Abstract
We investigated inbreeding depression and genetic load in a small (Ne ∼ 100) population of the Glanville fritillary butterfly (Melitaea cinxia), which has been completely isolated on a small island [Pikku Tytärsaari (PT)] in the Baltic Sea for at least 75 y. As a reference, we studied conspecific populations from the well-studied metapopulation in the Åland Islands (ÅL), 400 km away. A large population in Saaremaa, Estonia, was used as a reference for estimating genetic diversity and Ne. We investigated 58 traits related to behavior, development, morphology, reproductive performance, and metabolism. The PT population exhibited high genetic load (L = 1 − WPT/WÅL) in a range of fitness-related traits including adult weight (L = 0.12), flight metabolic rate (L = 0.53), egg viability (L = 0.37), and lifetime production of eggs in an outdoor population cage (L = 0.70). These results imply extensive fixation of deleterious recessive mutations, supported by greatly reduced diversity in microsatellite markers and immediate recovery (heterosis) of egg viability and flight metabolic rate in crosses with other populations. There was no significant inbreeding depression in most traits due to one generation of full-sib mating. Resting metabolic rate was significantly elevated in PT males, which may be related to their short lifespan (L = 0.25). The demographic history and the effective size of the PT population place it in the part of the parameter space in which models predict mutation accumulation. This population exemplifies the increasingly common situation in fragmented landscapes, in which small and completely isolated populations are vulnerable to extinction due to high genetic load.
Keywords: fixation load, segregation load, hybrid vigor, persisting population, inbreeding avoidance
Rampant loss and fragmentation of natural habitats is a prime cause of population and species extinctions. At their final stage, extinctions are typically ascribed to ecological processes (1), especially to demographic and environmental stochasticities, which increase the risk of extinction for remaining small populations (2–4). However, in the absence of gene flow, the viability of isolated remnant populations may be compromised also by genetic processes, such as inbreeding depression due to segregation of deleterious recessive mutations and overdominant alleles and mutational meltdown due to fixation of old and new deleterious mutations of smaller effect by random genetic drift (5–7). On the other hand, increased homozygosity due to inbreeding and drift may expose strongly deleterious recessive mutations to selection (8), thereby increasing mean fitness (9, 10), and local adaptation may further enhance fitness in the absence of disruptive gene flow (11–13). Complete isolation and reduction in population size, which are increasingly common predicaments for populations in human-dominated landscapes (14–16), can thus have contrasting effects on population viability. There are scores of studies focusing on the effects of drift or inbreeding or local adaptation (7, 17, 18), but their joint effects remain little studied and therefore poorly understood (19).
Increasing frequency and fixation of deleterious recessive mutations in isolated populations via inbreeding and drift are a particularly serious concern for conservation as they may erode population viability gradually, in an initially imperceptible but irreversible way. Theoretical models have been constructed to analyze the factors influencing the likelihood of mutational meltdown. These models have addressed the effects of effective population size (5) and the rate of appearance of new deleterious mutations and their effects (5), as well as compensatory beneficial mutations (20) and epistatic interactions (21). What is in short supply are empirical studies of genetic load and its consequences in natural populations. Previous studies have rarely documented the history of isolation and past population sizes. In the absence of this information, researchers have merely assumed that small current population size combined with reduced genetic variation is indicative of long-term isolation and that such populations potentially suffer from the accumulation of deleterious mutations (22). In the case of naturally isolated insect populations, the focus of research has been on reduced genetic diversity (23–27). Strong support for genetic load due to drift is provided by demonstration of fitness recovery (heterosis) in crosses between isolated populations (28, 29). Studies of natural populations have demonstrated fitness recovery due to a small number of unrelated immigrants entering an isolated population (14, 30–32) and leading to genetic rescue (33, 34), regardless of whether reduced fitness was due to inbreeding depression (segregation load) or drift (fixation load) (35, 36).
Here, we report on inbreeding depression, genetic load (fitness), and local adaptation in a population of the Glanville fritillary butterfly (Melitaea cinxia) on the small island of Pikku Tytärsaari (PT) in the northern Baltic Sea. This small population has been completely isolated for at least 75 y. As a reference, we use the large conspecific metapopulation in the Åland Islands (ÅL), located 400 km away, which has been intensively studied for the past 20 y (37, 38). The amount of neutral genetic variation lost by the small isolated population is consistent with model predictions, given its demographic history. We found no convincing evidence for local adaptation and only limited inbreeding depression due to one generation of full-sib mating, supporting a high degree of relatedness between individuals and purging of strongly deleterious alleles. In contrast, the isolated PT population exhibits large fitness reduction in comparison with the reference population in a range of traits including body size, flight metabolic rate, egg viability, adult longevity, and lifetime production of eggs and larvae. Finally, we observed complete fitness recovery (heterosis) in crosses between PT and other regional populations, implying high genetic load in this isolated population, which may be undergoing a genetic meltdown to extinction.
Results
Population Origin and Genetic Diversity.
The Glanville fritillary was first recorded on the small uninhabited island of PT in the middle of the Gulf of Finland in 1936. The island has a single meadow of ca. 10 ha with the larval host plant Veronica spicata along the shore. The nearest mainland (Estonia) is ca. 30 km away. The Glanville fritillary was most likely unintentionally introduced from the Estonian coast by people who used the island as a base for fishing and travel. It is practically certain that there has been no further gene flow to the island because it has been visited very seldom by anyone since World War II (WWII) and the butterfly is a poor disperser (39). Exhaustive surveys of the entire habitat area on the island revealed 111 and 198 larval family groups (full siblings) in 2009 and 2011, respectively. Using the formula in Hartl and Clark 2007 (40) and assuming that the expected heterozygosities in the initial and current populations are 0.73 and 0.43, respectively (Table 1) and that the age of the population is 100 y (=100 generations), we obtain Ne = 95, which is consistent with the above-cited numbers of larval family groups (some females produce two or even more larval groups that survive until the autumn) (41).
Table 1.
Summary of genetic results for the Pikku Tytärsaari (PT, Russia), the Saaremaa (SA, Estonia), and the Åland Islands (ÅL, Finland) populations
| PT | SA | ÅL | |
| Island area, km2 | 1.5 | 2,673 | 13,517 |
| Sample size | 69 | 59 | 28 |
| No. of alleles (SD) | 3.57 (0.54) | 10.57 (2.44) | 8.14 (3.0) |
| Expected heterozygosity (SD) | 0.43 (0.09) | 0.73 (0.07) | 0.72 (0.21) |
| Observed heterozygosity (SD) | 0.47 (0.12) | 0.58 (0.14)* | 0.53 (0.16)* |
| Gene diversity (SD)† | 0.37 (0.22) | 0.65 (0.36) | 0.62 (0.34) |
| Shared alleles with PT only (total) | 7 (20) | 1 (13) | |
| Fst with PT | 0.28 | 0.41 | |
| Inbreeding DyadML (variance)‡ | 0.36 (0.06) | 0.31 (0.06) | 0.40 (0.05) |
| Relatedness DyadML (variance)‡ | 0.80 (0.10) | 0.27 (0.07) | 0.39 (0.05) |
*All seven markers were included in the analysis although four markers showed indication of null alleles in SA and ÅL populations, probably due to a very large size range of alleles.
†Gene diversity refers to expected heterozygosity over all data and is calculated with Arlequin (94).
‡Dyadic maximum-likelihood inbreeding (F; tests for deviation from random mating) and relatedness values are calculated with the Coancestry (95) program. Variance refers to the 95% confidence interval for 1,000 bootstrapping samples.
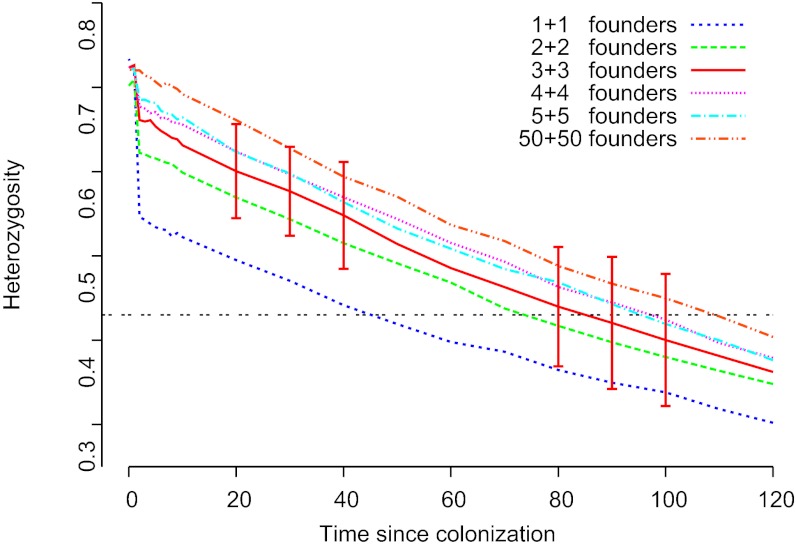
Data for seven microsatellites indicate closer similarity of the PT population to the Estonian population from Saaremaa (SA) than to the Finnish population from the ÅL (Table 1). The PT population shares seven alleles in the seven microsatellites with only the Estonian population, but only one allele with only the ÅL population (P < 0.05). The pairwise Fst values between PT and Estonia and between PT and ÅL are 0.28 and 0.41, respectively, whereas Fst between Estonia and ÅL is 0.22 (all values statistically significant, P < 0.05). The PT population is genetically clearly less diverse than the two other populations (Table 1). The number of alleles in each microsatellite is maximally four, suggesting that the offspring of a single mated female may have established the population (taking into account that the introduction most likely took place at the larval stage; SI Materials and Methods). However, modeling the reduction in genetic diversity over time (SI Materials and Methods) is more consistent with colonization by the offspring of two or more females (Fig. 1). The modeling results are consistent with the knowledge that the population is at least 75 y old, and the results suggest that the population is unlikely to be older than 100 y (Fig. 1). We sequenced the mitochondrial DNA gene COI, which had only one haplotype in PT, but this is not informative because the same haplotype is common in both SA and ÅL, which had five and three haplotypes, respectively.
Fig. 1.
Model-predicted reduction in expected heterozygosity in the PT population with time since the colonization. The results are shown for six different propagule sizes, from a single mated female to 50 females mated with 50 males. Bars giving 1 SD are shown for the propagule size 3+3 and are very similar to all other propagule sizes. The propagule is assumed to have the genetic parameters of the Estonian population on the island of Saaremaa (Table 1). The horizontal line gives the expected heterozygosity in the PT population.
The DyadML inbreeding coefficients in Table 1 are nearly identical in the three populations, whereas the value of relatedness is very much higher in PT than in the two other populations (Table 1). These results, and taking into account that the observed (0.47) and expected heterozygosities (0.43) are nearly equal, indicate that there is no excess of nonrandom matings in the PT population compared with the other populations. Estimates using other models implemented in Coancestry (Materials and Methods) were compatible with those of DyadML. The overall positive inbreeding values are at least partly due to null alleles in the microsatellite data.
Life-History Traits and Fitness.
We measured life-history traits and fitness components of individuals mated with full sibs vs. unrelated individuals in a common garden laboratory experiment, separately for the PT and ÅL populations (Table 2). Additionally, we recorded comparable data for other life-history traits, morphology, and metabolic rates in the laboratory and in a large outdoor population cage (Table 3 and Materials and Methods).
Table 2.
Life-history traits and fitness components in outcrossed (between-family) and full-sibling matings in butterflies originating from the old isolated PT population and the large ÅL reference population and the corresponding estimates of inbreeding depression and genetic load
| ÅL |
PT |
PT vs. ÅL§ |
|||||||||||
| Average trait value* |
t test† |
Inbreeding depression:‡ |
Average trait value* |
t test† |
Inbreeding depression:‡ |
t test |
Genetic load: |
||||||
| Trait | Outcrossed | Sib- mated | t | P | 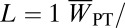 |
Outcrossed | Sib- mated | t | P | 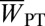 |
t | P | LPT |
| Prezygotic | |||||||||||||
| Timing of reproduction | |||||||||||||
| Mating rate, /d, sqrt | 1.900 | 1.650 | 1.93 | 0.06 | 0.13 | 1.237 | 1.239 | −0.02 | 0.99 | 0.00 | 3.41 | 0.00 | 0.35 |
| Oviposition rate, first egg clutch, /d, log | 0.343 | 0.319 | 1.26 | 0.22 | 0.07 | 0.507 | 0.539 | −0.16 | 0.87 | −0.06 | −1.75 | 0.09 | −0.48 |
| Oviposition rate, egg clutches/d, sqrt | 0.161 | 0.138 | 1.13 | 0.27 | 0.14 | 0.228 | 0.194 | 0.89 | 0.38 | 0.15 | −1.99 | 0.06 | −0.42 |
| Egg clutch size | |||||||||||||
| Size of first egg clutch | 241 | 201 | 1.69 | 0.10 | 0.17 | 148 | 153 | −0.29 | 0.78 | −0.03 | 4.65 | 0.00 | 0.39 |
| Average size of all egg clutches | 192 | 169 | 1.20 | 0.24 | 0.12 | 118 | 112 | 0.34 | 0.74 | 0.04 | 4.75 | 0.00 | 0.39 |
| Postzygotic | |||||||||||||
| Survival | |||||||||||||
| Hatching rate of first egg clutch, %, asin | 82 | 56 | 2.60 | 0.01 | 0.32 | 64 | 41 | 2.40 | 0.02 | 0.36 | 2.36 | 0.02 | 0.22 |
| Average hatching rate of egg clutches, %, asin | 73 | 53 | 2.05 | 0.05 | 0.28 | 46 | 28 | 1.85 | 0.08 | 0.39 | 4.81 | 0.00 | 0.37 |
| Larval survival before diapause, %, asin | 95 | 73 | 4.28 | 0.00 | 0.23 | 53 | 59 | −0.21 | 0.83 | −0.11 | 4.36 | 0.00 | 0.44 |
| Development | |||||||||||||
| Average embryo development time, /d | 0.075 | 0.073 | 1.61 | 0.12 | 0.02 | 0.069 | 0.067 | 1.85 | 0.08 | 0.03 | 6.90 | 0.00 | 0.08 |
| Average larval growth rate, /d | 0.045 | 0.042 | 3.04 | 0.01 | 0.07 | 0.046 | 0.048 | −0.61 | 0.55 | −0.04 | −0.48 | 0.64 | −0.02 |
| Average larval weight, mg | 4.5 | 4.7 | −0.56 | 0.58 | −0.04 | 2.2 | 2.2 | 0.38 | 0.71 | 0.00 | 11.84 | 0.00 | 0.51 |
| Reproductive fitness | |||||||||||||
| Larval group size at diapause, sqrt | 190 | 104 | 3.38 | 0.00 | 0.45 | 54 | 43 | −0.13 | 0.90 | 0.20 | 5.35 | 0.00 | 0.72 |
| Lifetime larval production, sqrt | 443 | 284 | 1.97 | 0.06 | 0.36 | 139 | 103 | 1.30 | 0.21 | 0.26 | 4.13 | 0.00 | 0.69 |
The traits are divided into prezygotic and postzygotic. Transformations of variables: sqrt, square-root transformation; log, logarithmic transformation; asin, arcsine transformation.
*The average trait values are calculated from nontransformed data.
†Average trait values for the cross types were tested with a t test (using transformed data if a transformation was used).
‡Inbreeding depression coefficients  , where
, where  and
and 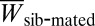 are the average values for sib-mated and outcrossed females.
are the average values for sib-mated and outcrossed females.
§The last three columns give a t test for the difference between the average trait values for outcrossed females from PT and ÅL,  and
and 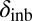 , and the genetic load L for PT in comparison with ÅL (
, and the genetic load L for PT in comparison with ÅL (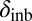 ).
).
Table 3.
Larval development and other life-history traits and adult morphology and metabolic rate in butterflies originating from the old isolated PT population and the large ÅL reference population
| Significance† |
Female average‡ |
Male average‡ |
||||||
| Method | Pop | Sex | Pop:sex | ÅL | PT | ÅL | PT | |
| Life history and fitness | ||||||||
| Larval weight at fifth instar, mg | CG | ** | NS | NS | 5.37 | 4.55 | 5.06 | 4.54 |
| Pupal weight, mg | CG | **** | *** | * | 187.83 | 173.78 | 156.99 | 150.34 |
| Pupal period, d | CG | **** | NS | NS | 9.5 | 10.5 | 9.4 | 10.5 |
| Adult longevity, mated, d | CG | ** | * | * | 23.0 | 22.3 | 22.9 | 17.2 |
| Adult longevity, not mated, d | CG | **** | *** | NS | 22.9 | 13.4 | 17.0 | 10.3 |
| Probability of flight behavior | PC | *** | ** | NS | 0.11 | 0.09 | 0.23 | 0.1 |
| Mobility | PC | NS | *** | ** | −1.33 | −0.27 | 0.95 | 0.06 |
| Time of day at start of oviposition, h | PC | **** | — | — | 13.42 | 15.05 | — | — |
| Lifetime-corrected number of eggs laid | PC | *** | — | — | 52.15 | 15.41 | — | — |
| Adult morphology | ||||||||
| Adult weigt, mg | CG | **** | **** | NS | 93.8 | 81.59 | 55.7 | 49.68 |
| Thorax weight, age residuals | CG | **** | *** | NS | 0.52 | −0.31 | 0.69 | −0.38 |
| Log abdomen weight, age/mating/ovip residual | CG | NS | ** | NS | 0.22 | −0.23 | 0.33 | −0.18 |
| Wing area, mm2 | CG | * | **** | NS | 119.33 | 113.5 | 97.82 | 99.03 |
| Wing loading, adult weight/wing area | CG | *** | **** | NS | 7.77 | 7.16 | 5.71 | 5.05 |
| Aspect ratio, 4 × wing length2/wing area | CG | NS | NS | ** | 1.13 | 1.16 | 1.18 | 1.13 |
| Outer wing ratio | CG | *** | NS | * | 0.44 | 0.427 | 0.434 | 0.433 |
| Metabolic rate (weight residual) | Pop (F) | Pop (M) | Pop:Sex | |||||
| Peak FMR, CO2 mL/h | CG, MR | **** | *** | NS | 1.68 | 0.97 | 1.78 | 0.95 |
| Peak FMRend, CO2 mL/h | CG, MR | **** | **** | NS | 0.73 | 0.32 | 1.01 | 0.37 |
| Integrated FMR, CO2 mL | CG, MR | **** | **** | NS | 0.22 | 0.11 | 0.26 | 0.11 |
| RMR, CO2 mL/h | CG, MR | NS | **** | **** | 0.11 | 0.12 | 0.05 | 0.1 |
*P < 0.1; **P < 0.05; ***P < 0.01; ****P < 0.001; CG, common garden experiment; MR, metabolic rate measurement; NS, nonsignificant; ovip, oviposition; PC, population cage experiment; Pop, population.
†Shown are the results for models with the fixed effects of population, sex, and their interaction. For metabolic rates (FMR corrected for adult weight, RMR corrected for adult age), the results are shown for models with the fixed effect of population separately for females and males.
‡The last four columns give the average trait values for females and males. The average FMR and RMR values are absolute values, not residuals.
For most traits, butterflies from the isolated PT population showed significantly lower trait value than butterflies from ÅL (Tables 2 and 3; full results in Tables S1–S4). The reduction in the PT population due to genetic load was measured as 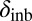 , where
, where  and
and 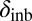 are the average trait values for outcrossed females in the PT and ÅL populations, respectively. For 9 of the 13 traits L > 0.2, for 4 traits L > 0.4, and for lifetime larval production, the best composite measure of fitness, L = 0.69 (Table 2), implying high genetic load in the PT population. The exceptional trait was the rate of laying egg clutches: PT females laid their first egg clutch at a younger age than ÅL females, and the average time interval between subsequent clutches was shorter in PT females (Table 2).
are the average trait values for outcrossed females in the PT and ÅL populations, respectively. For 9 of the 13 traits L > 0.2, for 4 traits L > 0.4, and for lifetime larval production, the best composite measure of fitness, L = 0.69 (Table 2), implying high genetic load in the PT population. The exceptional trait was the rate of laying egg clutches: PT females laid their first egg clutch at a younger age than ÅL females, and the average time interval between subsequent clutches was shorter in PT females (Table 2).
The rate of development was lower in the PT population. Thus, the length of the fifth larval instar following diapause was ca. 20% longer, and similarly the embryo (Table 2) and pupal development times (Table 3) were about 1 d (ca. 10%) longer in the PT than the ÅL population. One exception is the final seventh larval instar, which was about 1 d (ca. 10%) shorter in female PT larvae (Table S1). Adult longevity was significantly shorter in PT butterflies, especially in males (25% shorter; Table 3). Among the butterflies that did not succeed to mate in small cages in the laboratory, most likely because of their inferior condition, lifespan of PT butterflies was about 40% shorter than the lifespan of ÅL butterflies (both sexes; Table 3).
Reproductive performance was very poor in PT males. In the laboratory mating cages, PT males took significantly longer to mate than ÅL males (Table 2). In the large outdoor population cage with competition for mates among free-flying males from several regional populations (Materials and Methods), only 1 of 18 PT males succeeded to mate, in contrast to 15 of 20 ÅL males that mated at least once (χ2 = 22.2, df = 1, P < 0.0001). The low mating success of PT males in the outdoor cage was probably partly due to their limited flight activity. During regular censuses of butterflies in the cage (Materials and Methods and Table 3, full results in Table S1), PT males were observed to fly with probability 0.10, compared with 0.23 in ÅL males (χ2 = 6.13, df = 1, P = 0.013).
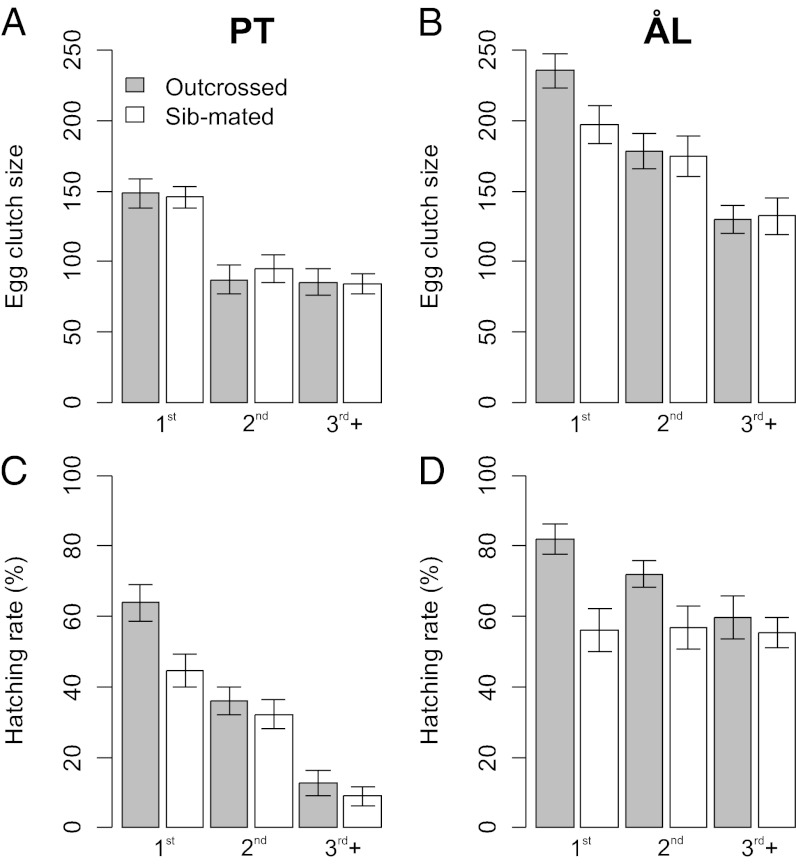
The reproductive performance of PT females was inferior to that of ÅL females (Tables 2 and 3 and Tables S2 and S4), with the above-mentioned exception of shorter interval between consecutive egg clutches in PT females. This exception is likely to be related to the much smaller average egg clutch size in PT females (39% smaller; Table 2 and Fig. 2). Consequently, although PT females laid more egg clutches under the common garden conditions in the laboratory, their lifetime egg production was much smaller (∼350 eggs) than that of ÅL females (∼600 eggs). In the outdoor population cage, the lifetime-corrected number of eggs laid by PT females was about 30% (L = 0.70) of that in ÅL females (Table 3). Finally, there was an intriguing difference in the timing of oviposition, PT females starting to oviposit on average 1.5 h later in the day than ÅL females (Table 3). This difference was not affected by ambient temperature.
Fig. 2.
(A–D) Clutch size (A and B) and egg-hatching rate (%) (C and D) in the first two clutches and as the average in the subsequent clutches in PT (A and C) and ÅL females (B and D). Clutches from outcrossed matings (between family, nPT = 17, nÅL = 20) are shown as shaded bars and clutches from sibling matings (nPT = 12, nÅL = 20) as open bars. Error bars show the SD.
Lifetime (prediapause) larval production is our best composite measure of fitness, as it includes several components of female reproductive success, such as the number of egg clutches, egg clutch size, hatching rate, and larval survival until diapause. On the basis of this measure, the fitness of the PT population is reduced to 31% of the fitness of the reference population. This fitness is, however, likely to be an underestimate, as it does not take into account the smaller body mass of PT prediapause larvae (49% of the mass of ÅL larvae) or larval development following diapause.
Inbreeding Depression.
Inbreeding depression was measured as  , where
, where 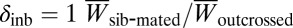 and
and  are the average trait values for sib-mated and outcrossed females. The traits are divided into prezygotic and postzygotic traits in Table 2. In the case of prezygotic traits, where the female is not inbred, there is a trend suggesting higher inbreeding depression in ÅL (average
are the average trait values for sib-mated and outcrossed females. The traits are divided into prezygotic and postzygotic traits in Table 2. In the case of prezygotic traits, where the female is not inbred, there is a trend suggesting higher inbreeding depression in ÅL (average  is 0.126 and 0.020 in ÅL and PT, respectively). Mating with a brother reduced the trait values of ÅL females: They took longer to mate, oviposited at a lower rate, and laid smaller egg clutches than females mated to a nonrelative (Table 2 and Fig. 2B). In contrast, there was no such inbreeding depression in PT females in the prezygotic traits (Table 2 and Fig. 2A). Among the postzygotic traits, egg viability was significantly affected by inbreeding in both populations, but prediapause larval survival showed significant inbreeding depression only in the ÅL population (Table 2). Traits related to the rates of embryo and larval development showed no inbreeding depression in either population, except for larval growth rate, which showed low but significant inbreeding depression in ÅL (Table 2). Unlike the other traits in Table 2, in which larger values increase lifetime reproductive performance, rates of embryo and prediapause larval development may be under stabilizing selection, with no advantage from higher rates, and hence these traits do not show inbreeding depression (smaller values) in the present analysis. Finally, considering larval group size at diapause and lifetime larval production, two compound traits related to lifetime reproductive success, there was high and significant inbreeding depression in the ÅL population (the latter was nearly significant with P = 0.06 on the basis of a t test on the square-root transformed variable and significant with P = 0.03 on the basis of a nonparametric Mann–Whitney U test;
is 0.126 and 0.020 in ÅL and PT, respectively). Mating with a brother reduced the trait values of ÅL females: They took longer to mate, oviposited at a lower rate, and laid smaller egg clutches than females mated to a nonrelative (Table 2 and Fig. 2B). In contrast, there was no such inbreeding depression in PT females in the prezygotic traits (Table 2 and Fig. 2A). Among the postzygotic traits, egg viability was significantly affected by inbreeding in both populations, but prediapause larval survival showed significant inbreeding depression only in the ÅL population (Table 2). Traits related to the rates of embryo and larval development showed no inbreeding depression in either population, except for larval growth rate, which showed low but significant inbreeding depression in ÅL (Table 2). Unlike the other traits in Table 2, in which larger values increase lifetime reproductive performance, rates of embryo and prediapause larval development may be under stabilizing selection, with no advantage from higher rates, and hence these traits do not show inbreeding depression (smaller values) in the present analysis. Finally, considering larval group size at diapause and lifetime larval production, two compound traits related to lifetime reproductive success, there was high and significant inbreeding depression in the ÅL population (the latter was nearly significant with P = 0.06 on the basis of a t test on the square-root transformed variable and significant with P = 0.03 on the basis of a nonparametric Mann–Whitney U test;  = 0.45 and 0.36, respectively), but weaker and nonsignificant inbreeding depression in the PT population (
= 0.45 and 0.36, respectively), but weaker and nonsignificant inbreeding depression in the PT population ( = 0.20 and 0.26; Table 2).
= 0.20 and 0.26; Table 2).
The inbreeding experiment could not be continued after larval diapause (fifth larval instar) due to very low survival of postdiapause PT larvae. Before the diapause, there were signs of disease in many groups of PT larvae, and their overall prediapause mortality was much higher (50%) than in ÅL larvae (5%; Table 2). In terms of lifetime production of diapause larvae the fitness of PT females was only 31% of the fitness of ÅL females (Table 2), but in reality the difference was much greater as the PT larvae were only half of the size of the ÅL larvae (Table 2) and apparently in poor condition.
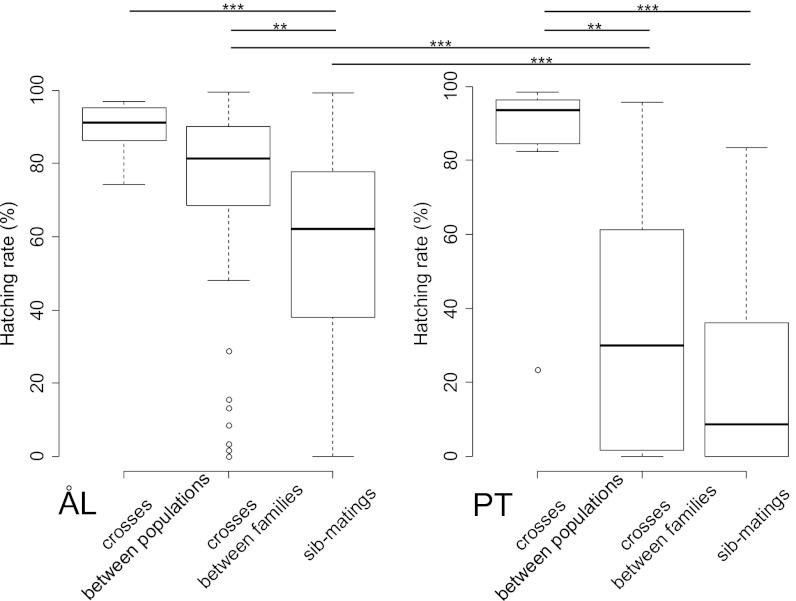
In the course of the outdoor cage experiment, we obtained naturally occurring crosses within and between PT and four other regional populations (Materials and Methods). In the case of egg viability, we compared three types of matings for both PT and ÅL females, namely mating (i) with a full sib, (ii) with an unrelated male from the same population, and (iii) with a male from another regional population (Fig. 3). In the PT population, egg viability was very low in females mated with a sib or a same-population male, but very high in females mated with a male from a different population, indicating very strong heterosis and hence high genetic load. In the ÅL population, sib-mated females had lower egg viability than females mated with an unrelated same-population male or with a male from another population, indicating inbreeding depression.
Fig. 3.
Egg-hatching rate (%) in three types of matings in ÅL and PT populations. The mating types are crosses between five regional populations, including ÅL and PT (Materials and Methods; nPT = 11, nÅL = 14 egg clutches), crosses between different families from the same regional population (nPT = 57, nÅL = 85 egg clutches), and sib mating (nPT = 60, nÅL = 58 egg clutches). The lines above the graphs indicate significant differences between pairs of treatments: **P < 0.01; ***P < 0.001.
Adult Morphology.
PT larvae were significantly lighter than ÅL larvae in all stages of development (Tables 2 and 3), and adult PT butterflies were ca. 10% lighter (L = 0.12, average for females and males) than ÅL butterflies (Table 3). Apart from the overall size, there were substantial differences in adult morphology between the two populations (Table 3, full results in Table S3). Age at dissection affected thorax weight negatively in males (PAGE < 0.001) and females (PAGE < 0.001), whereas in the case of abdomen weight mating had an effect in males (PMATING = 0.02) and age and oviposition event had an effect in females (PAGE = 0.01, POVIPOSITION < 0.001). We corrected for these effects while comparing the two populations. Both uncorrected and corrected thorax weights were smaller, especially in PT males (Table 3, Tables S3 and S5, and Fig. S1).
Although PT butterflies were smaller than ÅL butterflies, the front wing area was similar in both populations and hence wing loading was significantly smaller in PT butterflies [wing loading is defined as the ratio of adult weight/wing area, and it is generally positively correlated with acceleration capacity but negatively correlated with sustained flight (42)] (Table 3 and Table S6, PCA C). Comparing wing area with age-corrected thorax weight, there was a significant population–sex interaction, indicating that thorax weight in relation to front wing area was more reduced in PT males than females (Table S3). Average front wing length, width, and perimeter and the areas of large and small triangles within the front wing did not differ between the populations, but there were subtle significant differences in wing shape. PT females had a higher aspect ratio [leaner wings, generally associated with fast flight (42)], whereas in males the aspect ratio was lower in PT butterflies (significant population–sex interaction, Table 3). In a principal component analysis (Table S6, PCA D), the component that is analogous to aspect ratio differed significantly between the populations in both females and males.
Flight and Resting Metabolic Rates.
The flight metabolic rate (FMR) was positively correlated with adult weight in both sexes, whereas butterfly age had a slight negative effect on resting metabolic rate (RMR). While comparing the two populations below, we used residuals from linear regressions accounting for these effects.
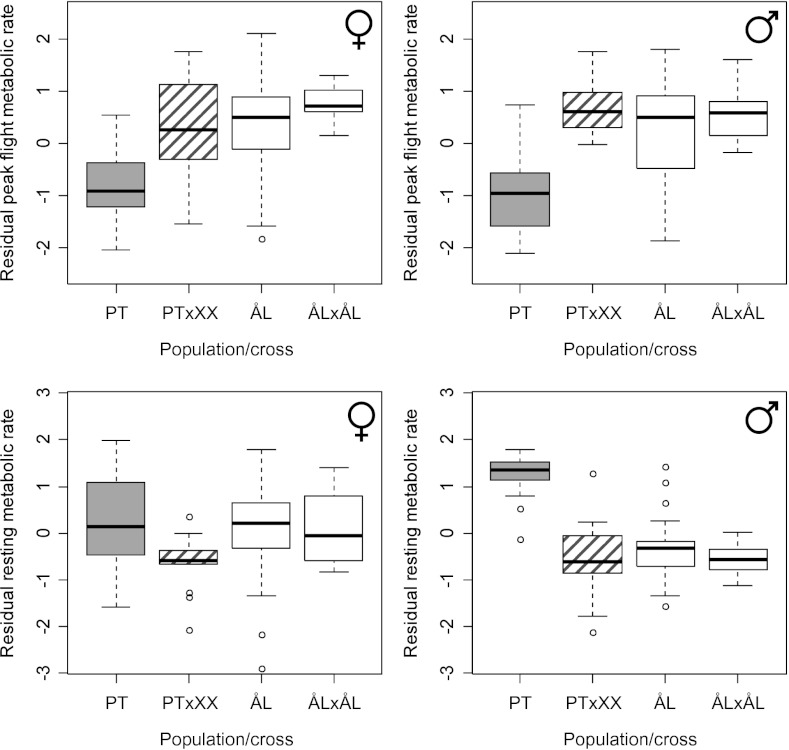
The FMR of PT butterflies was only half of that of ÅL butterflies (average genetic load of FMR measures L = 0.53; Table 3 and Figs. 4 and 5). The difference was evident in both peak and integrated FMR, but was especially notable at the end of the 15-min flight period (Fig. 4 and Table 3), when the PT butterflies became exhausted sooner and more severely than ÅL butterflies. The ÅL males typically exhibited multiple peaks of CO2 emission during the 15-min experiment, but in PT males CO2 emission peaked only in the beginning of the experiment. A principal component analysis supports these conclusions (Table S6, PCA B). There was no difference in the RMR between the populations in females, but in males the difference was highly significant and the reversed of that in FMR, PT males showing almost twice as high RMR than ÅL males (Fig. 5). The ratio FMR/RMR (a measure of metabolic scope) was much higher in ÅL females (15.3) and especially males (35.6) than in PT females (8.1) and males (9.5).
Fig. 4.

The CO2 emission curves during the 15-min flight experiment (including the base line) for five randomly selected females and males from the two populations. Butterflies from PT are represented by thick solid lines and butterflies from ÅL by thin dashed lines.
Fig. 5.
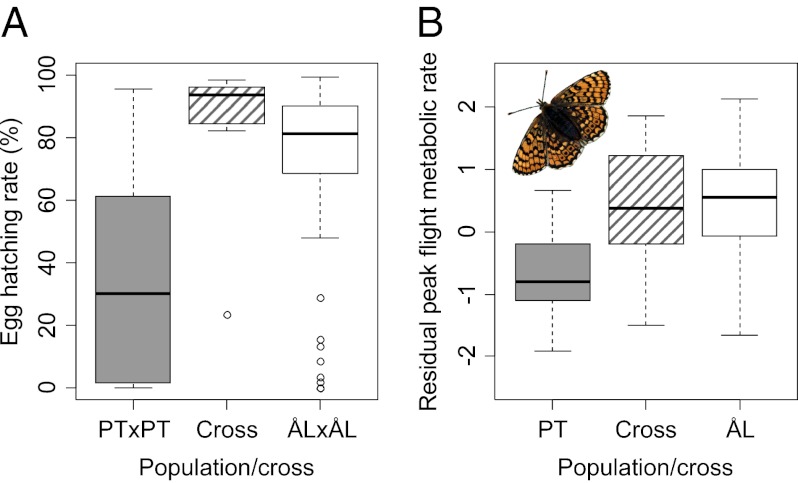
Peak flight metabolic rate (Upper, residual from a linear model including adult weight) in females and males and resting metabolic rate (Lower, residual from a linear model including butterfly age at measurement) in females and males in four lines of butterflies: PT, butterflies reared from field-collected PT larvae (shaded boxes, nfemales = 25, nmales = 25); PT×XX, crosses between PT females and males from other regional populations (boxes with shaded hatched lines, nfemales = 16, nmales = 18); ÅL, butterflies reared from field-collected ÅL larvae (open boxes, nfemales = 25, nmales = 25); and ÅL×ÅL, crosses between different families from the Åland metapopulation (open boxes, nfemales = 9, nmales = 12). The PT×XX and ÅL×ÅL crosses were obtained from the outdoor population cage (Materials and Methods).
Between-population PT crosses showed strong heterosis in metabolic rates (Fig. 5). The offspring of PT females crossed with a male from another regional population [ÅL, Saaremaa, Uppland (Sweden), or Öland (Sweden)] had significantly higher peak FMR than pure-bred PT butterflies [Tukey’s honestly significant difference (HSD), P < 0.001 for both males and females], which was not significantly different from the FMR of ÅL butterflies. The significantly elevated RMR in PT males (Table 3) entirely disappeared in the crosses (Tukey’s HSD, P < 0.001), and the hybrid males had similar RMR to that of ÅL males (Fig. 5).
Discussion
When an isolated population remains small for an extended period, some of the mutations present at the time of isolation become fixed, whereas others become purged (5, 9, 43). At the same time, new detrimental mutations enter the population, and the smaller their effect the higher is the probability of their fixation (44, 45). Given enough time, such a population is likely to show reduced fitness due to high load of fixed mutations, but no reduced fitness upon further inbreeding. The reduction in inbreeding depression is due to increased homozygosity: Fixed loci show no inbreeding depression, whereas a part of the mutations with the strongest deleterious effects have been purged (35, 46, 47). The old isolated population of the Glanville fritillary on the island of PT in the Baltic Sea represents a prime example. First, the fitness of the PT population is reduced to <30% of the fitness of the large reference population in the ÅL (on the basis of lifetime larval production, our best composite measure of fitness, and the expected performance of the larvae following diapause), representing one of the greatest fitness declines reported for natural populations. Second, the PT population has much less genetic variation in microsatellite loci than the ÅL population, consistent with large-scale fixation of alleles by genetic drift. Third, in the PT population most traits were little or not at all affected by one generation of full-sib mating, in contrast to substantial inbreeding depression (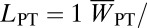 ∼ 0.25) in the ÅL population. Fourth, hybrid offspring between the PT butterflies and individuals from other regional populations showed complete fitness recovery in the two traits studied, egg viability and flight metabolic rate. Finally, we found no clear evidence for local adaptation, which is discussed further in Female Performance. Reduced genetic variation in the isolated population is likely to constrain local adaptation (48).
∼ 0.25) in the ÅL population. Fourth, hybrid offspring between the PT butterflies and individuals from other regional populations showed complete fitness recovery in the two traits studied, egg viability and flight metabolic rate. Finally, we found no clear evidence for local adaptation, which is discussed further in Female Performance. Reduced genetic variation in the isolated population is likely to constrain local adaptation (48).
Examples of fitness reduction in small isolated populations include studies on mammals [Weddell seals (16)], reptiles and amphibians [adders (15, 49), timber rattlesnakes (50), European tree frogs (28, 51), and natterjack toads (52)], fish [Japanese rosy bitterling (53)], aquatic snails (54), and plants [Carex davalliana (55)]. In these studies, the fitness of the isolated population ranged from 30% to 90% of the fitness of the corresponding nonisolated conspecific population, with an average of 65%, although it should be noted that the fitness components and the methods used varied greatly between the studies. These studies examined on average 3.5 fitness components (range from 2 to 6), whereas our study has reported 58 traits (Table S7) for natural populations of the Glanville fritillary. Not all traits studied here are independent of each other, for which reason we summarized variation in morphometric and metabolic traits with principal component analyses. Studying only a part of the life cycle is likely to lead to an underestimate of the true fitness effect (56, 57). In some studies, for instance in the case of the Florida panther (32, 58, 59) and the greater prairie chicken (14), researchers have demonstrated fitness recovery via genetic rescue when individuals from other populations were introduced to isolated populations.
Models predict that mutation accumulation may generate a significant risk of extinction in small populations (5, 44). Lynch et al. (5) concluded that sexual populations with an effective size smaller than 100 individuals are unlikely to persist for more than a few hundred generations, assuming mutation parameters based on Drosophila studies. The theory and empirical results on mutational meltdown are, however, controversial (60), and the impacts depend on the nature of the genetic load (61). Mutational meltdown is more likely to take place with a predominance of mutations of small effect (35). Our results are in agreement with the prediction of mutational meltdown within an order of 100 generations in populations with an effective size of 100 or less. It remains an open question to what extent the high genetic load in our study population is due to fixation (or drift to high frequency) of deleterious mutations present in the population at the time of establishment and to what extent it is due to fixation of newly arisen mutations, although given the relatively short time since colonization, it is likely that the majority of fixed deleterious mutations arise from standing detrimental variation.
Female Performance.
In the case of female reproductive performance, both fecundity (lifetime egg production) and offspring viability were significantly reduced in the PT population in comparison with the reference ÅL population. There was limited inbreeding depression in the PT population, with one notable exception: Inbreeding depression in egg viability was even greater in the PT (δinb = 0.39) than in the ÅL population (δinb = 0.28; average for all clutches, Table 2), although there was also a significant reduction in egg viability in outcrossed females in the PT compared with the ÅL population (L = 0.37), indicating fixation load. Mutations affecting egg viability operate early in the life cycle, and often such mutations are more deleterious than mutations expressed later in the life cycle (62). As purging of highly deleterious mutations is more efficient than purging of mildly deleterious mutations, other things being equal, the high inbreeding depression of egg viability in the PT population is surprising. Other studies have previously reported that purging may not effectively reduce inbreeding depression in all traits (63, 64).
Noninbred females in the ÅL reference population took longer to mate and oviposit and laid smaller egg clutches, when mated to a brother as opposed to an unrelated male. In contrast, there was no such effect in PT females. The parsimonious explanation of these results is kin recognition and inbreeding avoidance in the ÅL population but not in the PT population, in which all individuals are highly related. One previous study found no evidence for kin recognition in the Glanville fritillary (65) but it used different methods from those of the present study.
We did not find any clear evidence for local adaptation in the PT population, although this result does not rule out the possibility of local adaptation in traits not investigated in the present study. Here, we define local adaptation as a trait that is expected to increase, on the basis of biological knowledge, the fitness of its carrier. An adaptive trait may be specific to a certain environment (e.g., low dispersal rate in an isolated island population) or a trait may be adaptive more generally (e.g., high fecundity). There are two female traits in the PT population that at first could be interpreted as indicating local adaptation: reduced FMR and high rate of laying egg clutches. In the Glanville fritillary, FMR and dispersal rate in the field are strongly correlated (66), and hence reduced FMR in PT could be interpreted as being an adaptation to an isolated island, where high dispersal rate is most likely selected against (67, 68). However, the local adaptation hypothesis is not supported by the observation that high FMR has contrasting effects on dispersal propensity in the two sexes; only females with high FMR are more likely to disperse, whereas males with high FMR are actually less dispersive than males with low FMR, but the former benefit from high FMR through their superior capacity to acquire mates in the natal population (69). In the PT population, FMR was similarly reduced in both sexes. Second, in crosses between PT and other populations FMR was at the level of that of the reference ÅL butterflies, which is most parsimoniously explained by heterosis. Third, flight morphology of PT females does not indicate reduced flight ability. PT females had a smaller thorax than ÅL females, but they had a higher wing aspect ratio [associated with fast flight (42)] and smaller wing loading [generally associated with low acceleration capacity but higher capacity for sustained flight (42)] than ÅL females. We therefore suggest that low FMR in PT butterflies is another consequence of large genetic load (see Male Performance for resting metabolic rate in males).
Turning to the larger number of smaller egg clutches laid by PT than by ÅL females, this difference could possibly be an adaptation to spread the risk of entire clutches being parasitized by the specialist parasitoid Cotesia melitaearum, which occurs on PT (Materials and Methods), especially because parasitism is known to increase with the size of larval groups (70). The average rate of parasitism must be higher on PT than in ÅL, because most small local populations in ÅL lack the parasitoid (71). On the other hand, apart from the effect of parasitism by Cotesia, larval survival increases with group size (72), which should select for larger groups. Another hypothesis is that PT butterflies have evolved a faster adult life history, but it is not clear why that would be especially advantageous in their environment, and in fact it is not, as the lifetime number of eggs produced by PT females was only 30% of that produced by ÅL females. Finally, a nonadaptive explanation of the oviposition pattern is that PT females are constrained to lay small egg clutches, which take a shorter time to mature than large clutches, and hence PT females are able to lay the next egg clutch sooner than ÅL females.
Male Performance.
In the case of males, fitness reduction in the PT population is dramatic and leaves little scope for alternative interpretations. At the physiological level, the flight metabolic rate was greatly reduced, as in females. Additionally, the RMR of PT males was twice the rate of ÅL males. Increased RMR may increase the oxidative stress that is associated with elevated energy use. Males in the PT population had an especially short lifespan (Table 3), which can be related to their high RMR (73, 74), although we realize that association between RMR and lifespan is still under debate (75). Studies examining the effect of increased homozygosity (inbreeding) on metabolic rate are scarce (76–78). A study on the cricket Gryllodes sigillatus (only males) found an 80% increase in RMR of inbred crickets over that in outcrossed individuals, along with a slight decrease in metabolic rate during forced exercise (79). Elevated levels of oxidative stress have been reported in inbred Drosophila males (80), and other studies have shown reduced lifespan in populations suffering from inbreeding or genetic load (81, 82).
Reduced mass-specific FMR combined with reduced mass of flight muscles go some way toward explaining the reduced flight activity of PT males in the population cage. Reduced flight activity, in turn, may explain at least partly the reduced mating success of PT males while in competition with males from other populations. Previous studies on insects have demonstrated that reduced flight activity of males decreases their mating success (83, 84). The dramatically reduced egg-hatching rate of the second and subsequent egg clutches in the PT population is probably another indication of low performance of PT males, which may transfer small amounts of or low-quality sperm during matings. In Drosophila simulans, males suffering from inbreeding depression transfer smaller amounts of sperm (80), whereas in the butterfly Bicyclus anynana, egg-hatching rate is more affected by the inbreeding of the male than of the female (85). The general adverse effect of inbreeding on male reproductive performance in insects has been reported for Drosophila melanogaster (80, 86, 87), the butterfly B. anynana (88), and the Glanville fritillary (89).
Conclusion
Our results on the Glanville fritillary indicate a high load of fixed or near-fixed deleterious recessive mutations in an old isolated population. The demographic history and the size of the population place it in the part of the parameter space where models predict the possibility of mutational meltdown. Given the relatively short time since colonization, it is likely that the majority of fixed deleterious mutations originate from standing detrimental variation, whereas the role of newly arisen mutations would increase with time. Our results suggest that the PT population exemplifies the increasingly common situation in human-fragmented landscapes in which small and completely isolated populations are vulnerable to extinction due to fitness decline caused by high genetic load.
Materials and Methods
Study Species and Populations.
PT is a small (1 × 2 km) and very isolated Russian island in the Gulf of Finland, located ∼30 km north of the Estonian coast. The island has a single shoreline meadow of about 10 ha with V. spicata, the larval host plant of the Glanville fritillary. The Glanville fritillary was recorded on the island in 1936 and again in 1994, and it has persisted until the present. The island has been uninhabited by people, but it was frequently visited before WWII by fishermen coming from the nearby larger island of Tytärsaari. Material has been brought to the island from the Estonian coast, most likely leading to accidental introduction of larvae. Introduction at the larval stage is supported by the presence of the specialist braconid parasitoid C. melitaearum, which is an even worse disperser than the butterfly (71) but could have arrived as parasitized host larvae. The PT population was surveyed thoroughly in late August 2009, and 111 larval family groups were detected. Five larvae were sampled per group. For comparison, we sampled three larvae per larval family group from ca. 100 different local populations in the well-studied large metapopulation in the ÅL in Finland (38, 90). The total census size of the ÅL metapopulation is some tens of thousands of individuals (38), but the effective metapopulation size is much smaller, probably of the order of 1,000 individuals. Postdiapause larvae were reared in 2 groups, taken out of diapause either in February (group 1) or in April 2010 (group 2). Details of the rearing conditions are given in SI Materials and Methods. After eclosion, butterflies were sexed and marked individually with a number on the underside of the hind wing. Butterflies from group 1 were assigned to either measurement of flight and resting metabolic rates or an inbreeding experiment. Group 2 butterflies were released 24 h after eclosion into a large outdoor population cage.
Microsatellite Genotyping and Analyses.
DNA was extracted with the NucleoMag 96 Tissue system (Macherey-Nagel) according to the manufacturer’s protocol from 156 unrelated butterflies from the PT and ÅL populations and a population from the large island Saaremaa in Estonia. Samples were genotyped for seven microsatellites, including AM0, AM1, AM2, AM3, and AM4 described by Sarhan (91) and two new ones selected from the genome assembly of the Glanville fritillary (details in SI Materials and Methods). Design of robust microsatellite primers is difficult for Lepidoptera (92), and the microsatellites have often null alleles, due to loss of the longer allele (93). The allele distributions and Fst values were calculated with Arlequin (94) and the within-population dyadic maximum-likelihood inbreeding (F) and relatedness values (denoted as DyadML estimates) with Coancestry (95). The nine-parameter model of inbreeding based on within-population pairwise identity-by-descent (IBD) estimates (96) was used as originally implemented by Wang (97). The DyadML relatedness method can be used in the presence of inbreeding and genotyping errors (96). In Table 1, “gene diversity” refers to the expected heterozygosity over all data, whereas the “expected heterozygosity” is the mean for individual loci [see Arlequin 3.5 documentation, section 8.1. (94)].
Decline in Heterozygosity Since the Colonization.
We conducted simulations to estimate the number of founding individuals of the PT population, using a simulation software similar to that in Gasbarra et al. (98). The simulation is done in two phases. In the first phase, a random pedigree is constructed backward in time, from the youngest generation toward the founders. In the second phase, neutral genetic data for each individual in the pedigree are simulated forward in time. Generations are assumed to be nonoverlapping, consistent with the biology of the Glanville fritillary. The simulation was repeated 100 times and the expected heterozygosity over the seven markers was computed for generations t = 0, 10, … , 120. The mean values for different propagule sizes were compared with the expected heterozygosities for the PT sample (Fig. 1 and Table 1). For details see SI Materials and Methods.
Larval Development and Adult Morphology.
Postdiapause larval development was studied using larvae from group 2. The dates of the larval molts and their weights in the beginning of each molt were recorded individually (Mettler-Toledo XS 105 analytical balance, accuracy 0.01 mg). Pupae were weighed on the day after pupation and the length of the pupal period was recorded. Adult and abdomen weights were measured for 51 PT and 50 ÅL butterflies, adult weight immediately following the measurement of flight metabolic rate (below). Thorax weight was measured for all PT adults (n = 176) and for most ÅL butterflies (n = 106). For details see SI Materials and Methods.
Wings were detached from the thorax, using tweezers (n = 51 and n = 50 for PT and ÅL, respectively). One forewing was chosen to represent each individual. The forewings were photographed against a white background, using a digital camera (Canon PowerShot A710 IS) and a ruler for scale. The image analysis software ImageJ 1.37v (Wayne Rasband, http://rsb.info.nih.gov/ij/) was used for wing measurements (accuracy 0.01 mm). Forewing length, width, perimeter, area, areas within the small and large triangles within the wing, wing loading, and aspect ratio were measured as described in SI Materials and Methods.
Population Cage Experiment.
The large outdoor population cage (32 × 26 × 3 m) covered a dry meadow rich in flowering nectar plants and gardened to resemble the natural habitat of the Glanville fritillary (99, 100). The experimental setup was the same as used by Saastamoinen and Hanski (99). Altogether 207 butterflies (group 2) were released into the cage in June 2010, representing the following regional populations: 13 females and 18 males from PT, 14 and 21 from ÅL, 31 and 27 from Saaremaa, 18 and 26 from Uppland, and 17 and 22 from Öland. Matings and ovipositions occurred naturally by free-flying butterflies and were constantly monitored, along with regular censuses of butterflies in the cage (SI Materials and Methods). Following oviposition, the leaf supporting the egg clutch was collected and the eggs were reared in the laboratory (SI Materials and Methods). The eggs were counted at the age of 3 d. Following egg hatching, the larvae were counted.
Inbreeding Experiment.
An inbreeding experiment was conducted with butterflies from the PT and ÅL populations (group 1) by comparing within-family crosses (full-sib mating, nPT = 12, nÅL = 20) with between-family crosses (nPT = 17, nÅL = 20). Butterflies were mated in small cylindrical net cages (13 × 30 cm) in the laboratory. After mating, the females were isolated and allowed to oviposit on individual potted host plants (SI Materials and Methods). Oviposition rate refers to the number of egg clutches laid per day during the lifetime of a female, including the females that laid no egg clutches. The numbers of eggs and larvae per group were counted. For each female, the first larval group was reared until diapause (SI Materials and Methods). The average larval weight at diapause was measured, and development time and mortality of larvae were recorded. Adult longevity was recorded separately for butterflies that mated in the experiment, those that did not succeed to mate, and those that did not participate in the mating experiment (no opportunity to mate).
Flight and Resting Metabolic Rates.
FMR and RMR were measured for 25 females and 25 males from both populations in 2010. Each individual was chosen from a different family. In the following generation, we measured the metabolic rates of the offspring of PT females mated with males from another regional population (ÅL, Saaremaa, Uppland, or Öland) (16 females and 18 males) and of the offspring of ÅL × ÅL crosses (9 females and 12 males). We could not obtain any PT × PT offspring for these measurements. The flight metabolic rate was measured using flow-through respirometry in the constant temperature of 30 °C [see SI Materials and Methods and Niitepõld et al. (66) for details]. We used the average CO2 emission rate during 60 s of stable baseline before the 15-min flight experiment as a measure of RMR. Peak FMR refers to the maximum rate of CO2 emission during the flight, integrated FMR is the total volume of CO2 produced during the 15-min experiment, and peak FMREND and integrated FMREND represent the peak CO2 emission and the total CO2 volume produced during the last 5 min of the experiment, respectively. Measures of FMR and RMR were corrected for variation in body mass and age, respectively, by using residuals from linear regressions.
Statistical Analyses.
Statistical analyses related to the inbreeding experiment, the population cage experiment, and adult morphology and metabolism were carried out using PASW Statistics 17.0, R Version 64, and R Version 2.10.1 (101), respectively. We used two-way ANOVA to test for the effects of the population of origin (PT or ÅL) and sex on the morphological, physiological, developmental, life-history, and fitness-related traits (details in SI Materials and Methods). Resting metabolic rate and peak flight metabolic rate were compared in the first (field-collected)- and second (laboratory-reared)-generation butterflies (PT × “another population” and ÅL × ÅL crosses, as explained above), using ANOVA and Tukey’s HSD tests. Many of the traits that were included in this study are correlated, and we therefore performed several principal component analyses on traits related to metabolism and morphology. The methods are given in SI Materials and Methods and results in Tables S5 and S6. For traits included in the inbreeding experiment, the difference between average trait values of between-family vs. within-family crosses was tested with a t test. Oviposition rate of the first egg clutch was log transformed; egg hatching rate and larval survival rate were arcsine transformed; and oviposition rate, larval production, larval group size, and mating rate were square-root transformed to make the variables normally distributed. Nonparametric Mann–Whitney U tests gave qualitatively similar results.
Supplementary Material
Acknowledgments
We thank Elena Glazkova for surveying and sampling the PT population; Suvi Ikonen, Coong Lo, Elli Lappalainen, Annukka Ruokolainen, and Toshka Nyman for technical assistance; Christoph Haag, Ilik Saccheri, and two anonymous referees for comments on the manuscript; Kristjan Niitepõld for advice on insect respirometry; Marjo Saastamoinen and Maaike De Jong for discussions; the many research assistants who took part in the population cage experiment and helped to collect and rear the larvae; and the staff of the Lammi Biological Station. A.L.M. was funded by the Finnish School in Wildlife Biology, Conservation, and Management. This study was supported by European Research Council Grant AdG 232826 (to I.H.), the Academy of Finland (Finnish Center of Excellence Programme Grants 131155, 38604, and 44887; to I.H.), and the Lammi Biological Station Foundation of Environmental Research (M.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 14744 (volume 109, number 37).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205789109/-/DCSupplemental.
References
- 1.Caughley G. Directions in conservation biology. J Anim Ecol. 1994;63:215–244. [Google Scholar]
- 2.Lande R. Risks of population extinction from demographic and environmental stochasticity and random catastrophes. Am Nat. 1993;142:911–927. doi: 10.1086/285580. [DOI] [PubMed] [Google Scholar]
- 3.Hanski I. Metapopulation dynamics. Nature. 1998;396:41–49. [Google Scholar]
- 4.Beissinger S, Westphal M. On the use of demographic models of population viability in endangered species management. J Wildl Manage. 1998;62:821–841. [Google Scholar]
- 5.Lynch M, Conery J, Burger R. Mutation accumulation and the extinction of small populations. Am Nat. 1995;146:489–518. [Google Scholar]
- 6.Frankham R. Conservation genetics. Annu Rev Genet. 1995;29:305–327. doi: 10.1146/annurev.ge.29.120195.001513. [DOI] [PubMed] [Google Scholar]
- 7.Lande R. Mutation and conservation. Conserv Biol. 1995;9:782–791. [Google Scholar]
- 8.Glémin S. How are deleterious mutations purged? Drift versus nonrandom mating. Evolution. 2003;57:2678–2687. doi: 10.1111/j.0014-3820.2003.tb01512.x. [DOI] [PubMed] [Google Scholar]
- 9.Hedrick PW. Purging inbreeding depression and the probability of extinction: Full-sib mating. Heredity (Edinb) 1994;73:363–372. doi: 10.1038/hdy.1994.183. [DOI] [PubMed] [Google Scholar]
- 10.Leberg PL, Firmin BD. Role of inbreeding depression and purging in captive breeding and restoration programmes. Mol Ecol. 2008;17:334–343. doi: 10.1111/j.1365-294X.2007.03433.x. [DOI] [PubMed] [Google Scholar]
- 11.Slatkin M. Gene flow in natural populations. Annu Rev Ecol Syst. 1985;16:393–430. [Google Scholar]
- 12.Slatkin M. Gene flow and the geographic structure of natural populations. Science. 1987;236:787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- 13.Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecol Lett. 2004;7:1225–1241. [Google Scholar]
- 14.Westemeier RL, et al. Tracking the long-term decline and recovery of an isolated population. Science. 1998;282:1695–1698. doi: 10.1126/science.282.5394.1695. [DOI] [PubMed] [Google Scholar]
- 15.Madsen T, Stille B, Shine R. Inbreeding depression in an isolated population of adders Vipera berus. Biol Conserv. 1996;75:113–118. [Google Scholar]
- 16.Gelatt TS, et al. History and fate of a small isolated population of Weddell seals at White island, Antarctica. Conserv Genet. 2010;11:721–735. [Google Scholar]
- 17.Crnokrak P, Roff DA. Inbreeding depression in the wild. Heredity (Edinb) 1999;83:260–270. doi: 10.1038/sj.hdy.6885530. [DOI] [PubMed] [Google Scholar]
- 18.Aardema ML, Scriber JM, Hellmann JJ. Considering local adaptation in issues of Lepidopteran conservation-a review and recommendations. Am Midl Nat. 2011;165:294–303. [Google Scholar]
- 19.Lopez S, Rousset F, Shaw FH, Shaw RG, Ronce O. Joint effects of inbreeding and local adaptation on the evolution of genetic load after fragmentation. Conserv Biol. 2009;23:1618–1627. doi: 10.1111/j.1523-1739.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- 20.Whitlock MC, Griswold CK, Peters AD. Compensating for the meltdown: The critical effective size of a population with deleterious and compensatory mutations. Ann Zool Fenn. 2003;40:169–183. [Google Scholar]
- 21.Keightley PD, Otto SP. Interference among deleterious mutations favours sex and recombination in finite populations. Nature. 2006;443:89–92. doi: 10.1038/nature05049. [DOI] [PubMed] [Google Scholar]
- 22.Frankham R. Genetics and extinction. Biol Conserv. 2005;126:131–140. [Google Scholar]
- 23.Meglecz E, Neve G, Pecsenye K, Varga Z. Genetic variations in space and time in Parnassius mnemosyne (L.) (Lepidoptera) populations in north-east Hungary: Implications for conservation. Biol Conserv. 1999;89:251–259. [Google Scholar]
- 24.Joyce DA, Pullin AS. Conservation implications of the distribution of genetic diversity at different scales: A case study using the marsh fritillary butterfly (Euphydryas aurinia) Biol Conserv. 2003;114:453–461. [Google Scholar]
- 25.Williams BL, Brawn JD, Paige KN. Landscape scale genetic effects of habitat fragmentation on a high gene flow species: Speyeria idalia (Nymphalidae) Mol Ecol. 2003;12:11–20. doi: 10.1046/j.1365-294x.2003.01700.x. [DOI] [PubMed] [Google Scholar]
- 26.Vandewoestijne S, Baguette M. Genetic population structure of the vulnerable bog fritillary butterfly. Hereditas. 2004;141:199–206. doi: 10.1111/j.1601-5223.2004.01849.x. [DOI] [PubMed] [Google Scholar]
- 27.Cassel A, Tammaru T. Allozyme variability in central, peripheral and isolated populations of the scarce heath (Coenonympha hero:Lepidoptera, Nymphalidae): Implications for conservation. Conserv Genet. 2003;4:83–93. [Google Scholar]
- 28.Luquet E, et al. Heterozygosity-fitness correlations among wild populations of European tree frog (Hyla arborea) detect fixation load. Mol Ecol. 2011;20:1877–1887. doi: 10.1111/j.1365-294X.2011.05061.x. [DOI] [PubMed] [Google Scholar]
- 29.Ebert D, et al. A selective advantage to immigrant genes in a Daphnia metapopulation. Science. 2002;295:485–488. doi: 10.1126/science.1067485. [DOI] [PubMed] [Google Scholar]
- 30.Vilà C, et al. Rescue of a severely bottlenecked wolf (Canis lupus) population by a single immigrant. Proc Biol Sci. 2003;270:91–97. doi: 10.1098/rspb.2002.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madsen T, Ujvari B, Olsson M. Novel genes continue to enhance population growth in adders (Vipera berus) Biol Conserv. 2004;120:145–147. [Google Scholar]
- 32.Johnson WE, et al. Genetic restoration of the Florida panther. Science. 2010;329:1641–1645. doi: 10.1126/science.1192891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hedrick PW, Fredrickson R. Genetic rescue guidelines with examples from Mexican wolves and Florida panthers. Conserv Genet. 2010;11:615–626. [Google Scholar]
- 34.Tallmon DA, Luikart G, Waples RS. The alluring simplicity and complex reality of genetic rescue. Trends Ecol Evol. 2004;19:489–496. doi: 10.1016/j.tree.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Glémin S, Ronfort J, Bataillon T. Patterns of inbreeding depression and architecture of the load in subdivided populations. Genetics. 2003;165:2193–2212. doi: 10.1093/genetics/165.4.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paland S, Schmid B. Population size and the nature of genetic load in Gentianella germanica. Evolution. 2003;57:2242–2251. doi: 10.1111/j.0014-3820.2003.tb00236.x. [DOI] [PubMed] [Google Scholar]
- 37.Hanski I. Eco-evolutionary spatial dynamics in the Glanville fritillary butterfly. Proc Natl Acad Sci USA. 2011;108:14397–14404. doi: 10.1073/pnas.1110020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanski I. Metapopulation Ecology. Oxford: Oxford University Press; 1999. pp. 205–265. [Google Scholar]
- 39.Ovaskainen O, et al. Tracking butterfly movements with harmonic radar reveals an effect of population age on movement distance. Proc Natl Acad Sci USA. 2008;105:19090–19095. doi: 10.1073/pnas.0802066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartl DL, Clark AG. Principles of Population Genetics. 4th Ed. Sunderland, MA: Sinauer; 2007. pp. 121–128. [Google Scholar]
- 41.Austin A, Ovaskainen O, Hanski I. Size and genetic composition of the colonizing propagules in a butterfly metapopulation. Oikos. 2011;120:1357–1365. [Google Scholar]
- 42.Berwaerts K, Van Dyck H, Aerts P. Does flight morphology relate to flight performance? An experimental test with the butterfly Pararge aegeria. Funct Ecol. 2002;16:484–491. [Google Scholar]
- 43.Wang JL. Effects of population structures and selection strategies on the purging of inbreeding depression due to deleterious mutations. Genet Res. 2000;76:75–86. doi: 10.1017/s0016672399004450. [DOI] [PubMed] [Google Scholar]
- 44.Lande R. Risk of population extinction from fixation of new deleterious mutations. Evolution. 1994;48:1460–1469. doi: 10.1111/j.1558-5646.1994.tb02188.x. [DOI] [PubMed] [Google Scholar]
- 45.Whitlock MC, Ingvarsson PK, Hatfield T. Local drift load and the heterosis of interconnected populations. Heredity (Edinb) 2000;84:452–457. doi: 10.1046/j.1365-2540.2000.00693.x. [DOI] [PubMed] [Google Scholar]
- 46.Keller LF, Waller DM. Inbreeding effects in wild populations. Trends Ecol Evol. 2002;17:230–241. [Google Scholar]
- 47.Roze D, Rousset F. Joint effects of self-fertilization and population structure on mutation load, inbreeding depression and heterosis. Genetics. 2004;167:1001–1015. doi: 10.1534/genetics.103.025148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitlock MC. Selection, load and inbreeding depression in a large metapopulation. Genetics. 2002;160:1191–1202. doi: 10.1093/genetics/160.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Madsen T, Shine R, Olsson M, Wittzell H. Conservation biology - restoration of an inbred adder population. Nature. 1999;402:34–35. [Google Scholar]
- 50.Clark RW, Marchand MN, Clifford BJ, Stechert R, Stephens S. Decline of an isolated timber rattlesnake (Crotalus horridus) population: Interactions between climate change, disease, and loss of genetic diversity. Biol Conserv. 2011;144:886–891. [Google Scholar]
- 51.Luquet E, et al. Consequences of genetic erosion on fitness and phenotypic plasticity in European tree frog populations (Hyla arborea) J Evol Biol. 2011;24:99–110. doi: 10.1111/j.1420-9101.2010.02138.x. [DOI] [PubMed] [Google Scholar]
- 52.Rowe G, Beebee TJ. Population on the verge of a mutational meltdown? Fitness costs of genetic load for an amphibian in the wild. Evolution. 2003;57:177–181. doi: 10.1111/j.0014-3820.2003.tb00228.x. [DOI] [PubMed] [Google Scholar]
- 53.Kawamura K. Low genetic variation and inbreeding depression in small isolated populations of the Japanese rosy bitterling, Rhodeus ocellatus kurumeus. Zoolog Sci. 2005;22:517–524. doi: 10.2108/zsj.22.517. [DOI] [PubMed] [Google Scholar]
- 54.Puurtinen M, Knott KE, Suonpää S, van Ooik T, Kaitala V. Genetic variability and drift load in populations of an aquatic snail. Evolution. 2004;58:749–756. doi: 10.1111/j.0014-3820.2004.tb00408.x. [DOI] [PubMed] [Google Scholar]
- 55.Hooftman DAP, van Kleunen M, Diemer M. Effects of habitat fragmentation on the fitness of two common wetland species, Carex davalliana and Succisa pratensis. Oecologia. 2003;134:350–359. doi: 10.1007/s00442-002-1096-0. [DOI] [PubMed] [Google Scholar]
- 56.Charlesworth D, Charlesworth B. Inbreeding depression and its evolutionary consequences. Annu Rev Ecol Syst. 1987;18:237–268. [Google Scholar]
- 57.Szulkin M, Garant D, McCleery RH, Sheldon BC. Inbreeding depression along a life-history continuum in the great tit. J Evol Biol. 2007;20:1531–1543. doi: 10.1111/j.1420-9101.2007.01325.x. [DOI] [PubMed] [Google Scholar]
- 58.Barone MA, et al. Reproductive characteristics of male Florida panthers - Comparative-studies from Florida, Texas, Colorado, Latin-America, and North-American zoos. J Mammal. 1994;75:150–162. [Google Scholar]
- 59.Hedrick PW. Gene flow and genetic restoration - The Florida panther as a case-study. Conserv Biol. 1995;9:996–1007. doi: 10.1046/j.1523-1739.1995.9050988.x-i1. [DOI] [PubMed] [Google Scholar]
- 60.Charlesworth D, Morgan M, Charlesworth B. Mutation accumulation in finite outbreeding and inbreeding populations. Genet Res. 1993;61:39–56. [Google Scholar]
- 61.Garcia-Dorado A. Tolerant versus sensitive genomes: The impact of deleterious mutation on fitness and conservation. Conserv Genet. 2003;4:311–324. [Google Scholar]
- 62.Husband B, Schemske D. Evolution of the magnitude and timing of inbreeding depression in plants. Evolution. 1996;50:54–70. doi: 10.1111/j.1558-5646.1996.tb04472.x. [DOI] [PubMed] [Google Scholar]
- 63.Boakes EH, Wang J, Amos W. An investigation of inbreeding depression and purging in captive pedigreed populations. Heredity (Edinb) 2007;98:172–182. doi: 10.1038/sj.hdy.6800923. [DOI] [PubMed] [Google Scholar]
- 64.Grueber CE, Laws RJ, Nakagawa S, Jamieson IG. Inbreeding depression accumulation across life-history stages of the endangered Takahe. Conserv Biol. 2010;24:1617–1625. doi: 10.1111/j.1523-1739.2010.01549.x. [DOI] [PubMed] [Google Scholar]
- 65.Haikola S, Singer MC, Pen I. Has inbreeding depression led to avoidance of sib mating in the Glanville fritillary butterfly (Melitaea cinxia)? Evol Ecol. 2004;18:113–120. [Google Scholar]
- 66.Niitepõld K, et al. Flight metabolic rate and Pgi genotype influence butterfly dispersal rate in the field. Ecology. 2009;90:2223–2232. doi: 10.1890/08-1498.1. [DOI] [PubMed] [Google Scholar]
- 67.Ronce O. How does it feel to be like a rolling stone? Ten questions about dispersal evolution. Annu Rev Ecol Evol Syst. 2007;38:231–253. [Google Scholar]
- 68.Hanski I, Mononen T. Eco-evolutionary dynamics of dispersal in spatially heterogeneous environments. Ecol Lett. 2011;14:1025–1034. doi: 10.1111/j.1461-0248.2011.01671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Niitepõld K, Mattila ALK, Harrison PJ, Hanski I. Flight metabolic rate has contrasting effects on dispersal in the two sexes of the Glanville fritillary butterfly. Oecologia. 2011;165:847–854. doi: 10.1007/s00442-010-1886-8. [DOI] [PubMed] [Google Scholar]
- 70.Lei GC, Camara MD. Behaviour of a specialist parasitoid, Cotesia melitaearum: From individual behaviour to metapopulation processes. Ecol Entomol. 1999;24:59–72. [Google Scholar]
- 71.van Nouhuys S, Hanski I. Colonization rates and distances of a host butterfly and two specific parasitoids in a fragmented landscape. J Anim Ecol. 2002;71:639–650. [Google Scholar]
- 72.Kuussaari M, van Nouhuys S, Hellmann JJ, Singer MC. In: On the Wings of Checkerspots: A Model System for Population Biology. Ehrlich PR, Hanski I, editors. New York: Oxford Univ Press; 2004. pp. 138–160. [Google Scholar]
- 73.Nilsson JA. Metabolic consequences of hard work. Proc Biol Sci. 2002;269:1735–1739. doi: 10.1098/rspb.2002.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alonso-Alvarez C, et al. Increased susceptibility to oxidative stress as a proximate cost of reproduction. Ecol Lett. 2004;7:363–368. [Google Scholar]
- 75.Hulbert AJ, Pamplona R, Buffenstein R, Buttemer WA. Life and death: Metabolic rate, membrane composition, and life span of animals. Physiol Rev. 2007;87:1175–1213. doi: 10.1152/physrev.00047.2006. [DOI] [PubMed] [Google Scholar]
- 76.Rantala MJ, Roff DA. Analysis of the importance of genotypic variation, metabolic rate, morphology, sex and development time on immune function in the cricket, Gryllus firmus. J Evol Biol. 2006;19:834–843. doi: 10.1111/j.1420-9101.2005.01048.x. [DOI] [PubMed] [Google Scholar]
- 77.Mikkelsen K, Loeschcke V, Kristensen TN. Trait specific consequences of fast and slow inbreeding: Lessons from captive populations of Drosophila melanogaster. Conserv Genet. 2010;11:479–488. [Google Scholar]
- 78.Clare MR. 1925 A study of oxygen metabolism in Drosophila melanogaster (ProQuest Dissertations and Theses) University of Pennsylvania, Philadelphia, Pennsylvania. [Google Scholar]
- 79.Ketola T, Kotiaho JS. Inbreeding, energy use and condition. J Evol Biol. 2009;22:770–781. doi: 10.1111/j.1420-9101.2009.01689.x. [DOI] [PubMed] [Google Scholar]
- 80.Okada K, Blount JD, Sharma MD, Snook RR, Hosken DJ. Male attractiveness, fertility and susceptibility to oxidative stress are influenced by inbreeding in Drosophila simulans. J Evol Biol. 2011;24:363–371. doi: 10.1111/j.1420-9101.2010.02170.x. [DOI] [PubMed] [Google Scholar]
- 81.van Oosterhout C, Zijlstra WG, van Heuven MK, Brakefield PM. Inbreeding depression and genetic load in laboratory metapopulations of the butterfly Bicyclus anynana. Evolution. 2000;54:218–225. doi: 10.1111/j.0014-3820.2000.tb00022.x. [DOI] [PubMed] [Google Scholar]
- 82.Fox CW, Stillwell RC. Environmental effects on sex differences in the genetic load for adult lifespan in a seed-feeding beetle. Heredity (Edinb) 2009;103:62–72. doi: 10.1038/hdy.2009.31. [DOI] [PubMed] [Google Scholar]
- 83.Watt WB, Donohue K, Carter PA. Adaptation at specific loci. 6. Divergence vs parallelism of polymorphic allozymes in molecular function and fitness-component effects among Colias species (Lepidoptera, Pieridae) Mol Biol Evol. 1996;13:699–709. [Google Scholar]
- 84.De Block M, Stoks R. Flight-related body morphology shapes mating success in a damselfly. Anim Behav. 2007;74:1093–1098. [Google Scholar]
- 85.Saccheri IJ, Lloyd HD, Helyar SJ, Brakefield PM. Inbreeding uncovers fundamental differences in the genetic load affecting male and female fertility in a butterfly. Proc Biol Sci. 2005;272:39–46. doi: 10.1098/rspb.2004.2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sharp PM. The effect of inbreeding on competitive male-mating ability in Drosophila melanogaster. Genetics. 1984;106:601–612. doi: 10.1093/genetics/106.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Enders LS, Nunney L. Sex-specific effects of inbreeding in wild-caught Drosophila melanogaster under benign and stressful conditions. J Evol Biol. 2010;23:2309–2323. doi: 10.1111/j.1420-9101.2010.02085.x. [DOI] [PubMed] [Google Scholar]
- 88.Joron M, Brakefield PM. Captivity masks inbreeding effects on male mating success in butterflies. Nature. 2003;424:191–194. doi: 10.1038/nature01713. [DOI] [PubMed] [Google Scholar]
- 89.Haikola S, et al. Inbreeding depression and the maintenance of genetic load in Melitaea cinxia metapopulations. Conserv Genet. 2001;2:325–335. [Google Scholar]
- 90.Nieminen M, Siljander M, Hanski I. In: On the Wings of Checkerspots: A Model System for Population Biology. Ehrlich PR, Hanski I, editors. New York: Oxford Univ Press; 2004. pp. 63–91. [Google Scholar]
- 91.Sarhan A. Isolation and characterization of five microsatellite loci in the Glanville fritillary butterfly (Melitaea cinxia) Mol Ecol Notes. 2006;6:163–164. [Google Scholar]
- 92.Tay WT, Behere GT, Batterham P, Heckel DG. Generation of microsatellite repeat families by RTE retrotransposons in lepidopteran genomes. BMC Evol Biol. 2010;10:144–159. doi: 10.1186/1471-2148-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Orsini L, Corander J, Alasentie A, Hanski I. Genetic spatial structure in a butterfly metapopulation correlates better with past than present demographic structure. Mol Ecol. 2008;17:2629–2642. doi: 10.1111/j.1365-294X.2008.03782.x. [DOI] [PubMed] [Google Scholar]
- 94.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 95.Wang J. COANCESTRY: A program for simulating, estimating and analysing relatedness and inbreeding coefficients. Mol Ecol Resour. 2011;11:141–145. doi: 10.1111/j.1755-0998.2010.02885.x. [DOI] [PubMed] [Google Scholar]
- 96.Milligan BG. Maximum-likelihood estimation of relatedness. Genetics. 2003;163:1153–1167. doi: 10.1093/genetics/163.3.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang J. Triadic IBD coefficients and applications to estimating pairwise relatedness. Genet Res. 2007;89:135–153. doi: 10.1017/S0016672307008798. [DOI] [PubMed] [Google Scholar]
- 98.Gasbarra D, Sillanpää MJ, Arjas E. Backward simulation of ancestors of sampled individuals. Theor Popul Biol. 2005;67:75–83. doi: 10.1016/j.tpb.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 99.Saastamoinen M, Hanski I. Genotypic and environmental effects on flight activity and oviposition in the Glanville fritillary butterfly. Am Nat. 2008;171:701–712. doi: 10.1086/587531. [DOI] [PubMed] [Google Scholar]
- 100.Hanski I, Saastamoinen M, Ovaskainen O. Dispersal-related life-history trade-offs in a butterfly metapopulation. J Anim Ecol. 2006;75:91–100. doi: 10.1111/j.1365-2656.2005.01024.x. [DOI] [PubMed] [Google Scholar]
- 101.R Development Core Team R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna) 2009. Available at http://www.R-project.org. Accessed Dec. 14, 2009.