SUMMARY
Culturing methods are the primary approach for microbiological analysis of plaque-biofilms in rodent models of dental caries. In this study, we developed strategies for isolation of DNA and RNA from in vivo formed plaque-biofilms to analyze the viable bacterial population and gene expression. Plaque-biofilm samples from rats were treated with propidium monoazide to isolate DNA from viable cells, and the purified DNA was used to quantify total bacteria and S. mutans population via qPCR and specific primers; the same samples were also analyzed by colony forming unit (CFU) counting. In parallel, RNA was isolated from plaque-biofilm samples (from same animals) and used for transcriptional analyses via RT-qPCR. The viable population of both S. mutans and total bacteria assessed by qPCR were positively correlated with the CFU data (P<0.001; r>0.8). However, the qPCR data showed higher bacterial cell counts, particularly for total bacteria (vs. CFU). Moreover, S. mutans proportion in the plaque-biofilm determined by qPCR analysis showed strong correlation with incidence of smooth-surface caries (P=0.0022, r=0.71). The purified RNAs presented high RNA integrity numbers (>7), which allowed measurement of the expression of genes that are critical for S. mutans virulence (e.g. gtfB and gtfC). Our data show that the viable microbial population and the gene expression can be analyzed simultaneously, providing a global assessment of the infectious aspect of the disease dental caries. Our approach could enhance the value of the current rodent model in further understanding the pathophysiology of this disease and facilitating the exploration of novel anti-caries therapies.
Keywords: in vivo biofilms, Streptococcus mutans, DNA, RNA, viable bacterial population, gene expression
INTRODUCTION
Dental caries continues to be the single most common and costly biofilm-dependent oral disease in the US and worldwide (Marsh, 2003; Dye et al., 2007). Dental biofilms, clinically known as plaque, are complex biofilms formed on susceptible tooth surfaces. The interactions of specific microorganisms (and their products), salivary constituents and dietary carbohydrates (e.g. sucrose) are responsible for the establishment of virulent-cariogenic biofilms. The major controlling virulence factors modulating the pathogenesis of dental caries disease are the formation of an extracellular polysaccharide (EPS) matrix by mutans streptococii (especially Streptococcus mutans) and acidification of the dental biofilm milieu (Quivey et al., 2000, Bowen and Koo, 2011).
Oral biofilms in vivo harbor a mixed flora (Dewhirst et al., 2010). In cariogenic biofilms, the microorganisms are embedded and/or covered by EPS, are usually potent producers of acid, and are acid tolerant; which creates a highly adhesive, cohesive and acidic milieu. Among oral bacteria, mutans streptococci (mainly S. mutans) are prime EPS-rich matrix formers via synthesis of glucan by glucosyltransferases (S. mutans Gtf B, C and D) and fructan by fructosyltransferases. These exoenzymes can be incorporated in the pellicle and bacteria surfaces, and utilize sucrose as substrate for EPS matrix formation (as reviewed by Bowen and Koo, 2011). Therefore, the development of cariogenic biofilms is a result of complex diet-host-pathogen interactions. Several approaches have been employed to study the dental caries development process, including analyses of clinical samples (Loesche, 1986; Marsh, 2003; Dewhirst et al., 2010), in situ caries models (Zero, 1995), and a rodent model of dental caries (Keyes, 1960; Fitzgerald and Keyes, 1960; 1963).
The rodent model of dental caries using cariogenic streptococci is well established (Keyes, 1960; Fitzgerald and Keyes, 1960; 1963; Tanzer, 1979; Bowen et al., 1988a; 1988b; 1991), which was fundamental for advancing our understanding about the complex pathophysiological aspects of the disease. For example, studies using the animal model provided critical information on (i) the infectious character of the disease (Keyes, 1960; Fitzgerald and Keyes, 1960; 1963; Tanzer, 1979; Bowen et al., 1988a; 1991), (ii) the contribution of specific genes in expression of virulence via colonization, transmissibility and carious lesions evaluations (Yamashita et al, 1993; Fozo et al., 2007) and, (iii) investigation of host factors such salivary proteins on dental caries development (Bowen et al., 1988a; 1988b;Catalánet al., 2011). Moreover, it has been valuable for investigating the potential therapeutic effects of cariostatic agents (Koo et al., 2005; 2010).
The animal model has at least two key evaluation components, scoring of carious lesions and microbiological assessment of plaque-biofilms. The caries scoring system (Keyes’ method) has been thoroughly tested and is well established and standardized (Larson, 1981). In contrast, the plaque-biofilm analyses are still based on determination of microbial population via culturing methods and colony forming unit (CFU) counting, which have inherent limitations (e.g. uniform and consistent biofilm dispersion, specificity of culture media, oxygen tension, pH, plating variability). With the availability of modern genomics and transcriptomic tools, additional and improved information could be extracted to understand the infectious aspects of the disease. For example, precise measurement of the viable microbial population and gene expression profile of the microorganisms inhabiting within biofilms formed in vivo could be achieved. However, to the best of our knowledge, there is a lack of methods for such analyses in rodent models. Therefore, the aim of this study was to develop molecular approaches to (i) isolate genomic DNA for quantification of viable bacteria population and (ii) purify RNA to determine gene expression from in vivo plaque-biofilm samples using a well-established rat model of dental caries.
MATERIALS AND METHODS
Animal study
The animal experiment was performed according to methods described previously (Bowen et al., 1988; 1991; Koo et al., 2005). Pups aged 19 and 20 days, from 3 litters, were infected by mouth using an actively growing biofilms of S. mutans UA159 (Koo et al., 2005). These biofilms were 4-days old, whichhave not reached stationary phase. The stationary phase is characterized by reduced metabolic activity, detachment of the biofilm from the apatitic surface, and lack of further growth (and accumulation) even when continuously fed with fresh medium. At age 21 days, the animals were weaned and randomly housed in pairs. A total of 16 animals was provided with a cariogenic diet (diet 2000, which contains 56% sucrose; Harlan Teklab, Madison, WI) and 5% sucrose water ad libitum (Bowen et al., 1988). Establishment of infection was confirmed by platting oral swab directly on Mitis Salivarius Agar (Difco). The experiment proceeded for 3 weeks; the rats were killed by CO2 asphyxiation. The standard microbiological assessment (via plating) and caries evaluation were carried out using previously described methods (Koo et al., 2005). Molecular approaches were developed to evaluate viable bacterial content and S. mutans gene expression within plaque-biofilms as depicted in Figure 1.
Figure 1. Experimental design.

Stepwise approach used to evaluate planktonic culture, in vitro and in vivo biofilm samples.
Briefly, the lower left and right jaws were aseptically dissected (including incisors) and gently rinsed three times withsaline solution (0.9% NaCl). The left jaw was suspended in 5.0 ml of sterile saline solution, and sonicated on ice using a Branson Sonifier 450 (three 10-second pulses with 15-second intervals at 7 W; Branson Ultrasonics Co., Conn., USA), to remove biofilms formed on tooth surfaces. One ml of the biofilm suspension was serially diluted with saline solution and the dilutions plated on Mitis Salivarius Agar (Difco) containing bacitracin (Sigma) to estimate S. mutans ppopulation, and Blood Agar (Blood Agar base plus 5% sheep blood) to determine the total cultivable microbiota (i.e. CFU data; Bowen et al., 1988a, 1991). The remaining 4 ml of biofilm suspension were centrifuged and the pellet stored at −20°C, and later submitted to DNA extraction for quantification of bacterial cells via qPCR. The lower right jaw was incubated in 5 ml of RNALater® solution (Ambion, Austin, TX, USA) for 1 h at room temperature. This incubation time is enough to preserve the integrity of the RNA and avoid dental erosion due to the low pH of this solution as tested experimentally (data not shown). After the incubation, sonication was performed (1 pulse of 10 seconds; 7 Watts) and the biofilm suspensions were transferred to new 15 ml tubes. Additional 5 ml of 1X ice cold PBS was added to each tube containing the jaws (vortexed briefly), followed by sonication (1 pulse of 10 seconds; 7 Watts). The suspension was then transferred to the 15 ml tubes containing the initial 5 ml of each biofilm suspension, and centrifuged (5500g at 4 °C for 20 min). This step was repeated once, and the biofilms were submitted immediately to RNA extraction for further gene expression analysis. Fast processing time is critical to maintain RNA integrity. Blood and epithelial contamination was not a problem for the DNA and RNA analyses as shown in the following sections. The animals’ jaws were transferred to new containers and stored for caries score evaluation. All jaws were defleshed, and the teeth prepared for caries scoring by means of Larson’s modification of Keyes’ system (Larson, 1981). This study was reviewed and approved by the University of Rochester Committee on Animal Resources (Protocol number UCAR #2003-124, University of Rochester Medical Center, Rochester, NY, USA).
Isolation of DNA and quantification of bacteria population via quantitative real-time PCR (qPCR)
Optimization of procedure in vitro
A comprehensive and stepwise procedure was performed to optimize genomic DNA extraction and purification from viable bacterial cells with intact membrane and further quantification via qPCR (see details in Appendix 1). First, planktonic cultures were used to test the correlations of optical density (OD600nm), CFU (viable cells), and number of cells by counting chamber method and by qPCR. Propidium monoazide (PMA) prior DNA isolation was then used to determine whether PMA would interfere with accurate quantification of bacterial cells (Nocker et al., 2007a). Second, the methodology combining PMA, DNA isolation and qPCR was explored using in vitro formed biofilms. The results of these analyses demonstrated that addition of PMA prior to DNA isolation is the most optimal condition for quantification of viable bacterial cells via qPCR (Appendix 1), and thereby used for our in vivo biofilm samples. The combination of PMA and qPCR will quantify only the cells with intact membrane (i.e. viable cells). Because the PMA cross-linked to DNA of dead cells and extracellular DNA (eDNA, present on the extracellular matrix of biofilms) modify the DNA and inhibits the PCR amplification of the extracted DNA (Nocker et al., 2006; 2007a,b; Nocker and Camper, 2009).
In vivo analyses
The in vivo biofilm pellets (stored at −20°C) were resuspended with 500 μl of TE (50 mM Tris, 10 mM EDTA, pH 8.0). Using a pipette, the biofilm suspensions were transferred to light transparent 1.5 ml microcentrifuge tubes; then mixed with PMA as described by Nocker et al. (2007a). Briefly, 1.5 ul of PMA (20 mM in 20% dimethyl sulfoxide; Biotium, Hayward, CA) were added to the biofilm suspensions. The tubes were incubated in the dark for 5 minutes, at room temperature, with occasionally mixing. Next, the samples were exposed to light for 3 minutes (600W halogen light source). After photo-induced cross-linking, the biofilm suspensions were centrifuged (13,000 × g / 10 minutes/ 4 °C) and the supernatant discarded. The pellet was resuspended with 100 μl of TE (50 mM Tris, 10 mM EDTA, pH 8.0), followed by incubation with 10.9 μl lysozyme (100 mg/mL stock) and 5 μl mutanolysin (5 U/μL stock) (37°C / 30 min). Genomic DNA was then isolated using the MasterPure DNA purification kit (see Figure 2). The total bacteria and S. mutans contents, in each in vivo biofilm sample, were determined using qPCR. Ten nanograms of genomic DNA per sample and negative controls (without DNA) were amplified by a MyiQ real-time PCR detection system with iQ SYBR Green supermix (Bio-Rad Laboratories, Inc., CA, USA) and specific primers. For S. mutans quantification a 16S rRNA primer was used (Klein et al., 2010). A standard curve based on the genome size of S. mutans (2.03 Mb) was prepared as described previously (Doležel et al., 2003). Briefly, one genome copy represents one S. mutans cell; and the range tested was 108 to 102 cells (PCR efficiency: 98 to 102%). This standard curve was used to transform the quantification cycle (Cq) values to the relative number of S. mutans cells. To determine the number of S. mutans cells in the original sample, the numbers of cells detected in the qPCR runs were multiplied by the dilution factor from the DNA dilution step. For the total bacteria content determination a universal primer was designed (5-GGTTAAGTCCCGCAACGAGC-3 and 5-AGGGGCATGATGATTTGACG-3) using Beacon Designer 2.0 software (Premier Biosoft International, Palo Alto, CA, USA). The standard curve was based on average of oral bacteria genome size of sequenced oral bacteria (2.45 Mb), as available on http://www.oralgen.lanl.gov/ (at the time that our experiments were performed). This curve was tested by preparing it with 3 distinct templates: (i) S. mutans genomic DNA, (ii) a pool of DNA from 4 oral bacteria S. mutans UA159, Actinomyces naeslundii ATCC 12104, Streptococcus gordonii DL-1, Streptococcus oralis ATCC 35037 (from planktonic cultures at mid exponential phase), and (iii) DNA extracted from an in vivo plaque-biofilm sample selected arbitrarily. These 3 curves were evaluated based on detection of S. mutans proportion to the total bacteria content in samples (in vitro planktonic and biofilm) with known cell numbers and CFU data. The standard curve prepared with DNA extracted from in vivo plaque-biofilm was selected because it provided the best correlation with the cell numbers/CFU data.
Figure 2. DNA isolated from in vitro and in vivo biofilm samples.
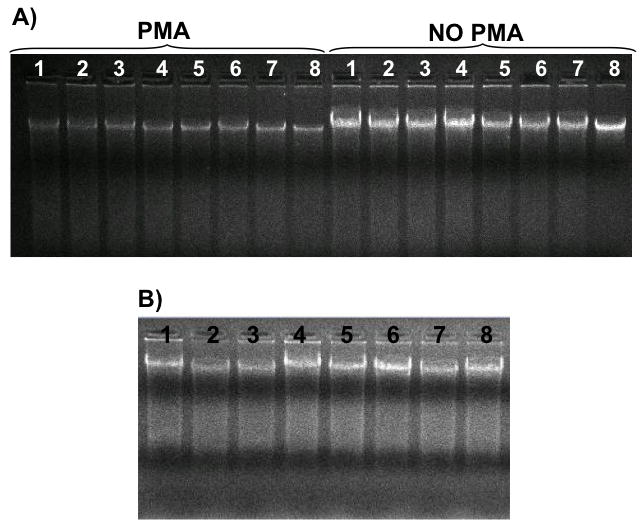
A) DNA purified from in vitro S. mutans biofilms (68 h-old). Lanes 1 to 4: biofilms grown with 1% sucrose; lanes 5 to 8: biofilms grown with 1% sucrose + 1% starch. NO PMA: samples not mixed with PMA prior DNA isolation; PMA: samples mixed and incubated with PMA prior DNA isolation. B) DNA isolated from in vivo biofilms (Lanes 1 to 8), all samples were incubated with PMA.
RNA extraction and purification from in vivo biofilms
The RNA extraction and purification from plaque-biofilm samples was conducted according to the optimized protocol for in vitro biofilms as detailed elsewhere (Cury and Koo, 2007). The pellets from 5 in vivo formed biofilm (pre-incubated in RNALater® solution) were combined and resuspended with 750 μl of NAES (50 mM sodium acetate pH 5.0, 10 mM EDTA, 1% SDS in molecular grade water), followed by the recommended DNAses treatments for RNA purification (Cury and Koo, 2007). The quality of the purified RNA was examined by an Agilent 2100 Electrophoresis Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) (Mueller et al., 2004), which provides the RNA integrity number (RIN). A RIN greater than 5 is considered to be a good total RNA quality for quantitative RT-PCR (Fleige & Pfaffl, 2006). Biofilm samples were pooled to isolate enough RNA for downstream analysis (i.e. RNA yield for cDNA synthesis and RT-qPCR), as tested experimentally. We also tested whether MICROBEnrich™ kit (Ambion), which is designed to enrich bacterial RNA from mixtures containing human, mouse, or rat RNA and bacterial RNA, could improve our RNA extraction and purification procedure (following the manufacturer’s instructions). However, the use of the kit resulted in significant loss of RNA without improving the overall quality of the RNA (i.e. RIN number), and therefore was not used in this study.
Reverse transcription quantitative real-time PCR (RT-qPCR)
RT-qPCR was performed to evaluate expression of genes gtfB, gtfC and gtfD (and internal control 16S rRNA). These genes were selected because they are critical virulence genes associated with the pathogenesis of dental caries as determined in rodent models (Yamashita et al., 1993). Briefly, 200 ng of RNA and specific primers (Koo et al., 2006) were used to generate cDNA in duplicate (SuperScript III Reverse Transcriptase, Invitrogen, Carlsbad, CA). For each sample, a mock reaction without addition of reverse transcriptase was performed (negative RT control) to check for DNA contamination. Superscript III Reverse Transcriptase in combination with specific primers was used to improve efficiency of cDNA synthesis (Ståhlberg et al., 2004). The resulting cDNAs and negative controls were amplified by a MyiQ real-time PCR detection system with iQ SYBR Green supermix (Bio-Rad Laboratories, Inc., CA, USA) and specific primers for gtfB, gtfC, gtfD and 16S rRNA as described previously (Koo et al., 2006; Aires et al., 2008). Standard curves for each gene were prepared as described elsewhere (Yin et al., 2001). The standard curves were used to transform the quantification cycle (Cq) values to the relative number of cDNA molecules. Total cDNA abundance was normalized using S. mutans specific 16S rRNA gene as an internal control (Koo et al., 2006; Klein et al., 2009; 2010).
Statistical analyses
An exploratory data analysis was performed to determine the most appropriate statistical test; the assumptions of equality of variances and normal distribution of errors were also checked. Spearman correlation tests were carried out using original data to evaluate the relationship between (1) bacterial population determined by CFU and number of cells by qPCR, (2) CFU (OR number of cells) and caries scores (total smooth or sulcal caries lesions, including severity of lesions). Statistical software JMP version 8.0 (SAS Institute, Cary, NC, USA) was used to perform the analyses. The level of significance was set at 5%.
RESULTS
In this study we established a methodology for quantification of viable bacterial cells using in vitro biofilm samples. The details of the optimization of the methodology are described in the Appendix 1. Then, we used the rodent modelfor dental caries development for analyses of in vivo biofilms. After 3 weeks of experiment, all animal presented caries lesions. The incidence and severity of smooth-surface and sulcal-surface caries are shown in the Table 1. The in vivo cariogenic biofilms were further evaluated, as follow.
Table 1.
Development of dental caries in rats (Larson’s modification of Keyes’ score).
| Caries | Smooth-surface caries | Sulcal-surface caries |
|---|---|---|
| (E) | 59.8 (17.0) | 45.5 (10.2) |
| Severity | ||
| (Ds) | 19.9 (10.6) | 42.6 (3.0) |
| (Dm) | 7.8 (5.6) | 14.1 (5.1) |
| (Dx) | 6.2 (4.8) | 8.0 (5.3) |
Values denote means with SD in parentheses (n = 16). E = enamel; Ds = Dentin exposed; Dm = 3/4 of the dentin affected; Dx = whole dentin affected.
DNA isolation from in vivo biofilms for assessment of viable bacteria populations
The DNA yield from animal’s plaque ranged from 8.40 to 48.22 μg of DNA per biofilm (per jaw: 20.00 ±13.62 μg of DNA). These DNA samples were used for downstream analysis using qPCR. The number of viable bacteria cells correlate positively with the CFU data, for both total bacteria and S. mutans (r and P values were 0.87 and <0.0001 for total bacteria, and 0.82 and 0.0007 for S. mutans, respectively; see Figure 3). The proportion of S. mutans (in relation to total bacteria) within plaque-biofilm is higher for plating than for qPCR (Table 2). One explanation is that qPCR may quantify total bacteria more precisely than standard culturing, as plating-CFU counting could miss those organisms that are strict anaerobes or not cultivable in the conditions performed in our study (e.g. lower amounts or unavailability of an essential nutrient in the culture media used). There were no qPCR inhibitors in the DNA samples recovered from in vivo plaque-biofilms (Figure 4). Moreover, the proportion of S. mutans population detected in the plaque-biofilms by qPCR was superior to CFU in correlating with the incidence smooth caries (E) (r=0.7 and P=0.0022 for qPCR vs. r=0.28 and P>0.05 for CFU). Furthermore, the qPCR data also correlated with incidence of sulcal caries (r=0.52; P=0.0405). However, S. mutans population detected by both culture and qPCR methods do not correlate with neither severity of smooth surface caries or with severity of sulcal caries.
Figure 3. Bacteria population detected in animal’s plaque at 3 weeks.
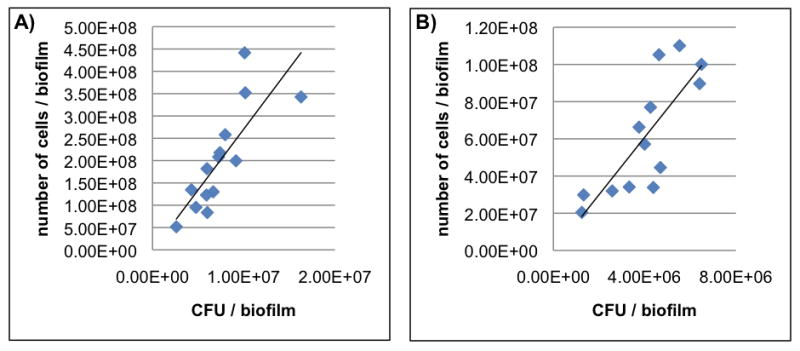
The relationship between number of bacteria cells with intact membranes (cells/biofilm) and CFU data are shown for both total bacteria (A) and S. mutans (B) in biofilms formed in vivo. Spearman Correlation Coefficient r and P value were r =0.87 and P <0.0001 for total bacteria (A), and r = 0.82 and P = 0.0007 for S. mutans (B).
Table 2.
Bacterial population detected in the animals’ plaque.
| Data | CFU×106/ml | cells×107/ml |
|---|---|---|
| Total counts | 7.58 (3.13) | 23.1 (12.3) |
| S. mutans counts | 4.32 (2.09) | 6.6 (3.2) |
| % of S. mutans | 56.99 | 28.57 |
Values denote means with SD in parentheses (n = 16).
Figure 4. qPCR amplification plot and standard curve data for S. mutans quantification.

There are no qPCR inhibitors in the plaque-biofilm samples and the values of expression detected for the tested samples (unknowns) are within the standard curve range.
RNA isolation from in vivo biofilms for gene expression analysis by RT-qPCR
Based on a RIN value, our RNA was of good quality and integrity for in vivo formed biofilms (Fleige & Pfaffl, 2006). The RIN values were 9.2 and 7.7 (Figure 5); but one sample presented RIN lower than 5 (RIN= 4.1) due to prolonged processing time (emphasizing the importance of fast sample manipulation to avoid RNA degradation), and was not included in the RT-qPCR assay. The yield of purified RNA ranged from 3.25 to 8.45 μg per pooled biofilms from 5 jaws (5.62 ± 2.63 μg), which are sufficient to synthesize cDNAs and perform gene expression assays. There were no RT-qPCR inhibitors, as shown by the amplification plot depicted in Figure 6. We successfully measured the expression of genes gtfB, gtfC and gtfD, which is known to generate low transcripts abundance (Figure 7).
Figure 5. RNA samples isolated from in vitro (A) and in vivo (B)biofilms.
A1) Bioanalyzer gel visualization. RNA samples extracted and purified from in vitro biofilms: sample 1 (RIN 10.0) and sample 2 (RIN 9.8). A2) Electropherogram summary for sample 1. B1) Bioanalyzer gel visualization. RNA samples extracted and purified from in vivo biofilms: 1 Pink R (RIN 9.2) and 2 Pink 1L (RIN 7.7). B2) Electropherogram summary for sample 1 Pink R.
Figure 6. RT-qPCR amplification plot and standard curve data for gtfB.
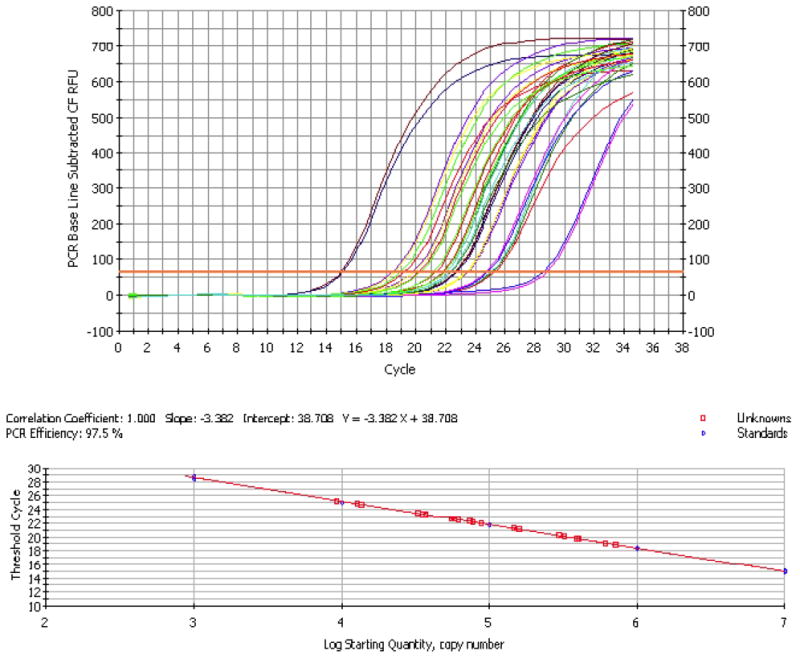
There are no RT-qPCR inhibitors in the plaque-biofilm samples and the values of expression detected for the tested samples (unknowns) are within the standard curve range.
Figure 7. Expression of S. mutans genes gtfC, gtfB, and gtfD.

The mRNA level of gtfB, gtfC and gtfD in each sample was normalized to that of 16S rRNA. Data are expressed as means ± standard deviations (n= 2; samples with RIN > 5.0; each samples was run twice).
DISCUSSION
Dental caries is a highly complex biofilm-dependent infectious disease. Moreover, this disease is a major public health concern worldwide. The rodent model of dental caries offers an unparalleled approach to further understand the complexity of diet-pathogen-host interactions involved in the pathophysiology of the disease, and to explore new preventive approaches. A major limitation with the model is the lack of any improvement of methodologies to examine the plaque-biofilms formed on the animals’ teeth. Of fundamental value is the ability to precisely determine the content of viable microbial populations. More importantly, the level of gene expression by microorganisms within biofilms can be determined to evaluate virulence in biofilms formed in vivo. In this study, we established two methods to: 1) measure the number of viable cells via combination of PMA with qPCR); and 2) purify RNA from in vivo biofilms with good RIN for downstream gene expression evaluation.
Improving quantification of microbial viable cell populations
An inherent characteristic of in vitro and in vivo biofilms is the presence of intact live bacterial cells at distinct growth phases and dead cells or cells with damaged membrane, which are considered ‘dead’ cells (Nocker and Camper, 2009). Moreover, biofilm matrices present extracellular DNA (eDNA) as a result of mainly bacteria autolysis (Steinberger and Holden, 2005; Kreth et al., 2009; Perry et al., 2009; Klein et al., 2010). Therefore, an extra step prior to genomic DNA isolation was introduced to allow differentiation of live and dead bacteria by the selective removal of DNA from dead cells and eDNA by using propidium monoazide (PMA) and ethidium monoazide (for details about the principle see Nocker et al., 2006). This strategy was successful for both Gram positive and Gram negative organisms grown in vitro, including oral bacteria species (Nocker et al., 2006; 2007a,b; Nocker and Camper, 2009; Loozen et al., 2011).
Here, we demonstrated that the approach of combining PMA incubation prior to DNA isolation, followed by qPCR amplification works well for characterization of viable population from animal’s plaque. The quantification of viable cells by qPCR correlates well with the traditional CFU counting method, which is considered the gold standard method to assess microbial population in the animal model of dental caries. Furthermore, the number of viable cells in vivo detected by qPCR correlated well with incidence of smooth- and sulcal-surface caries lesions. Future studies using a longer experimental period (e.g. 5 weeks) may clarify whether there could be a correlation of number of cells and severity of smooth and sulcal caries lesions.
One great advantage of using PMA and qPCR in combination is the flexibility to design multiple species-specific probes to analyze different proportions and types of microorganisms within the same sample. This would be nearly impossible with standard culturing methods based on combination of various selective media. Because any genus-, species-, sub-species-specific probes can be designed, the options are limitless for assessing the microbiota, dominance of a specific organism or group of organisms that are not only associated with caries but also other oral infectious diseases (e.g. periodontitis). Thus, microbial population and composition assessment via qPCR could be a more sensitive and versatile alternative to culturing-based techniques for evaluation of in vivo samples, including in human plaque studies. Several studies using human dental plaque samples have quantified microorganisms using DNA isolated from both live and dead cells (including eDNA), without using any approachto select out DNA from viable cells only (Gross et al., 2010; Tanner et al., 2011). Therefore, the PMA-based strategy would provide more reliable estimation of the viable oral microorganisms within plaque.
The use of PMA may have some disadvantages. The main drawbacks are the penetration of PMA in viable cells with reversibly damaged membranes, and the limited access of this chemical to all dead microorganisms (Fittipaldi et al., 2011). Thus, particular attention should be given during sample homogenization when adding PMA, to allow optimum access of this chemical to DNA from dead cells, from membrane compromised cells and eDNA. Nevertheless, the approach described in this study clearly improved the detection of viable cells in both our in vitro and, more importantly in vivo biofilms.
Assessing gene expression in biofilms formed in vivo
The virulence of biofilms can be determined by quantification of physiological and metabolic processes of (potential) pathogens in their natural habitat. Assessment of transcriptomic responses by these pathogens within biofilms could provide critical information about their expression of virulence in vivo. Furthermore, we could identify the molecular mechanisms of action of therapeutic agents that modulate development of cariogenic biofilm. However, information about gene expression from in vivo samples is sparse (Aires et al., 2008; Toro et al., 2010). The methodology used here can yield excellent RNA quality from animals’ dental plaque, which is not only important for RT-qPCR (particularly for assessing low abundance transcripts) but also for other transcriptome analyses such as RNAseq.
We used S. mutans genes gtfB, gtfC and gtfD to demonstrate the utility of the method. These genes were selected because they are (i) associated with sucrose-mediated bacterial binding and S. mutans colonization in vivo, (ii) responsible for EPS-rich matrix formation, and (iii) proven virulence factors associated with cariogenesis (as reviewed in Koo and Bowen, 2011). The detection of gtfBCD genes transcripts is relevant from technical standpoint because they are detected in low abundance under most biofilm growth conditions in vitro. These genes rarely meet the minimum intensity filters for cDNA microarrays (Klein et al., 2010), thus it is critical to optimize the gtfBCD expression via RT-qPCR. This successful methodology has important implications because the number of S. mutans detected within plaque-biofilm may be less important than the expression of the genes involved in the cariogenic biofilm formation.
Additional microorganisms may also be involved in the pathogenesis of dental caries, particularly through their ability to produce acids and tolerate acidic environment within the plaque-biofilm milieu (Kanasi et al., 2010; Gross et al., 2010; Palmer et al., 2010). Most oral bacteria do not participate in the assembly of the insoluble exopolysaccharide (EPS) matrix, which is associated with cariogenicity (Koo and Bowen, 2011). In contrast, S. mutans are the prime developers of the EPS-rich matrix in cariogenic biofilms though the actions of Gtfs enzymes. The construction of a complex three-dimensional extracellular matrix enmeshing the bacterial cells provides a highly adherent and cohesive biofilms (Bowen and Koo, 2011). The presence of EPS surrounding bacterial clusters appears to be essential for the assembly of protective (against antimicrobials) and acidic microenvironments facilitating S. mutans and other aciduric organisms to survive and thrive, contributing to the onset of a cariogenic biofilm (Bowen and Koo, 2011).
Thus, the use of the methodology presented here for the evaluation of the expression of known virulence factors of this bacterium (i.e. gtfBCD), and possibly novel genes that could be related to virulence in vivo, may open new ways to characterize potential novel therapeutic targets. We are currently assessing expression of S. mutans genes in animals’ plaque treated or not with novel and well-established (e.g. fluoride) therapeutic agents. Moreover, our approach may have applications for further investigation of the role of other organisms (via expression of gene products in vivo) in the pathogenesis of the disease.
It is noteworthy that there are limitations of this methodology. The data generated is an average of expression of all cells present throughout the in vivo plaque-biofilm; not all S. mutans cells may be expressing gtfs genes within the biofilm. The microorganisms growing in biofilms present physiological heterogeneity. Some cells may be going under cellular division, others may not be metabolically active, others may present alteration in the membrane composition, and some cells may be dead (Stewart and Franklin, 2008). The RNA yield is another limitation of evaluation of in vivo formed biofilms. To have enough purified RNA, it is necessary to combine 5 plaque-biofilm samples. In addition, rapid sample processing is warranted to avoid RNA degradation. Additional optimization of the method is warranted to improve RNA yield, so that RNA can be isolated from a single plaque-biofilm sample instead of pooled samples.
Clearly, the simultaneous assessment of microbial population, expression of virulence genes (e.g. linked to colonization and development of cariogenic biofilms) and caries scores improved the quantity and quality of information obtained from the rodent model. These approaches may be more time consuming than traditional methods used in animal studies, but on other hand the yield of biological relevant data is greatly enhanced, which may have direct application in clinical/translational research. The inclusion of advanced molecular biology tools brings a novel dimension to a classical animal model of dental caries that may advance this well-established scientific model.
Supplementary Material
Acknowledgments
The authors are thankful to Dr. Gene E. Watson for technical assistance during the animal experiment. We also gratefully acknowledge Dr. William H. Bowen for helpful discussions during the course of this study and for critical reading of the manuscript. This work was supported in part by NIH grant R01 DE016139-05.
Footnotes
CONFLINCT OF INTERST: None.
Appendix 1. Detailed strategy for viable cell quantification using qPCR.
References
- Aires CP, Del Bel Cury AA, Tenuta LMA, Klein MI, Koo H, Duarte S, Cury JA. Effect of starch and sucrose on dental biofilm formation and on root dentine demineralization. Caries Res. 2008;42:380–386. doi: 10.1159/000154783. [DOI] [PubMed] [Google Scholar]
- Bowen WH, Koo H. Biology of Streptococcus mutans Derived Glucosyltransferases: Role in Extracellular Matrix formation of Cariogenic Biofilms. Caries Res. 2011;45:69–86. doi: 10.1159/000324598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen WH, Madison KM, Pearson SK. Influence of desalivation in rats on incidence of caries in intact cagemates. J Dent Res. 1988a;67:1316–1318. doi: 10.1177/00220345880670101401. [DOI] [PubMed] [Google Scholar]
- Bowen WH, Pearson SK, Young DA. The Effect of Desalivation on Coronal and Root-surface Caries in Rats. J Dent Res. 1988b;67:21–23. doi: 10.1177/00220345880670010301. [DOI] [PubMed] [Google Scholar]
- Bowen WH, Schilling K, Giertsen E, Pearson S, Lee SF, Bleiweis A, Beeman D. Role of a cell surface-associated protein in adherence and dental caries. Infect Immun. 1991;59:4606–4609. doi: 10.1128/iai.59.12.4606-4609.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalán MA, Scott-Anne K, Klein MI, Koo H, Bowen WH, Melvin JE. Elevated Incidence of Dental Caries in a Mouse Model of Cystic Fibrosis. PLoS One. 2011;6:e16549. doi: 10.1371/journal.pone.0016549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cury JA, Koo H. Extraction and purification of total RNA from Streptococcus mutans biofilms. Anal Biochem. 2007;365:208–214. doi: 10.1016/j.ab.2007.03.021. [DOI] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J, Bartoš J, Voglmayr H, Greilhuber J. Nuclear DNA content and genome size of trout and human. Cytometry. 2003;51A:127–128. doi: 10.1002/cyto.a.10013. [DOI] [PubMed] [Google Scholar]
- Dye BA, Tan S, Smith V, Lewis BG, Barker LK, Thornton-Evans G, Eke PI, Beltrán-Aguilar ED, Horowitz AM, Li CH. Trends in oral health status: United States, 1988–1994 and 1999–2004. Vital Health Stat. 2007;11:1–92. [PubMed] [Google Scholar]
- Fittipaldi M, Codony F, Adrados B, Camper AK, Morató J. Viable real-time PCR in environmental samples: can all data be interpreted directly? Microb Ecol. 2011;61:7–12. doi: 10.1007/s00248-010-9719-1. [DOI] [PubMed] [Google Scholar]
- Fitzgerald RJ, Keyes PH. Demonstration of the etiologic role of streptococci in experimental caries in the hamster. J Am Dent Assoc. 1960;61:9–19. doi: 10.14219/jada.archive.1960.0138. [DOI] [PubMed] [Google Scholar]
- Fitzgerald RJ, Keyes PH. Ecologic factors in dental caries: The fate of antibiotic-resistant cariogenic streptococci in hamsters. Am J Pathol. 1963;42:759–772. [PMC free article] [PubMed] [Google Scholar]
- Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT–PCR performance. Mol Aspects Med. 2006;27:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Fozo EM, Scott-Anne K, Koo H, Quivey RG., Jr Role of unsaturated fatty acid biosynthesis in virulence of Streptococcus mutans. Infect Immun. 2007;75:1537–1539. doi: 10.1128/IAI.01938-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross EL, Leys EJ, Gasparovich SR, Firestone ND, Schwartzbaum JA, Janies DA, Asnani K, Griffen AL. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J Clin Microbiol. 2010;48:4121–4128. doi: 10.1128/JCM.01232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanasi E, Dewhirst FE, Chalmers NI, Kent R, Jr, Moore A, Hughes CV, Pradhan N, Loo CY, Tanner AC. Clonal analysis of the microbiota of severe early childhood caries. Caries Res. 2010;44:485–497. doi: 10.1159/000320158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes PH. The infectious and transmissible nature of experimental dental caries. Findings and implications. Arch Oral Biol. 1960;1:304–320. doi: 10.1016/0003-9969(60)90091-1. [DOI] [PubMed] [Google Scholar]
- Klein MI, DeBaz L, Agidi S, Lee H, Xie G, Lin AH, Hamaker BR, Lemos JA, Koo H. Dynamics of Streptococcus mutans transcriptome in response to starch and sucrose during biofilm development. PLoS One. 2010;5:e13478. doi: 10.1371/journal.pone.0013478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein MI, Duarte S, Xiao J, Mitra S, Foster TH, Koo H. Structural and molecular basis of the role of starch and sucrose in Streptococcus mutans biofilms development. Appl Environ Microbiol. 2009;75:837–841. doi: 10.1128/AEM.01299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H, Duarte S, Murata RM, Scott-Anne K, Gregoire S, Watson GE, Singh AP, Vorsa N. Influence of cranberry proanthocyanidins on formation of biofilms by Streptococcus mutans on saliva-coated apatitic surface and on dental caries development in vivo. Caries Res. 2010;44:116–126. doi: 10.1159/000296306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H, Schobel B, Scott-Anne K, Watson G, Bowen WH, Cury JA, Rosalen PL, Park YK. Apigenin and tt -farnesol with fluoride effects on S. mutans biofilms and dental caries. J Dent Res. 2005;84:1016–1020. doi: 10.1177/154405910508401109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H, Seils J, Abranches J, Burne RA, Bowen WH, Quivey RG., Jr Influence of apigenin on gtf gene expression in Streptococcus mutans UA159. Antimicrob Agents Chemother. 2006;50:542–546. doi: 10.1128/AAC.50.2.542-546.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Vu H, Zhang Y, Herzberg MC. Characterization of hydrogen peroxide-induced DNA release by Streptococcus sanguinis and Streptococcus gordonii. J Bacteriol. 2009;191:6281–6291. doi: 10.1128/JB.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson RM. Merits and modifications of scoring rat dental caries by Keyes’ method. In: Tanzer JM, editor. Animal Models in Cariology. Abstracts special suppl. Washington, DC: Microbiology; 1981. pp. 195–203. [Google Scholar]
- Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loozen G, Boon N, Pauwels M, Quirynen M, Teughels W. Live/dead real-time polymerase chain reaction to assess new therapies against dental plaque-related pathologies. Mol Oral Microbiol. 2011;26:253–261. doi: 10.1111/j.2041-1014.2011.00615.x. [DOI] [PubMed] [Google Scholar]
- Marsh PD. Are dental diseases examples of ecological catastrophes? Microbiology. 2003;149:279–294. doi: 10.1099/mic.0.26082-0. [DOI] [PubMed] [Google Scholar]
- Mueller O, Lightfoot S, Schroeder A. RNA integrity number (RIN): Standardization of RNA quality control. Agilent Application Note. 2004:1–8. pub. 5989-1165EN. [Google Scholar]
- Nocker A, Camper AK. Novel approaches toward preferential detection of viable cells using nucleic acid amplification techniques. FEMS Microbiol Lett. 2009;291:137–142. doi: 10.1111/j.1574-6968.2008.01429.x. [DOI] [PubMed] [Google Scholar]
- Nocker A, Cheung CY, Kamper AK. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J Microbio Meth. 2006;67:310–320. doi: 10.1016/j.mimet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- Nocker A, Sossa K, Camper AK. Molecular monitoring of disinfection efficacy. J Microbiol Methods. 2007a;70:252–260. doi: 10.1016/j.mimet.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Nocker A, Sossa P, Burr M, Camper AK. Use of propidium monoazide for live dead distinction in microbial ecology. Appl Environ Microbiol. 2007b;73:5111–5117. doi: 10.1128/AEM.02987-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CA, Kent R, Jr, Loo CY, Hughes CV, Stutius E, Pradhan N, Dahlan M, Kanasi E, Arevalo Vasquez SS, Tanner AC. Diet and caries-associated bacteria in severe early childhood caries. J Dent Res. 2010;89:1224–1229. doi: 10.1177/0022034510376543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JA, Cvitkovitch DG, Lévesque CM. Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol Lett. 2009;299:261–266. doi: 10.1111/j.1574-6968.2009.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivey RG, Jr, Kuhnert WL, Hahn K. Adaptation of oral streptococci to low pH. Adv Microb Physiol. 2000;42:239–274. doi: 10.1016/s0065-2911(00)42004-7. [DOI] [PubMed] [Google Scholar]
- Ståhlberg A, Kubista M, Pfaffl M. Comparison of reverse transcriptases in gene expression analysis. Clin Chem. 2004;50:1678–1680. doi: 10.1373/clinchem.2004.035469. [DOI] [PubMed] [Google Scholar]
- Steinberger RE, Holden PA. Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl Environ Microbiol. 2005;71:5404–5410. doi: 10.1128/AEM.71.9.5404-5410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- Tanner AC, Kent RL, Jr, Holgerson PL, Hughes CV, Loo CY, Kanasi E, Chalmers NI, Johansson I. Microbiota of severe early childhood caries before and after therapy. J Dent Res. 2011;90:1298–1305. doi: 10.1177/0022034511421201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzer JM. Essential dependence of smooth surface caries on, and augmentation of fissure caries by, sucrose and Streptococcus mutans infection. Infect Immun. 1979;25:526–531. doi: 10.1128/iai.25.2.526-531.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro E, Nascimento MM, Suarez-Perez E, Burne RA, Elias-Boneta A, Morou-Bermudez E. The effect of sucrose on plaque and saliva urease levels in vivo. Arch Oral Biol. 2010;55:249–254. doi: 10.1016/j.archoralbio.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y, Bowen WH, Burne RA, Kuramitsu HK. Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infect Immun. 1993;61:3811–3817. doi: 10.1128/iai.61.9.3811-3817.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin JL, Shackel NA, Zekry A, McGuinness PH, Richards C, Putten KV, McCaughan GW, Eris JM, Bishop GA. Real-time reverse transcriptase-polymerase chain reaction (RT-PCR) for measurement of cytokine and growth factor mRNA expression with fluorogenic probes or SYBR Green I. Immunol Cell Biol. 2001;79:213–221. doi: 10.1046/j.1440-1711.2001.01002.x. [DOI] [PubMed] [Google Scholar]
- Zero DT. In situ caries models. Adv Dent Res. 1995;9:214–234. doi: 10.1177/08959374950090030501. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



