Abstract
In contrast to the prevailing dogma in the 1990s, recent studies have suggested that an evolutionary history of segregation distortion within species may contribute to sterility in species hybrids. However, this recent work identified segregation distortion exclusively in species hybrids which may never have had an evolutionary history of segregation distortion in either parent species. We expand on previous work by using a strain of Drosophila persimilis exhibiting segregation distortion within species to generate QTL maps for segregation distortion and hybrid sterility in crosses between D. persimilis and D. pseudoobscura. The maps localize regions along the XR contributing to both phenotypes, and they indicate one region of overlap between the two maps. This overlap could provide preliminary evidence for an association between segregation distortion within species and hybrid sterility, but the localizations are currently too broad to have confidence in this conclusion. This work is a first step towards possibly supporting a genetic conflict model of speciation in this system.
Keywords: Drosophila, Speciation, Reproductive Isolation, Meiotic Drive, Hybrid Incompatibility
INTRODUCTION
Sex chromosome segregation distortion, genetic conflict, and speciation
Diploid taxa typically transmit the two alleles from each locus to their offspring in equal Mendelian proportions. In contrast, “segregation distortion” (SD) describes the phenomenon wherein alleles are not found in equal proportions among the offspring. Meiotic drive is a form of SD characterized by the loss of gametes carrying a certain allele, such that by the end of meiosis, the favored allele is passed on more often than its homolog (Sandler and Novitski 1957, Werren 2011). Many Drosophila species exhibit SD of the entire X chromosome in males (reviewed in Jaenike 2001), but it is sometimes unclear whether this SD occurs during meiosis (Policansky 1974, Pardo-Manuel de Villena and Sapienza 2001), so we conservatively refer to this phenomenon as X chromosome segregation distortion or the “sex-ratio” (SR) trait. The SR trait is expressed during either meiosis or after, during sperm maturation (reviewed in Presgraves 2008) and biases transmission of the male's X chromosome, causing a higher ratio of female relative to male progeny. Since the distorter is located on the X chromosome, the resultant females produce male offspring of which about half exhibit SR. Continuing the cycle, those SR male offspring then produce a disproportionately large number of female SR-carrier offspring relative to male non-carrier offspring. While some SR factors are associated with fertility reduction, and are not under direct selection, SR factors can rise in frequency quickly due to the transmission advantage alone.
The spread of factors causing segregation distortion, such as SR, can initiate a genetic conflict, where selfish genetic elements are detrimental to their host or decrease transmission of other genetic elements, while increasing transmission of themselves (Werren 2011). Due to the spread of SR X chromosomes, selection will favor other chromosomes that bear loci suppressing SR, hence recovering transmission and/or recovering fertility. After SR suppression, new X chromosome distorters (or new modifiers that restore the original distorter) that arise will again spread through the population, reinitiating selection for new suppressors. As these elements jockey for higher frequency, an arms race is set off – potentially accumulating modifiers that suppress distortion, as well as modifiers that enhance distortion, and likely increasing divergence between populations (Carvalho et al. 1997, Hall 2004). A suppressor may be specific to a particular distorter, and therefore may not function in hybrids when confronted with different distorters. Suppression malfunction in hybrids could lead to defects in chromosomal segregation, for example, mutual distortion from two unsuppressed homologs may prevent both homologs from entering gametes, resulting in aneuploidy, gamete failure, and hybrid sterility (Frank 1991, Hurst and Pomiankowski 1991). Since hybrid sterility prevents gene exchange between taxa, genetic conflict can possibly contribute to speciation via arms races and the resultant formation of reproductive incompatibilities.
If this type of genetic conflict leads to speciation, then hybrid sterility between species should sometimes be associated with segregation distortion (reviewed in: Johnson 2010, McDermott and Noor 2010, Presgraves 2010). Indeed, studies in Drosophila simulans/D. mauritiana and D. pseudoobscura USA/D. pseudoobscura Bogota have identified that genes causing hybrid sterility also cause segregation distortion in hybrids (Tao et al. 2001, Phadnis and Orr 2009). However, the distorter/sterility alleles from these two studies have only been detected in systems of “cryptic distortion,” where the distortion was not identified within species – only in species hybrids. Such cases may suggest an evolutionary history of segregation distortion within species causing hybrid sterility, or merely a hybrid defect, acting simultaneously with hybrid sterility but without causation. In contrast, the model system used here, Drosophila persimilis, has alleles causing SR within species. Observing an association between segregation distortion within species and hybrid sterility between species could provide a new type of support for the genetic conflict models of speciation.
Drosophila persimilis “sex-ratio” and hybrid sterility alleles on the X chromosome
Here, we investigate a strain of Drosophila persimilis which exhibits SR (SR D. persimilis), and carries the distorter(s) within an inversion on the right arm of the X chromosome (XR) (Sturtevant and Dobzhansky 1936). The XR of SR D. persimilis has a different inversion arrangement than Standard (non-sex-ratio) D. persimilis, but the XR of SR D. persimilis shares the chromosomal arrangement with Standard D. pseudoobscura (used to map hybrid sterility—see Figure 1). D. pseudoobscura is the sister species of D. persimilis, and when they hybridize, male hybrid offspring are sterile, while female hybrid offspring are fertile. Some strains of D. pseudoobscura also exhibit the SR trait, but those strains carry a chromosomal arrangement that differs from both Standard D. pseudoobscura and SR D. persimilis (Sturtevant and Dobzhansky 1936). Since the XR of SR D. persimilis and Standard D. pseudoobscura are homosequential, recombination along the XR can proceed in hybrid female gametes, unlike in inversion heterozygotes where recombination products from within the inversion are not recovered in the offspring. Historically, fine-mapping hybrid sterility loci between D. persimilis and D. pseudoobscura has been difficult because Standard lines of both species (which are heterozygous for large inversions) are often used. Since hybrid sterility loci in these lines are located within inversions (Noor et al. 2001), the only progeny in Standard hybrids are non-recombinant, making mapping within the inversion impossible.
Figure 1.
Orientation of inversions and location of markers along XR. (A) Arrows indicate the orientation of the inversion, located from band 26D to band 38 on the D. pseudoobscura polytene chromosome (images of chromosomes adapted from Figures 8 and 9 of Schaeffer et al. (2008), with permission from Genetics Society of America). SR D. persimilis and Standard (nonSR) D. pseudoobscura are homosequential for the XR inversion, while Standard D. persimilis carries a unique arrangement. (B) The XR of D. pseudoobscura was used to map the marker locations, given its better coverage and annotation (Schaeffer et al. 2008). Markers represented only three of the eight scaffolds (group 6, group 8a, and group8b), since our work focused on the region carrying the inversion. The marker locations are indicated to scale based on the physical location; the recombination distance (cM) indicated between each marker is based on our sterility map results.
Comparing the results from various published studies, we find circumstantial evidence of an association between segregation distortion and hybrid sterility in these species, associated with factors on the XR chromosome arm. Previous studies mapped sterility factors between D. persimilis and D. pseudoobscura (Dobzhansky 1936, Wu and Beckenbach 1983, Orr 1989, Noor et al. 2001), and distorters in SR D. persimilis (Wu and Beckenbach 1983) to the XR chromosome arm. Wu and Beckenbach (1983) were the first to describe a possible association between segregation distortion and hybrid sterility in D. persimilis and D. pseudoobscura. They introgressed sections of the SR XR from D. persimilis into D. pseudoobscura, and observed that certain segments (at least three loci) of the SR D. persimilis XR marked by visible mutations resulted in hybrid sterility in a D. pseudoobscura background. Orr (1989) also observed that the SR XR from D. persimilis increased the proportion of sterile hybrids. In contrast, when the Standard XR from D. persimilis is crossed into a D. pseudoobscura background, it reduces the proportion of sterile hybrids (Noor et al. 2001, McDermott and Noor 2011). This consistently greater sterility associated with the XR chromosome arm in crosses using distorting rather than non-distorting strains of D. persimilis strongly suggests an association between segregation distortion and hybrid sterility in these species. Here, we expand on Wu and Beckenbach's (1983) work in this system by using more, and genome-sequence-localized, markers to map the responsible regions more precisely.
MATERIALS AND METHODS
Fly stocks
The D. persimilis MSH1993 line (Standard XR chromosome arrangement) was derived from females collected at Mount Saint Helena, California, in 1993 (Noor 1995). D. persimilis Santa Cruz Island (SCI) was obtained from the Drosophila Species Stock Center (stock number 14011-0111.50). This line contained both SR and Standard flies – sublines were established of Standard D. persimilis SCI, and strongly distorting SR D. persimilis SCI. The strongly distorting subline was created by selecting individuals each generation that produced a strong female-biased sex ratio. This selection was continued for 11 generations. The strongly distorting subline was used here for the crosses to map both hybrid sterility and segregation distortion. The D. pseudoobscura white line was obtained from the Drosophila Species Stock Center (stock number 14011-0121.12).
Fly crosses
For the cross used to map sterility (Figure 2A): D. pseudoobscura white (hereafter psw) females were collected as virgin adults and maintained for at least six days to reach sexual maturity and receptivity. After six days, psw females were crossed to SR D. persimilis males from the strongly distorting subline of D. persimilis SCI. F1 females were backcrossed to psw males to generate backcross male progeny (hereafter “BCps males”) for fertility assays and preliminary tests of linkage between sterility and distortion factors. Single pair crosses of seven day old BCps males and psw females were set up, and after two days, the male was collected, assayed for sperm motility, and genotyped. The female remained in the vial for another 7-8 days to lay eggs. If larvae are present after a total of 9-10 days, the female would be discarded, and the cross would be kept to later count the sex ratio of the progeny.
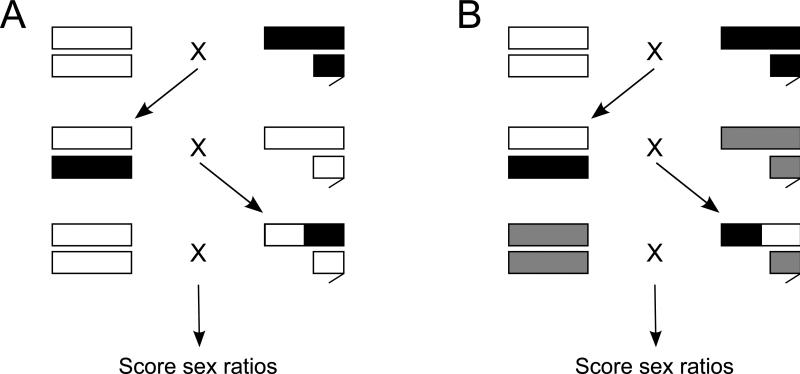
Figure 2.
Crossing scheme to map sterility and segregation distortion on the X chromosome. Long boxes indicate X chromosomes, and short boxes indicate the Y chromosome (autosomes are not shown). The crosses for sterility (A) and segregation distortion (B) both start by crossing D. pseudoobscura white (white) to SR D. persimilis SCI (black). F1 hybrid females are then either backcrossed to D. pseudoobscura white for the sterility cross, or crossed to D. persimilis MSH1993 (gray) for the distortion cross. The resultant males are then backcrossed to their respective paternal species, and sex ratios of their progeny are counted.
As we were only interested in mapping hybrid sterility to the XR chromosome arm (where distortion factor(s) are located), we sought to limit the flies used for analysis to those without other regions known to contribute to hybrid male sterility. We limited the dataset in two ways. First, since the red eye allele is associated with hybrid sterility factors on the D. persimilis XL (Orr 1987, Noor et al. 2001), we only assayed white-eyed BCps male progeny, which would bear the fertile XL alleles in a D. pseudoobscura background. Therefore any observed hybrid sterility can be attributed to factors on the XR without interference from the XL. Second, prior to scoring markers on the XR chromosome arm, we scored microsatellite markers on the XL and sterility-conferring 2nd chromosome, and limited the dataset to just those flies bearing the D. pseudoobscura (non-sterility-conferring) allele at both loci (see below).
For the cross used to map segregation distortion (Figure 2B): The F1 females from the sterility cross were crossed to D. persimilis MSH1993 males to generate backcross male progeny (hereafter “BCper males”). Red-eyed BCper males were selected to minimize hybrid sterility effects, since the XL inversion would be of D. persimilis origin. For this cross, all white-eyed BCper males are fully sterile, thus interfering with mapping segregation distortion, which requires fertility to observe. Single-pair crosses of BCper males and D. persimilis MSH1993 females were set up (both seven days old), and if larvae were present in the vial after 9-10 days, the male was collected for genotyping, and the female was discarded. Numbers of male and female progeny were counted for each fertile cross to obtain sex ratios. All crosses were performed on standard sugar/yeast/agar medium at 20°C ± 1° and 80% relative humidity.
Fertility assays
BCps adult males were collected two days after setting up the cross to psw females, and maintained alone in food-containing vials. One to three days later, the fertility of each backcross male was assessed by dissection of the testes in Ringer's solution following Coyne (1984). A male was scored as “fertile” if at least one motile sperm was observed and “sterile” if no motile sperm were observed. Treating fertility as a binary trait is conservative (Campbell and Noor 2001), although other methods of scoring sperm motility exist (Davis and Wu 1996, Moehring et al. 2006, Reed et al. 2008). All dissected BCps males were labeled and stored at -20°C.
Microsatellite genotyping
DNA was isolated from all dissected BCps and BCper males following the Gloor and Engels (1992) protocol. Microsatellite genotyping was performed in two steps. First, all BCps males were genotyped for markers associated with each inversion that distinguishes D. pseudoobscura from D. persimilis. The markers used for this initial screen were DPSX046 (XL), DPSX014 (XR), and DPS2026 (2). Primer sequences for all markers are available in Supplementary Table 1. Since we are interested in mapping traits located on the XR, two additional XR markers were added: DPSX063, and DPSX021B2. These markers denoted the species identity of the inversion arrangement on these chromosome arms. Second, only those BCps males that were hemizygous or homozygous for the D. pseudoobscura allele at the XL and 2nd chromosome inversion markers were further genotyped at additional markers on the XR. The markers used on this limited set of flies were DPSX029, DPSX007, DPSX048, DPSX024, DPSX030, DPSX012, and DPSX062. Locations of these and the preliminary XR markers are shown in Figure 1B. Markers were chosen in an attempt to narrow down large peaks, causing an uneven concentration of markers along the XR. PCR amplification protocol and touchdown cycle details are described in Noor et al. (2001) and Palumbi (1996), respectively. PCR products were visualized on acrylamide gels on LiCor 4200 and 4300 DNA sequencer/analyzers.
Data Analysis
To analyze hybrid sterility in BCps males, all males included in the limited set were used for mapping (a total of 1898 males). All males that produced 20 or greater progeny were also used for a preliminary test of linkage between sterility and distorter loci. Scoring at least 20 progeny enhances detection of moderate segregation distortion, so crosses producing fewer than 20 progeny were not scored for this phenotype. Numbers of females and male progeny from each BCps male were counted and tested for distortion using a binomial test, comparing the observed adult offspring sex ratio to an expectation of 55% adult female offspring (the sex ratio of adult progeny from males with no D. persimilis genotypes on their XR). The number of estimated false positives was determined using sex ratio data collected from Standard males that produced 20 or greater progeny, and equals the number of sex ratios (fraction of females) greater than two standard deviations above the mean. To map segregation distortion in BCper males, only males that produced 15 or greater progeny were used to ensure any segregation distortion would be detectable. A larger cutoff for progeny number was used to map hybrid sterility because the large sample size of BCps males producing over 20 progeny allowed us to be more conservative. This also explains why we used a cutoff of 20 progeny for our false positive estimate for the segregation distortion cross.
For BCps and BCper analyses, recombination distances were obtained using OneMap (Margarido et al. 2007) via R 2.13.1 (Ihaka and Gentleman 1996) (see Figure 1B). Both traits were mapped using Composite Interval Mapping (CIM) in Windows QTL Cartographer 2.5_009 (Wang et al. 2011), and permuted 1000 times to establish a significance threshold. While CIM does not account for binary data (e.g. our sterility data), Categorical Trait Multiple Interval Mapping (CT-MIM) in Windows QTL Cartographer does. However, CT-MIM fails when using sex-linked and hemizygous data (Shengchu Wang, personal communication), and using CIM to analyze binary data appears comparable to CT-MIM (Moehring et al. 2004). The BCper analysis to map segregation distortion used the fraction of the total progeny that are female, and therefore CIM is appropriate.
RESULTS
Characterization of D. persimilis SR
Unlike previous reports for homozygous SR D. pseudoobscura (Wallace 1948, Curtsinger and Feldman 1980, Beckenbach 1983, Beckenbach 1996, but see Gebhardt and Anderson 1993), our homozygous SR D. persimilis females from Santa Cruz Island (SCI) were fertile, and therefore stocks of SR were easily made. The D. persimilis SCI line from the Drosophila Species Stock Center was polymorphic for the SR inversion, so we created sublines homozygous for either the SR or Standard inversion. The subline used in the crosses exhibited an average of 73% female progeny – lower than Wu and Beckenbach's (1983) observation of 95-99% female progeny, indicating variation between lines of SR D. persimilis. The presence of the D. persimilis SR inversion was confirmed by polytene squash, and genetic markers were used later that differentiated between the inversion types.
QTL Mapping segregation distortion in SR D. persimilis
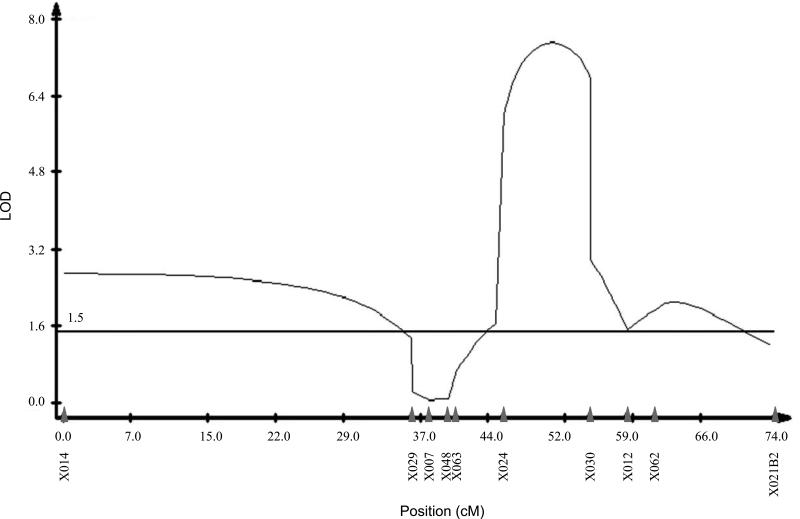
To create the QTL map for segregation distortion, 10,683 BCper males were mated, but only 288 produced at least one offspring. These 288 BCper males were genotyped at 10 XR markers: DPSX014, DPSX029, DPSX007, DPSX048, DPSX063, DPSX024, DPSX030, DPSX012, DPSX062, and DPSX021B2 (locations shown in Figure 1B). Only BCper males producing over 15 progeny (109) were used for QTL mapping. We used CIM in Windows QTL Cartographer to generate the QTL map for segregation distortion shown in Figure 3. Between the markers DPSX014 and DPSX029 is a 3.69 Mb minor peak. However, both the absence of a marker to the left of DPSX014, and the absence of a marker between DPSX014 and DPSX029 prevents certainty of the exact QTL effect and position. It is possible that the addition of another marker to the left of DPSX014 (proximal to the centromere) could further extend the QTL, or addition of another marker between DPSX014 and DPSX029 could reduce the size of the QTL. Between the markers DPSX024 and DPSX012 (2.06 Mb) is a peak of major effect. Our segregation distortion map also has a third minor peak starting at DPSX012, but no conclusions can be drawn for the region distal to DPSX062. The presence of multiple peaks suggests multiple factors may be required for segregation distortion, as initially observed by Wu and Beckenbach (1983).
Figure 3.
QTL map of segregation distortion on the D. persimilis SCI XR. Map was generated using Composite Interval Mapping (CIM) of BCper males producing 15 or greater progeny. Significance threshold (LOD 1.5) was calculated by 1000 permutations, shown as the horizontal line. Marker locations are indicated with solid gray arrowheads.
QTL mapping hybrid male sterility between SR D. persimilis SCI and D. pseudoobscura white
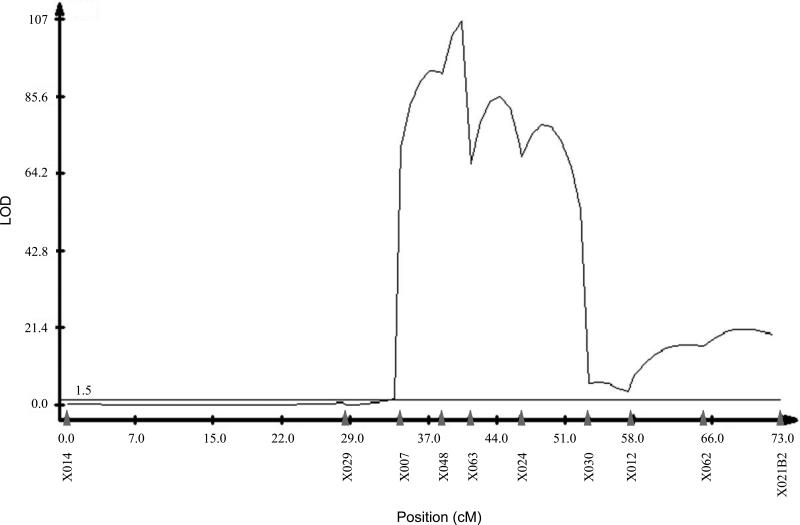
To create the QTL map for hybrid sterility, 5329 BCps males were mated, tested for fertility, genotyped at 2 initial markers: DPSX046 (chromosome XL), DPS2026 (chromosome 2). Of these 5329, 1898 exhibited D. pseudoobscura genotypes at the XL and 2nd chromosome and were genotyped at the same markers used in the segregation distortion map. We used CIM in Windows QTL Cartographer to generate a QTL map, shown in Figure 4, of hybrid sterility between Drosophila persimilis SCI and Drosophila pseudoobscura white. Our QTL map shows a large proximal region of no significant effect, followed by one broad significant region with possibly three peaks, and a less significant minor peak near the distal end of the inversion. The broad region starts at marker DPSX007 and ends at DPSX030, and is approximately 5.23 Mb long. The second region is between DPSX012 and DPSX062. For the region beyond DPSX062, no conclusions can be drawn, because adding another marker to the right of DPSX021B2 (distal to the centromere) could further extend the QTL, or addition of another marker between DPSX062 and DPSX021B2 could reduce the size of the QTL. Unfortunately, each of these regions is too large to identify candidate loci. Maps of both traits appear to overlap at two peaks: the major peak between markers DPSX024 and DPSX012 (2.06 Mb), and the minor peak between markers DPSX012 and DPSX021B2.
Figure 4.
QTL map along XR of hybrid sterility between SR D. persimilis SCI and D. pseudoobscura white. Map was generated using Composite Interval Mapping (CIM) of BCps males with ps genotypes at XL and 2nd chromosome inversions. Significance threshold (LOD 1.5) was calculated by 1000 permutations, shown as the horizontal line. Marker locations are indicated with solid gray arrowheads.
Preliminary test of linkage between hybrid sterility and segregation distortion
Using the results of the hybrid sterility cross, we conducted a one-way test of whether loci causing hybrid sterility and segregation distortion are linked. If the loci causing hybrid sterility and segregation distortion are incompletely linked, recombination should be able to separate them. In this case, we may observe sterile backcross hybrid (BCps) males inheriting only the sterility factor, but also BCps males inheriting only the segregation distortion factor, who will be fertile with biased sex ratios. However, absence of such recombinant males is uninformative, given that we are not certain if the D. pseudoobscura Y chromosome is sensitive to the D. persimilis SR XR distortion factor(s) (see Discussion). Therefore, the presence of biased sex ratios in the progeny of BCps males serves as a one-way test of linkage of the two traits. Of the 350 BCps males that produced progeny, 20 exhibited significantly female-biased sex ratios. However, 21 BCps males had progeny with male-biased sex ratios. If the loci causing hybrid sterility and segregation distortion were unlinked, we should expect to see a greater number of female-biased sex ratios than male-biased (since the presence of male-biased sex ratios are assumed to be arising by chance). We cannot exclude that a subset of the BCps males producing offspring with female-biased sex ratios are recombinants between separate segregation distortion- and sterility-conferring factors, but a more parsimonious conclusion is most (or perhaps all) are false positives. Hence, we failed to reject the hypothesis that hybrid sterility and segregation distortion are completely linked.
DISCUSSION
In this study, we characterized and mapped SR Drosophila persimilis SCI segregation distortion, and mapped hybrid sterility of this species with its sister species, D. pseudoobscura. Additionally, we found possible overlap between the two maps, indicating a potential for shared loci causing both segregation distortion within D. persimilis and hybrid sterility between these two species that can be explored in future work.
Characterization of Segregation Distortion in D. persimilis
Despite their overall genetic similarity, D. persimilis and D. pseudoobscura each maintain distinct segregation distortion systems. The SR XR of D. pseudoobscura contains three non-overlapping inversions (Dobzhansky 1939, Beckenbach 1996), while D. persimilis only contains one (Sturtevant and Dobzhansky 1936). D. pseudoobscura females homozygous for the SR inversion have reduced fertility compared to both Standard homozygotes, and inversion heterozygotes (Wallace 1948, Curtsinger and Feldman 1980, Beckenbach 1983, Beckenbach 1996, but see Gebhardt and Anderson 1993). The SR D. persimilis from Santa Cruz Island does not appear to inflict reduced fertility in female SR inversion homozygotes allowing the strain to be easily used as a stock.
Critical for this study, SR D. persimilis and ST D. pseudoobscura share the same XR chromosomal arrangement, allowing mapping of SR factors, unlike SR D. pseudoobscura which carries unique inversion arrangements, preventing recovery of recombinant progeny. Many SR systems in Drosophila exhibit a fertility reduction within species, so it is possible that “hybrid” sterility is merely the within-species sterility acting across species. However, SR males in Drosophila exhibit a range of fertility reduction, where many only see a slight reduction in fertility when rapid remating takes place (reviewed in Price and Wedell 2008). In hybrids, there must be additional factors causing hybrid sterility, or the same factors but with a stronger effect. Since our line of SR D. persimilis did not show any detectable within-species sterility, our observation of sterility in our crosses is unlikely to be a within-species fertility reduction, though we cannot completely exclude that such a reduction is overexpressed in a hybrid background. A related argument is that since hybrids are sterile with or without segregation distortion, there is no reason to expect the two traits to be associated. However, the causes of sterility in hybrids with an SR D. persimilis parent are different than in nonSR D. persimilis parents, as indicated by the difference in effects of the XR chromosome between SR and non-SR D. persimilis (Noor et al. 2001). While more is known about the within-species fertility effects of SR in D. pseudoobscura (Wallace 1948, Beckenbach 1978, Curtsinger and Feldman 1980, Beckenbach 1981, Wu 1983a, Wu 1983b, Beckenbach 1996, Price et al. 2008a, Price et al. 2008b, Price et al. 2009), SR D. persimilis actually serves as a better model system for studying the genetic basis of SR given that it is homosequential with the Standard D. pseudoobscura XR.
QTL Mapping
Wu and Beckenbach (1983) failed to observe the SR trait in any of their isolated segments of the SR XR of D. persimilis when in a D. pseudoobscura background, suggesting either that the presence of all four segments were required for segregation distortion to be expressed, or that the D. pseudoobscura Y chromosome is not sensitive to the D. persimilis SR distorter. They mapped a region causing hybrid sterility linked to the sepia (se) gene, located about 0.5 Mb proximal to our marker DPSX029, which shows a significant effect on segregation distortion (Figure 3) but not hybrid sterility (Figure 4) in our study. Other work has demonstrated the strong effect of the SR D. persimilis XR on hybrid sterility with D. pseudoobscura (Orr 1989), while the Standard XR from D. persimilis reduces the proportion of sterile hybrids (Noor et al. 2001). Results from these previous studies provide circumstantial evidence of an association of factors located on the XR with segregation distortion and hybrid sterility in these species.
Our segregation distortion QTL map would be more precise with a larger sample size; however, given the success rate of crosses, a larger sample size would be extremely difficult (e.g. 100,000 crosses to generate 1000 individuals with 15 or greater progeny). A lower cutoff value would increase the sample size, but even with a cutoff value of 5 progeny, our sample size only increases from 109 to 205. Our cutoff of 15 progeny is within the range used by recent studies: Phadnis (2011) used 5 progeny, Dyer (2011) used 20 progeny, and Bastide et al. (2011) used 70 progeny (though D. simulans, used by Bastide et al., is more fecund than D. pseudoobscura or D. persimilis).
Association of hybrid sterility and segregation distortion
We failed to identify clear recombinants (barring twenty possible cases in 350 that may be false positives) between XR factors conferring distortion and factors conferring sterility, indicating that the two traits are caused by the same factor or factors very close together. We cannot estimate the actual recombination rate without including the sterile individuals, in whom recombination is undetectable since the phenotype cannot be studied– they could have inherited both sterility and segregation distortion factors (nonrecombinant), or inherited only sterility factor(s) (recombinant). However, another plausible reason we saw so few backcross males produce biased sex ratios is that the D. pseudoobscura Y chromosome may not be sensitive to the D. persimilis SR distorter. Wu and Beckenbach (1983) also did not see evidence of sex ratio bias in backcross males carrying a D. persimilis X and a D. pseudoobscura Y. In Drosophila mediopunctata, D. quinaria, and D. simulans (Carvalho et al. 1997, Jaenike 1999, Montchamp-Moreau et al. 2001), within-species variation does exist for Y chromosome sensitivity, so such a difference between D. persimilis and D. pseudoobscura is possible. With this caveat, we are unable to conclude whether our mapping results demonstrate linkage of the segregation distortion and hybrid sterility factors, or merely a loss of drive due to insensitivity of the Y chromosome in our cross. However, had we observed a sex ratio bias in the progeny of the backcross males, it would have been evidence against linkage of the segregation distortion and hybrid sterility factors. Since we did not observe a sex ratio bias, the possibility remains that the factors are linked.
The major peaks of both hybrid sterility and segregation distortion maps colocalize, despite the large width of the sterility map. They also share a minor peak towards the telomere of the XR, starting at marker DPSX012. Combined with the data from the preliminary linkage test, this evidence suggests a possible association between the two traits. However, the broad peak of the hybrid sterility map could be masking multiple factors contributing to hybrid sterility between D. persimilis and D. pseudoobscura. Multiple peaks in our segregation distortion map also suggest multiple required factors (Figure 3). A similar finding suggested at least three segregation distortion factors on the XR (Wu and Beckenbach 1983). We also could be missing segregation distortion peaks because so few BCper males were fertile enough to produce greater than 15 progeny. Many of the sterile males may also be harboring segregation distorter alleles linked to the hybrid sterility allele, but we cannot detect this sex ratio bias because of the linked sterility allele.
Using a species like D. persimilis that exhibits segregation distortion within species to understand the possible link between segregation distortion, hybrid sterility, and speciation provides for more direct testing than using cryptic hybrid distortion – which may merely be a hybrid incompatibility itself. This work built on studies done by Tao et al. (2001) and Phadnis and Orr (2009) by using a species exhibiting within-species segregation distortion, and expanded on Wu and Beckenbach (1983) by adding more individuals to map hybrid sterility and more (and genome-sequence-localized) markers for localizing both traits. Future work could include developing a molecular assay to try to measure X vs. Y-bearing gametes directly, hence allowing semi-sterile individuals to be scored for meiotic drive.
The proposed association between hybrid incompatibilities and segregation distortion continues to strengthen as more genes are discovered that influence both phenotypes: tmy (Tao et al. 2001), Ovd (Phadnis and Orr 2009), nad6/PPR gene (Case and Willis 2008, Barr and Fishman 2010), and other hybrid incompatibility genes suggested to be influenced by segregation distortion (reviewed in Johnson 2010). This association also strengthens the link to speciation, providing more evidence for Frank's (1991) and Hurst and Pomiankowski's (1991) hypothesis that segregation distortion can cause hybrid sterility in some cases. In further support of their hypothesis, Bastide et al. (2011) provides evidence that a possible arms race between SR segregation distorters and suppressors in the D. simulans Paris system is ongoing.
In summary, we characterized and developed lines of SR D. persimilis, mapped segregation distortion within D. persimilis, and mapped hybrid sterility between D. persimilis and D. pseudoobscura. Our maps suggest a possible association between both traits. This work expands on previous mapping studies and opens opportunities for other research using SR D. persimilis. Future work in all SR systems will hopefully provide answers to the various questions surrounding segregation distortion, including its association with hybrid sterility and speciation.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank A. Graham and B. Manzano-Winkler for technical assistance, and Ö. Kayikçi, K. Ferris, members of the Noor laboratory, and two anonymous reviewers for critical comments on the manuscript. This work was funded by National Institutes of Health grants GM076051 and GM086445.
WORKS CITED
- Barr CM, Fishman L. The nuclear component of a cytonuclear hybrid incompatibility in Mimulus maps to a cluster of pentatricopeptide repeat genes. Genetics. 2010;184:455–465. doi: 10.1534/genetics.109.108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastide H, Cazemajor M, Ogereau D, Derome N, Hospital F, Montchamp-Moreau C. Rapid rise and fall of selfish sex-ratio X chromosomes in Drosophila simulans: spatiotemporal analysis of phenotypic and molecular data. Mol Biol Evol. 2011;28:2461–2470. doi: 10.1093/molbev/msr074. [DOI] [PubMed] [Google Scholar]
- Beckenbach A. Multiple mating and the sex-ratio trait in Drosophila pseudoobscura. Evolution. 1981;35:275–281. doi: 10.1111/j.1558-5646.1981.tb04886.x. [DOI] [PubMed] [Google Scholar]
- Beckenbach A. Fitness analysis of the sex-ratio polymorphism in experimental populations of Drosophila pseudoobscura. Am Nat. 1983;121:630–648. [Google Scholar]
- Beckenbach AT. The “sex-ratio” trait in Drosophila pseudoobscura: fertility relations of males and meiotic drive. Am Nat. 1978;112:97–117. [Google Scholar]
- Beckenbach AT. Selection and the “'sex-ratio” polymorphism in natural populations of Drosophila pseudoobscura. Evolution. 1996;50:787–794. doi: 10.1111/j.1558-5646.1996.tb03888.x. [DOI] [PubMed] [Google Scholar]
- Campbell R, Noor MAF. Assessing hybrid male fertility in Drosophila species: Correlation between sperm motility and production of offspring. Dros. Inf. Serv. 2001;84:6–9. [Google Scholar]
- Carvalho AB, Vaz SC, Klaczko LB. Polymorphism for Y-linked suppressors of sex-ratio in two natural populations of Drosophila mediopunctata. Genetics. 1997;146:891–902. doi: 10.1093/genetics/146.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case AL, Willis JH. Hybrid male sterility in Mimulus (Phrymaceae) is associated with a geographically restricted mitochondrial rearrangement. Evolution. 2008;62:1026–39. doi: 10.1111/j.1558-5646.2008.00360.x. [DOI] [PubMed] [Google Scholar]
- Coyne JA. Genetic basis of male sterility in hybrids between 2 closely related species of Drosophila. P Natl Acad Sci-Biol. 1984;81:4444–4447. doi: 10.1073/pnas.81.14.4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtsinger JW, Feldman MW. Experimental and theoretical analysis of the “sex-ratio” polymorphism in Drosophila pseudoobscura. Genetics. 1980;94:445–466. doi: 10.1093/genetics/94.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AW, Wu CI. The broom of the sorcerer's apprentice: The fine structure of a chromosomal region causing reproductive isolation between two sibling species of Drosophila. Genetics. 1996;143:1287–1298. doi: 10.1093/genetics/143.3.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics. 1936;21:113–135. doi: 10.1093/genetics/21.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Facts and problems of the “sex-ratio” condition in Drosophila. Scientia Genetica. 1939;11:67–75. [Google Scholar]
- Dyer KA. Local selection underlies the geographic distribution of sex-ratio drive in Drosophila neotestacea. Evolution. 2011;66:973–984. doi: 10.1111/j.1558-5646.2011.01497.x. [DOI] [PubMed] [Google Scholar]
- Frank SA. Divergence of meiotic drive-suppression systems as an explanation for sex-biased hybrid sterility and inviability. Evolution. 1991;45:262–267. doi: 10.1111/j.1558-5646.1991.tb04401.x. [DOI] [PubMed] [Google Scholar]
- Gebhardt MD, Anderson WW. Temperature-related fertility selection on body-size and the sex-ratio gene arrangement in Drosophila pseudoobscura. Genet Res. 1993;62:63–75. doi: 10.1017/s0016672300031578. [DOI] [PubMed] [Google Scholar]
- Gloor GB, Engels WR. Single-fly DNA preps for PCR. Dros. Inf. Serv. 1992;71:148–149. [Google Scholar]
- Hall DW. Meiotic drive and sex chromosome cycling. Evolution. 2004;58:925–931. doi: 10.1111/j.0014-3820.2004.tb00426.x. [DOI] [PubMed] [Google Scholar]
- Hurst LD, Pomiankowski A. Causes of sex-ratio bias may account for unisexual sterility in hybrids - a new explanation of Haldane's rule and related phenomena. Genetics. 1991;128:841–858. doi: 10.1093/genetics/128.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 1996;5:299–314. [Google Scholar]
- Jaenike J. Suppression of sex-ratio meiotic drive and the maintenance of Y-chromosome polymorphism in Drosophila. Evolution. 1999;53:164–174. doi: 10.1111/j.1558-5646.1999.tb05342.x. [DOI] [PubMed] [Google Scholar]
- Jaenike J. Sex chromosome meiotic drive. Annu Rev Ecol Syst. 2001;32:25–49. [Google Scholar]
- Johnson NA. Hybrid incompatibility genes: remnants of a genomic battlefield? Trends Genet. 2010;26:317–325. doi: 10.1016/j.tig.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Margarido GRA, Souza AP, Garcia AAF. OneMap: software for genetic mapping in outcrossing species. Hereditas. 2007;144:78–79. doi: 10.1111/j.2007.0018-0661.02000.x. [DOI] [PubMed] [Google Scholar]
- McDermott SR, Noor MA. The role of meiotic drive in hybrid male sterility. Philos Trans R Soc Lond B Biol Sci. 2010;365:1265–1272. doi: 10.1098/rstb.2009.0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott SR, Noor MAF. Genetics of hybrid male sterility among strains and species in the Drosophila pseudoobscura species group. Evolution. 2011;65:1969–1978. doi: 10.1111/j.1558-5646.2011.01256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehring AJ, Li J, Schug MD, Smith SG, deAngelis M, Mackay TF, Coyne JA. Quantitative trait loci for sexual isolation between Drosophila simulans and D. mauritiana. Genetics. 2004;167:1265–1274. doi: 10.1534/genetics.103.024364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehring AJ, Llopart A, Elwyn S, Coyne JA, Mackay TF. The genetic basis of postzygotic reproductive isolation between Drosophila santomea and D. yakuba due to hybrid male sterility. Genetics. 2006;173:225–233. doi: 10.1534/genetics.105.052985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montchamp-Moreau C, Ginhoux V, Atlan A. The Y chromosomes of Drosophila simulans are highly polymorphic for their ability to suppress sex-ratio drive Evolution. 2001;55:728–737. doi: 10.1554/0014-3820(2001)055[0728:tycods]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Noor MA. Speciation driven by natural selection in Drosophila. Nature. 1995;375:674–675. doi: 10.1038/375674a0. [DOI] [PubMed] [Google Scholar]
- Noor MAF, Grams KL, Bertucci LA, Almendarez Y, Reiland J, Smith KR. The genetics of reproductive isolation and the potential for gene exchange between Drosophila pseudoobscura and D. persimilis via backcross hybrid males. Evolution. 2001;55:512–521. doi: 10.1554/0014-3820(2001)055[0512:tgoria]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Orr HA. Genetics of male and female sterility in hybrids of Drosophila pseudoobscura and Drosophila persimilis. Genetics. 1987;116:555–563. doi: 10.1093/genetics/116.4.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. Localization of genes causing postzygotic isolation in 2 hybridizations involving Drosophila pseudoobscura. Heredity. 1989;63:231–237. doi: 10.1038/hdy.1989.96. [DOI] [PubMed] [Google Scholar]
- Palumbi SR. Molecular Systematics. Sinauer; Sunderland, MA: 1996. Nucleic Acids II. the polymerase chain reaction. [Google Scholar]
- Pardo-Manuel de Villena F, Sapienza C. Nonrandom segregation during meiosis: the unfairness of females. Mamm Genome. 2001;12:331–339. doi: 10.1007/s003350040003. [DOI] [PubMed] [Google Scholar]
- Phadnis N. Genetic architecture of male sterility and segregation distortion in Drosophila pseudoobscura Bogota-USA Hybrids. Genetics. 2011;189:1001–1009. doi: 10.1534/genetics.111.132324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis N, Orr HA. A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science. 2009;323:376–379. doi: 10.1126/science.1163934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Policansky D. Sex-ratio, meiotic drive, and group selection in Drosophila pseudoobscura. Am Nat. 1974;108:75–90. [Google Scholar]
- Presgraves DC. Sperm biology: an evolutionary perspective. Elsevier; Burlington, MA: 2008. Drive and sperm: evolution and genetics of male meiotic drive. [Google Scholar]
- Presgraves DC. The molecular evolutionary basis of species formation. Nat Rev Genet. 2010;11:175–180. doi: 10.1038/nrg2718. [DOI] [PubMed] [Google Scholar]
- Price TA, Bretman AJ, Avent TD, Snook RR, Hurst GD, Wedell N. Sex ratio distorter reduces sperm competitive ability in an insect. Evolution. 2008a;62:1644–1652. doi: 10.1111/j.1558-5646.2008.00386.x. [DOI] [PubMed] [Google Scholar]
- Price TA, Hodgson DJ, Lewis Z, Hurst GD, Wedell N. Selfish genetic elements promote polyandry in a fly. Science. 2008b;322:1241–1243. doi: 10.1126/science.1163766. [DOI] [PubMed] [Google Scholar]
- Price TA, Lewis Z, Smith DT, Hurst GD, Wedell N. Sex ratio drive promotes sexual conflict and sexual coevolution in the fly Drosophila pseudoobscura. Evolution. 2009;64:1504–1509. doi: 10.1111/j.1558-5646.2009.00896.x. [DOI] [PubMed] [Google Scholar]
- Price TA, Wedell N. Selfish genetic elements and sexual selection: their impact on male fertility. Genetica. 2008;132:295–307. doi: 10.1007/s10709-007-9173-2. [DOI] [PubMed] [Google Scholar]
- Reed LK, LaFlamme BA, Markow TA. Genetic architecture of hybrid male sterility in Drosophila: analysis of intraspecies variation for interspecies isolation. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler L, Novitski E. Meiotic drive as an evolutionary force. Am Nat. 1957;91:105–110. [Google Scholar]
- Sturtevant AH, Dobzhansky T. Geographical distribution and cytology of “sex ratio” in Drosophila pseudoobscura and related species. Genetics. 1936;21:473–490. doi: 10.1093/genetics/21.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Hartl DL, Laurie CC. Sex-ratio segregation distortion associated with reproductive isolation in Drosophila. Proc Natl Acad Sci U S A. 2001;98:13183–13188. doi: 10.1073/pnas.231478798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace B. Studies on sex-ratio in Drosophila pseudoobscura. I. Selection and sex-ratio. Evolution. 1948;2:189–217. [PubMed] [Google Scholar]
- Wang S, Basten CJ, Zeng ZB. Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University; Raleigh, NC: 2011. ( http://statgen.ncsu.edu/qtlcart/WQTLCart.htm) [Google Scholar]
- Werren JH. Selfish genetic elements, genetic conflict, and evolutionary innovation. P Natl Acad Sci USA. 2011;108:10863–10870. doi: 10.1073/pnas.1102343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI. Virility deficiency and the sex-ratio trait in Drosophila pseudoobscura .II. Multiple mating and overall virility selection. Genetics. 1983a;105:663–679. doi: 10.1093/genetics/105.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI. Virility deficiency and the sex-ratio trait in Drosophila pseudoobscura. .I. Sperm displacement and sexual selection. Genetics. 1983b;105:651–662. doi: 10.1093/genetics/105.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI, Beckenbach AT. Evidence for extensive genetic differentiation between the sex-ratio and the standard arrangement of Drosophila pseudoobscura and Drosophila persimilis and identification of hybrid sterility Factors. Genetics. 1983;105:71–86. doi: 10.1093/genetics/105.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.