection of pCMV-HA alone, wild-type vector 1 alone or with mutations M103I, K122R, or G145A, were calculated for supernatants (A,C) and cell lysates (B,D) from Huh7 (A,B) and HepG2 (C,D) cells in duplicate and compared at days 3 (grey bars), 5 (white bars), and 7 (black bars) post-transfection. * p<0.05; ** p<0.01; + p<0.001; ++ p<0.0001.
Effects of Single Point Mutations on HBsAg Expression
HBsAg expression was also assessed for the M103I, K122R, or G145A mutations. Supernatant levels of HBsAg were similar between wild-type, M103I, and K122R in both Huh7 and HepG2 cells (Figures 1A,C). In lysates, HBsAg levels were slightly lower for expression vectors containing the M103I and K122R mutations compared to wild-type (Figures 1B,D). In contrast, the G145A mutation resulted in significantly lower HBsAg levels in both cell supernatants and lysates. These trends were also observed for the second wild-type expression vector (Supplemental Figure 2).
Effects of Multiple Mutations on HBsAg Expression
When examining the combined effects of these mutations, M103I+K122R resulted in decreased HBsAg levels in both supernatants (Figures 2A,C) and lysates (Figures 2B,D), but remained well above the ELISA detection limit. In contrast, combinations of mutations containing G145A resulted in significantly lower HBsAg levels – some below detectable limits. The most pronounced reductions in HBsAg synthesis was observed with M103I+K122R+G145A (Figure 2, Supplemental Figure 3).
Figure 2.
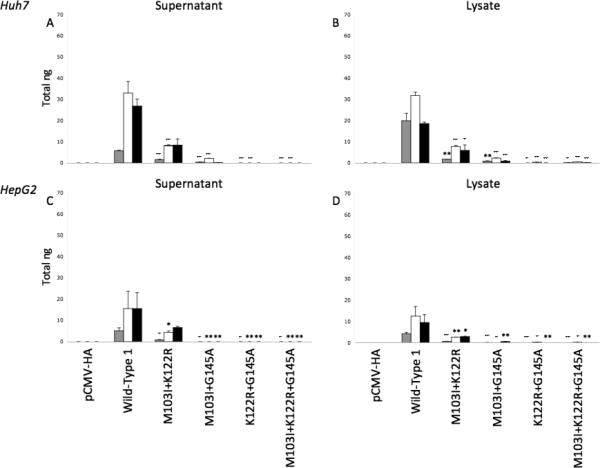
Total ng of HBsAg produced by transfection of pCMV-HA alone, wild-type vector 1 alone or with mutation combinations M103I+K122R, M103I+G145A, K122R+G145A, or M103I+K122R+G145A, were calculated for supernatants (A,C) and cell lysates (B,D) from Huh7 (A,B) and HepG2 (C,D) cells in duplicate and compared for days 3 (grey bars), 5 (white bars), and 7 (black bars) post-transfection. * p<0.05; ** p<0.01; + p<0.001; ++ p<0.0001.
Effects of Mutations on HBsAg Secretion
Ratios of lysate (intracellular) to supernatant (extracellular) HBsAg were calculated to compare HBsAg secretion versus retention. At day 3 post-transfection, the wild-type vector resulted in higher intracellular levels of HBsAg in Huh7 cells (ratios >1 in bold, Table 3). Over time, HBsAg secretion increased, resulting in ratios <1. The G145A, M103I+G145A, K122R+G145A, and M103I+K122R+G145A vectors continued to have higher intracellular HBsAg levels at all time points in Huh7 cells, when above the detection limit. Similar results were observed in HepG2 cells, in which the only constructs with ratios >1 were those containing the G145A mutation alone or in combination. However, these findings were not observed with the second wild-type vectors (Supplemental Table 1), since lysate HBsAg levels were lower overall with these constructs.
Table 3. HBsAglysate:HBsAgsupernatant Ratios.
Ratios comparing lysate HBsAg to supernatant HBsAg (HBsAglysate:HBsAgsupernatant) were calculated at days 3, 5, and 7 post-transfection for all constructs with the wild-type 1 backbone in both Huh7 and HepG2 cells. Bold values indicate constructs with higher intracellular/lysate levels of HBsAg (values >1). UD – indicates undetectable levels of HBsAg (below the lower limit of detection for the ELISA, <0.5ng/mL) in the lysate and/or supernatant.
| Day 3 | Day 5 | Day 7 | ||||
|---|---|---|---|---|---|---|
| Construct | Huh7 | HepG2 | Huh7 | HepG2 | Huh7 | HepG2 |
| Wild Type 1 | 3.44 | 0.83 | 0.98 | 0.85 | 0.70 | 0.63 |
| M103I | 2.95 | 0.68 | 0.90 | 0.83 | 0.51 | 0.52 |
| K122R | 1.88 | 0.76 | 0.90 | 0.60 | 0.60 | 0.50 |
| G145A | UD/0.43 | UD/0.29 | 1.27 | UD/1.09 | 1.61 | 1.11 |
| M103I+K122R | 1.18 | 1.12 | 0.96 | 0.62 | 0.69 | 0.44 |
| M103I+G145A | 2.64 | UD/0.17 | 1.09 | UD/UD | UD/0.85 | UD/0.62 |
| K122R+G145A | UD/UD | UD/UD | UD/0.31 | UD/0.35 | UD/UD | UD/UD |
| M103I+K122R+G145A | UD/0.08 | UD/UD | UD/0.44 | UD/0.26 | UD/0.08 | UD/UD |
| pCMV-HA | UD/UD | UD/UD | UD/UD | UD/UD | UD/UD | UD/UD |
Discussion
To date, most O-HBV studies have focused on prevalence(16,19) and/or identification of mutations associated with O-HBV(22,23). In contrast, functional analysis of specific HBV mutations on HBsAg synthesis has rarely been reported(26-28). Thus, to determine whether mutations associated with O-HBV infection may play a role in the lack of detectable HBsAg in these individuals, additional investigation is required. The current in vitro system specifically evaluates HBsAg protein expression, since the CMV-IE promoter drives transcription with little variation in mRNA expression. Thus, we examined the effects of three previously identified O-HBV mutations in the S region – M103I, K122R, and G145A – on HBsAg expression(21).
Of the three mutations characterized, G145A contributes the most to decreased HBsAg synthesis in vitro. Additional mutations at position 145 of HBsAg have been previously characterized, although not always in O-HBV infections(29-31). The G145R mutation has been described as an immune escape mutant(31), as well as a potential contributor to lamivudine resistance(29); therefore, G145A may also represent a potential vaccine escape mutant(32). The G145R mutation has consistently resulted in decreased HBsAg levels and increased replication in lamivudine resistant mutants, especially among those with additional polymerase mutations, such as L180M and M204V(30). In this study, extracellular levels of HBsAg were slightly above detectable levels at times with the G145A mutation, indicating that the HBsAg secreted from infected hepatocytes would likely be below the limit of detection for commercial assays once diluted in the blood. Although the lack of detectable HBsAg could represent a diagnostic error(33-37), HBsAg synthesis was consistently detectable for G145A with the commercial ELISA kit utilized even at low levels. Western Blot to detect HBsAg was also attempted but was found to be much less sensitive than ELISA (data not shown). In addition, one study compared several commercial ELISA kits to assess detection of amino acid changes at position 145, including G145A(38). Although G145A had not been identified in vivo, it was detectable with all kits tested; thus, a false-negative HBsAg result in vivo seems unlikely.
The M103I or K122R mutations alone did not significantly affect HBsAg expression, although it is interesting to note that when these two mutations were combined, HBsAg expression was decreased. Although the K122R mutation results in a serotype change from ‘d’ to ‘y’, both K122R and M103I may have minor effects with respect to protein folding, antigenicity, detection by anti-HBs, and protein expression, due to their location within the antigenic determinant region of HBsAg(20,22,28,34,37,39).
For one wild-type construct, the small amount of HBsAg produced with the G145A mutation, alone or in combination, was retained within hepatocytes, as indicated by higher intracellular HBsAg ratios in Table 3. While this could represent an artifact of low overall HBsAg levels and was only observed for one of the two wild-type constructs, it is intriguing given that certain HBsAg mutations may result in a drastic increase in retention of viral proteins due to misfolding(40-42). However, the low levels of intracellular HBsAg observed indicate that if HBsAg misfolding is occurring, then proper proteasomal degradation is likely still taking place. Nonetheless, future investigations should evaluate the folding and sub-cellular localization of mutant forms of HBsAg.
The current investigation focused on the impact of HBsAg mutations on protein expression without influence from potential mutations in other HBV regions. While full-length HBV PCR was attempted, the concentration of PCR product was too low to permit subsequent generation of replication-competent vectors. A small number of prior studies have utilized full-length, replication-competent O-HBV genomes. For example, one polymerase mutation (terminal protein mutation T165P)(43) or several amino acid mutations throughout the genome(44,45) have resulted in defective replication or overall decreases in HBsAg and HBV DNA levels. Other studies identified a single nucleotide(26) or amino acid mutation(34) that lead to decreased PS2/S mRNA levels, resulting in decreased HBsAg. Additional investigations studied heavily mutated viral sequences within the PS1 and PS2/S promoters(46) or S region(27) that lead to altered HBsAg secretion. However, to date, only three studies have focused specifically on individual mutations within the S region and their effects on HBsAg synthesis. One study evaluated the Y100C mutation. Surprisingly, HBsAg levels were higher than wild-type; thus, why HBsAg was not detectable in the serum remains unclear(47). In the second study, HBsAg expression was decreased by 40% with the P142A mutation and by 60% with the D144V mutation; however, when both mutations were included, HBsAg expression was decreased by only 20%(28). Finally, in the third study, the I110M, G119E, and R169P mutations were each capable of impairing virion secretion(48). These studies, along with the current investigation, demonstrate that multiple mutations within HBsAg have the potential to impact HBsAg expression and secretion, although the specific mutations may differ among O-HBV-infected patients.
Several aspects of the current investigation are unique compared to previously published studies. First, the specific mutations assessed in this study – M103I, K122R, and G145A – had not been included in previous functional analyses of HBsAg expression during O-HBV infection and, therefore, extend the small body of previously available research that utilized in vitro models to evaluate specific instances of undetectable HBsAg in vivo. The current study also examined changes in HBsAg expression over 7 days, since HBsAg synthesis and secretion are dynamic processes that may fluctuate over time. Additionally, both Huh7 and HepG2 cell lines were utilized to assess potential differences between these cell lines in vitro, since hepatocyte cell lines may not recapitulate infection in humans. Finally, two chronic/wild-type HBV sequences were used as backbones for the constructed HBsAg expression vectors to assess the consistency of the findings.
In summary, the S gene mutations M103I, K122R, and G145A were evaluated for their impact on HBsAg synthesis given that a lack of detectable HBsAg in the serum is a hallmark of O-HBV infection. The G145A mutation, alone and in combination, resulted in significantly decreased extracellular and intracellular HBsAg expression. Interestingly, the G145A mutation, also results in a W153C mutation in the reverse transcriptase region of the HBV polymerase. Previous mutations at this position, although not W153C, have been shown to decrease HBV replication(30). Additional cohorts will be utilized in the future to generate full-length, replication-competent viruses to further evaluate the simultaneous effects of these and other mutations on HBsAg synthesis and viral replication.
Supplementary Material
Acknowledgements
Funding was provided in part by a Cincinnati Digestive Health Center pilot award (P30 DK078392) to JTB and by an NIDDK K24 award (DK070528) to KES. This publication was also supported by an Institutional Clinical and Translational Science Award, NIH/NCRR Grant (5UL1RR026314-03). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
This work was presented at the American Society for Virology 30th Annual Meeting in Minneapolis, MN 7/16/2011 - 7/20/2011.
The authors report no conflicts of interest.
References
- 1.Lavanchy D. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. Journal of Viral Hepatitis. 2004;11(2):97–107. doi: 10.1046/j.1365-2893.2003.00487.x. [DOI] [PubMed] [Google Scholar]
- 2.Lok ASF, McMahon BJ. Chronic hepatitis B. Hepatology. 2007;45(2):507–39. doi: 10.1002/hep.21513. [DOI] [PubMed] [Google Scholar]
- 3.Bodsworth NJ, Cooper DA, Donovan B. The influence of human immunodeficiency virus type 1 infection on the development of the hepatitis B virus carrier state. Journal of Infectious Diseases. 1991;163(5):1138–40. doi: 10.1093/infdis/163.5.1138. [DOI] [PubMed] [Google Scholar]
- 4.Jilg W, Sieger E, Zachoval R, Schatzl H. Individuals with antibodies against hepatitis B core antigen as the only serological marker for hepatitis B infection: high percentage of carriers of hepatitis B and C virus. Journal of Hepatology. 1995;23(1):14–20. doi: 10.1016/0168-8278(95)80305-x. [DOI] [PubMed] [Google Scholar]
- 5.Shire NJ, Rouster SD, Stanford SD, et al. The prevalence and significance of occult hepatitis B virus in a prospective cohort of HIV-infected patients. Journal of Acquired Immune Deficiency Syndromes. 2007;44(3):309–14. doi: 10.1097/QAI.0b013e31802e29a9. [DOI] [PubMed] [Google Scholar]
- 6.Lok AS, Everhart JE, Di Bisceglie AM, et al. Occult and previous hepatitis B virus infection are not associated with hepatocellular carcinoma in United States patients with chronic hepatitis C. Hepatology. 2011;54(2):434–42. doi: 10.1002/hep.24257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikeda K, Kobayashi M, Someya T, et al. Occult hepatitis B virus infection increases hepatocellular carcinogenesis by eight times in patients with non-B, non-C liver cirrhosis: a cohort study. Journal of Viral Hepatitis. 2009;16(6):437–43. doi: 10.1111/j.1365-2893.2009.01085.x. [DOI] [PubMed] [Google Scholar]
- 8.Kannangai R, Molmenti E, Arrazola L, et al. Occult hepatitis B viral DNA in liver carcinomas from a region with a low prevalence of chronic hepatitis B infection. Journal of Viral Hepatitis. 2004;11(4):297–301. doi: 10.1111/j.1365-2893.2004.00502.x. [DOI] [PubMed] [Google Scholar]
- 9.Matsuoka S, Nirei K, Tamura A, et al. Influence of occult hepatitis B virus coinfection on the incidence of fibrosis and hepatocellular carcinoma in chronic hepatitis C. Intervirology. 2008;51(5):352–61. doi: 10.1159/000187720. [DOI] [PubMed] [Google Scholar]
- 10.Squadrito G, Pollicino T, Cacciola I, et al. Occult hepatitis B virus infection is associated with the development of hepatocellular carcinoma in chronic hepatitis C patients. Cancer. 2006;106(6):1326–30. doi: 10.1002/cncr.21702. [DOI] [PubMed] [Google Scholar]
- 11.Tamori A, Nishiguchi S, Kubo S, et al. Sequencing of human-viral DNA junctions in hepatocellular carcinoma from patients with HCV and occult HBV infection. Journal of Medical Virology. 2003;69(4):475–81. doi: 10.1002/jmv.10334. [DOI] [PubMed] [Google Scholar]
- 12.Baginski I, Chemin I, Hantz O, et al. Transmission of serologically silent hepatitis B virus along with hepatitis C virus in two cases of posttransfusion hepatitis. Transfusion. 1992;32(3):215–20. doi: 10.1046/j.1537-2995.1992.32392213803.x. [DOI] [PubMed] [Google Scholar]
- 13.Campe H, Hillebrand GF, Mairhofer H, Nitschko H, Jager G. Undetected chronic hepatitis B virus infection of a vaccinated dialysis patient after liver transplantation. Nephrology, Dialysis, Transplantation. 2005;20(7):1492–4. doi: 10.1093/ndt/gfh823. [DOI] [PubMed] [Google Scholar]
- 14.Thiers V, Nakajima E, Kremsdorf D, et al. Transmission of hepatitis B from hepatitis-B-seronegative subjects. Lancet. 1988;2(8623):1273–6. doi: 10.1016/s0140-6736(88)92891-7. [DOI] [PubMed] [Google Scholar]
- 15.Yuen M-F, Ka-Ho Wong D, Lee C-K, et al. Transmissibility of Hepatitis B Virus (HBV) Infection through Blood Transfusion from Blood Donors with Occult HBV Infection. Clinical Infectious Diseases. 2011;52(5):624–32. doi: 10.1093/cid/ciq247. [DOI] [PubMed] [Google Scholar]
- 16.Hofer M, Joller-Jemelka HI, Grob PJ, Luthy R, Opravil M. Frequent chronic hepatitis B virus infection in HIV-infected patients positive for antibody to hepatitis B core antigen only. Swiss HIV Cohort Study. European Journal of Clinical Microbiology & Infectious Diseases. 1998;17(1):6–13. doi: 10.1007/BF01584356. [DOI] [PubMed] [Google Scholar]
- 17.Nunez M, Rios P, Perez-Olmeda M, Soriano V. Lack of ‘occult’ hepatitis B virus infection in HIV-infected patients. AIDS. 2002;16(15):2099–101. doi: 10.1097/00002030-200210180-00024. [DOI] [PubMed] [Google Scholar]
- 18.Rodríguez-Torres M, Gonzalez-Garcia J, Bräu N, et al. Occult hepatitis B virus infection in the setting of hepatitis C virus (HCV) and human immunodeficiency virus (HIV) co-infection: Clinically relevant or a diagnostic problem? Journal of Medical Virology. 2007;79(6):694–700. doi: 10.1002/jmv.20836. [DOI] [PubMed] [Google Scholar]
- 19.Torbenson M, Kannangai R, Astemborski J, Strathdee SA, Vlahov D, Thomas DL. High prevalence of occult hepatitis B in Baltimore injection drug users. Hepatology. 2004;39(1):51–7. doi: 10.1002/hep.20025. [DOI] [PubMed] [Google Scholar]
- 20.Hu K-Q. Occult hepatitis B virus infection and its clinical implications. Journal of Viral Hepatitis. 2002;9(4):243–57. doi: 10.1046/j.1365-2893.2002.00344.x. [DOI] [PubMed] [Google Scholar]
- 21.Martin CM, Welge JA, Shire NJ, et al. Genomic variability associated with the presence of occult hepatitis B virus in HIV co-infected individuals. Journal of Viral Hepatitis. 2010;17(8):588–97. doi: 10.1111/j.1365-2893.2009.01214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weinberger KM, Bauer T, Bohm S, Jilg W. High genetic variability of the group-specific a-determinant of hepatitis B virus surface antigen (HBsAg) and the corresponding fragment of the viral polymerase in chronic virus carriers lacking detectable HBsAg in serum. Journal of General Virology. 2000;81(5):1165–74. doi: 10.1099/0022-1317-81-5-1165. [DOI] [PubMed] [Google Scholar]
- 23.Kao J-H, Chen P-J, Lai M-Y, Chen D-S. Sequence analysis of pre-S/surface and pre-core/core promoter genes of hepatitis B virus in chronic hepatitis C patients with occult HBV infection. Journal of Medical Virology. 2002;68(2):216–20. doi: 10.1002/jmv.10188. [DOI] [PubMed] [Google Scholar]
- 24.Chu C-J, Keeffe EB, Han S-H, et al. Hepatitis B virus genotypes in the United States: results of a nationwide study. Gastroenterology. 2003;125(2):444–51. doi: 10.1016/s0016-5085(03)00895-3. [DOI] [PubMed] [Google Scholar]
- 25.Günther S, Li BC, Miska S, Kruger DH, Meisel H, Will H. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. Journal of Virology. 1995;69(9):5437–44. doi: 10.1128/jvi.69.9.5437-5444.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hass M, Hannoun C, Kalinina T, Sommer G, Manegold C, Günther S. Functional analysis of hepatitis B virus reactivating in hepatitis B surface antigen-negative individuals. Hepatology. 2005;42(1):93–103. doi: 10.1002/hep.20748. [DOI] [PubMed] [Google Scholar]
- 27.Jeantet D, Chemin I, Mandrand B, Zoulim F, Trepo C, Kay A. Characterization of two hepatitis B virus populations isolated from a hepatitis B surface antigen-negative patient. Hepatology. 2002;35(5):1215–24. doi: 10.1053/jhep.2002.32710. [DOI] [PubMed] [Google Scholar]
- 28.Schories M, Peters T, Rasenack J. Isolation, characterization and biological significance of hepatitis B virus mutants from serum of a patient with immunologically negative HBV infection. Journal of Hepatology. 2000;33(5):799–811. doi: 10.1016/s0168-8278(00)80313-x. [DOI] [PubMed] [Google Scholar]
- 29.Amini-Bavil-Olyaee S, Alavian S-M, Adeli A, et al. Hepatitis B virus genotyping, core promoter, and precore/core mutations among Afghan patients infected with hepatitis B: A preliminary report. Journal of Medical Virology. 2006;78(3):358–64. doi: 10.1002/jmv.20547. [DOI] [PubMed] [Google Scholar]
- 30.Bock CT, Tillmann HL, Torresi J, et al. Selection of hepatitis B virus polymerase mutants with enhanced replication by lamivudine treatment after liver transplantation. Gastroenterology. 2002;122(2):264–73. doi: 10.1053/gast.2002.31015. [DOI] [PubMed] [Google Scholar]
- 31.Echevarría JM, Avellón A. Hepatitis B virus genetic diversity. Journal of Medical Virology. 2006;78(S1):S36–S42. doi: 10.1002/jmv.20605. [DOI] [PubMed] [Google Scholar]
- 32.Seddigh-Tonekaboni S, Waters JA, Jeffers S, et al. Effect of variation in the common “a” determinant on the antigenicity of hepatitis B surface antigen. Journal of Medical Virology. 2000;60(2):113–21. doi: 10.1002/(sici)1096-9071(200002)60:2<113::aid-jmv2>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 33.El Chaar M, Candotti D, Crowther RA, Allain JP. Impact of hepatitis B virus surface protein mutations on the diagnosis of occult hepatitis B virus infection. Hepatology. 2010;52(5):1600–10. doi: 10.1002/hep.23886. [DOI] [PubMed] [Google Scholar]
- 34.Jeantet D, Chemin I, Mandrand B, et al. Cloning and expression of surface antigens from occult chronic hepatitis B virus infections and their recognition by commercial detection assays. Journal of Medical Virology. 2004;73(4):508–15. doi: 10.1002/jmv.20119. [DOI] [PubMed] [Google Scholar]
- 35.Louisirirotchanakul S, Khupulsup K, Akraekthalin S, et al. Comparison of the technical and clinical performance of the Elecsys® HBsAg II assay with the Architect®, AxSym®, and Advia® Centaur HBsAg screening assays. Journal of Medical Virology. 2010;82(5):755–62. doi: 10.1002/jmv.21706. [DOI] [PubMed] [Google Scholar]
- 36.Mizuochi T, Okada Y, Umemori K, Mizusawa S, Yamaguchi K. Evaluation of 10 commercial diagnostic kits for in vitro expressed hepatitis B virus (HBV) surface antigens encoded by HBV of genotypes A to H. Journal of Virological Methods. 2006;136(1-2):254–6. doi: 10.1016/j.jviromet.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 37.Moerman B, Moons V, Sommer H, Schmitt Y, Stetter M. Evaluation of sensitivity for wild type and mutant forms of hepatitis B surface antigen by four commercial HBsAg assays. Clinical Laboratory. 2004;50(3-4):159–62. [PubMed] [Google Scholar]
- 38.Coleman PF, Chen YC, Mushahwar IK. Immunoassay detection of hepatitis B surface antigen mutants. Journal of Medical Virology. 1999;59(1):19–24. doi: 10.1002/(sici)1096-9071(199909)59:1<19::aid-jmv4>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 39.Wu C, Zhang X, Tian Y, et al. Biological significance of amino acid substitutions in hepatitis B surface antigen (HBsAg) for glycosylation, secretion, antigenicity and immunogenicity of HBsAg and hepatitis B virus replication. Journal of General Virology. 2010;91(2):483–92. doi: 10.1099/vir.0.012740-0. [DOI] [PubMed] [Google Scholar]
- 40.Bruss V, Ganem D. Mutational analysis of hepatitis B surface antigen particle assembly and secretion. Journal of Virology. 1991;65(7):3813–20. doi: 10.1128/jvi.65.7.3813-3820.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalinina T, Riu A, Fischer L, Will H, Sterneck M. A dominant hepatitis B virus population defective in virus secretion because of several S-gene mutations from a patient with fulminant hepatitis. Hepatology. 2001;34(2):385–94. doi: 10.1053/jhep.2001.26516. [DOI] [PubMed] [Google Scholar]
- 42.Khan N, Guarnieri M, Ahn SH, et al. Modulation of hepatitis B virus secretion by naturally occurring mutations in the S gene. Journal of Virology. 2004;78(7):3262–70. doi: 10.1128/JVI.78.7.3262-3270.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blum HE, Galun E, Liang TJ, von Weizsacker F, Wands JR. Naturally occurring missense mutation in the polymerase gene terminating hepatitis B virus replication. Journal of Virology. 1991;65(4):1836–42. doi: 10.1128/jvi.65.4.1836-1842.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blum HE, Liang TJ, Galun E, Wands JR. Persistence of hepatitis B viral DNA after serological recovery from hepatitis B virus infection. Hepatology. 1991;14(1):56–63. doi: 10.1002/hep.1840140110. [DOI] [PubMed] [Google Scholar]
- 45.Miyagawa M, Minami M, Fujii K, et al. Molecular characterization of a variant virus that caused de novo hepatitis B without elevation of hepatitis B surface antigen after chemotherapy with rituximab. Journal of Medical Virology. 2008;80(12):2069–78. doi: 10.1002/jmv.21311. [DOI] [PubMed] [Google Scholar]
- 46.Sengupta S, Rehman S, Durgapal H, Acharya SK, Panda SK. Role of surface promoter mutations in hepatitis B surface antigen production and secretion in occult hepatitis B virus infection. Journal of Medical Virology. 2007;79(3):220–8. doi: 10.1002/jmv.20790. [DOI] [PubMed] [Google Scholar]
- 47.Mello FC, Martel N, Gomes SA, Araujo NM. Expression of hepatitis B virus surface antigen containing Y100C variant frequently detected in occult HBV infection. Hepatitis Research and Treatment. 2011 doi: 10.1155/2011/695859. Epub 2011 Feb 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ito K, Qin Y, Guarnieri M, et al. Impairment of Hepatitis B Virus Virion Secretion by Single-Amino-Acid Substitutions in the Small Envelope Protein and Rescue by a Novel Glycosylation Site. Journal of Virology. 2010;84(24):12850–61. doi: 10.1128/JVI.01499-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


