Abstract
Background
Klf5 plays an important role in maturation and maintenance of the mouse ocular surface. Here, we quantify WT and Klf5-conditional null (Klf5CN) corneal gene expression, identify Klf5-target genes and compare them with the previously identified Klf4-target genes to understand the molecular basis for non-redundant functions of Klf4 and Klf5 in the cornea.
Methodology/Principal Findings
Postnatal day-11 (PN11) and PN56 WT and Klf5CN corneal transcriptomes were quantified by microarrays to compare gene expression in maturing WT corneas, identify Klf5-target genes, and compare corneal Klf4- and Klf5-target genes. Whole-mount corneal immunofluorescent staining was employed to examine CD45+ cell influx and neovascularization. Effect of Klf5 on expression of desmosomal components was studied by immunofluorescent staining and transient co-transfection assays. Expression of 714 and 753 genes was increased, and 299 and 210 genes decreased in PN11 and PN56 Klf5CN corneas, respectively, with 366 concordant increases and 72 concordant decreases. PN56 Klf5CN corneas shared 241 increases and 98 decreases with those previously described in Klf4CN corneas. Xenobiotic metabolism related pathways were enriched among genes decreased in Klf5CN corneas. Expression of angiogenesis and immune response-related genes was elevated, consistent with neovascularization and CD45+ cell influx in Klf5CN corneas. Expression of 1574 genes was increased and 1915 genes decreased in WT PN56 compared with PN11 corneas. Expression of ECM-associated genes decreased, while that of solute carrier family members increased in WT PN56 compared with PN11 corneas. Dsg1a, Dsg1b and Dsp were down-regulated in Klf5CN corneas and their corresponding promoter activities were stimulated by Klf5 in transient co-transfection assays.
Conclusions/Significance
Differences between PN11 and PN56 corneal Klf5-target genes reveal dynamic changes in functions of Klf5 during corneal maturation. Klf5 contributes to corneal epithelial homeostasis by regulating the expression of desmosomal components. Klf4- and Klf5-target genes are largely distinct, consistent with their non-redundant roles in the mouse cornea.
Introduction
The transparent and refractive cornea plays a central role in vision. Abnormal development and/or maintenance of the cornea result in severe defects in vision [1], [2]. Molecular and cellular events involved in corneal development, maturation and maintenance have been studied in great detail [3]–[9]. Members of different transcription factor families including Krüppel-like factors (KLF) influence corneal morphogenesis [10]–[25]. More than 17 members of the KLF family have been identified in mammals [26], [27], many of which are expressed in the ocular surface in varying amounts [17], [28], [29]. Among them, structurally related Klf4 and Klf5 are two of the most highly expressed transcription factors in the mouse cornea [29], [30]. Our previous studies demonstrated that both Klf4 and Klf5 are essential for normal maturation and maintenance of the mouse ocular surface [22], [31].
Klf4 and Klf5 exert tissue-dependent and non-redundant influences on the mouse ocular surface in spite of possessing an identical DNA-binding domain. Conditional disruption of Klf4 in the developing mouse ocular surface resulted in numerous defects including corneal epithelial fragility, stromal edema, altered stromal collagen fibril organization, endothelial vacuolation and loss of mucin producing conjunctival goblet cells [21], [22], [32]. Similar conditional disruption of Klf5 also resulted in multiple defects including translucent cornea, abnormal eyelids with malformed meibomian glands and a conjunctiva devoid of goblet cells [31]. Microarray comparison of WT and Klf4CN corneal and conjunctival transcriptomes identified significant differences in Klf4-target genes in these adjacent tissues, suggesting tissue-dependent functions for Klf4 [21], [33].
Here, we test the hypothesis that the basis for non-redundant functions of structurally related Klf4 and Klf5 lies in their distinct target genes in the mouse cornea. As most of the Klf5CN ocular surface defects appeared in post-eyelid opening stages [31], we identified the corneal Klf5-target genes before eyelid opening at PN11 and in young adults at PN56. This study design also enabled us to examine the changes in gene expression accompanying WT corneal maturation between PN11 and PN56. We report that Klf5 regulates a wide array of genes associated with a diverse spectrum of functions such as cell adhesion, barrier function, maintenance of hydration, and xenobiotic metabolism. We also show that the corneal Klf5- and Klf4-target genes are largely distinct, consistent with their non-redundant roles in the mouse cornea. Furthermore, we identified significant differences in Klf5-target genes between PN11 and PN56, revealing dynamic changes in Klf5 functions in the maturing cornea.
Results
Microarray analysis and validation of results
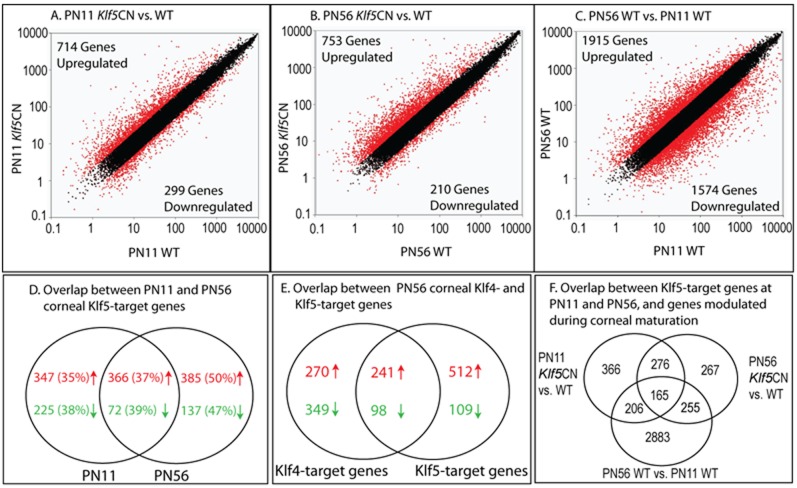
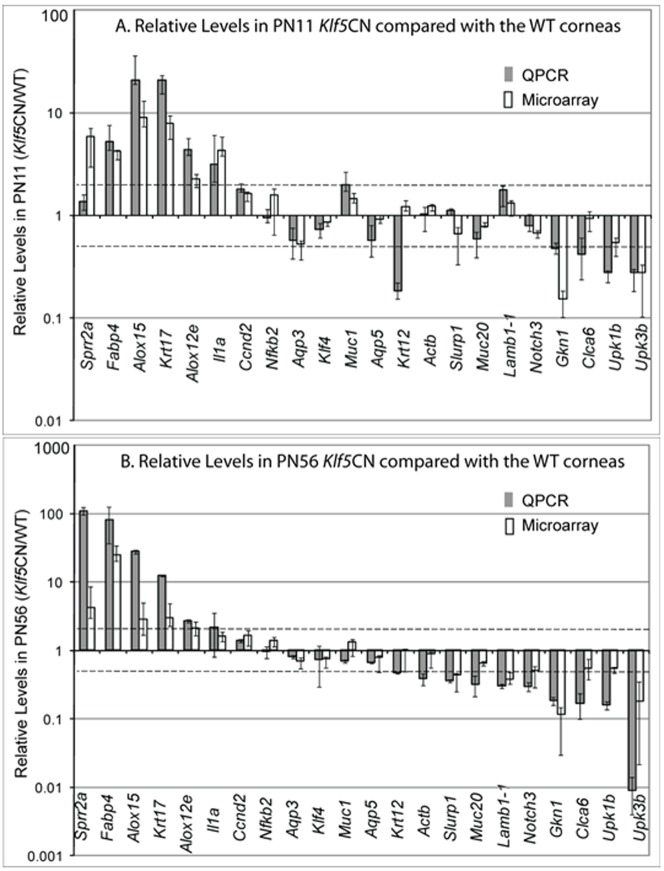
We compared the WT and Klf5CN corneal transcriptomes in immature PN11 corneas just before eyelid opening and in young adult PN56 corneas to identify the changes in gene expression associated with post-eyelid opening Klf5CN corneal phenotype [31]. We also compared the PN56 Klf5-target genes with those reported previously for Klf4 [21] to determine the extent of overlap between Klf4- and Klf5-target genes. Scatter plots of the WT vs. Klf5CN comparisons at PN11 (Fig. 1A) and PN56 (Fig. 1B), and the PN11 WT vs. PN56 WT comparison (Fig. 1C) show overall distribution of the panels measured by these microarrays. A large number of genes with distinct or overlapping expression were identified between (i) corneal Klf5-target genes at PN11 and PN56 (Fig. 1D), (ii) corneal Klf4- [21] and Klf5-target genes at PN56 (Fig. 1E), and (iii) the genes modulated during WT corneal maturation compared with the Klf5-target genes at PN11 and PN56 (Fig. 1F). Microarray results were validated by QPCR comparison of selected genes whose expression was increased, decreased or relatively unaffected in PN11 or PN56 Klf5CN corneas (Fig. 2). There was a general conformity between the microarray and QPCR results, indicating that the microarray results accurately represent the changes in Klf5CN corneal gene expression at these two stages (Fig. 2).
Figure 1. Comprehensive view of the changes in PN11 and PN56 Klf5CN corneal gene expression.
A–C. Scatter plots showing the significantly affected genes in (A) PN11 Klf5CN compared with the WT corneas, (B) PN56 Klf5CN compared with the WT corneas, and (C) PN56 WT compared with PN11 WT corneas. (D). Venn representation of numbers of unique characterized genes which are differentially expressed in Klf5CN corneas vs. WT corneas at PN11 or PN56. In parentheses is the percentage of genes showing valid >2-fold changes between PN11 WT and PN56 WT samples. (E). Venn representation of the overlap between Klf4- and Klf5-target genes in PN56 corneas. (F). Venn representation of the overlap between aggregate Klf5-target genes at PN11 and PN56, and genes modulated during WT corneal maturation.
Figure 2. Validation of microarray results by QPCR analysis of the expression of selected genes.
Note that the relative levels are plotted on a log scale.
Changes in Klf5-target genes during corneal maturation
Table 1 gives a complete breakdown of the changes in 21,815 unique characterized genes represented on the microarray. Corneal ablation of Klf5 resulted in decreased expression of 299 and 210 genes at PN11 and PN56 respectively (with 72 concordant decreases), and increased expression of 714 and 753 genes (with 366 concordant increases; Fig. 1D; Table 1). About 41% of the genes modulated in Klf5CN corneas were also modulated during WT corneal maturation between PN11 and PN56, compared with only 15% for those unaffected by disruption of Klf5 (Table 1), providing evidence for the important role of Klf5 in regulating post-eyelid opening corneal maturation. The top 50 most affected genes in PN11 and PN56 Klf5CN corneas are listed in Tables 2, 3, 4, and 5 (See Tables S2, S3, S4, S5 for the complete list).
Table 1. Distribution of modulated genes.
| Developmental changes PN56 vs. PN11 | PN11: Effects in Klf5CN vs. WT corneas | ||||
| Increased | Decreased | Unchanged | |||
| PN56: Effects in Klf5CN vs. WT corneas | Increased | Increased | 37 (22) | 0 (0) | 16 (6) |
| Decreased | 99 (42) | 1 (0) | 175 (48) | ||
| Unchanged | 230 (79) | 1 (0) | 194 (44) | ||
| Decreased | Increased | 0 (0) | 25 (17) | 47 (22) | |
| Decreased | 0 (0) | 3 (1) | 17 (5) | ||
| Unchanged | 1 (0) | 44 (29) | 73 (26) | ||
| Unchanged | Increased | 58 (18) | 31 (11) | 1360 (128) | |
| Decreased | 62 (7) | 55 (7) | 1503 (80) | ||
| Unchanged | 227 (16) | 139 (48) | 17,417 (304) | ||
Numbers of unique characterized genes which are differentially expressed in Klf5CN vs. WT corneas at PN11 or at PN56 (rows) are shown. Rows are broken down according to developmental changes, i.e., differences between PN56 WT and PN11 WT corneas, as designated in the shaded column. Data sets discordant between PN11 and PN56 (i.e., increased in PN11 Klf5CN but decreased in PN56 Klf5CN, or vice versa) are too small to be meaningful. Number of genes which also show valid >2-fold changes in PN56 Klf4CN cornea are shown in parentheses.
Table 2. Top 50 genes whose expression is most decreased in PN11 Klf5CN compared with the WT corneas.
| Gene symbol | Description | Mean log intensity in WT | Mean log intensity in Klf5CN | Fold Difference |
| Klf5 | Krüppel-like factor 5 | 9.10 | 3.87 | 0.03 |
| Aqp3 | aquaporin 3 | 11.11 | 7.54 | 0.08 |
| Folr1 | folate receptor 1 (adult) | 8.52 | 5.01 | 0.09 |
| Krt4 | keratin 4 | 7.33 | 4.12 | 0.11 |
| Serpinb3a | serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3A | 7.10 | 3.90 | 0.11 |
| Ces3 | carboxylesterase 3 | 8.71 | 5.60 | 0.12 |
| Gkn1 | gastrokine 1 | 6.21 | 3.35 | 0.14 |
| Snx31 | sorting nexin 31 | 8.59 | 5.84 | 0.15 |
| Adh6b | alcohol dehydrogenase 6B (class V) | 9.35 | 6.60 | 0.15 |
| Ppp1r3c | protein phosphatase 1, regulatory (inhibitor) subunit 3C | 7.14 | 4.50 | 0.16 |
| Txnip | thioredoxin interacting protein | 9.25 | 6.63 | 0.16 |
| Scd2 | stearoyl-Coenzyme A desaturase 2 | 8.88 | 6.27 | 0.16 |
| Cyp2b19 | cytochrome P450, family 2, subfamily b, polypeptide 19 | 6.29 | 3.68 | 0.16 |
| Cnpy1 | canopy 1 homolog (zebrafish) | 5.95 | 3.40 | 0.17 |
| Pax6 | paired box gene 6 | 9.08 | 6.60 | 0.18 |
| Acaa2 | acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) | 9.39 | 6.94 | 0.18 |
| Syt7 | synaptotagmin VII | 7.12 | 4.73 | 0.19 |
| Sfrp1 | secreted frizzled-related protein 1 | 9.60 | 7.21 | 0.19 |
| Paqr5 | progestin and adipoQ receptor family member V | 8.32 | 5.98 | 0.20 |
| Rapgef3 | Rap guanine nucleotide exchange factor (GEF) 3 | 5.97 | 3.65 | 0.20 |
| Fgf21 | fibroblast growth factor 21 | 6.96 | 4.66 | 0.20 |
| Acsm1 | acyl-CoA synthetase medium-chain family member 1 | 8.38 | 6.10 | 0.21 |
| Jakmip1 | janus kinase and microtubule interacting protein 1 | 7.64 | 5.40 | 0.21 |
| Es22 | esterase 22 | 5.56 | 3.33 | 0.21 |
| Gldc | glycine decarboxylase | 7.03 | 4.83 | 0.22 |
| Ripply3 | ripply3 homolog (zebrafish) | 6.99 | 4.83 | 0.22 |
| Bre | brain and reproductive organ-expressed protein | 9.45 | 7.28 | 0.22 |
| Sox15 | SRY-box containing gene 15 | 8.13 | 5.97 | 0.22 |
| Gpt | glutamic pyruvic transaminase, soluble | 8.29 | 6.18 | 0.23 |
| Tkt | transketolase | 9.66 | 7.55 | 0.23 |
| Stk35 | serine/threonine kinase 35 | 6.18 | 4.07 | 0.23 |
| Ngef | neuronal guanine nucleotide exchange factor | 6.49 | 4.39 | 0.23 |
| Akr1b7 | aldo-keto reductase family 1, member B7 | 9.73 | 7.63 | 0.23 |
| Slc16a12 | solute carrier family 16 (monocarboxylic acid transporters), member 12 | 8.60 | 6.55 | 0.24 |
| Pmm1 | phosphomannomutase 1 | 8.68 | 6.64 | 0.24 |
| Sdc1 | syndecan 1 | 8.01 | 5.99 | 0.25 |
| Mamdc2 | MAM domain containing 2 | 8.05 | 6.03 | 0.25 |
| Ckmt1 | creatine kinase, mitochondrial 1, ubiquitous | 9.50 | 7.49 | 0.25 |
| Tm9sf2 | transmembrane 9 superfamily member 2 | 8.91 | 6.92 | 0.25 |
| Bnc1 | basonuclin 1 | 10.00 | 8.03 | 0.26 |
| Cps1 | carbamoyl-phosphate synthetase 1 | 6.29 | 4.34 | 0.26 |
| Shmt1 | serine hydroxymethyltransferase 1 (soluble) | 5.82 | 3.88 | 0.26 |
| Angptl7 | angiopoietin-like 7 | 11.61 | 9.68 | 0.26 |
| Ascl2 | achaete-scute complex homolog 2 (Drosophila) | 5.35 | 3.44 | 0.27 |
| Padi4 | peptidyl arginine deiminase, type IV | 6.06 | 4.16 | 0.27 |
| Fjx1 | four jointed box 1 (Drosophila) | 8.25 | 6.35 | 0.27 |
| Slc22a18 | solute carrier family 22 (organic cation transporter), member 18 | 6.68 | 4.79 | 0.27 |
| Lemd1 | LEM domain containing 1 | 5.71 | 3.83 | 0.27 |
| Prss32 | protease, serine, 32 | 6.95 | 5.11 | 0.28 |
| Etv4 | ets variant gene 4 (E1A enhancer binding protein, E1AF) | 5.51 | 3.68 | 0.28 |
Genes whose expression is also decreased in the PN56 Klf5CN corneas are shown in bold.
Table 3. Top 50 genes whose expression is most increased in PN11 Klf5CN compared with the WT corneas.
| Gene symbol | Description | Mean log intensity in WT | Mean log intensity in Klf5CN | Fold Difference |
| Sftpd | surfactant associated protein D | 4.11 | 11.36 | 151.84 |
| Ccl8 | chemokine (C-C motif) ligand 8 | 5.31 | 12.04 | 105.55 |
| Expi | extracellular proteinase inhibitor | 6.16 | 12.55 | 83.66 |
| Lcn2 | lipocalin 2 | 6.72 | 12.06 | 40.53 |
| Retnla | resistin like alpha | 5.62 | 10.95 | 40.27 |
| Ppbp | pro-platelet basic protein | 3.44 | 8.69 | 37.93 |
| Ltf | lactotransferrin | 6.85 | 12.06 | 36.88 |
| Cd209e | CD209e antigen | 3.32 | 8.17 | 28.77 |
| Cxcl3 | chemokine (C-X-C motif) ligand 3 | 3.32 | 8.13 | 27.94 |
| Hp | haptoglobin | 3.63 | 8.36 | 26.49 |
| S100a9 | S100 calcium binding protein A9 (calgranulin B) | 5.64 | 10.36 | 26.43 |
| Ear11 | eosinophil-associated, ribonuclease A family, member 11 | 3.32 | 8.01 | 25.81 |
| Spink12 | serine peptidase inhibitor, Kazal type 11 | 3.45 | 7.99 | 23.23 |
| Cxcl17 | chemokine (C-X-C motif) ligand 17 | 5.25 | 9.51 | 19.20 |
| Cxcl2 | chemokine (C-X-C motif) ligand 2 | 3.35 | 7.37 | 16.20 |
| Cytip | cytohesin 1 interacting protein | 5.55 | 9.54 | 15.84 |
| Flt1 | FMS-like tyrosine kinase 1 | 4.11 | 8.09 | 15.80 |
| Cxcl5 | chemokine (C-X-C motif) ligand 5 | 6.62 | 10.56 | 15.41 |
| Krt16 | keratin 16 | 6.74 | 10.68 | 15.31 |
| Sprr1a | small proline-rich protein 1A | 5.86 | 9.68 | 14.12 |
| Nrsn1 | neurensin 1 | 3.56 | 7.24 | 12.83 |
| Ms4a6d | membrane-spanning 4-domains, subfamily A, member 6D | 5.46 | 9.07 | 12.27 |
| Socs3 | suppressor of cytokine signaling 3 | 6.08 | 9.66 | 11.96 |
| Slfn4 | schlafen 4 | 3.87 | 7.43 | 11.75 |
| Slco1a5 | solute carrier organic anion transporter family, member 1a5 | 3.54 | 7.07 | 11.58 |
| S100a8 | S100 calcium binding protein A8 (calgranulin A) | 7.25 | 10.73 | 11.13 |
| Ccl9 | chemokine (C-C motif) ligand 9 | 6.52 | 9.99 | 11.11 |
| Spink5 | serine peptidase inhibitor, Kazal type 5 | 5.38 | 8.81 | 10.83 |
| Cd14 | CD14 antigen | 6.11 | 9.53 | 10.71 |
| Cp | ceruloplasmin | 7.60 | 10.98 | 10.40 |
| Clec4d | C-type lectin domain family 4, member d | 3.72 | 7.08 | 10.26 |
| Stfa2l1 | stefin A2 like 1 | 5.88 | 9.22 | 10.13 |
| Ccl6 | chemokine (C-C motif) ligand 6 | 5.42 | 8.76 | 10.06 |
| Ifi203 | interferon activated gene 203 | 5.11 | 8.30 | 9.11 |
| Chi3l1 | chitinase 3-like 1 | 7.81 | 10.98 | 9.00 |
| Ccl5 | chemokine (C-C motif) ligand 5 | 3.32 | 6.47 | 8.89 |
| Gcnt2 | glucosaminyl (N-acetyl) transferase 2, I-branching enzyme | 5.21 | 8.32 | 8.65 |
| Sprr2a | small proline-rich protein 2A | 3.32 | 6.43 | 8.62 |
| Alox15 | arachidonate 15-lipoxygenase | 6.68 | 9.74 | 8.34 |
| Acpp | acid phosphatase, prostate | 4.14 | 7.19 | 8.30 |
| Ccl7 | chemokine (C-C motif) ligand 7 | 3.65 | 6.70 | 8.30 |
| Tm4sf1 | transmembrane 4 superfamily member 1 | 7.39 | 10.43 | 8.22 |
| Cxcl1 | chemokine (C-X-C motif) ligand 1 | 5.99 | 9.01 | 8.08 |
| Htra4 | HtrA serine peptidase 4 | 3.32 | 6.33 | 8.02 |
| Il1b | interleukin 1 beta | 4.95 | 7.95 | 8.01 |
| Nckap1l | NCK associated protein 1 like | 3.44 | 6.42 | 7.90 |
| Krt23 | keratin 23 | 5.27 | 8.24 | 7.84 |
| Plb1 | phospholipase B1 | 4.05 | 7.01 | 7.77 |
| Pmaip1 | phorbol-12-myristate-13-acetate-induced protein 1 | 4.71 | 7.67 | 7.74 |
| Slc26a4 | solute carrier family 26, member 4 | 3.39 | 6.33 | 7.71 |
Genes whose expression is also increased in the PN56 Klf5CN corneas are shown in bold.
Table 4. Top 50 genes whose expression is most decreased in PN56 Klf5CN compared with the WT corneas.
| Gene symbol | Description | Mean log intensity in WT | Mean log intensity in Klf5CN | Fold Difference |
| Klf5 | Kruppel-like factor 5 | 10.22 | 4.75 | 0.02 |
| Gldc | glycine decarboxylase | 7.88 | 4.73 | 0.11 |
| Ppp1r3c | protein phosphatase 1, regulatory (inhibitor) subunit 3C | 10.56 | 7.63 | 0.13 |
| Gkn1 | gastrokine 1 | 7.50 | 4.59 | 0.13 |
| Cyp24a1 | cytochrome P450, family 24, subfamily a, polypeptide 1 | 7.75 | 5.06 | 0.15 |
| Enpep | glutamyl aminopeptidase | 9.38 | 6.72 | 0.16 |
| Klk11 | kallikrein related-peptidase 11 | 6.71 | 4.11 | 0.16 |
| Folr1 | folate receptor 1 (adult) | 7.79 | 5.43 | 0.20 |
| Cd55 | CD55 antigen | 6.58 | 4.25 | 0.20 |
| Dsg1a | desmoglein 1 alpha | 11.33 | 9.06 | 0.21 |
| Trpm3 | transient receptor potential cation channel, subfamily M, member 3 | 7.20 | 4.98 | 0.21 |
| Il1f5 | interleukin 1 family, member 5 (delta) | 7.78 | 5.58 | 0.22 |
| Pygl | liver glycogen phosphorylase | 9.93 | 7.74 | 0.22 |
| Ldlr | low density lipoprotein receptor | 6.63 | 4.51 | 0.23 |
| Lect1 | leukocyte cell derived chemotaxin 1 | 7.83 | 5.72 | 0.23 |
| Car3 | carbonic anhydrase 3 | 6.11 | 4.01 | 0.23 |
| Ces3 | carboxylesterase 3 | 10.11 | 8.04 | 0.24 |
| Fam184b | family with sequence similarity 184, member B | 7.56 | 5.48 | 0.24 |
| Mab21l1 | mab-21-like 1 (C. elegans) | 8.51 | 6.45 | 0.24 |
| Paqr5 | progestin and adipoQ receptor family member V | 8.08 | 6.03 | 0.24 |
| Dlg2 | discs, large homolog 2 (Drosophila) | 7.79 | 5.75 | 0.24 |
| Sorbs2 | sorbin and SH3 domain containing 2 | 9.45 | 7.41 | 0.24 |
| Myo6 | myosin VI | 7.24 | 5.21 | 0.24 |
| Tkt | transketolase | 9.65 | 7.62 | 0.25 |
| Slc14a1 | solute carrier family 14 (urea transporter), member 1 | 9.34 | 7.32 | 0.25 |
| Col4a3 | collagen, type IV, alpha 3 | 8.91 | 6.89 | 0.25 |
| Aldh1a1 | aldehyde dehydrogenase family 1, subfamily A1 | 10.12 | 8.11 | 0.25 |
| Vgll3 | vestigial like 3 (Drosophila) | 7.81 | 5.89 | 0.27 |
| Abcc9 | ATP-binding cassette, sub-family C (CFTR/MRP), member 9 | 6.78 | 4.86 | 0.27 |
| Mmab | methylmalonic aciduria (cobalamin deficiency) type B homolog (human) | 5.80 | 3.90 | 0.27 |
| Tox | thymocyte selection-associated high mobility group box | 6.12 | 4.24 | 0.27 |
| Crygb | crystallin, gamma B | 8.29 | 6.43 | 0.27 |
| Ano4 | anoctamin 4 | 5.57 | 3.71 | 0.28 |
| Capsl | calcyphosine-like | 6.06 | 4.21 | 0.28 |
| Dkk1 | dickkopf homolog 1 (Xenopus laevis) | 7.11 | 5.26 | 0.28 |
| Usp19 | ubiquitin specific peptidase 19 | 6.12 | 4.27 | 0.28 |
| Ano9 | anoctamin 9 | 6.62 | 4.77 | 0.28 |
| Mt3 | metallothionein 3 | 5.17 | 3.32 | 0.28 |
| Glb1l2 | galactosidase, beta 1-like 2 | 5.75 | 3.91 | 0.28 |
| Kif26a | kinesin family member 26A | 6.71 | 4.87 | 0.28 |
| Prss32 | protease, serine, 32 | 8.46 | 6.67 | 0.29 |
| Cyp2b19 | cytochrome P450, family 2, subfamily b, polypeptide 19 | 5.36 | 3.59 | 0.29 |
| Slc4a11 | solute carrier family 4, sodium bicarbonate transporter-like, member 11 | 9.40 | 7.64 | 0.29 |
| Cryaa | crystallin, alpha A | 8.39 | 6.65 | 0.30 |
| Scrn1 | secernin 1 | 5.67 | 3.93 | 0.30 |
| Phlda2 | pleckstrin homology-like domain, family A, member 2 | 6.20 | 4.47 | 0.30 |
| Osbpl6 | oxysterol binding protein-like 6 | 6.50 | 4.77 | 0.30 |
| Prkcb | protein kinase C, beta | 8.48 | 6.77 | 0.31 |
| Fxyd4 | FXYD domain-containing ion transport regulator 4 | 7.29 | 5.59 | 0.31 |
| Ipw | imprinted gene in the Prader-Willi syndrome region | 6.07 | 4.38 | 0.31 |
Genes whose expression is also decreased in the PN11 Klf5CN corneas are shown in bold.
Table 5. Top 50 genes whose expression is most increased in PN56 Klf5CN compared with the WT corneas.
| Gene symbol | Description | Mean log intensity in WT | Mean log intensity in Klf5CN | Fold Difference |
| Ppbp | pro-platelet basic protein | 5.28 | 10.79 | 45.32 |
| Sprr2d | small proline-rich protein 2D | 4.73 | 10.14 | 42.29 |
| Cxcl3 | chemokine (C-X-C motif) ligand 3 | 4.82 | 10.13 | 39.83 |
| Clca2 | chloride channel calcium activated 2 | 3.32 | 8.40 | 33.76 |
| Fabp4 | fatty acid binding protein 4, adipocyte | 4.37 | 9.44 | 33.53 |
| Cd163 | CD163 antigen | 3.74 | 8.57 | 28.62 |
| Sprr2f | small proline-rich protein 2F | 6.65 | 11.49 | 28.60 |
| Chi3l3 | chitinase 3-like 3 | 3.67 | 8.38 | 26.19 |
| Gsdmc | gasdermin C | 5.18 | 9.70 | 22.88 |
| Ccl8 | chemokine (C-C motif) ligand 8 | 6.09 | 10.59 | 22.68 |
| S100a8 | S100 calcium binding protein A8 (calgranulin A) | 6.60 | 11.03 | 21.60 |
| Cxcl5 | chemokine (C-X-C motif) ligand 5 | 6.93 | 11.10 | 17.96 |
| Igj | immunoglobulin joining chain | 5.48 | 9.64 | 17.95 |
| Pip | prolactin induced protein | 4.55 | 8.69 | 17.72 |
| Ear11 | eosinophil-associated, ribonuclease A family, member 11 | 3.50 | 7.64 | 17.65 |
| Sprr2a | small proline-rich protein 2A | 7.55 | 11.69 | 17.65 |
| Srgn | serglycin | 7.01 | 11.15 | 17.63 |
| Saa3 | serum amyloid A 3 | 5.00 | 9.04 | 16.45 |
| Ms4a6d | membrane-spanning 4-domains, subfamily A, member 6D | 3.92 | 7.81 | 14.76 |
| Clca1 | chloride channel calcium activated 1 | 3.84 | 7.61 | 13.71 |
| Chi3l4 | chitinase 3-like 4 | 7.05 | 10.80 | 13.50 |
| Mrc1 | mannose receptor, C type 1 | 5.27 | 9.00 | 13.25 |
| Aqp4 | aquaporin 4 | 3.99 | 7.70 | 13.03 |
| Fcgr2b | Fc receptor, IgG, low affinity IIb | 4.66 | 8.29 | 12.43 |
| Ccl24 | chemokine (C-C motif) ligand 24 | 3.32 | 6.96 | 12.42 |
| Serpina3g | serine (or cysteine) peptidase inhibitor, clade A, member 3G | 4.63 | 8.21 | 11.96 |
| Nlrp10 | NLR family, pyrin domain containing 10 | 4.43 | 7.98 | 11.74 |
| Igl-V1 | immunoglobulin lambda chain, variable 1 | 3.32 | 6.80 | 11.15 |
| S100a9 | S100 calcium binding protein A9 (calgranulin B) | 7.47 | 10.92 | 10.97 |
| Ccr1 | chemokine (C-C motif) receptor 1 | 3.96 | 7.41 | 10.92 |
| Chi3l1 | chitinase 3-like 1 | 7.83 | 11.27 | 10.91 |
| Csf3r | colony stimulating factor 3 receptor (granulocyte) | 4.01 | 7.43 | 10.71 |
| Il1b | interleukin 1 beta | 5.71 | 9.11 | 10.56 |
| Clec4d | C-type lectin domain family 4, member d | 3.84 | 7.23 | 10.51 |
| Mfap4 | microfibrillar-associated protein 4 | 3.94 | 7.30 | 10.33 |
| C1qa | complement component 1, q subcomponent, alpha polypeptide | 6.22 | 9.58 | 10.25 |
| Mmp3 | matrix metallopeptidase 3 | 8.19 | 11.50 | 9.89 |
| H2-Ab1 | histocompatibility 2, class II antigen A, beta 1 | 5.92 | 9.20 | 9.72 |
| Igh-6 | immunoglobulin heavy chain 6 (heavy chain of IgM) | 5.26 | 8.51 | 9.49 |
| Clec7a | C-type lectin domain family 7, member a | 5.31 | 8.54 | 9.39 |
| Tyrobp | TYRO protein tyrosine kinase binding protein | 5.16 | 8.37 | 9.25 |
| Lcn2 | lipocalin 2 | 8.71 | 11.92 | 9.24 |
| Cxcl13 | chemokine (C-X-C motif) ligand 13 | 3.85 | 7.04 | 9.12 |
| Mmp13 | matrix metallopeptidase 13 | 4.73 | 7.92 | 9.09 |
| Cytip | cytohesin 1 interacting protein | 5.98 | 9.16 | 9.06 |
| Nrsn1 | neurensin 1 | 3.77 | 6.93 | 8.96 |
| Gatm | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 4.46 | 7.62 | 8.95 |
| S1pr3 | sphingosine-1-phosphate receptor 3 | 4.00 | 7.15 | 8.91 |
| Mcpt2 | mast cell protease 2 | 3.40 | 6.53 | 8.77 |
| Ccl9 | chemokine (C-C motif) ligand 9 | 5.93 | 9.06 | 8.74 |
Genes whose expression is also increased in the PN11 Klf5CN corneas are shown in bold.
Changes in gene expression during WT corneal maturation
Comparison of the WT corneal transcriptomes at PN11 and PN56 revealed that the expression of 1574 genes decreased and 1915 genes increased by more than 2-fold between PN11 and PN56 (Fig. 1C). The 50 most affected genes in the WT PN56 compared with PN11 corneas are listed in Tables 6–7 (See Tables S6 and S7 for the complete list). Transcripts encoding different collagens and other major extracellular matrix (ECM)-related proteins were significantly decreased between PN11 and PN56, suggesting that most of the ingredients for stromal ECM are produced before or around eyelid opening (Tables 7 and 8). Similarly, expression of Adam family proteinases and other MMPs that play significant roles in remodeling ECM [34] also was sharply decreased between PN11 and PN56 (Table 9), providing further evidence that most of the stromal ECM is in place by eyelid opening stage and little stromal remodeling occurs in the adult cornea [35].
Table 6. Top 50 genes whose expression is most increased during post-eyelid opening WT corneal maturation between PN11 and PN56.
| Gene symbol | Description | Mean log intensity in PN11 | Mean log intensity in PN56 | Fold Difference |
| Dsp | desmoplakin | 3.56 | 10.89 | 160.99 |
| Lce3a | late cornified envelope 3A | 3.32 | 10.19 | 116.53 |
| Scd2 | stearoyl-Coenzyme A desaturase 2 | 4.05 | 10.28 | 75.18 |
| Psca | prostate stem cell antigen | 6.24 | 11.98 | 53.45 |
| Clca4 | chloride channel calcium activated 4 | 3.44 | 8.65 | 37.16 |
| Dsc3 | desmocollin 3 | 4.79 | 9.89 | 34.31 |
| Il1f9 | interleukin 1 family, member 9 | 4.88 | 9.96 | 33.83 |
| Lce3c | late cornified envelope 3C | 3.32 | 8.12 | 27.78 |
| Oasl1 | oligoadenylate synthetase-like 1 | 5.47 | 10.26 | 27.52 |
| Vps35 | vacuolar protein sorting 35 | 3.95 | 8.67 | 26.32 |
| Piga | phosphatidylinositol glycan anchor biosynthesis, class A | 4.69 | 9.41 | 26.29 |
| Prkci | protein kinase C, iota | 4.15 | 8.79 | 25.04 |
| Expi | extracellular proteinase inhibitor | 6.16 | 10.76 | 24.20 |
| Ggta1 | glycoprotein galactosyltransferase alpha 1, 3 | 3.32 | 7.91 | 24.00 |
| Il1f6 | interleukin 1 family, member 6 | 3.91 | 8.45 | 23.34 |
| Hif1a | hypoxia inducible factor 1, alpha subunit | 3.80 | 8.24 | 21.65 |
| Met | met proto-oncogene | 3.75 | 8.15 | 21.23 |
| Mobkl1b | MOB1, Mps One Binder kinase activator-like 1B (yeast) | 4.60 | 9.00 | 21.15 |
| Sema4d | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D | 3.32 | 7.70 | 20.75 |
| Pdcd6ip | programmed cell death 6 interacting protein | 4.61 | 8.97 | 20.44 |
| Mxd1 | MAX dimerization protein 1 | 4.03 | 8.38 | 20.43 |
| Pdlim5 | PDZ and LIM domain 5 | 4.51 | 8.80 | 19.54 |
| Sprr2a | small proline-rich protein 2A | 3.32 | 7.55 | 18.73 |
| Lin7c | lin-7 homolog C (C. elegans) | 4.44 | 8.58 | 17.56 |
| Sftpd | surfactant associated protein D | 4.11 | 8.24 | 17.49 |
| Adam10 | a disintegrin and metallopeptidase domain 10 | 3.58 | 7.70 | 17.47 |
| Vcl | Vinculin | 3.32 | 7.44 | 17.42 |
| Trp63 | transformation related protein 63 | 3.71 | 7.82 | 17.28 |
| Tgfbr1 | transforming growth factor, beta receptor I | 3.32 | 7.38 | 16.67 |
| Slc1a1 | solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 | 4.61 | 8.54 | 15.26 |
| Spink5 | serine peptidase inhibitor, Kazal type 5 | 5.38 | 9.30 | 15.21 |
| Sdcbp2 | syndecan binding protein (syntenin) 2 | 4.54 | 8.46 | 15.07 |
| Gja1 | gap junction protein, alpha 1 | 6.03 | 9.91 | 14.68 |
| Obfc2a | oligonucleotide/oligosaccharide-binding fold containing 2A | 4.96 | 8.82 | 14.50 |
| Cxadr | coxsackie virus and adenovirus receptor | 3.75 | 7.61 | 14.50 |
| Mff | mitochondrial fission factor | 7.34 | 11.19 | 14.37 |
| Sprr1a | small proline-rich protein 1A | 5.86 | 9.69 | 14.26 |
| Clic5 | chloride intracellular channel 5 | 4.48 | 8.26 | 13.78 |
| Nr1d2 | nuclear receptor subfamily 1, group D, member 2 | 3.32 | 7.10 | 13.74 |
| Pax6 | paired box gene 6 | 4.54 | 8.30 | 13.60 |
| Gch1 | GTP cyclohydrolase 1 | 4.66 | 8.43 | 13.56 |
| Il1a | interleukin 1 alpha | 4.09 | 7.84 | 13.45 |
| Dnm1l | dynamin 1-like | 3.32 | 7.07 | 13.39 |
| Ide | insulin degrading enzyme | 4.54 | 8.28 | 13.39 |
| Slc5a1 | solute carrier family 5 (sodium/glucose cotransporter), member 1 | 7.41 | 11.14 | 13.28 |
| Dock1 | dedicator of cytokinesis 1 | 3.39 | 7.10 | 13.10 |
| Sgpl1 | sphingosine phosphate lyase 1 | 3.32 | 7.03 | 13.06 |
| Malat1 | metastasis associated lung adenocarcinoma transcript 1 (non-coding RNA) | 7.96 | 11.66 | 13.05 |
| Slc19a2 | solute carrier family 19 (thiamine transporter), member 2 | 4.74 | 8.44 | 13.00 |
| Atrx | alpha thalassemia/mental retardation syndrome X-linked homolog (human) | 3.32 | 7.00 | 12.79 |
Genes encoding cell junctional complex components or markers of epithelial stratification are shown in bold.
Table 7. Top 50 genes whose expression is most decreased during post-eyelid opening WT corneal maturation between PN11 and PN56.
| Gene symbol | Description | Mean log intensity in PN11 | Mean log intensity in PN56 | Fold Difference |
| Mfap4 | microfibrillar-associated protein 4 | 11.77 | 3.94 | 0.00 |
| Cpz | carboxypeptidase Z | 10.50 | 3.59 | 0.01 |
| Col11a1 | collagen, type XI, alpha 1 | 10.77 | 4.05 | 0.01 |
| Col9a1 | collagen, type IX, alpha 1 | 11.51 | 4.81 | 0.01 |
| Col5a2 | collagen, type V, alpha 2 | 12.06 | 5.95 | 0.01 |
| Meg3 | maternally expressed 3 | 11.22 | 5.14 | 0.02 |
| Col5a1 | collagen, type V, alpha 1 | 10.43 | 4.50 | 0.02 |
| Dlk1 | delta-like 1 homolog (Drosophila) | 10.15 | 4.23 | 0.02 |
| Agtr2 | angiotensin II receptor, type 2 | 10.19 | 4.28 | 0.02 |
| Fmod | fibromodulin | 11.95 | 6.08 | 0.02 |
| Rian | RNA imprinted and accumulated in nucleus | 9.47 | 3.61 | 0.02 |
| H19 | H19 fetal liver mRNA | 11.32 | 5.48 | 0.02 |
| Adamts2 | a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 2 | 9.48 | 3.68 | 0.02 |
| Matn4 | matrilin 4 | 11.63 | 5.95 | 0.02 |
| Col3a1 | collagen, type III, alpha 1 | 11.26 | 5.81 | 0.02 |
| Pik3cd | phosphatidylinositol 3-kinase catalytic delta polypeptide | 12.06 | 6.73 | 0.03 |
| Thbs2 | thrombospondin 2 | 11.78 | 6.49 | 0.03 |
| Twist2 | twist homolog 2 (Drosophila) | 9.03 | 3.82 | 0.03 |
| Clec3b | C-type lectin domain family 3, member b | 8.77 | 3.57 | 0.03 |
| Aplnr | apelin receptor | 9.21 | 4.04 | 0.03 |
| Cryga | crystallin, gamma A | 9.02 | 3.89 | 0.03 |
| Mfap2 | microfibrillar-associated protein 2 | 11.15 | 6.07 | 0.03 |
| Fbn2 | fibrillin 2 | 9.06 | 4.00 | 0.03 |
| Ctsk | cathepsin K | 11.39 | 6.39 | 0.03 |
| Dpep1 | dipeptidase 1 (renal) | 10.50 | 5.53 | 0.03 |
| Col6a2 | collagen, type VI, alpha 2 | 12.35 | 7.49 | 0.03 |
| Kazald1 | Kazal-type serine peptidase inhibitor domain 1 | 9.89 | 5.05 | 0.04 |
| Aspn | Aspirin | 9.25 | 4.41 | 0.04 |
| Mmp23 | matrix metallopeptidase 23 | 8.04 | 3.32 | 0.04 |
| Thbs4 | thrombospondin 4 | 11.04 | 6.35 | 0.04 |
| Camk4 | calcium/calmodulin-dependent protein kinase IV | 8.28 | 3.60 | 0.04 |
| C1qtnf2 | C1q and tumor necrosis factor related protein 2 | 9.40 | 4.80 | 0.04 |
| Mcpt4 | mast cell protease 4 | 8.04 | 3.44 | 0.04 |
| Itih5 | inter-alpha (globulin) inhibitor H5 | 8.06 | 3.47 | 0.04 |
| Pi16 | peptidase inhibitor 16 | 8.09 | 3.51 | 0.04 |
| Lox | lysyl oxidase | 11.18 | 6.60 | 0.04 |
| Mfap5 | microfibrillar associated protein 5 | 8.13 | 3.60 | 0.04 |
| Col6a1 | collagen, type VI, alpha 1 | 12.49 | 7.97 | 0.04 |
| Col24a1 | collagen, type XXIV, alpha 1 | 7.84 | 3.32 | 0.04 |
| Aebp1 | AE binding protein 1 | 9.73 | 5.22 | 0.04 |
| Pycr1 | pyrroline-5-carboxylate reductase 1 | 8.34 | 3.84 | 0.04 |
| Mmp2 | matrix metallopeptidase 2 | 11.37 | 6.87 | 0.04 |
| Pdgfrl | platelet-derived growth factor receptor-like | 11.01 | 6.54 | 0.05 |
| B3gnt9 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 | 8.44 | 3.98 | 0.05 |
| Tac1 | tachykinin 1 | 8.86 | 4.41 | 0.05 |
| Angptl7 | angiopoietin-like 7 | 11.61 | 7.17 | 0.05 |
| Creb3l1 | cAMP responsive element binding protein 3-like 1 | 8.40 | 3.96 | 0.05 |
| Cgref1 | cell growth regulator with EF hand domain 1 | 7.74 | 3.32 | 0.05 |
| Gpx7 | glutathione peroxidase 7 | 9.69 | 5.27 | 0.05 |
| Gpr124 | G protein-coupled receptor 124 | 8.53 | 4.13 | 0.05 |
| Igfbp4 | insulin-like growth factor binding protein 4 | 11.62 | 7.24 | 0.05 |
ECM-associated genes are shown in bold.
Table 8. Expression of ECM-related genes between PN11 and PN56.
| Gene symbol | Description | Mean log intensity for PN11 | Mean log intensity for PN56 | Fold Difference |
| A. Collagens | ||||
| Col11a1 | collagen, type XI, alpha 1 | 10.77 | 4.05 | 0.01 |
| Col9a1 | collagen, type IX, alpha 1 | 11.51 | 4.81 | 0.01 |
| Col5a2 | collagen, type V, alpha 2 | 12.06 | 5.95 | 0.01 |
| Col5a1 | collagen, type V, alpha 1 | 10.43 | 4.50 | 0.02 |
| Col3a1 | collagen, type III, alpha 1 | 11.26 | 5.81 | 0.02 |
| Col6a2 | collagen, type VI, alpha 2 | 12.35 | 7.49 | 0.03 |
| Col6a1 | collagen, type VI, alpha 1 | 12.49 | 7.97 | 0.04 |
| Col24a1 | collagen, type XXIV, alpha 1 | 7.84 | 3.32 | 0.04 |
| Col1a2 | collagen, type I, alpha 2 | 13.04 | 8.71 | 0.05 |
| Col14a1 | collagen, type XIV, alpha 1 | 9.76 | 5.75 | 0.06 |
| Col27a1 | collagen, type XXVII, alpha 1 | 8.78 | 5.03 | 0.07 |
| Col1a1 | collagen, type I, alpha 1 | 12.83 | 9.08 | 0.08 |
| Col2a1 | collagen, type II, alpha 1 | 7.02 | 3.32 | 0.08 |
| Col16a1 | collagen, type XVI, alpha 1 | 10.92 | 7.52 | 0.10 |
| Col11a2 | collagen, type XI, alpha 2 | 7.33 | 4.04 | 0.10 |
| Col6a3 | collagen, type VI, alpha 3 | 12.48 | 9.36 | 0.12 |
| Col5a3 | collagen, type V, alpha 3 | 7.56 | 4.47 | 0.12 |
| Col13a1 | collagen, type XIII, alpha 1 | 7.10 | 4.11 | 0.13 |
| Col15a1 | collagen, type XV, alpha 1 | 6.78 | 3.85 | 0.13 |
| Col20a1 | collagen, type XX, alpha 1 | 6.01 | 3.32 | 0.16 |
| Col4a2 | collagen, type IV, alpha 2 | 10.76 | 8.37 | 0.19 |
| Col8a1 | collagen, type VIII, alpha 1 | 10.90 | 8.51 | 0.19 |
| Col4a1 | collagen, type IV, alpha 1 | 11.14 | 8.99 | 0.23 |
| Col12a1 | collagen, type XII, alpha 1 | 12.32 | 10.45 | 0.28 |
| Col18a1 | collagen, type XVIII, alpha 1 | 7.48 | 5.64 | 0.28 |
| Col4a5 | collagen, type IV, alpha 5 | 11.08 | 9.81 | 0.42 |
| Col23a1 | collagen, type XXIII, alpha 1 | 6.44 | 5.22 | 0.43 |
| Col7a1 | collagen, type VII, alpha 1 | 9.90 | 8.74 | 0.45 |
| B. Other ECM-related proteins | ||||
| Lamb1-1 | laminin B1 subunit 1 | 8.68 | 7.56 | 0.46 |
| Lama1 | laminin, alpha 1 | 5.30 | 3.32 | 0.25 |
| Lama2 | laminin, alpha 2 | 8.86 | 5.40 | 0.09 |
| Lama4 | laminin, alpha 4 | 7.65 | 3.41 | 0.05 |
| Lamb2 | laminin, beta 2 | 8.33 | 6.32 | 0.25 |
| Lamc1 | laminin, gamma 1 | 9.09 | 7.01 | 0.24 |
| Lgals1 | lectin, galactose binding, soluble 1 | 11.61 | 8.00 | 0.08 |
| Lgals7 | lectin, galactose binding, soluble 7 | 8.13 | 6.13 | 0.25 |
| Lman1 | lectin, mannose-binding, 1 | 9.95 | 8.89 | 0.48 |
| Lum | Lumican | 12.81 | 10.12 | 0.16 |
| Sdc2 | syndecan 2 | 7.75 | 6.68 | 0.48 |
| Sdc3 | syndecan 3 | 8.86 | 6.07 | 0.15 |
| Sdc4 | syndecan 4 | 10.70 | 8.21 | 0.18 |
| Kera | keratocan | 12.81 | 9.27 | 0.09 |
| Vim | vimentin | 11.64 | 8.46 | 0.11 |
| Fndc1 | fibronectin type III domain containing 1 | 9.31 | 6.74 | 0.17 |
| Fndc3b | fibronectin type III domain containing 3B | 9.45 | 8.13 | 0.40 |
| Fndc4 | fibronectin type III domain containing 4 | 6.29 | 4.22 | 0.24 |
| Fndc5 | fibronectin type III domain containing 5 | 6.34 | 3.69 | 0.16 |
| Chad | chondroadherin | 7.63 | 4.18 | 0.09 |
| Chpf | chondroitin polymerizing factor | 8.01 | 5.53 | 0.18 |
| Chpf2 | chondroitin polymerizing factor 2 | 8.41 | 6.42 | 0.25 |
| Chsy3 | chondroitin sulfate synthase 3 | 8.48 | 5.71 | 0.15 |
| Chst1 | carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 | 5.81 | 4.76 | 0.48 |
| Chst14 | carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 14 | 7.87 | 6.29 | 0.33 |
| Chst5 | carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5 | 10.46 | 9.15 | 0.40 |
| Chst7 | carbohydrate (N-acetylglucosamino) sulfotransferase 7 | 6.16 | 3.62 | 0.17 |
| Chst11 | carbohydrate sulfotransferase 11 | 7.89 | 5.63 | 0.21 |
| Chst12 | carbohydrate sulfotransferase 12 | 7.30 | 6.03 | 0.41 |
| Chst2 | carbohydrate sulfotransferase 2 | 5.93 | 3.71 | 0.22 |
| Has2 | hyaluronan synthase 2 | 7.34 | 5.83 | 0.35 |
Table 9. Expression of metalloproteinases and other genes associated with matrix remodeling between PN11 and PN56.
| Gene symbol | Description | Mean log intensity for PN11 | Mean log intensity for PN56 | Fold Difference |
| A. Decreased Expression | ||||
| Adamts2 | a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 2 | 9.48 | 3.68 | 0.02 |
| Adam33 | a disintegrin and metallopeptidase domain 33 | 7.08 | 3.96 | 0.12 |
| Adamts12 | a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 12 | 6.55 | 3.52 | 0.12 |
| Adam22 | a disintegrin and metallopeptidase domain 22 | 6.96 | 4.46 | 0.18 |
| Adamts10 | a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 10 | 8.05 | 5.83 | 0.22 |
| Adamts9 | a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 9 | 6.34 | 4.76 | 0.33 |
| Adamts3 | a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 3 | 6.68 | 5.61 | 0.48 |
| Mmp23 | matrix metallopeptidase 23 | 8.04 | 3.32 | 0.04 |
| Mmp2 | matrix metallopeptidase 2 | 11.37 | 6.87 | 0.04 |
| Mmp14 | matrix metallopeptidase 14 (membrane-inserted) | 9.59 | 5.52 | 0.06 |
| Mmp16 | matrix metallopeptidase 16 | 5.85 | 3.40 | 0.18 |
| Mmp15 | matrix metallopeptidase 15 | 7.38 | 5.83 | 0.34 |
| Mxra8 | matrix-remodeling associated 8 | 10.66 | 8.48 | 0.22 |
| Mxra7 | matrix-remodeling associated 7 | 7.65 | 6.15 | 0.35 |
| B. Increased Expression | ||||
| Adam10 | a disintegrin and metallopeptidase domain 10 | 3.58 | 7.70 | 17.47 |
| Adam17 | a disintegrin and metallopeptidase domain 17 | 5.07 | 7.41 | 5.06 |
| Adamts1 | a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 1 | 6.69 | 8.25 | 2.96 |
Expression of several cell-junctional complexes and late markers of stratified squamous epithelial cells increased significantly between PN11 and PN56 (shown in bold in Table 6), when much of the corneal epithelial stratification occurs. In addition, expression of several members of the solute carrier family was significantly elevated between PN11 and PN56 (Table 10), reflecting the elevated need for solute transport in metabolically active adult corneas. While specific corneal functions of many of these solute carrier family members are not known, it is noteworthy that mutations in SLC4A11 and SLC16A12 are associated with congenital hereditary endothelial dystrophy (CHED) [36] and microcornea [37], respectively. Another important change that takes place between PN11 and PN56 corneas is the increased expression of several oxidative stress related genes including ceruloplasmin, an antioxidant enzyme upregulated in different neurodegenerative disorders including glaucoma [38], [39], Arachidonate lipoxygenase-12 and -15, which promote epithelial wound healing and host defense [40], carbonic anhydrase-2, -12, and -13, overexpressed in human glaucoma [41]–[43], and calcium binding proteins S100A8 and A9 (Table 6), suggesting an increase in oxidative stress in the adult compared with the PN11 corneas.
Table 10. WT corneal expression of solute carrier family members increased between PN11 and PN56.
| Gene symbol | Description | Mean log intensity in PN11 | Mean log intensity in PN56 | Fold Difference |
| Slc1a1 | solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 | 4.61 | 8.54 | 15.26 |
| Slc5a1 | solute carrier family 5 (sodium/glucose cotransporter), member 1 | 7.41 | 11.14 | 13.28 |
| Slc19a2 | solute carrier family 19 (thiamine transporter), member 2 | 4.74 | 8.44 | 13.00 |
| Slc6a14 | solute carrier family 6 (neurotransmitter transporter), member 14 | 7.93 | 11.48 | 11.70 |
| Slc28a3 | solute carrier family 28 (sodium-coupled nucleoside transporter), member 3 | 5.34 | 8.72 | 10.40 |
| Slc39a8 | solute carrier family 39 (metal ion transporter), member 8 | 4.95 | 8.22 | 9.63 |
| Slc4a11 | solute carrier family 4, sodium bicarbonate transporter-like, member 11 | 6.43 | 9.40 | 7.84 |
| Slc35a3 | solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 | 4.36 | 7.02 | 6.31 |
| Slc22a4 | solute carrier family 22 (organic cation transporter), member 4 | 3.65 | 6.08 | 5.41 |
| Slc12a2 | solute carrier family 12, member 2 | 7.94 | 10.26 | 4.99 |
| Slc39a6 | solute carrier family 39 (metal ion transporter), member 6 | 5.16 | 7.47 | 4.97 |
| Slc25a15 | solute carrier family 25 (mitochondrial carrier ornithine transporter), member 15 | 3.62 | 5.76 | 4.42 |
| Slc10a7 | solute carrier family 10 (sodium/bile acid cotransporter family), member 7 | 3.48 | 5.60 | 4.35 |
| Slc6a6 | solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | 3.32 | 5.35 | 4.09 |
| Slc14a1 | solute carrier family 14 (urea transporter), member 1 | 6.62 | 8.54 | 3.76 |
| Slc25a24 | solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 24 | 6.85 | 8.68 | 3.55 |
| Slc38a9 | solute carrier family 38, member 9 | 4.24 | 5.96 | 3.29 |
| Slc22a5 | solute carrier family 22 (organic cation transporter), member 5 | 5.20 | 6.64 | 2.71 |
| Slc25a30 | solute carrier family 25, member 30 | 4.72 | 6.15 | 2.69 |
| Slc11a2 | solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 | 6.50 | 7.92 | 2.68 |
| Slc37a1 | solute carrier family 37 (glycerol-3-phosphate transporter), member 1 | 6.91 | 8.32 | 2.66 |
| Slco4a1 | solute carrier organic anion transporter family, member 4a1 | 7.36 | 8.75 | 2.61 |
| Slc4a7 | solute carrier family 4, sodium bicarbonate cotransporter, member 7 | 4.65 | 5.99 | 2.54 |
| Slc44a4 | solute carrier family 44, member 4 | 7.12 | 8.44 | 2.50 |
| Slc38a10 | solute carrier family 38, member 10 | 5.19 | 6.44 | 2.38 |
| Slc5a8 | solute carrier family 5 (iodide transporter), member 8 | 8.21 | 9.45 | 2.36 |
| Slc20a2 | solute carrier family 20, member 2 | 6.11 | 7.35 | 2.36 |
| Slc44a2 | solute carrier family 44, member 2 | 6.08 | 7.31 | 2.34 |
| Slc25a36 | solute carrier family 25, member 36 | 6.30 | 7.51 | 2.32 |
| Slc38a2 | solute carrier family 38, member 2 | 8.38 | 9.57 | 2.27 |
| Slc4a4 | solute carrier family 4 (anion exchanger), member 4 | 6.91 | 8.09 | 2.27 |
| Slc16a12 | solute carrier family 16 (monocarboxylic acid transporters), member 12 | 8.60 | 9.76 | 2.24 |
| Slc25a13 | solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 | 7.49 | 8.62 | 2.20 |
| Slc35b4 | solute carrier family 35, member B4 | 5.66 | 6.79 | 2.19 |
| Slc6a20a | solute carrier family 6 (neurotransmitter transporter), member 20A | 5.95 | 7.02 | 2.10 |
Differences in PN56 corneal Klf4- and Klf5-target genes
Comparison of the PN56 corneal Klf5-target genes with those described previously for Klf4 [21] identified 260 common targets (204 increased and 56 decreased; Fig. 1E; Tables S8 and S9), with many more modulated exclusively in the Klf4CN (270 increased and 349 decreased) or Klf5CN (512 increased and 109 decreased) corneas. Most of the common increases in Klf4CN and Klf5CN corneas are associated with immune response, reflecting enhanced inflammatory conditions in those corneas. Regulation of largely distinct sets of target genes by Klf4 and Klf5 is consistent with their non-redundant functions in the mouse cornea [22], [31].
Elevated immune response in Klf5CN corneas
Canonical pathway analysis of the aggregate Klf5-target genes identified 26 significantly (p<0.001) enriched pathways, predominantly associated with immune function (Table S10). Expression of most of the genes associated with these pathways was increased, increasing the likelihood that these pathways represent indirect response to disruption of Klf5. To overcome this limitation, we repeated pathway analyses selectively for the genes with decreased expression in Klf5CN corneas. Several xenobiotic stress response-related pathways were predominantly enriched in genes with decreased expression upon disruption of Klf5, suggesting that Klf5 plays a role in xenobiotic stress response in the cornea (Table 11).
Table 11. Canonical pathways enriched among genes with reduced expression in Klf5CN corneas.
| PN11 Klf5CN | PN56 Klf5CN | Aggregate | ||||
| Canonical Pathway (Total number of genes in pathway) | −log(p) | Number of genes | −log(p) | Number of genes | −log(p) | Number of genes |
| LPS/IL-1 Mediated Inhibition of RXR Function (223) | 4.5 | 11 | 3.62 | 8 | 5.67 | 15 |
| Metabolism of Xenobiotics by Cytochrome P450 (197) | 4.82 | 8 | 2.16 | 4 | 5.38 | 10 |
| Glutathione Metabolism (92) | 4.29 | 6 | 2 | 4.35 | 7 | |
| Xenobiotic Metabolism Signaling (296) | 2.33 | 9 | 4.2 | 10 | 3.68 | 14 |
| Arachidonic Acid Metabolism (208) | 4.02 | 8 | 2 | 2.97 | 8 | |
| Fatty Acid Metabolism (185) | 3.31 | 7 | 3 | 3.75 | 9 | |
| Glycerolipid Metabolism (154) | 2.21 | 5 | 3 | 3.58 | 8 | |
| Mechanisms of Viral Exit from Host Cells (45) | 2.84 | 4 | 1 | 3.18 | 5 | |
| Butanoate Metabolism (129) | 3 | 3 | 3.08 | 6 | ||
| Eicosanoid Signaling (77) | 3.05 | 5 | 1 | 2.36 | 5 | |
| Valine, Leucine and Isoleucine Degradation (108) | 3 | 3 | 2.82 | 6 | ||
| PXR/RXR Activation (89) | 2 | 2.78 | 4 | 2.36 | 5 | |
| Cysteine Metabolism (90) | 2.6 | 4 | 2 | 2.04 | 4 | |
| Retinol Metabolism (61) | 1 | 2.56 | 3 | 2.56 | 4 | |
| Propanoate Metabolism (122) | 2 | 3 | 2.36 | 5 | ||
| NRF2-mediated Oxidative Stress Response (192) | 6 | 5 | 2.27 | 9 | ||
| Chondroitin Sulfate Biosynthesis (68) | 3 | 2.19 | 3 | 2.08 | 4 | |
| Aryl Hydrocarbon Receptor Signaling (159) | 4 | 2.18 | 5 | 7 | ||
| Urea Cycle and Metabolism of Amino Groups (78) | 2.14 | 3 | 3 | |||
| β-alanine Metabolism (93) | 1 | 2.11 | 3 | 4 | ||
Pathways were selected where at least one group was significantly enriched at p<0.01 (i.e., −log(p)>2). For clarity, −log(p) cells are left blank if −log(p)<2.
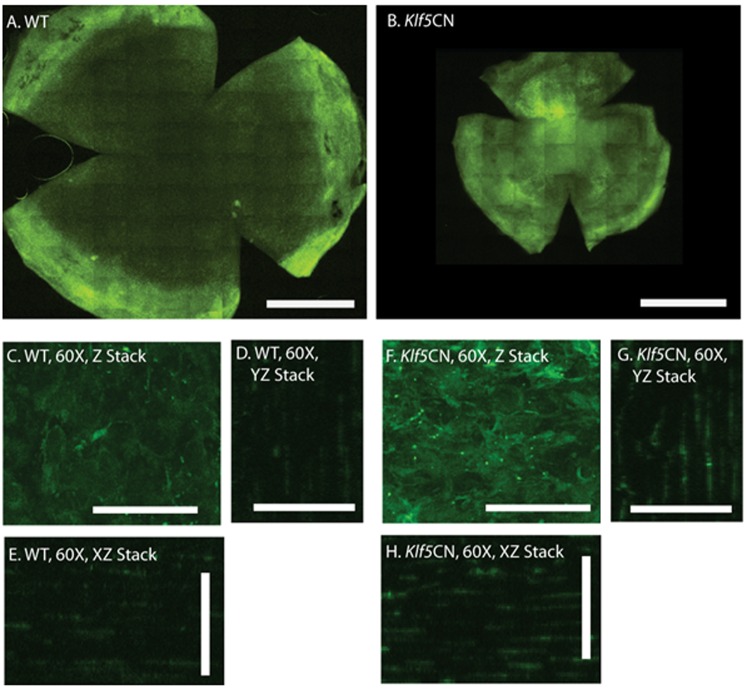
Expression of 107 of 368 genes (29.1%) containing “immun” or “inflamm” in the GO Biological Process notation was increased, while expression of only seven of these genes decreased in PN11 or PN56 Klf5CN corneas (Table S11). Increased expression of the immune response related genes, together with the previously reported hypercellularity of the Klf5CN corneal stroma [31] suggested a robust increase in immune-response in the Klf5CN corneas. Immunofluorescent staining of corneal flat mounts with anti-CD45 antibody demonstrated increased influx of CD45+ cells distributed throughout the Klf5CN compared with the WT corneal stroma sparsely populated with CD45+ cells (Fig. 3).
Figure 3. Enhanced influx of CD45+ cells into Klf5CN corneas.
Flat mounts of PN56 WT (A) and Klf5CN (B) corneas were stained with FITC-conjugated anti-CD45 antibody and examined by confocal microscopy. Representative stacked images of the central corneal stroma are shown at 60× magnification (Panels C–H). Compared with the WT stroma, enhanced influx of clusters of CD45+ cells is observed throughout the depth of Klf5CN stromas. Scale bars: 1 mm in Panels A and B; 40 µm in Panels C–H. Data are representative of 4 independent experiments. Klf5CN corneas are smaller than the WT, consistent with their small eye size reported previously.
Klf5CN corneal neovascularization (CNV)
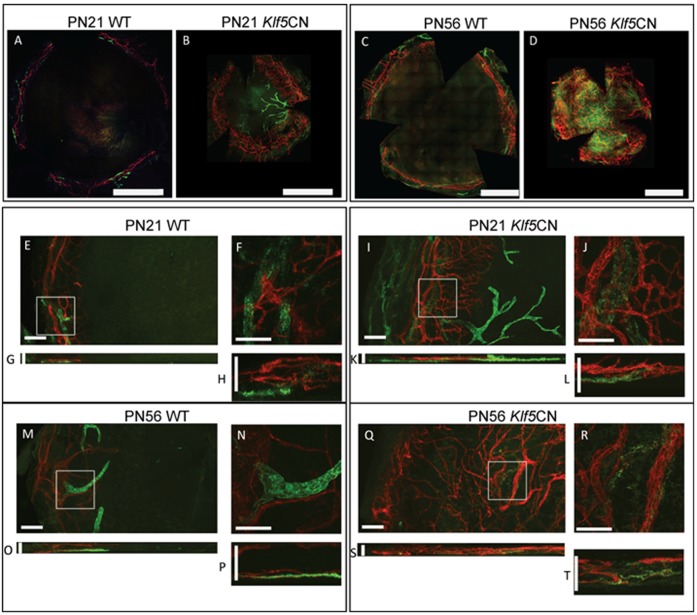
Whole-mount corneal immunofluorescent staining with anti-CD31 and anti-Lyve1 antibody revealed that the enhanced inflammatory environment in Klf5CN corneas is accompanied by extensive CNV (Fig. 4). Klf5CN CNV was apparent as early as PN21, when the Lyve1+ lymph vessels were much more pronounced and penetrated deeper into the central cornea, unlike the CD31+ blood vessels that remained in the peripheral region without reaching the central cornea (Fig. 4). By PN56, CNV was observed throughout the Klf5CN cornea, with blood vessels overtaking lymph vessels, which appeared to have regressed (Fig. 4). Examination of the XY-stack of confocal images revealed that CD31+ blood vessels are present in the anterior of the Klf5CN corneal stroma, unlike the Lyve1+ lymph vessels located in the posterior (Fig. 4).
Figure 4. Neovascularization in Klf5CN corneas.
Flat mounts of PN21 and PN56 WT and Klf5CN corneas were subjected to immunofluorescent staining with anti-CD31 (red) and anti-Lyve1 (green) antibody to detect blood vessels and lymph vessels, respectively. A–D, images of whole corneas generated by stitching together individual images from adjacent areas (Panels A–D; Scale bars = 1 mm). Vessels were blocked from entering the corneas at the limbus in WT but not Klf5CN corneas. Z-stack and XY-stacks of confocal images collected at 20× (panels E, G, I, K, M, O, Q and S; Scale bars = 100 µm) and 60× magnification (panels F, H, J, L, N, P, R and T; Scale bars = 50 µm) are shown. Data are representative of 4 independent experiments. Klf5CN corneas are smaller than the WT, consistent with their small eye size reported previously.
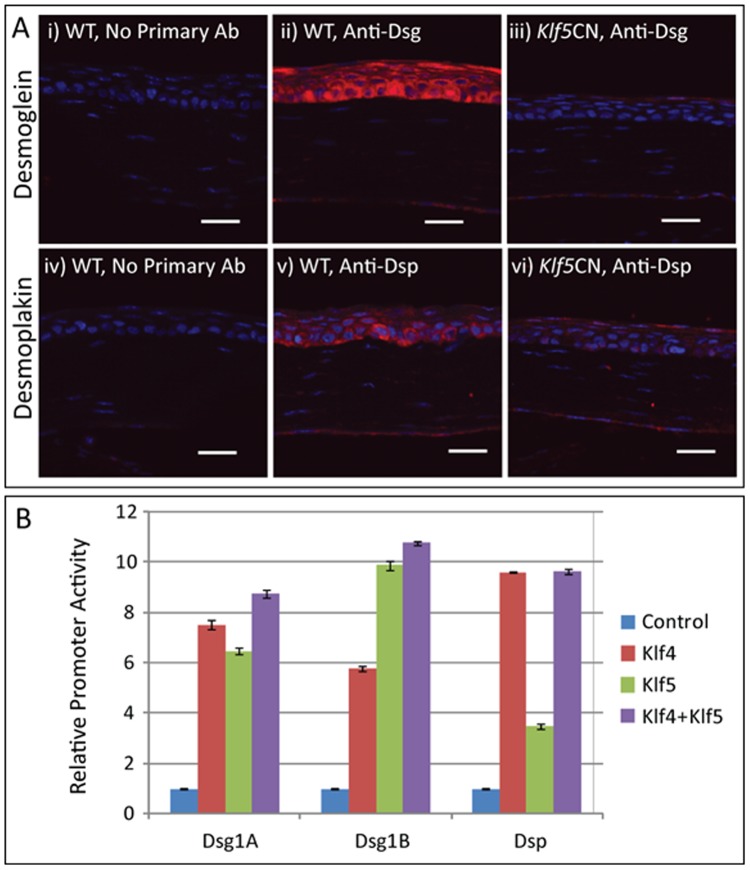
Klf5 regulates the expression of desmosomal components Dsg1a, Dsg1b and Dsp
Desmosomes are essential for corneal epithelial homeostasis [44]–[46]. Previously, we reported that Klf4 contributes to the formation and maintenance of corneal epithelial permeability barrier by regulating the expression of desmosomal components [47]. Microarray data presented here revealed that desmosomal components Dsg1a and Dsg1b are decreased in Klf5CN corneas (Table 4, Tables S2 and S4). Consistent with the microarray data, immunofluorescence revealed a sharp decrease in the epithelial expression of desmogleins, and a moderate decrease in desmoplakin in the Klf5CN corneas (Fig. 5A). Next, we tested the effect of Klf4 and/or Klf5 on Dsg1a, Dsg1b and Dsp promoter activities by transient co-transfection assays in NCTC cells using the previously described reporter vectors [47]. Dsg1a, Dsg1b, and Dsp promoter activities were stimulated by 7.5-, 6.5- and 8.7-fold, 5.8-, 9.9- and 10.8-fold, and 9.6-, 3.5- and 9.6-fold, respectively, when co-transfected with Klf4, Klf5, or both (Fig. 5B). Relative to Klf4, Klf5 had a comparable effect on Dsg1a, stronger stimulatory effect on Dsg1b and weaker stimulatory effect on Dsp promoter activities (Fig. 5B). Co-transfection with both Klf4 and Klf5 did not result in an additive or synergistic stimulation of these promoter activities (Fig. 5B), suggesting that Klf4 and Klf5 function through the same cis- elements within the Dsg1a, Dsg1b, and Dsp promoters [47]. Taken together with our previous report [47], these results demonstrate that one of the ways by which Klf4 and Klf5 contribute to corneal epithelial homeostasis is by regulating the expression of desmosomal components Dsg1a, Dsg1b and Dsp.
Figure 5. Klf5 contributes to corneal epithelial homeostasis by regulating the expression of desmogleins and desmoplakin.
(A) Immunofluorescence shows abundant expression of desmogleins (red, panel ii) and desmoplakin (red, panel v) in the WT but not in the Klf5CN corneal epithelium (panels iii and vi, respectively). Sections processed in a similar manner but without the primary antibody served as negative controls (panels i and iv). Scale bars = 25 µm. (B) Relative activities of 2 kb Dsg1a, Dsg1b and Dsp promoter fragments measured by transient co-transfection assays with or without Klf4, Klf5, or both in NCTC cells. Error bars indicate Standard Error of Mean (SEM). Data are representative of three independent experiments.
Influence of Klf4 and Klf5 on gene regulatory networks in the cornea
In order to determine the influence of Klf4 and Klf5 on gene regulatory networks in the cornea, we examined the expression of other transcription factors in PN11 and PN56 WT and Klf5CN corneas and compared them with the previous results from PN56 Klf4CN corneas [21]. Comparative analysis of the transcription factors decreased in more than one dataset (i.e., (a) PN56 Klf4CN vs. WT, (b) PN56 Klf5CN vs. WT, (c) PN11 Klf5CN vs. WT, and (d) PN56 WT vs. PN11 WT) identified Pax6, Bnc1, Cux1, Tox and Satb1 as common targets of Klf4 and Klf5 that were also modulated during corneal maturation (Table 12). Pathway analysis of the affected transcription factors revealed distinct networks predominantly associated with development and tissue homeostasis (Figures S2 and S3). The differences in these associated networks in spite of the five common transcription factor targets for Klf4 and Klf5 are consistent with their non-redundant functions in the mouse cornea. Among the common transcription factor targets of Klf4 and Klf5, while Pax6 and Bnc1 are known to regulate corneal epithelial homeostasis [5], [14], [48]–[52], corneal functions of Cux1, Tox and Satb1 are not yet known. Increased expression of Cux1, which suppresses collagen synthesis [53], is consistent with the decreased expression of collagens during WT corneal maturation. The transcription factors whose expression increased in PN56 Klf4CN and both PN11 and PN56 Klf5CN corneas (Atf3, Litaf, Runx1, Nfkbie and Fhl2; Table 12) are known to be upregulated during inflammation [54]–[57], raising a possibility that their increased expression reflects pro-inflammatory conditions in these corneas and may not be due to direct de-repression in the absence of Klf4 or Klf5.
Table 12. Transcription factors (TFs) differentially expressed in more than one comparison.
| Transcription factor | Klf4CN vs. WT (PN56) | Klf5CN vs. WT (PN56) | Klf5CN vs. WT (PN11) | PN56 WT vs. PN11 WT |
| Klf5 | 1.14 | 0.028 | 0.027 | 2.20 |
| Bnc1 | 0.36 | 0.45 | 0.27 | (1.58) |
| Pax6 | 0.38 | (0.64) | 0.16 | 12.95 |
| Cux1 | 0.37 | (0.60) | 0.47 | 3.56 |
| Satb1 | 0.24 | 0.43 | (0.56) | 0.46 |
| Tox | 0.23 | 0.29 | 0.45 | 0.47 |
| Sox4 | 8.19 | 4.16 | 2.10 | 0.32 |
| Irf8 | 2.38 | 4.68 | 2.47 | 0.46 |
| Mecom | 1.8 | 2.30 | (1.41) | 0.33 |
| Ar | 1.24 | 2.85 | (1.65) | 0.38 |
| Atf3 | 3.77 | 2.28 | 3.41 | (1.55) |
| Fhl2 | 2.80 | 2.98 | 3.47 | (0.77) |
| Litaf | 2.04 | 2.52 | 2.78 | (1.05) |
| Nfkbie | 2.22 | 3.92 | 3.59 | 0.41 |
| Runx1 | 2.70 | 2.05 | 2.60 | 2.49 |
In the first group, Bnc1, Pax6 and Cux1 have expression profiles most similar to Klf5, and therefore are strong candidates for direct control by Klf5 or for co-modulation with Klf5 by an upstream modulator. Members of the second group (Sox4, Irf8, Mecom and Ar) have expression profiles which are the inverse of Klf5. The third group shows increased expression in PN11 and PN56 Klf5CN, and PN56 Klf4CN corneas without a convincing change during WT corneal maturation. When the differences were less than 2-fold, those values are shown in parenthesis.
In order to identify if a regulatory relationship exists between different Klfs, we examined the effect of disruption of Klf5 on expression of other Klfs in the cornea. While transcripts for Klfs 1, 14 and 15 were essentially absent in the cornea, those for Klfs 2, 4, 6, 7, 10, 11, 12, 16 and 17 were present in all samples but showed no robust changes. During WT corneal maturation, the expression of Klfs 3, 5 and 6 was increased, while that of Klfs 2, 12 and 13 was decreased in PN56 compared with the PN11 corneas. Klf4 and Klf5 remained unaffected in Klf5CN and Klf4CN corneas, respectively, suggesting a lack of regulatory relationship between these two Klfs highly expressed in the cornea. The closely related Klf9 and Klf13 [27] were both increased in both Klf4CN and Klf5CN corneas, suggesting that Klf9 and Klf13 show similar compensatory changes whether Klf5 or Klf4 is ablated. Whether this represents a true regulatory relationship among Klfs 4, 5, 9 and 13 in the mouse cornea remains to be established.
Discussion
Previously, we demonstrated that disruption of Klf5 resulted in defective maturation of the mouse cornea in post-eyelid opening stages [31]. Here, we have employed microarray analysis to obtain a comprehensive view of the changes in corneal gene expression upon deletion of Klf5 in immature corneas around eyelid opening (PN11) and in adult (PN56) corneas. Our findings reveal the molecular basis of the wide ranging influence of Klf5 on corneal homeostasis and identify candidate target genes of Klf5 in the mouse cornea. In addition, the design of our study allowed us to identify the changes in gene expression between PN11 and PN56 WT corneas.
Earlier reports of Klf5-target gene profiling used chromatin immunoprecipitation followed by microarray (ChIP-Array) in embryonic stem cells [58], or microarray analysis following selective disruption of Klf5 in the mouse lung [59] or bladder urothelium [60]. Our results are largely consistent with those findings and identify additional Klf5-target genes such as Pax6 and Dsg1a, which play critical roles in the cornea. Differences in Klf5-target genes in the cornea (this study), ESCs [58], lung [59] and bladder urothelium [60] provide evidence for tissue-dependent functions of Klf5. For example, disruption of Klf5 resulted in increased expression of surfactant protein-D (Sftpd) in corneas (Table 2), in contrast to decreased expression in the lung [59], highlighting tissue-dependent functions of Klf5.
A striking feature of our results is the large number of genes whose expression is influenced by the absence of Klf5 in the mouse cornea. By comparison with similar studies of transcription factors such as FoxP2 [61], Sox2 [62], Myb [63] and Bcl11b [64], we predict that Klf5 is likely to directly regulate only a fraction of those genes whose expression is modulated in the Klf5CN corneas. The remaining genes are expected to be indirect targets of Klf5, through other transcription factors such as Pax6 [48]–[50], [65], whose expression is reduced in Klf5CN corneas (Table 12). Alternatively, they may represent physiological responses to the phenotype brought about by disruption of Klf5. For example, a large number of genes upregulated in Klf5CN corneas are immune response related, consistent with the massive infiltration of CD45+ cells (Fig. 3), and may not be directly regulated by Klf5. Though it is likely that the Klf5CN corneal neovascularization and influx of CD45+ cells indirectly contributed to increased expression of many of the immune response related genes, a similar effect may not be extended to a majority of the negatively regulated genes, which are more likely to represent true targets for Klf5.
Klf4 and Klf5 are both abundantly expressed in the mouse cornea [29], where they play critical, non-redundant roles [22], [31]. In order to understand the molecular basis for their corneal functions, it is necessary to identify and distinguish their target genes. Comparison of the Klf5CN corneal gene expression profile (this study) with that of Klf4CN corneas [21] revealed that roughly 2/3 of the corneal Klf4- and Klf5-target genes are Klf4- or Klf5-specific, with the rest being common targets. Canonical pathway analysis of genes exclusively modulated by Klf4 yielded “human embryonic stem cell pluripotency” as the most significantly enriched (p<10−5) pathway, in agreement with the importance of KLF4 in inducing pluripotency [66]. In contrast, “hepatic fibrosis/hepatic stellate cell activation” was the most significantly enriched (p<10−12) pathway among the genes exclusively modulated by Klf5 (Table S10), indicative of a general fibrotic response such as that observed in cultured human keratocytes exposed to TGF-β [67]. Molecular basis for the ability of Klf4 and Klf5 to regulate distinct sets of target genes in spite of possessing identical DNA-binding domain remains to be understood. Genome-wide identification of the nucleotide sequence of Klf4- and Klf5-bound cis-elements by chromatin immunoprecipitation followed by large scale sequencing (ChIP-Seq) is necessary to better understand target site selection by Klf4 and Klf5.
Being environmentally exposed, the cornea is frequently exposed to xenobiotic stress. Also, when the eyelids are closed during sleep, the avascular cornea is subjected to almost 75% drop in oxygen partial pressure [68], [69]. Thus, hypoxic and xenobiotic response pathways are expected to play an active role in corneal homeostasis. The important role of inhibitory PAS domain protein (IPAS) - a hypoxia repressor protein- in maintaining corneal avascularity [70], [71] further supports this contention. Moreover, corneal crystallin genes are induced by hypoxia or xenobiotics, further implicating hypoxic and xenobiotic stress in corneal gene expression [16], [72], [73]. Our data demonstrated significant enrichment of xenobiotic metabolism-related pathways among genes whose expression is decreased in Klf5CN corneas (Table 11). Thus, we suggest that Klf5 serves an important role in detoxification of the environmentally exposed avascular cornea, by supporting the expression of xenobiotic metabolism related genes.
Important changes take place in the mouse cornea as it matures following eyelid opening around PN12 [8], [74]. This study identified the changes in corneal gene expression between PN11 and PN56, revealing the molecular events underlying post-eyelid opening corneal maturation. Specifically, we demonstrate that during WT corneal maturation between PN11 and PN56, transcripts encoding (i) ECM components are sharply decreased, (ii) epithelial barrier-related proteins are sharply increased, and (iii) members of the solute carrier family proteins are elevated, consistent with (a) the active formation of stromal ECM around eyelid opening, with little remodeling taking place in adult corneal stroma, (b) rapid stratification of squamous epithelium in post-eyelid opening stages and (c) elevated demand for solute transport in the metabolically active adult cornea, respectively.
We compared the present data with a previous analysis of changes in corneal gene expression associated with post-eyelid opening maturation [75], which used Affymetrix MG74Av2 chips targeting a subset (8,666) of the 21,815 unique characterized genes examined here. Applying the present selection rules to their data [75] yielded 442 genes differentially expressed between their immature (PN10) and adult (PN49 to PN56) groups. Though there were differences between the two datasets (which could be attributed to several factors including differences in the mouse strains used, the nature of microarray chips used, and the parameters employed in filtering and analyzing the microarray data), our study confirmed 202 (45.7%) of these genes as differentially expressed between PN11 and PN56 corneas, with 36 concordant increases and 149 concordant decreases (7.0 and 5.1 times the number expected by contingency analysis, respectively, with a χ2 test yielding p = 6.8×10−161). Thus, the current study confirmed and expanded our knowledge of the changes in gene expression associated with post-eyelid opening corneal maturation [29], [75].
In summary, this report identifies dynamic changes in gene expression accompanying post-eyelid opening corneal maturation, and the role of Klf5 in this process. Our results show that Klf5 contributes to maturation and maintenance of cornea by regulating the expression of subsets of genes involved in specific functions such as cell-cell adhesion, epithelial barrier formation, maintenance of proper level of hydration and xenobiotic metabolism. The changes in Klf5CN corneal gene expression are consistent with the elevated immune response and CNV. These results also revealed significant differences between Klf4- and Klf5-target genes, consistent with their non-redundant roles in the mouse cornea. Taken together with our previous report [22], the present studies establish Klf5 as another important node in the genetic network of transcription factors required for corneal maturation and maintenance.
Materials and Methods
Ethics Statement
Mice used in these studies were maintained in accordance with the guidelines set forth by the Institutional Animal Care and Use Committee (IACUC) of the University of Pittsburgh, and the ARVO statement on the use of animals in ophthalmic and vision research. All procedures performed on mice reported in this study were approved by the University of Pittsburgh IACUC.
Conditional disruption of Klf5
Klf5CN mice were generated on a mixed background by mating Klf5loxP/loxP, Le-Cre/- mice with Klf5loxP/loxP mice as described before [31]. This mating scheme generated equal numbers of Klf5loxP/loxP, Le-Cre/- (Klf5CN) and Klf5loxP/loxP (control) offspring. Genomic DNA isolated from tail clippings was assayed for the presence of the Klf5-LoxP and Le-Cre transgenes by PCR using specific primers.
Isolation of total RNA, quality control, labeling and microarray analysis
WT and Klf5CN littermates (4 each at PN56 and 3 each at PN11) were used for comparison of corneal gene expression by microarrays. All corneas used in these studies were microdissected from freshly harvested eyeballs under a surgical microscope. Corneas were dissected using a pair of fine scissors (RS-5611 Vannas Curved Spring Scissors; Roboz Surgical Company, Germany) around the limbus, ensuring that they are free of contamination from iris, ciliary body and/or trabecular meshwork. Two dissected corneas from each mouse were pooled for isolation of total RNA using the RNeasy Mini kit (Qiagen, Germantown, MD). The quality and integrity of the isolated total RNA was confirmed using an Agilent Bioanalyzer (Figure S1); 1.0 µg sample RNAs were subsequently amplified and labeled using a 3′ IVT Express Kit (Affymetrix Inc., Santa Clara, CA) and hybridized to Affymetrix MG 430 2 chips [33]. Utilization of the same amount of total RNA (1.0 µg) from WT and Klf5CN corneas for labeling and hybridization ensured that the smaller size of Klf5CN corneas did not skew the microarray results.
The raw data obtained from microarray analysis were processed using Affymetrix GeneChip Operating Software (GCOS v 1.4) using software defaults, to assess the presence or absence of each transcript target sequence, its expression level, and then to make all relevant pair-wise statistical comparisons among samples. Expression levels were scaled to a target value of 150 using the software default (2% trimmed mean). Prior to scaling, mean microarray expression levels were 449±38 (mean ± SD, n = 13 with one outlier of 255), and target sequences were detected (called Present) for 57.8%±2.5% (mean ± SD, n = 14) of panels on the chip. After scaling, three redundant panels for the housekeeping gene GAPDH reported coefficients of variation of 12.3%, 10.9% and 13.1% respectively (n = 13 for each panel since each had one outlier value).
Processed data were sorted and inspected in an Excel spreadsheet (Microsoft, Redmond, WA) with BRB-ArrayTools (www.linus.nci.nih.gov/BRB-ArrayTools.html). Genes were considered differentially expressed between any two groups if they satisfied the following four criteria: (1) the average value for the high-expression group was >2 fold greater than the average value for the low expression group; (2) the high-expression group contained at least 3 detectable transcripts (called Present); (3) the differences between groups were significant at p≤0.057 in a two-tailed Mann-Whitney rank sum test. As this requirement could not be met in the PN11 WT vs. PN11 Klf5CN comparison, we required the minimum p value possible (0.1) instead; (4) for groups of m and n members, where m≥n, of the (m×n) pair-wise comparisons made by the GCOS software, at least ((m - 1)×n) show valid differences. We re-analyzed the previously published Klf4CN [21] data using current filters to compare PN56 WT corneas with Klf4CN corneas, at a cutoff of p = 0.063 in the Mann-Whitney test. MIAME-compliant microarray data were submitted to NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/; Accession Number GSE36229).
Validation of microarray results by real time QPCR
Applied Biosystems (ABI: Foster City, CA) was the source of the reagents, equipment and software for quantitative real time RT-PCR assays (QPCR). Total RNA isolated from pooled corneas of 10 WT or Klf5CN mice was quantified and the concentration adjusted with RNase-free water to 100 ng/µl. cDNA was generated using an ABI High Capacity cDNA Archive Kit and real-time QPCR assays were performed in a ABI StepOnePlus thermocycler using GAPDH as endogenous control for SYBR green assays and Pcx (pyruvate carboxylase) as endogenous control for FAM assays; the results were analyzed using ABI StepOnePlus software. Nucleotide sequence of different primers used is given in Table S1.
Immunofluorescent staining of corneal whole mounts
Corneas were dissected, flattened by three radial incisions, washed 3 times for 15 minutes each in PBS with 4% FBS and blocked in FC block (BD Pharmingen, San Jose, CA) for 20 minutes prior to incubation with FITC-conjugated anti-CD45 antibody, or FITC-conjugated anti-Lyve1 antibody and PE-conjugated anti-CD31 antibody (BD Pharmingen) overnight at 4°C. Corneas were then washed three times each for 30 minutes in PBS/4% FBS, fixed in 1% Paraformaldehyde for 2 hours at 4°C, rinsed 3 times again for 30 minutes each in PBS/4% FBS and mounted in Aqua Poly/Mount (Polysciences Inc, Warrington, PA). Images were acquired on a Nikon Ti-E and/or Olympus Fluoview 1000 confocal system with an Olympus IX81 microscope. Stacks were imaged at 20× (Numerical Aperture 0.85) and 60× (Numerical Aperture 1.42) through the cornea. Individual images were stitched together using Metamorph (Molecular Devices, Sunnyvale, CA) and Photoshop (Adobe Systems Inc., San Jose, CA) programs.
Immunofluorescence
Paraformaldehyde fixed, paraffin-embedded sections were deparaffinized with xylene, blocked with 10% goat serum in PBS for 1 h at room temperature (RT) in a humidified chamber, washed twice with PBS for 5 minutes each, incubated overnight with a 1∶50 dilution of anti-Dsg (recognizes both Dsg1a and Dsg1b) or anti-Dsp primary antibody raised in rabbit (Santa Cruz Biotechnology, Santa Cruz, CA) at 4°C, washed thrice with PBS for 5 minutes each, incubated with secondary antibody (AlexaFluor 546-coupled goat anti-rabbit IgG antibody; Molecular Probes, Carlsbad, CA) at a 1∶300 dilution for 1 h at RT, washed twice with PBS for 5 minutes each, incubated with DAPI for 10 minutes, mounted with Aqua Polymount (Polysciences, Inc), and observed with a fluorescence microscope. All images presented within each composite figure were acquired under identical settings and processed in a similar manner using Adobe Photoshop and Illustrator (Adobe, Mountain View, CA).
Reporter Vectors, Cell Culture, and Transient Transfection Assays
Different reporter vectors where approximately 2 kb Dsg1a, Dsg1b and Dsp promoter fragments drive the expression of luciferase reporter gene were described earlier [47]. Full-length Klf4 in pCI-Klf4 and full length Klf5 in pCMV-Sport-Klf5 was transiently expressed using the CMV promoter. Human skin keratinocyte NCTC cells were grown in Dulbecco's modified Eagle's medium(DMEM) supplemented with 10% fetal bovine serum, penicillin, and streptomycin at 37°C in a humidified chamber containing 5% CO2 in air. NCTC cells in 12-well plates were co-transfected with 0.5 µg pDsg1a-Luc, pDsg1b-Luc or pDsp-Luc, along with 20 ng pRL-SV40 (for normalization of transfection efficiency) and 0.5 µg pCI (control), or 0.25 µg each of pCI and pCI-Klf4, or pCI and pCMV-Sport6-Klf5, or pCI-Klf4 and pCMV-Sport6-Klf5 using 2.5 µL transfection reagent (Expressfect; Denville Scientific). After 2 days, cells were washed with PBS and lysed with 200 µL passive lysis buffer (Promega). 50 µL of lysate was analyzed using a dual-luciferase assay kit (Promega) and a microplate luminometer (Synergy-II; Biotek Instruments, Winooski, VT). The measurement was integrated over 10 seconds, with a delay of 2 seconds. Firefly luciferase activity normalized for transfection efficiency using the SV40 promoter-driven Renilla luciferase activity, were used to obtain mean promoter activities. Fold-activation was determined by dividing mean promoter activity by the promoter activity with only pCI. Error bars indicate Standard Error of Mean (SEM). Results presented are representative of three independent experiments.
Supporting Information
Quality of RNA used for microarray analysis. Total RNA isolated from PN11 and PN56 WT or Klf5CN corneas was subjected to Agilent Bioanalyzer analysis using nanoRNA chips. Resultant gel image confirming the RNA integrity is shown. Corresponding RNA Integrity Value (RIN) numbers are provided at the top of each lane.
(TIF)
Comparative analysis of the network of target genes influenced by transcription factors that are affected in Klf4 CN corneas. Lists of transcription factors downregulated by 1.5-fold in Klf4CN corneas (shaded yellow) were submitted to Ingenuity Pathway Analysis (IPA), where gene identifiers were mapped to their corresponding gene objects and overlaid onto a global molecular network in the Ingenuity Pathways Knowledge Base to generate the associated networks. Direct relationships are shown with solid arrows and indirect relationships with dashed arrows. Legends for different shapes used are shown.
(TIF)
Comparative analysis of the network of target genes influenced by transcription factors that are affected in Klf5 CN corneas. Lists of transcription factors downregulated by 1.5-fold in Klf5CN corneas (shaded yellow) were submitted to Ingenuity Pathway Analysis (IPA), where gene identifiers were mapped to their corresponding gene objects and overlaid onto a global molecular network in the Ingenuity Pathways Knowledge Base to generate the associated networks. Direct relationships are shown with solid arrows and indirect relationships with dashed arrows. Legends for different shapes used are shown.
(TIF)
Sequence of nucleotide primers used for QPCR.
(XLSX)
Most downregulated genes in PN11 Klf5 CN corneas.
(XLS)
Most upregulated genes in PN11 Klf5 CN corneas.
(XLS)
Most downregulated genes in PN56 Klf5 CN corneas.
(XLS)
Most upregulated genes in PN56 Klf5 CN corneas.
(XLS)
Most upregulated genes in PN56 WT compared with PN11 WT corneas.
(XLS)
Most downregulated genes in PN56 compared with PN11 WT corneas.
(XLS)
Genes whose expression is decreased at PN56 in both Klf4 CN and Klf5 CN corneas. Transcription factors are in bold. Genes are annotated by other effects as follows: (*) = developmental effect, (**) = PN11 Klf5CN effect, (***) = both. All 43 developmental effects are increases except Agpat3, Lamb1-1, Ntn1, Satb1 and Tox. All 48 PN56 Klf5CN effects are decreases except Vgll3.
(XLSX)
Genes whose expression is increased in both Klf4 CN and Klf5 CN corneas at PN56. Transcription factors are highlighted in bold. Genes are annotated by the other effects. (*) = developmental effect, (**) = Klf5CN effect in PN11 cornea also, (***) = both. All changes in the PN11 Klf5CN are increases and all developmental changes are decreases, except those shaded grey.
(XLSX)
Canonical pathways enriched in PN11 and PN56 corneal Klf5-target genes or in developmentally-modulated genes. Aggregates of differentially expressed genes in PN11or PN56 Klf5CN corneas, and during WT corneal maturation were subjected to canonical pathway analysis. Pathways for which at least one group showed significant (−log(p) >5) enrichment were selected. Where upregulated genes exceed downregulated genes by >2-fold, cells are colored red; where downregulated genes exceed upregulated genes by >2-fold, cells are colored green. Otherwise cells are colored yellow. For Klf5CN groups, significantly enriched pathways are dominated by upregulated genes. Developmentally modulated pathways contain a fair balance of up- and down-regulated genes.
(XLSX)
Immune-related genes whose expression changes in Klf5 CN corneas. 107 of 368 immun/inflam genes were upregulated in PN56 and/or PN11 Klf5CN corneas, with only 7 genes downregulated. Entries in bold show the smaller number of genes affected in PN56 Klf4CN corneas.
(XLSX)
Funding Statement
This work was supported by the National Eye Institute (NEI) K22 Career Development Award EY016875 (SKS), startup funds from the Department of Ophthalmology, Core Grant for Vision Research (5P30 EY08098-19), Research to Prevent Blindness and the Eye and Ear Foundation, Pittsburgh. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Klintworth GK (2003) The molecular genetics of the corneal dystrophies–current status. Front Biosci 8: d687–713. [DOI] [PubMed] [Google Scholar]
- 2. Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, et al. (2005) Kruppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res 15: 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nagasaki T, Zhao J (2003) Centripetal movement of corneal epithelial cells in the normal adult mouse. Invest Ophthalmol Vis Sci 44: 558–566. [DOI] [PubMed] [Google Scholar]
- 4. Cotsarelis G, Cheng SZ, Dong G, Sun TT, Lavker RM (1989) Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: implications on epithelial stem cells. Cell 57: 201–209. [DOI] [PubMed] [Google Scholar]
- 5. Collinson JM, Chanas SA, Hill RE, West JD (2004) Corneal development, limbal stem cell function, and corneal epithelial cell migration in the Pax6(+/−) mouse. Invest Ophthalmol Vis Sci 45: 1101–1108. [DOI] [PubMed] [Google Scholar]
- 6. Collinson JM, Morris L, Reid AI, Ramaesh T, Keighren MA, et al. (2002) Clonal analysis of patterns of growth, stem cell activity, and cell movement during the development and maintenance of the murine corneal epithelium. Dev Dyn 224: 432–440. [DOI] [PubMed] [Google Scholar]
- 7. Collinson JM, Quinn JC, Hill RE, West JD (2003) The roles of Pax6 in the cornea, retina, and olfactory epithelium of the developing mouse embryo. Dev Biol 255: 303–312. [DOI] [PubMed] [Google Scholar]
- 8. Zieske JD (2004) Corneal development associated with eyelid opening. Int J Dev Biol 48: 903–911. [DOI] [PubMed] [Google Scholar]
- 9. Hay ED (1979) Development of the vertebrate cornea. Int Rev Cytol 63: 263–322. [DOI] [PubMed] [Google Scholar]
- 10. Adhikary G, Crish JF, Bone F, Gopalakrishnan R, Lass J, et al. (2005) An involucrin promoter AP1 transcription factor binding site is required for expression of involucrin in the corneal epithelium in vivo. Invest Ophthalmol Vis Sci 46: 1219–1227. [DOI] [PubMed] [Google Scholar]
- 11. Adhikary G, Crish JF, Gopalakrishnan R, Bone F, Eckert RL (2005) Involucrin expression in the corneal epithelium: an essential role for Sp1 transcription factors. Invest Ophthalmol Vis Sci 46: 3109–3120. [DOI] [PubMed] [Google Scholar]
- 12. Chiambaretta F, Blanchon L, Rabier B, Kao WW, Liu JJ, et al. (2002) Regulation of corneal keratin-12 gene expression by the human Kruppel-like transcription factor 6. Invest Ophthalmol Vis Sci 43: 3422–3429. [PubMed] [Google Scholar]
- 13. Chiambaretta F, Nakamura H, De Graeve F, Sakai H, Marceau G, et al. (2006) Kruppel-like factor 6 (KLF6) affects the promoter activity of the alpha1-proteinase inhibitor gene. Invest Ophthalmol Vis Sci 47: 582–590. [DOI] [PubMed] [Google Scholar]
- 14. Davis J, Duncan MK, Robison WG Jr, Piatigorsky J (2003) Requirement for Pax6 in corneal morphogenesis: a role in adhesion. J Cell Sci 116: 2157–2167. [DOI] [PubMed] [Google Scholar]
- 15. Dwivedi DJ, Pontoriero GF, Ashery-Padan R, Sullivan S, Williams T, et al. (2005) Targeted deletion of AP-2alpha leads to disruption in corneal epithelial cell integrity and defects in the corneal stroma. Invest Ophthalmol Vis Sci 46: 3623–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hough RB, Piatigorsky J (2004) Preferential transcription of rabbit Aldh1a1 in the cornea: implication of hypoxia-related pathways. Mol Cell Biol 24: 1324–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakamura H, Chiambaretta F, Sugar J, Sapin V, Yue BY (2004) Developmentally regulated expression of KLF6 in the mouse cornea and lens. Invest Ophthalmol Vis Sci 45: 4327–4332. [DOI] [PubMed] [Google Scholar]
- 18. Nakamura H, Ueda J, Sugar J, Yue BY (2005) Developmentally regulated expression of Sp1 in the mouse cornea. Invest Ophthalmol Vis Sci 46: 4092–4096. [DOI] [PubMed] [Google Scholar]
- 19. Sivak JM, Mohan R, Rinehart WB, Xu PX, Maas RL, et al. (2000) Pax-6 expression and activity are induced in the reepithelializing cornea and control activity of the transcriptional promoter for matrix metalloproteinase gelatinase B. Dev Biol 222: 41–54. [DOI] [PubMed] [Google Scholar]
- 20. Sivak JM, West-Mays JA, Yee A, Williams T, Fini ME (2004) Transcription Factors Pax6 and AP-2alpha Interact To Coordinate Corneal Epithelial Repair by Controlling Expression of Matrix Metalloproteinase Gelatinase B. . Mol Cell Biol 24: 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Swamynathan SK, Davis J, Piatigorsky J (2008) Identification of candidate Klf4 target genes reveals the molecular basis of the diverse regulatory roles of Klf4 in the mouse cornea. Invest Ophthalmol Vis Sci 49: 3360–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Swamynathan SK, Katz JP, Kaestner KH, Ashery-Padan R, Crawford MA, et al. (2007) Conditional deletion of the mouse Klf4 gene results in corneal epithelial fragility, stromal edema, and loss of conjunctival goblet cells. Mol Cell Biol 27: 182–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ueta M, Hamuro J, Yamamoto M, Kaseda K, Akira S, et al. (2005) Spontaneous ocular surface inflammation and goblet cell disappearance in I kappa B zeta gene-disrupted mice. Invest Ophthalmol Vis Sci 46: 579–588. [DOI] [PubMed] [Google Scholar]
- 24. Francesconi CM, Hutcheon AE, Chung EH, Dalbone AC, Joyce NC, et al. (2000) Expression patterns of retinoblastoma and E2F family proteins during corneal development. Invest Ophthalmol Vis Sci 41: 1054–1062. [PubMed] [Google Scholar]
- 25. Lambiase A, Merlo D, Mollinari C, Bonini P, Rinaldi AM, et al. (2005) Molecular basis for keratoconus: lack of TrkA expression and its transcriptional repression by Sp3. Proc Natl Acad Sci U S A 102: 16795–16800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bieker JJ (2001) Kruppel-like factors: three fingers in many pies. J Biol Chem 276: 34355–34358. [DOI] [PubMed] [Google Scholar]
- 27. Swamynathan SK (2010) Kruppel-like factors: three fingers in control. Hum Genomics 2010/06/01 ed 263–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chiambaretta F, De Graeve F, Turet G, Marceau G, Gain P, et al. (2004) Cell and tissue specific expression of human Kruppel-like transcription factors in human ocular surface. Mol Vis 10: 901–909. [PubMed] [Google Scholar]
- 29. Norman B, Davis J, Piatigorsky J (2004) Postnatal gene expression in the normal mouse cornea by SAGE. Invest Ophthalmol Vis Sci 45: 429–440. [DOI] [PubMed] [Google Scholar]
- 30. McConnell BB, Ghaleb AM, Nandan MO, Yang VW (2007) The diverse functions of Kruppel-like factors 4 and 5 in epithelial biology and pathobiology. Bioessays 29: 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kenchegowda D, Swamynathan S, Gupta D, Wan H, Whitsett J, et al. (2011) Conditional Disruption of Mouse Klf5 Results in Defective Eyelids with Malformed Meibomian Glands, Abnormal Cornea and Loss of Conjunctival Goblet Cells. Developmental Biology [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Young RD, Swamynathan SK, Boote C, Mann M, Quantock AJ, et al. (2009) Stromal edema in klf4 conditional null mouse cornea is associated with altered collagen fibril organization and reduced proteoglycans. Invest Ophthalmol Vis Sci 50: 4155–4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gupta D, Harvey SA, Kaminski N, Swamynathan S (2011) Mouse conjunctival forniceal gene expression during postnatal development and its regulation by Kruppel-like factor 4. Invest Ophthalmol Vis Sci [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sivak JM, Fini ME (2002) MMPs in the eye: emerging roles for matrix metalloproteinases in ocular physiology. Prog Retin Eye Res 21: 1–14. [DOI] [PubMed] [Google Scholar]
- 35. Hausman RE (2007) Ocular extracellular matrices in development. Prog Retin Eye Res 26: 162–188. [DOI] [PubMed] [Google Scholar]
- 36. Vithana EN, Morgan P, Sundaresan P, Ebenezer ND, Tan DT, et al. (2006) Mutations in sodium-borate cotransporter SLC4A11 cause recessive congenital hereditary endothelial dystrophy (CHED2). Nat Genet 38: 755–757. [DOI] [PubMed] [Google Scholar]
- 37. Kloeckener-Gruissem B, Vandekerckhove K, Nurnberg G, Neidhardt J, Zeitz C, et al. (2008) Mutation of solute carrier SLC16A12 associates with a syndrome combining juvenile cataract with microcornea and renal glucosuria. Am J Hum Genet 82: 772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stasi K, Nagel D, Yang X, Ren L, Mittag T, et al. (2007) Ceruloplasmin upregulation in retina of murine and human glaucomatous eyes. Invest Ophthalmol Vis Sci 48: 727–732. [DOI] [PubMed] [Google Scholar]
- 39. Farkas RH, Chowers I, Hackam AS, Kageyama M, Nickells RW, et al. (2004) Increased expression of iron-regulating genes in monkey and human glaucoma. Invest Ophthalmol Vis Sci 45: 1410–1417. [DOI] [PubMed] [Google Scholar]
- 40. Gronert K, Maheshwari N, Khan N, Hassan IR, Dunn M, et al. (2005) A role for the mouse 12/15-lipoxygenase pathway in promoting epithelial wound healing and host defense. J Biol Chem 280: 15267–15278. [DOI] [PubMed] [Google Scholar]
- 41. Bonanno JA (2003) Identity and regulation of ion transport mechanisms in the corneal endothelium. Prog Retin Eye Res 22: 69–94. [DOI] [PubMed] [Google Scholar]
- 42. Bonanno JA, Nyguen T, Biehl T, Soni S (2003) Can variability in corneal metabolism explain the variability in corneal swelling? Eye Contact Lens 29: S7–9; discussion S26–29, S192–194. [DOI] [PubMed] [Google Scholar]
- 43. Liao SY, Ivanov S, Ivanova A, Ghosh S, Cote MA, et al. (2003) Expression of cell surface transmembrane carbonic anhydrase genes CA9 and CA12 in the human eye: overexpression of CA12 (CAXII) in glaucoma. J Med Genet 40: 257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gipson IK (1992) Adhesive mechanisms of the corneal epithelium. Acta Ophthalmol Suppl 13–17. [DOI] [PubMed] [Google Scholar]
- 45. Kottke MD, Delva E, Kowalczyk AP (2006) The desmosome: cell science lessons from human diseases. J Cell Sci 119: 797–806. [DOI] [PubMed] [Google Scholar]
- 46. Litjens SH, de Pereda JM, Sonnenberg A (2006) Current insights into the formation and breakdown of hemidesmosomes. Trends Cell Biol 16: 376–383. [DOI] [PubMed] [Google Scholar]
- 47. Swamynathan S, Kenchegowda D, Piatigorsky J, Swamynathan SK (2011) Regulation of Corneal Epithelial Barrier Function by Kruppel-like Transcription Factor 4. Invest Ophthalmol Vis Sci 52: 1762–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Davis J, Piatigorsky J (2011) Overexpression of Pax6 in mouse cornea directly alters corneal epithelial cells: changes in immune function, vascularization, and differentiation. Invest Ophthalmol Vis Sci 52: 4158–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dora N, Ou J, Kucerova R, Parisi I, West JD, et al. (2008) PAX6 dosage effects on corneal development, growth, and wound healing. Dev Dyn 237: 1295–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li W, Chen YT, Hayashida Y, Blanco G, Kheirkah A, et al. (2008) Down-regulation of Pax6 is associated with abnormal differentiation of corneal epithelial cells in severe ocular surface diseases. J Pathol 214: 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gage PJ, Zacharias AL (2009) Signaling “cross-talk” is integrated by transcription factors in the development of the anterior segment in the eye. Dev Dyn 238: 2149–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang X, Tseng H (2007) Basonuclin-null mutation impairs homeostasis and wound repair in mouse corneal epithelium. PLoS One 2: e1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fragiadaki M, Ikeda T, Witherden A, Mason RM, Abraham D, et al. (2011) High doses of TGF-beta potently suppress type I collagen via the transcription factor CUX1. Mol Biol Cell 22: 1836–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stucchi A, Reed K, O'Brien M, Cerda S, Andrews C, et al. (2006) A new transcription factor that regulates TNF-alpha gene expression, LITAF, is increased in intestinal tissues from patients with CD and UC. Inflamm Bowel Dis 12: 581–587. [DOI] [PubMed] [Google Scholar]
- 55. Tergaonkar V, Correa RG, Ikawa M, Verma IM (2005) Distinct roles of IkappaB proteins in regulating constitutive NF-kappaB activity. Nat Cell Biol 7: 921–923. [DOI] [PubMed] [Google Scholar]
- 56. Chu PH, Yeh LK, Lin HC, Jung SM, Ma DH, et al. (2008) Deletion of the FHL2 gene attenuating neovascularization after corneal injury. Invest Ophthalmol Vis Sci 49: 5314–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hai T, Wolford CC, Chang YS (2010) ATF3, a hub of the cellular adaptive-response network, in the pathogenesis of diseases: is modulation of inflammation a unifying component? Gene Expr 15: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Parisi S, Cozzuto L, Tarantino C, Passaro F, Ciriello S, et al. (2010) Direct targets of Klf5 transcription factor contribute to the maintenance of mouse embryonic stem cell undifferentiated state. BMC Biol 8: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wan H, Luo F, Wert SE, Zhang L, Xu Y, et al. (2008) Kruppel-like factor 5 is required for perinatal lung morphogenesis and function. Development 135: 2563–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bell SM, Zhang L, Mendell A, Xu Y, Haitchi HM, et al. (2011) Kruppel-like factor 5 is required for formation and differentiation of the bladder urothelium. Dev Biol 358: 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vernes SC, Oliver PL, Spiteri E, Lockstone HE, Puliyadi R, et al. (2011) Foxp2 regulates gene networks implicated in neurite outgrowth in the developing brain. PLoS Genet 7: e1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fang X, Yoon JG, Li L, Yu W, Shao J, et al. (2011) The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis. BMC Genomics 12: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhao L, Glazov EA, Pattabiraman DR, Al-Owaidi F, Zhang P, et al. (2011) Integrated genome-wide chromatin occupancy and expression analyses identify key myeloid pro-differentiation transcription factors repressed by Myb. Nucleic Acids Res 39: 4664–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tang B, Di Lena P, Schaffer L, Head SR, Baldi P, et al. (2011) Genome-wide identification of bcl11b gene targets reveals role in brain-derived neurotrophic factor signaling. PLoS One 6: e23691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Leiper LJ, Ou J, Walczysko P, Kucerova R, Lavery DN, et al. (2009) Control of patterns of corneal innervation by Pax6. Invest Ophthalmol Vis Sci 50: 1122–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lewitzky M, Yamanaka S (2007) Reprogramming somatic cells towards pluripotency by defined factors. Curr Opin Biotechnol 18: 467–473. [DOI] [PubMed] [Google Scholar]
- 67. Harvey SA, Guerriero E, Charukamnoetkanok N, Piluek J, Schuman JS, et al. (2010) Responses of cultured human keratocytes and myofibroblasts to ethyl pyruvate: a microarray analysis of gene expression. Invest Ophthalmol Vis Sci 51: 2917–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liesegang TJ (2002) Physiologic changes of the cornea with contact lens wear. CLAO J 28: 12–27. [PubMed] [Google Scholar]
- 69. Mandell RB, Fatt I (1965) Thinning of the human cornea on awakening. Nature 208: 292–293. [DOI] [PubMed] [Google Scholar]
- 70. Makino Y, Cao R, Svensson K, Bertilsson G, Asman M, et al. (2001) Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature 414: 550–554. [DOI] [PubMed] [Google Scholar]
- 71. Makino Y, Uenishi R, Okamoto K, Isoe T, Hosono O, et al. (2007) Transcriptional up-regulation of inhibitory PAS domain protein gene expression by hypoxia-inducible factor 1 (HIF-1): a negative feedback regulatory circuit in HIF-1-mediated signaling in hypoxic cells. J Biol Chem 282: 14073–14082. [DOI] [PubMed] [Google Scholar]
- 72. Nebert DW, Roe AL, Dieter MZ, Solis WA, Yang Y, et al. (2000) Role of the aromatic hydrocarbon receptor and [Ah] gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem Pharmacol 59: 65–85. [DOI] [PubMed] [Google Scholar]
- 73. Boesch JS, Miskimins R, Miskimins WK, Lindahl R (1999) The same xenobiotic response element is required for constitutive and inducible expression of the mammalian aldehyde dehydrogenase-3 gene. Arch Biochem Biophys 361: 223–230. [DOI] [PubMed] [Google Scholar]
- 74. Wolosin JM, Budak MT, Akinci MA (2004) Ocular surface epithelial and stem cell development. Int J Dev Biol 48: 981–991. [DOI] [PubMed] [Google Scholar]
- 75. Wu F, Lee S, Schumacher M, Jun A, Chakravarti S (2008) Differential gene expression patterns of the developing and adult mouse cornea compared to the lens and tendon. Exp Eye Res 87: 214–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quality of RNA used for microarray analysis. Total RNA isolated from PN11 and PN56 WT or Klf5CN corneas was subjected to Agilent Bioanalyzer analysis using nanoRNA chips. Resultant gel image confirming the RNA integrity is shown. Corresponding RNA Integrity Value (RIN) numbers are provided at the top of each lane.
(TIF)
Comparative analysis of the network of target genes influenced by transcription factors that are affected in Klf4 CN corneas. Lists of transcription factors downregulated by 1.5-fold in Klf4CN corneas (shaded yellow) were submitted to Ingenuity Pathway Analysis (IPA), where gene identifiers were mapped to their corresponding gene objects and overlaid onto a global molecular network in the Ingenuity Pathways Knowledge Base to generate the associated networks. Direct relationships are shown with solid arrows and indirect relationships with dashed arrows. Legends for different shapes used are shown.
(TIF)
Comparative analysis of the network of target genes influenced by transcription factors that are affected in Klf5 CN corneas. Lists of transcription factors downregulated by 1.5-fold in Klf5CN corneas (shaded yellow) were submitted to Ingenuity Pathway Analysis (IPA), where gene identifiers were mapped to their corresponding gene objects and overlaid onto a global molecular network in the Ingenuity Pathways Knowledge Base to generate the associated networks. Direct relationships are shown with solid arrows and indirect relationships with dashed arrows. Legends for different shapes used are shown.
(TIF)
Sequence of nucleotide primers used for QPCR.
(XLSX)
Most downregulated genes in PN11 Klf5 CN corneas.
(XLS)
Most upregulated genes in PN11 Klf5 CN corneas.
(XLS)
Most downregulated genes in PN56 Klf5 CN corneas.
(XLS)
Most upregulated genes in PN56 Klf5 CN corneas.
(XLS)
Most upregulated genes in PN56 WT compared with PN11 WT corneas.
(XLS)
Most downregulated genes in PN56 compared with PN11 WT corneas.
(XLS)
Genes whose expression is decreased at PN56 in both Klf4 CN and Klf5 CN corneas. Transcription factors are in bold. Genes are annotated by other effects as follows: (*) = developmental effect, (**) = PN11 Klf5CN effect, (***) = both. All 43 developmental effects are increases except Agpat3, Lamb1-1, Ntn1, Satb1 and Tox. All 48 PN56 Klf5CN effects are decreases except Vgll3.
(XLSX)
Genes whose expression is increased in both Klf4 CN and Klf5 CN corneas at PN56. Transcription factors are highlighted in bold. Genes are annotated by the other effects. (*) = developmental effect, (**) = Klf5CN effect in PN11 cornea also, (***) = both. All changes in the PN11 Klf5CN are increases and all developmental changes are decreases, except those shaded grey.
(XLSX)
Canonical pathways enriched in PN11 and PN56 corneal Klf5-target genes or in developmentally-modulated genes. Aggregates of differentially expressed genes in PN11or PN56 Klf5CN corneas, and during WT corneal maturation were subjected to canonical pathway analysis. Pathways for which at least one group showed significant (−log(p) >5) enrichment were selected. Where upregulated genes exceed downregulated genes by >2-fold, cells are colored red; where downregulated genes exceed upregulated genes by >2-fold, cells are colored green. Otherwise cells are colored yellow. For Klf5CN groups, significantly enriched pathways are dominated by upregulated genes. Developmentally modulated pathways contain a fair balance of up- and down-regulated genes.
(XLSX)
Immune-related genes whose expression changes in Klf5 CN corneas. 107 of 368 immun/inflam genes were upregulated in PN56 and/or PN11 Klf5CN corneas, with only 7 genes downregulated. Entries in bold show the smaller number of genes affected in PN56 Klf4CN corneas.
(XLSX)