Abstract
Background
The 235delC mutation of GJB2 gene is considered as a risk factor for the non-syndromic hearing loss (NSHL), and a significant difference in the frequency and distribution of the 235delC mutation has been described world widely.
Methods
A systematic review was performed by means of a meta-analysis to evaluate the influence of the 235delC mutation on the risk of NSHL. A literature search in electronic databases using keywords “235delC”, “GJB2” associated with “carrier frequency” was conducted to include all papers from January 1999 to June 2011. A total of 36 papers were included and there contained 13217 cases and 6521 controls derived from Oceania, American, Europe and Asian.
Results
A remarkable heterogeneity between these studies was observed. The combined results of meta-analysis showed that the 235delC mutant increased the risk of NSHL (OR = 7.9, 95%CI 4.77 ~ 13.11, P <0.00001). Meanwhile, heterogeneity of genetic effect was also observed due to the ethnic specificity and regional disparity. Therefore, the stratified meta-analysis was subsequently conducted and the results indicated that the 235delC mutation was significantly correlated with the risk of NHSL in the East Asian and South-east Asian populations (OR = 12.05, 95%CI 8.33~17.44, P <0.00001), but not significantly in the Oceania and European populations (OR = 10.36, 95%CI: 4.68~22.96, Z = 1.68, P >0.05).
Conclusions
The 235delC mutation of GJB2 gene increased the risk of NHSL in the East Asian and South-east Asian populations, but non-significantly associated with the NSHL susceptibility in Oceania and European populations, suggesting a significant ethnic specificity of this NSHL-associated mutation.
Keywords: GJB2, 235delC, Non-syndromic hearing loss, Meta-analysis
Introduction
Hearing impairment is the most common sensory disorder, present in 1 of every 1000 newborns, about half of which could be attributed to genetic factors [1]. There are two monogenic forms of hearing loss including syndromic (characterized by hearing loss in combination with other abnormalities) and non-syndromic (with only hearing loss) deafness [1]. Up to now, more than 70 loci for non-syndromic hearing loss (NSHL) had been identified (http://hereditaryhearingloss.org/). Among them, DFNB1 was considered to be the main cause of prelingual deafness. As one of the DFNB1 locus-linked genes, GJB2 was the first to be indentified which encoded the gap junction protein connexin 26 (Cx26). To date, more than a hundred different mutations of this gene has been implicated in NSHL [2]. The most common mutations of GJB2 gene comprised 35delG, 167delT and 235delC with the varied incidence among populations [2].
The 235delC mutation of GJB2 is the most frequently known mutation in some East Asian groups, with a carrier frequency of approximately 1% [3]. Recently, a substantial amount of research had been devoted to elucidate the influence of this mutation on the risk of NSHL, but conflicting results were obtained. Some studies reported that the 235delC mutation had only limited effect on the risk of NSHL [4,5], while others suggested that the 235delC mutation increased the risk of NSHL or was concerned with the specificity pathological type of NSHL [6,7]. In those reported studies, the genetic background of the ethnic specificity and the regional disparity were not completely considered, or small effects in human genetic association were difficult to be detected due to small sample size of case-control studies. To clarify the variable results, a further research on the association between ethnic specificity of the 235delC mutation and the NSHL susceptibility was essentially needed. In this study, a combined meta-analysis was performed by pooling data from all relevant case-control studies published in recent ten years. The association of the 235delC mutation with hearing loss and the heterogeneity of genetic effect were evaluated, based on which the stratified meta-analysis was subsequently conducted to assess the ethnic specificity of the 235delC mutation associated with the NSHL susceptibility.
Methods
Literature search
The electronic databases including PubMed, InterScience, British Library Direct, Embase, Sciencedirect, Chinese Biomedical Literature, Chinese WanFang database and China National Knowledge Infrastructure were searched from January 1999 to June 2011, fulfilling all case-control studies holding information on the epidemiology study in molecular genetics of hearing loss. The following terms were used in search strategies: GJB2, Connexin 26 (Cx26), mutation, variant, NSHL or NSHI (non-syndromic hearing impairment). All related citations were retrieved to find other relevant articles that were not initially identified. The entire literature search was performed by two independent researchers.
Study selection
The reference lists of all traced articles were examined manually and screened for eligibility using the following criteria: (1) the association of the 235delC mutation of GJB2 with NSHL was examined based on case-control design; (2) the selected papers contained complete information and represented Odds Ratio (OR) and 95% Confidence Interval (95%CI) directly or indirectly; (3) for the same studied populations, only the papers newly published or providing the complete data were adopted. All collected papers were scored and categorized according to the Newcastle-Ottawa Scale (NOS) for assessing the quality of case-control studies [8]. Studies with the low score (below 6) or the genetic type of control group inconsistent with the principle of Hardy-Weinberg Equilibrium (HWE) were categorized to Grade B, and the others were categorized to Grade A [9].
Data extraction
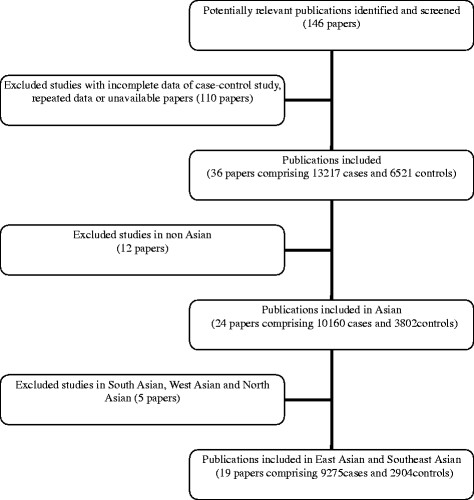
A total of 146 papers were obtained through the search of electronic databases. Among them, 36 papers associated with the 235delC mutation of GJB2 gene and the NSHL susceptibility were selected for the following data analysis. The flow chart of review process was shown as Figure 1.
Figure 1 .
Flow chart of review process.
Data analysis
The mean value of carrier frequency of the 235delC mutation in the individual studies was calculated. The included populations were divided into two groups (Asian and non-Asian groups) according to the distribution of carriers. The relationship between the 235delC mutation of GJB2 and hearing loss was evaluated for each group or continent. The statistical analysis was conducted using Review Manager 5.1 (RevMan) supplied by the Cochrane Collaboration (Oxford, UK). Heterogeneity among studies was assessed through Cochrane χ2 test and I2 statistic (P < 0.05 indicating the existence of between-study heterogeneity). Based on the test of heterogeneity, the combined meta-analysis and stratified meta-analysis were conducted respectively. The pooled OR was calculated using fixed or random effect model, along with the 95%CI to measure the strength of the genetic association. The publication bias was analyzed by STATA 11.0 and Begg’s test, and failsafe number (Nfs) was calculated (Nfs > 10, P > 0.05 means that the obtained conclusion was confident). The statistical analysis was performed using Software SPSS (Version 13, Chicago, USA). An online tool (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl/) was applied to test whether the genetic type of control group was consistent with Hardy-Weinberg equilibrium (HWE) for each study.
Results
The 235delC carrier frequency of GJB2 allele
Among 36 included papers, there were 24 papers containing the information on the 235delC carrier frequencies of GJB2 allele, as shown in Table 1.
Table 1.
235del mutation frequencies of ofGJB2allele in the individual studies
| No | Relevant Studies | TOTAL | HETRO | HOMO | TOTAL% |
|---|---|---|---|---|---|
| 1 |
Ohtsuka A, et al. 2003 [10] |
1227 |
92 |
16 |
5.05% |
| 2 |
Batissoco AC, et al. 2009 [11] |
300 |
1 |
0 |
0.17% |
| 3 |
Wattanasirichaigoon D, et al. 2004 [12] |
166 |
6 |
2 |
3.01% |
| 4 |
Padma G, et al. 2009 [13] |
456 |
0 |
1 |
0.22% |
| 5 |
Park HJ, et al. 2000 [14] |
147 |
5 |
5 |
5.10% |
| 6 |
Shalin H, et al. 2002 [15] |
48 |
0 |
1 |
2.08% |
| 7 |
Lee KY, et al. 2008 [16] |
29 |
0 |
2 |
6.90% |
| 8 |
Schimmenti LA, et al. 2008 [17] |
95 |
0 |
1 |
1.05% |
| 9 |
Posukh O, et al. 2005 [18] |
76 |
4 |
1 |
3.95% |
| 10 |
Abe S, et al. 2000 [19] |
35 |
4 |
1 |
8.57% |
| 11 |
Tang, HY et al. 2006 [20] |
610 |
1 |
0 |
0.08% |
| 12 |
Tekin M, et al. 2010 [21] |
534 |
12 |
2 |
1.50% |
| 13 |
Kudo T, et al. 2000 [22] |
63 |
1 |
3 |
5.56% |
| 15 |
Cheng X, et al. 2005 [23] |
740 |
1 |
1 |
0.20% |
| 16 |
Wang YC, et al. 2002 [24] |
169 |
6 |
8 |
6.51% |
| 17 |
Chen D, et al. 2009 [25] |
100 |
10 |
16 |
21.00% |
| 18 |
Xiao ZA, et al. 2004 [26] |
131 |
2 |
9 |
7.63% |
| 19 |
Wang SH, et al. 2009 [27] |
140 |
7 |
20 |
16.79% |
| 20 |
Hwa, HL et al. 2003 [28] |
324 |
16 |
11 |
5.86% |
| 21 |
Shi GZ, et al. 2004 [29] |
20 |
3 |
3 |
22.50% |
| 22 |
Dai P, et al. 2007 [30] |
3004 |
255 |
233 |
12.00% |
| 23 |
Liu ZX, et al. 2002 [31] |
118 |
20 |
14 |
20.34% |
| 24 | Liu YH, et al.2002 [32] | 210 | 13 | 27 | 15.95% |
(*χ2 = 6.667, P <0.05. A significant heterogeneity was found in the 235del mutation frequency of GJB2 allele).
Characteristics of included studies
A total of 36 case-control studies comprising 13217 patients’ cases with NSHL and 6521 controls were included in meta-analysis (Table 2). Each study was evaluated with a high score (not less than 5). Among them, there were 14 papers categorized to Grade A. All the included 36 case-control studies were consistent with the Hardy-Weinberg Equilibrium test, indicating a good representation of control populations.
Table 2.
Relevant studies on 235delC mutation and NSHL
| No | Author | Score/ Grade | No | Author | Score/ Grade |
|---|---|---|---|---|---|
| Studies in non-Asian populations | |||||
| 1 |
Dahl HM, et al. 2006 [4] |
6A |
7 |
Cheng X, et al. 2005 [23] |
5B |
| 2 |
Ramsebner R, et al. 2007 [5] |
5B |
8 |
Utrera R, et al. 2007 [33] |
5B |
| 3 |
Batissoco AC, et al. 2009 [11] |
5B |
9 |
Tang HY, et al. 2006 [20] |
6A |
| 4 |
Damalon V, et al. 2010 [34] |
5B |
10 |
Pollak A, et al. 2007 [35] |
5B |
| 5 |
Samanich J, et al. 2007 [36] |
5B |
11 |
Tóth T, et al. 2007 [37] |
5B |
| 6 |
Schimmenti LA, et al. 2008 [17] |
6A |
12 |
Neocleous V, et al. 2006 [38] |
5B |
| Studies in East Asian and Southeast Asian populations | |||||
| 13 |
Snoeckx RL, et al. 2005 [39] |
5B |
23 |
Xiao ZA, et al. 2004 [26] |
5B |
| 14 |
Wattanasirichaigoon D, et al. 2004 [12] |
5B |
24 |
Wang SH, et al. 2009 [ 27] |
6A |
| 15 |
Park HJ, et al. 2000 [14] |
6A |
25 |
Hwa HL, et al. 2003 [28] |
5B |
| 16 |
Lee KY, et al. 2008 [16] |
5B |
26 |
Shi GZ, et al. 2004 [29] |
6A |
| 17 |
Abe S, et al. 2000 [19] |
5B |
27 |
Dai P, et al. 2007 [30] |
6A |
| 18 |
Ohtsuka A, et al. 2003 [10] |
5B |
28 |
Liu ZX, et al. 2002 [31] |
5A |
| 19 |
Tekin M , et al. 2010 [21] |
5B |
29 |
Liu YH, et al. 2002 [32] |
6A |
| 20 |
Kudo T, et al. 2000 [22] |
5B |
30 |
Chen GM, et al. 2011 [40] |
5B |
| 21 |
Wang YC, et al. 2002 [24] |
6A |
31 |
Guo YF, et al. 2008 [41] |
6A |
| 22 |
Chen D, et al. 2009 [25] |
5B |
|
|
|
| Studies in South Asian, West Asian and North Asian populations | |||||
| 32 |
Padma G, et al. 2009 [13] |
6A |
35 |
Baysal E, et al. 2008 [42] |
6A |
| 33 |
Bhalla S, et al. 2009 [43] |
6A |
36 |
Posukh O, et al. 2005 [18] |
5B |
| 34 | Shalin H, et al. 2002 [15] | 5B | |||
Meta-analysis of overall data
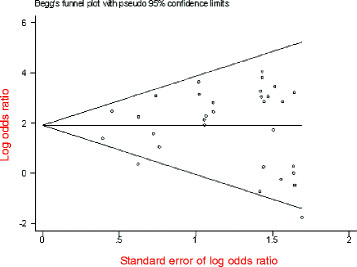
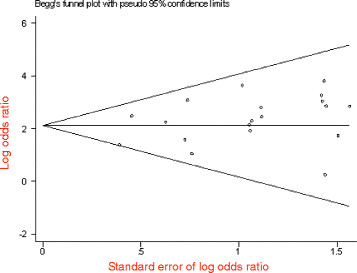
The 235delC mutation of GJB2 gene was not detected in five case-control studies, so the rest of 31 studies compromising 12437 patients’ cases and 5844 controls were included in meta-anslysis. The pooled OR was calculated using random effect model, and the forest plot on the association of mutation carriers with the risk of NSHL was fulfilled (OR = 7.9, 95%CI: 4.77~13.11, Z = 8.01, P<0.00001). As shown in Figure 2, the pooled OR and 95%CI values demonstrated that there was a higher prevalence of the 235delC mutation in the case group than in the control group. The publication bias was examined by use of funnel plot (Figure 3). The visual inspection of funnel plot seemed to be symmetrical, and most of the data points were located within 95%CI. However, an absence of corner was observed possibly due to the unpublished negative results. Additionally, the Begg’s test was conducted to analyze the symmetry of the funnel plot, and no significant publication bias was found (Nfs = 58.84>10, P = 0.341>0.05), which suggested that the publication bias had little effect on the result of meta-analysis and the conclusions obtained were confident.
Figure 2 .
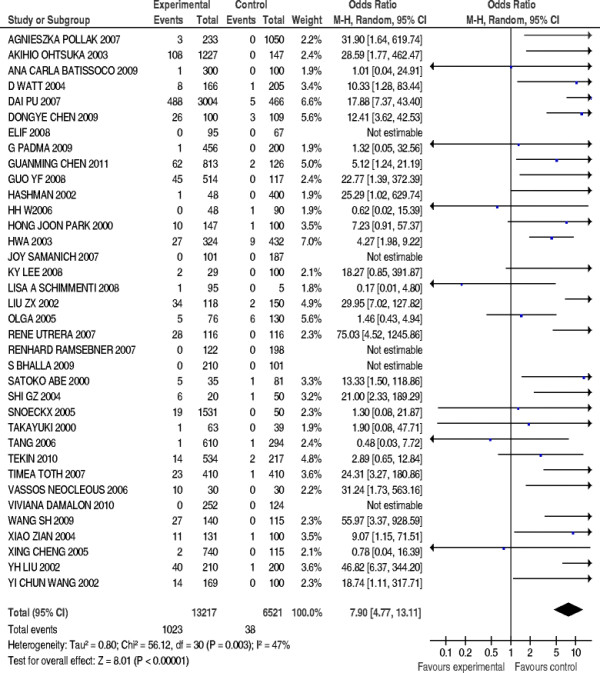
Forest plot on the association of 235delC mutation with the risk of NSHL.
Figure 3 .
Begg’s funnel plot assessing publication bias on all included studies.
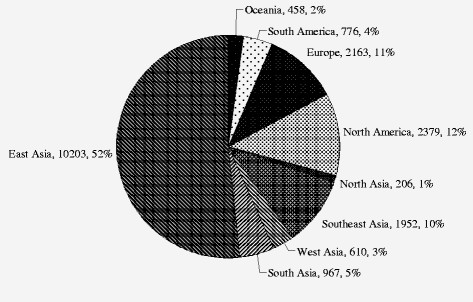
Heterogeneity of genetic effect was detected as shown in Figure 2 (χ2 = 56.12, df = 30, P = 0.003<0.05, I2 = 47%), which indicated that the association of the 235delC mutation with the risk of NSHL was varied due to the ethnic specificity or regional disparity. To elucidate the prevalence of the 235delC mutation related to the NSHL susceptibility more precisely, the stratified meta-analysis was subsequently performed according to the regional studies. It was observed that a high prevalence of the 235delC mutation related to hearing loss was reported in Asian (Figure 4). Therefore, the populations of the included studies were divided into Asian and Non-Asian population groups, and the stratified meta-analysis was conducted to each group.
Figure 4 .
Proportions of tested individuals in different continents.
Stratified meta-analysis of Asian population group
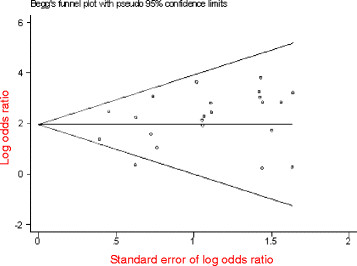
Two studies were excluded due to undetectable 235delC mutations in patients’ cases or incomplete statistical data in controls. And a total of 22 papers compromising 9855 patients’ cases and 3634 controls were included in the stratified meta-analysis. The pooled OR was calculated using random effect model, and forest plot on the association of mutation carriers with the risk of NSHL was fulfilled (OR = 8.95, 95%CI: 5.44 ~ 14.73, Z = 8.62, P < 0.00001). As shown in Figure 5, the pooled OR and 95%CI values demonstrated that the 235delC mutation of GJB2 gene was significantly associated with the risk of NSHL in Asian population. The visual inspection of funnel plot seemed to be symmetrical (Figure 6), and most of the data points were located within 95%CI. The Begg’s test was also used to analyze the symmetry of funnel plot, and no significant publication bias was found (Nfs = 35.46>10, P = 0.499>0.05), which suggested that and the publication bias had little effect on the result of meta-analysis and the conclusions obtained were confident.
Figure 5 .
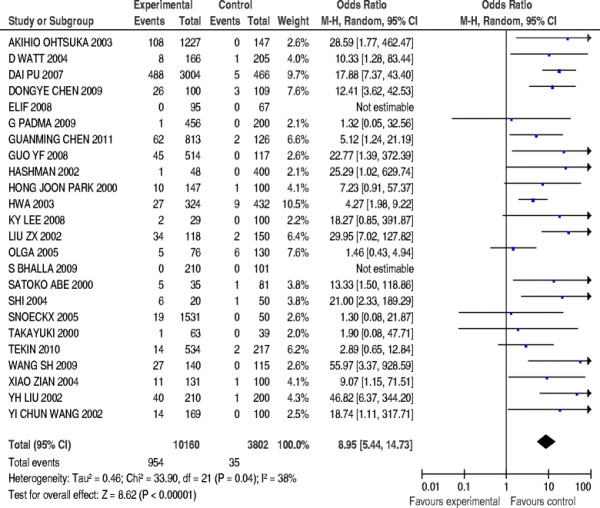
Forest plot on the association of 235delC mutation with the risk of NSHL in Asian populations.
Figure 6 .
Begg’s funnel plot assessing publication bias on included studies in Asian populations.
Heterogeneity of genetic effect was still detected as shown in Figure 5 (χ2 = 33.90, df = 21, P = 0.04<0.05, I2 = 38%), which demonstrated the presence of disparity in the prevalence of the 235delC mutation related to NSHL susceptibility among Asian populations suggesting the need of further stratified meta-analysis.
Stratified Meta-analysis of East Asian and Southeast Asian populations
The overall effect on included studies in East Asian and Southeast Asian populations was evaluated, and a total of 19 papers were included, compromising 9275 patients’ cases and 2904 controls. The forest plot on the association of mutation carriers with the risk of NSHL was fulfilled using random effect model (OR = 12.05, 95%CI: 8.33 ~ 17.44, Z = 13.21, P <0.00001). As shown in Figure 7, the pooled OR and 95%CI values demonstrated that the prevalence of the 235delC mutation was higher in the case group than in the control group. Publication bias was examined by use of funnel plot (Figure 8). The Begg’s test was also used to analyze the symmetry of funnel plot, and no significant publication bias was found (Nfs = 28.58>10, P = 0.576>0.05), which suggested that and the publication bias had little effect on the result of meta-analysis and the obtained conclusion was confident. In addition, no significant heterogeneity of genetic effect was found as shown in Figure 7 (χ2 = 27.16, df = 18, P = 0.08>0.05, I2 = 34%).
Figure 7 .
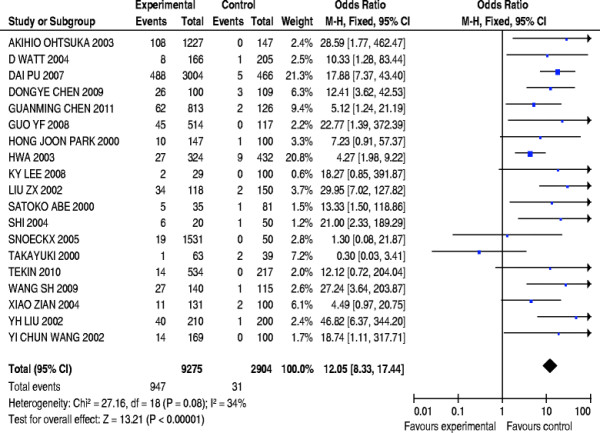
Forest plot on the association of 235delC mutation with the risk of NSHL in East Asian and Southeast Asian populations.
Figure 8 .
Begg’s funnel plot assessing publication bias on included studies in East Asian and Southeast Asian populations.
The above analytic results indicated that the 235delC mutation of GJB2 gene was significantly associated with the risk of NSHL in East Asian and Southeast Asian. Due to the insufficient searched studies, the meta-analysis was not performed to the regional studies in North Asian, West Asian and South Asian populations, respectively.
Stratified Meta-analysis of Non-Asian population group (Europe and Oceania populations)
Three case-control studies were excluded due to no 235delC mutation of GJB2 gene detected, and a total of 9 papers were included, compromising 2683 patients’ cases and 2397 controls. The pooled OR was calculated using random effect model, and forest plot on the association of mutation carriers with the risk of NSHL was fulfilled (Figure 9). The prevalence of the 235delC mutation seemed to be ethnic-specific, but was not significantly associated with the NSHL susceptibility in European and Oceania population (OR = 10.36, 95%CI: 4.68 ~ 22.96, Z = 1.68, P = 0.09>0.05).
Figure 9 .
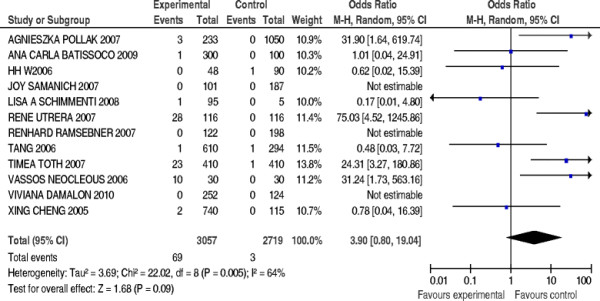
Forest plot on the association of 235delC mutation with the risk of NSHL in Europe and Oceania populations.
Discussion
The investigation on heterogeneity in genetic association were commonly leveled to the genetic effect and genetic model: the genetic effect focused on the difference in study method, potential interaction of gene-gene or gene-environment and linkage disequilibrium [13], while the genetic model was mainly concerned with the differential association between heterozygote (or homozygote) and the risk of human diseases. The development of hearing loss involved with numerous factors and experienced multiple steps, and the interaction of heredity (genes) and environmental factors played an important role, which could have a potential influence on the association of GJB2 mutations and the NSHL susceptibility with ethnic specificity or certain genetic backgrounds of affected families. Meanwhile, the same type of single nucleotide polymorphisms (SNP) could show various contributions to the genetic models derived from different studies due to the ethnic specificity.
By increasing the sample size, meta-analysis based on case-control studies has the potential to evaluate the between-study heterogeneity and detect small effects in genetic association. In this study, the combined results of meta-analysis showed that the 235delC mutant increased the risk of NSHL, but significant between-study heterogeneity and genetic-effect heterogeneity were detected, indicating that the ethnic specificity and regional disparity contributed to the association between the 235delC mutation of GJB2 gene and the NSHL susceptibility. Results of stratified meta-analysis showed that the 235delC mutation was prominently associated with the risk of NSHL in East Asian and Southeast Asian populations, but not significantly in European and Oceania populations. The 235delC mutation of GJB2 gene trended to be consistent with recessive model in Chinese population, while heterogeneity of genetic effect was also detected in studies of GJB2 mutation reported in Europe and Oceania, which suggested that multiple models could possibly be present in European and American populations, due to a complex interaction between GJB2 mutations and environmental factors. A further research on the prevalence of the 235delC mutation of GJB2 gene related to the NSHL susceptibility, as well as the regional disparity and ethnic specificity will be essentially need.
Conclusions
A systematic review was performed by means of a meta-analysis to evaluate the influence of the 235delC mutation on the NSHL susceptibility. The combined results of meta-analysis by pooling all relevant studies showed that the 235delC mutant increased the risk of NSHL. However, heterogeneity of genetic effect was observed due to the ethnic specificity and regional disparity. Therefore, stratified meta-analysis was conducted in different ethnic population group, and the results indicated that the 235delC mutation was prominently correlated with the risk of NHSL in the East Asian and South-east Asian populations, but non-significantly associated with the Oceania and European populations. Based on this study, a further research on the genetic mechanism of the NSHL susceptibility related to the 235delC mutation of GJB2 gene will be carried out in our future work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JY and YJL carried out the literature search and screening, performed the meta-analysis and drafted the manuscript. QJW participated in the design of the study and the statistical analysis. GQX and XC conceived of the design of the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Jun Yao, Email: joelyao@163.com.
Yajie Lu, Email: 13851543104@163.com.
Qinjun Wei, Email: wqj@njmu.edu.cn.
Xin Cao, Email: caoxin@njmu.edu.cn.
Guangqian Xing, Email: xing-gq@163.com.
Acknowledgements
This research was supported by the Grants from the National Natural Science Foundation of China (No. 31171217) and a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institution to XC, and the Grant from Jiangsu Health Administration of China (LJ201120) to GQX.
References
- Cohen MM, Gorlin RJ. In: Origins of Oxford monographs on medical genetics, No 28. Gorlin RJ, Toriello HV, Cohen MM, editor. Oxford University Press, New York; 1995. Hereditary hearing loss and its syndromes; pp. 9–21. [Google Scholar]
- Hilgert N, Smith RJH, Campa GV. Forty-six genes causing nonsyndromic hearing impairment: Which ones should be analyzed in DNA diagnostics. Mutat Res. 2009;681:189–196. doi: 10.1016/j.mrrev.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Park HJ, Ouyang XM. et al. Evidence of a founder effect for the 235delC mutation of GJB2 (connexin 26) in East Asians. Hum Genet. 2003;114:44–50. doi: 10.1007/s00439-003-1018-1. [DOI] [PubMed] [Google Scholar]
- Dahl HM, Tobin SE, Poulakis Z. et al. The contribution of GJB2 mutations to slight or mild hearing loss in Australian elementary school children. J Med Genet. 2006;43:850–855. doi: 10.1136/jmg.2006.042051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsebner R, Lucas T, Schoefer C. et al. Relevance of the A1555G mutation in the 12S rRNA gene for hearing impairment in Austria. BMC Med Genet. 2007;7:884–886. doi: 10.1097/MAO.0b013e3181461b26. [DOI] [PubMed] [Google Scholar]
- Li R, Greinwald JH, Yang L. et al. Molecular analysis of the mitochondrial 12S rRNA and tRNASer (UCN) genes in paediatric subjects with non-syndromic hearing loss. J Med Genet. 2004;41:615–620. doi: 10.1136/jmg.2004.020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabionet R, Gasparini P, Estivill X. Molecular genetics of hearing impairment due to mutations in gap junction genes encoding beta connexins. Hum Mutat. 2000;16:190–202. doi: 10.1002/1098-1004(200009)16:3<190::AID-HUMU2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Wells GA, Shea B, O’Connell D, The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. 3rd Symposium on Systematic Reviews, Oxford, UK; 2000. pp. 3–5. [ http://www.ohri.ca/programs/clinical_epidemiology/oxford.htm] [Google Scholar]
- Thakkinstian A, McElduff P, D’Este C. et al. A method for meta analysis of molecular association studies. Stat Med. 2005;24:1291–1306. doi: 10.1002/sim.2010. [DOI] [PubMed] [Google Scholar]
- Ohtsuka A, Yuge I, Kimura S. et al. GJB2 deafness gene shows a specificity spectrum of mutations in Japan, including a frequent founder mutation. Hum Genet. 2003;112:329–333. doi: 10.1007/s00439-002-0889-x. [DOI] [PubMed] [Google Scholar]
- Batissoco AC, Abreu-Silva RS, Braga MC. et al. Prevalence of GJB2 (Connexin-26) and GJB6 (Connexin-30) mutations in a cohort of 300 Brazilian hearing-impaired individuals: implications for diagnosis and genetic counseling. Ear Hearing. 2009;30:1–7. doi: 10.1097/AUD.0b013e31819144ad. [DOI] [PubMed] [Google Scholar]
- Wattanasirichaigoon D, Limwongse C, Jariengprasert C. et al. High prevalence of V37I genetic variant in the connexin-26 (GJB2) gene among non-syndromic hearing-impaired and control Thai individuals. Clin Genet. 2004;66:452–460. doi: 10.1111/j.1399-0004.2004.00325.x. [DOI] [PubMed] [Google Scholar]
- Padma G, Ramchander PV, Nandur UV. et al. GJB2 and GJB6 gene mutations found in Indian probands with congenital hearing impairment. J Genet. 2009;88:267–272. doi: 10.1007/s12041-009-0039-5. [DOI] [PubMed] [Google Scholar]
- Park HJ, Hahn S, Chun Y. et al. Connexin26 mutations associated with nonsyndromic hearing loss. Laryngoscope. 2000;110:1535–1538. doi: 10.1097/00005537-200009000-00023. [DOI] [PubMed] [Google Scholar]
- Shalin H, Walsh T, Sobe T. et al. Genetics of congenital deafness in the Palestinian population: multiple connexin 26 alleles with shared origins in the Middle East. Hum Genet. 2002;110:284–289. doi: 10.1007/s00439-001-0674-2. [DOI] [PubMed] [Google Scholar]
- Lee KY, Choi SY, Bae JW. et al. Molecular analysis of GJB2, GJB6 and SLC26A4 genes in Korean deafness patients. Int J Pediatr Otorhinolaryngol. 2008;72:1301–1309. doi: 10.1016/j.ijporl.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmenti LA, Martinez A, Telatar M. et al. Infant hearing loss and connexin texting in diverse population. Genet Med. 2008;10:517–524. doi: 10.1097/GIM.0b013e31817708fa. [DOI] [PubMed] [Google Scholar]
- Posukh O, Pallares-Ruiz N, Tadinova V. et al. First molecular screening of deafness in the Altai Republic population. BMC Med Genet. 2005;6:2–8. doi: 10.1186/1471-2350-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe S, Usami S, Shinkawa H. et al. Prevalent connexin 26 gene (GJB2) mutations in Japanese. J Med Genet. 2000;37:41–43. doi: 10.1136/jmg.37.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang HY, Fang P, Ward PA. et al. DNA sequence analysis of GJB2, encoding connexin 26: observations from a population of hearing impaired cases and variable carrier rates, complex genotypes, and ethnic stratification of alleles among controls. Am J Med Genet A. 2006;15:2401–2415. doi: 10.1002/ajmg.a.31525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekin M, Xia XJ, Erdenetungalag R. et al. GJB2 mutations in Mongolia: complex alleles, low frequency, and reduced fitness of the deaf. Ann Hum Genet. 2010;74:155–164. doi: 10.1111/j.1469-1809.2010.00564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo T, Ikeda K, Kure S. et al. Novel mutations in the connexin 26 gene (GJB2) responsible for childhood deafness in the Japanese population. Am J Med Genet. 2000;90:141–145. doi: 10.1002/(SICI)1096-8628(20000117)90:2<141::AID-AJMG10>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Cheng X, Li L, Brashears S. et al. Connexin26 variants and auditory neuropathy /dys-synchrony among children in schools for the deaf. Am J Med Genet A. 2005;139:13–18. doi: 10.1002/ajmg.a.30929. [DOI] [PubMed] [Google Scholar]
- Wang YC, Su MC, Su CC. et al. Mutations of Cx26 (GJB2) for prelingual deafness in Taiwan. Eur J Hum Genet. 2002;10:495–498. doi: 10.1038/sj.ejhg.5200838. [DOI] [PubMed] [Google Scholar]
- Chen D, Chen X, Cao K. et al. High prevalence of the connexin26 (GJB2) mutation in Chinese cochlear implant recipients. ORL J Otorhinolaryngol Relat Spec. 2009;71:212–215. doi: 10.1159/000229300. [DOI] [PubMed] [Google Scholar]
- Xiao ZA, Xie DH. GJB2 (Cx26) gene mutations in Chinese patients with congenital sensorineural deafness and a report of one novel mutation. Chin Med J. 2004;12:1797–1801. [PubMed] [Google Scholar]
- Wang SH, Hu ZM, Xiao Z. et al. GJB2 (connexin 26) gene mutation screen in patients with nonsydromic hearing loss in Human. J Cent South Univ (Med Sci) 2009;34:498–503. [PubMed] [Google Scholar]
- Hwa HL, Ko TM, Hsu CJ. et al. Mutation spectrum of the connexin 26 (GJB2) gene in Taiwanese patients with prelingual deafness. Genet Med. 2003;5:161–165. doi: 10.1097/01.GIM.0000066796.11916.94. [DOI] [PubMed] [Google Scholar]
- Shi GZ, Gong LX, Xu XH. GJB2 gene mutations in newborns with non-syndromic hearing impairment in Northern China. Hearing Res. 2004;197:19–23. doi: 10.1016/j.heares.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Dai P, Yu F, Han B. et al. Feature of nationwide distribution and frequency of a common gap junction beta-2 gene mutation in China. Chin J Otorhinolaryngol Head Neck Surg. 2007;42:804–808. [PubMed] [Google Scholar]
- Liu XZ, Xia XJ, Ke XM. et al. The prevalence of connexin 26 (GJB2) mutations in the Chinese population. Hum Genet. 2002;111:394–397. doi: 10.1007/s00439-002-0811-6. [DOI] [PubMed] [Google Scholar]
- Liu YH, Ke XM, Qi Y. et al. Connexin26 gene (GJB2): prevalence of mutations in the Chinese population. J Hum Genet. 2002;47:688–690. doi: 10.1007/s100380200106. [DOI] [PubMed] [Google Scholar]
- Utreta R, Angeli SI. Detection of the 35delG/GJB2 and del (GJB6-D13S1830) mutations in Venezuelan patients with autosomal recessive nonsyndromic hearing loss. Genet Test. 2007;11:347–352. doi: 10.1089/gte.2006.0526. [DOI] [PubMed] [Google Scholar]
- Damalon V, Lotersztein V, Béhèran A. et al. GJB2 and GJB6 genes: molecular study and identification of novel GJB2 mutations in the hearing-impaired Argentinean population. Audiol Neurotol. 2010;15:194–202. doi: 10.1159/000254487. [DOI] [PubMed] [Google Scholar]
- Pollak A, Ploski R, Murgia A. M34T and V37I Mutations in GJB2 associated hearing impairment: evidence for pathogenicity and reduced penetrance. Am J Med Genet A. 2007;143:2534–2543. doi: 10.1002/ajmg.a.31982. [DOI] [PubMed] [Google Scholar]
- Samanich J, Lowes C, Burk R. et al. Morrow Mutations in GJB2, GJB6, and mitochondrial DNA are rare in African, American and Caribbean Hispanic individuals with hearing impairment. Am J Med Genet A. 2007;143:830–838. doi: 10.1002/ajmg.a.31668. [DOI] [PubMed] [Google Scholar]
- Tóth T, Kupka S, Haack B. et al. Coincidence of mutations in different connexin genes in Hungarian patients. Int J Mol Med. 2007;20:315–321. [PubMed] [Google Scholar]
- Neocleous V, Aspris A, Shahpenterian V. et al. High Frequency of 35delG GJB2 mutation and absence of del(GJB6-D13S1830) in Greek Cypriot patients with nonsyndromic hearing loss. Genet Test. 2006;10:285–289. doi: 10.1089/gte.2006.10.285. [DOI] [PubMed] [Google Scholar]
- Snoeckx RL, Huygen PL, Feldmann D. et al. GJB2 mutations and degree of hearing loss: a multicenter study. Am J Hum Genet. 2005;77:945–957. doi: 10.1086/497996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GM, He F, Fu SQ. et al. GJB2 and mitochondrial DNA 1555A > G mutations in students with hearing loss in the Hubei Province of China. Int J Pediatr Otorhinolaryngol. 2011;75:1156–1159. doi: 10.1016/j.ijporl.2011.06.011. [DOI] [PubMed] [Google Scholar]
- Guo YF, Liu XW, Guan J. et al. GJB2, SLC26A4 and mitochondrial DNA A1555G mutations in prelingual deafness in Northern Chinese subjects. Acta Oto-Laryngologica. 2008;128:297–303. doi: 10.1080/00016480701767382. [DOI] [PubMed] [Google Scholar]
- Baysal E, Bayazit YA, Ceylaner S. et al. GJB2 and mitochondrial A1555G gene mutations in nonsyndromic profound hearing loss and carrier frequencies in healthy individuals. J Genet. 2008;87:53–57. doi: 10.1007/s12041-008-0007-5. [DOI] [PubMed] [Google Scholar]
- Bhalla S, Sharma R, Khandelwal G. et al. Low incidence of GJB2, GJB6 and mitochondrial DNA mutations in North Indian patients with nonsyndromic hearing impairment. Biochem Biophys Res Commun. 2009;385:445–448. doi: 10.1016/j.bbrc.2009.05.083. [DOI] [PubMed] [Google Scholar]