Abstract
Bisphenol A (BPA) is an industrial compound and a well known endocrine-disrupting chemical with estrogenic activity. The widespread exposure of individuals to BPA is suspected to affect a variety of physiological functions, including reproduction, development, and metabolism. Here we report that the mechanisms by which BPA and two congeners, bisphenol AF and bisphenol C (BPC), bind to and activate estrogen receptors (ER) α and β differ from that used by 17β-estradiol. We show that bisphenols act as partial agonists of ERs by activating the N-terminal activation function 1 regardless of their effect on the C-terminal activation function 2, which ranges from weak agonism (with BPA) to antagonism (with BPC). Crystallographic analysis of the interaction between bisphenols and ERs reveals two discrete binding modes, reflecting the different activities of compounds on ERs. BPA and 17β-estradiol bind to ERs in a similar fashion, whereas, with a phenol ring pointing toward the activation helix H12, the orientation of BPC accounts for the marked antagonist character of this compound. Based on structural data, we developed a protocol for in silico evaluation of the interaction between bisphenols and ERs or other members of the nuclear hormone receptor family, such as estrogen-related receptor γ and androgen receptor, which are two known main targets of bisphenols. Overall, this study provides a wealth of tools and information that could be used for the development of BPA substitutes devoid of nuclear hormone receptor-mediated activity and more generally for environmental risk assessment.
Keywords: crystal structure, endocrine disruptor, nuclear receptor, virtual screening
Bisphenols form a large family of chemicals that are commonly used in the manufacture of numerous consumer products. By far, the most widely used bisphenol (>3 million tons/y) is bisphenol A (BPA; 4-[2-(4-hydroxyphenyl)propan-2-yl]phenol), which is used in the manufacture of items such as plastics, food can linings, dentistry sealants, and thermal paper. Many other bisphenols are used in a variety of industrial applications, e.g., bisphenol AF (BPAF; 4-[1,1,1,3,3,3-hexafluoro-2-(4-hydroxyphenyl)propan-2-yl]phenol) in the fabrication of electronic materials, gas-permeable membranes, and plastic optical fibers, or bisphenol C (BPC; 4-[2,2-dichloro-1-(4-hydroxyphenyl)ethenyl]phenol) in the manufacture of fire-resistant polymers (Fig. 1A). Several studies have shown that BPA is released from consumer products, leading to its detection in food, drinking water, wastewater, air, and dust (1). Others studies have identified BPA in human serum, urine, adipose and placental tissues, and umbilical cord blood (2, 3). The major source of consumer exposure is likely to be through food and drinks in contact with BPA-containing materials (1), although a recent study has shown that BPA can be also absorbed by the skin (4). Finally, BPA is a significant contaminant of wastewater and biosolids from sewage treatment plants, which may affect wildlife at environmentally relevant concentrations (5). BPA has been shown to produce a range of adverse effects in laboratory animals, with major concerns regarding reproductive targets and embryonic development (6–8). More recently, it has been hypothesized that early exposure to BPA could also play a role in the onset of obesity and other metabolic syndromes (9). In this regard, a large body of data about endocrine-disrupting chemicals (EDCs) underlines the importance of exposure during early stages of development, which could result in numerous biological defects in adult life (10).
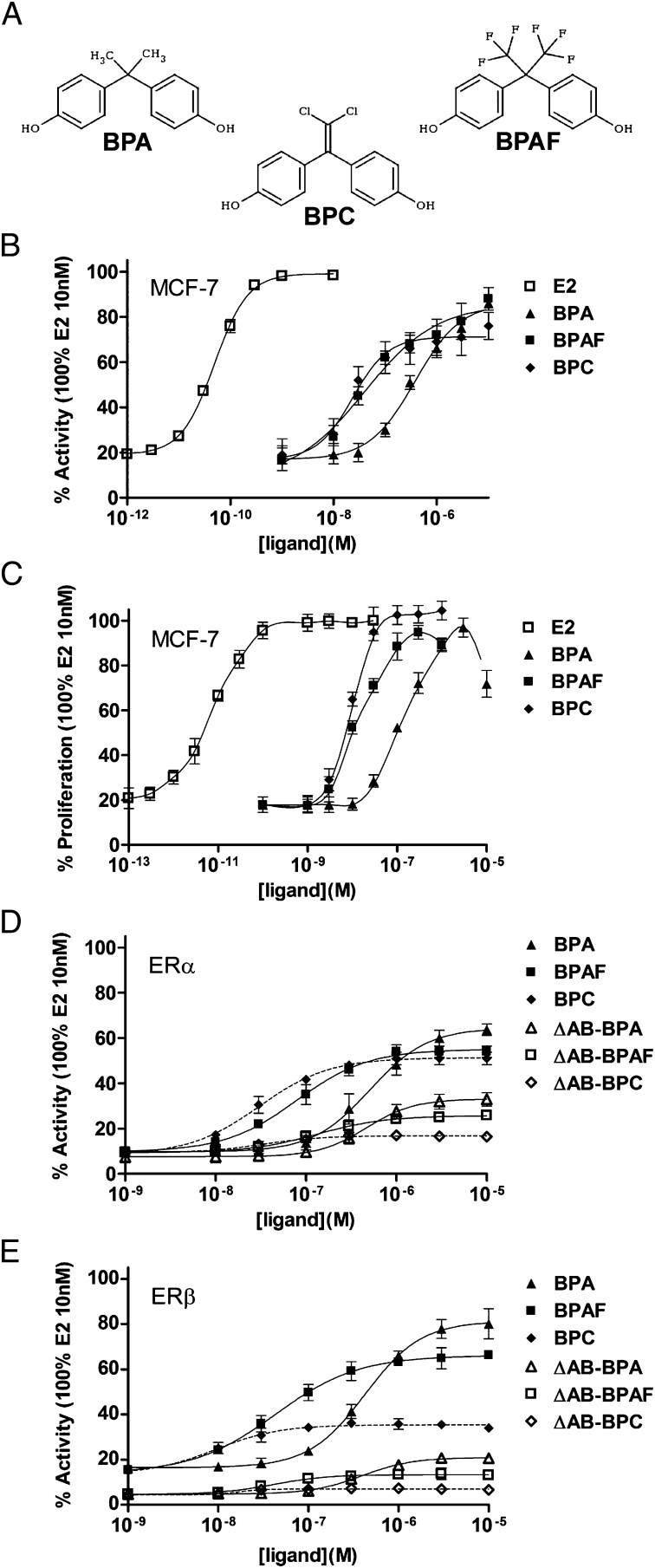
Fig. 1.
Dose–response curve for bisphenols in reporter cell lines. (A) Chemical structures of some bisphenols used in this study. (B) MELN, (D) HELN-ERα and -ΔAB-ERα, and (E) HELN-ERβ and -ΔAB-ERβ luciferase assays of BPA, BPAF, and BPC. (C) Proliferative response of BPA, BPAF, and BPC in MELN cells. The maximal luciferase and proliferation activity (100%) was obtained with 10 nM E2. Values are the mean ± SD from three separate experiments.
The molecular basis behind the deleterious effects of BPA is poorly understood, and a large controversy has been created within the field of endocrine disruption about low doses’ effects and the possible consequences of such exposures (11, 12). Although the two estrogen receptors (ERs), ERα and ERβ, are considered as the main targets of BPA (13, 14), several other cellular targets have been proposed for this compound. We and others have previously demonstrated that BPA or its halogenated derivatives also activate the pregnane X receptor (15, 16), the estrogen-related receptor γ (ERRγ) (17), or the peroxisome proliferator activated receptor γ (18, 19), and inhibit the androgen receptor (AR) (20) or the thyroid hormone receptor (21, 22). BPA has also been reported to interact with the G protein-coupled ER (23), so that the net effect of BPA could be caused by synergistic actions through different pathways.
ERα and ERβ are members of the nuclear hormone receptor (NR) family, acting as ligand-inducible transcription factors (24, 25). Their activity is regulated by 17β-estradiol (E2), which plays important roles in the growth and maintenance of a diverse range of tissues such as the mammary gland, uterus, bone, and cardiovascular and central nervous systems. The interaction of E2 with ERs initiates a series of molecular events, including the recruitment of members of the steroid receptor coactivator (SRC) family that culminate in the transcription of target genes (26). Like other members of this family, ERs contain three major functional domains, including an N-terminal A/B domain that harbors a transcriptional activation function (AF-1), a DNA-binding domain, and a C-terminal ligand-binding domain (LBD) hosting a ligand-dependent transcriptional activation function (AF-2). The LBD is crucially involved in most of the receptor functions because of its capacity of hormone binding, dimerization, and interaction with coregulatory complexes. The LBD also contributes to the modulation of the N-terminal AF-1 through interdomain crosstalk so that both AF-1 and AF-2 domains can recruit a range of coregulatory proteins and act individually or in a synergistic manner (27, 28). The precise structural basis of this interdomain communication is unknown, as no 3D structure of an entire NR has been obtained.
In contrast, many crystal structures of ER LBDs have been determined in complex with natural and synthetic ligands, revealing a conserved core of 12 α-helices (H1–H12) arranged into a three-layered sandwich fold (29–31). This arrangement generates a mostly hydrophobic cavity in the lower half of the domain to which hydrophobic ligands bind. In all hormone-bound structures, the ligand-binding cavity is sealed by the C-terminal helix H12. This conformation is specifically induced by the binding of hormones or synthetic agonists and is referred to as the active conformation because it favors the recruitment of coactivators to the so-called AF-2 surface. This surface formed by helices H3, H4, and H12 defines a hydrophobic binding groove for short LxxLL helical motifs found in coactivators.
Here we report on a study in which we combine biochemical, biophysical, structural, and cell-based assays to provide insights how BPA and two derivatives, BPAF and BPC, bind to and activate ERs. Based on these data, we have built a computational tool to predict the ER binding and activation properties of bisphenols and further extended this bioinformatic approach to ERRγ and AR, which are two known main targets of bisphenols.
Results and Discussion
Bisphenols Are Partial Activators of ERs and Potent Inducers of Cell Proliferation.
The agonistic potential of bisphenols (Fig. 1A) was monitored on ERα transcriptional activity and cell proliferation by using breast cancer ERα-positive MCF-7 reporter cells (MELN cell line) (32). In these cells, bisphenols exert a partial potency on luciferase reporter activity (Fig. 1B) but act as full ERα agonists on cell growth (Fig. 1C). We then monitored the effect of bisphenols on ERs transcriptional activity by using HeLa reporter cells stably expressing human ERα and ERβ (HELN ERα and ERβ lines) (32), allowing for a direct comparison of the effect of compounds on the two ER subtypes in a similar cellular context. As shown in Fig. 1D, bisphenols exhibit almost similar activation capabilities of ERα, inducing 60% to 70% of the transactivation seen with E2 (SI Appendix, Fig. S1). In contrast, the activation curves obtained with ERβ show different profiles, with BPA being the most potent (80% activity) and BPC inducing only a 35% activity (Fig. 1E). Accordingly, bisphenols act as partial antagonists in the presence of E2 (SI Appendix, Fig. S2). Transactivation assays (Fig. 1 D and E) suggested that BPAF and BPC bind more avidly to both receptors than BPA. This observation was validated by competitive binding assays with [3H]E2 (SI Appendix, Fig. S3). Together, these experiments show that bisphenols can be considered as selective ER modulators (SERMs) (24) that partially activate luciferase reporter in MCF-7 and HeLa cells while being fully active on MCF-7 cell proliferation.
AF-1 of ERs Is Indispensable for Bisphenol Activity.
Having characterized the estrogenic potential of BPA, BPAF, and BPC, we performed additional cell-based experiments aimed at assessing the relative contribution of the ERs AF-1 and AF-2 to this activity. We first examined the agonistic properties of bisphenols by using HELN cells stably transfected with ERs deleted of their N-terminal AB (AF-1) region (ΔAB-ERα and ΔAB-ERβ) (32). Interestingly, deletion of the AB domain strongly reduces the bisphenol-induced transcriptional activity of ERα and ERβ (Fig. 1 D and E), so that, in the presence of E2, BPC displays an almost full antagonistic activity (SI Appendix, Fig. S2). Next, we examined if the same phenomenon could be observed in a cellular response. For this purpose, we used the HELN ER cell lines, whose proliferation is known to decrease upon E2 treatment (32). In agreement with transcription data, bisphenols inhibit the proliferation of HELN ER cells as efficiently as E2 (SI Appendix, Fig. S4 A and B), but display a very weak efficacy in HELN ΔAB-ERs cells (SI Appendix, Fig. S4 C and D). This is in contrast with E2, whose inhibition properties remain unaffected upon deletion of the ER AB domains (SI Appendix, Fig. S4). The partial agonism of bisphenols is also observed in HeLa cells transiently transfected with another E2-regulated gene (pS2 promoter-luciferase; SI Appendix, Fig. S5), as well as on the expression of the ER target gene GREB1 in HELN ERs cells (SI Appendix, Fig. S6). In HeLa cells, bisphenols clearly act as partial agonists compared with E2. In contrast, in MCF-7 cells, expression of GREB1 and other endogenous E2-regulated genes (pS2, RIP140, and progesterone receptor) is fully activated by bisphenols (SI Appendix, Fig. S7). These data reveal that bisphenols act as SERMs whose activity relies mostly on the AF-1 and depends on the cellular context. The ranking order of potency, with BPA most potent, followed by BPAF and then BPC (Fig. 1 D and E), likely originates from the differential synergy between AF-1 and AF-2 created by the various bisphenols.
Bisphenols Render H12 Highly Dynamic and Disable AF-2.
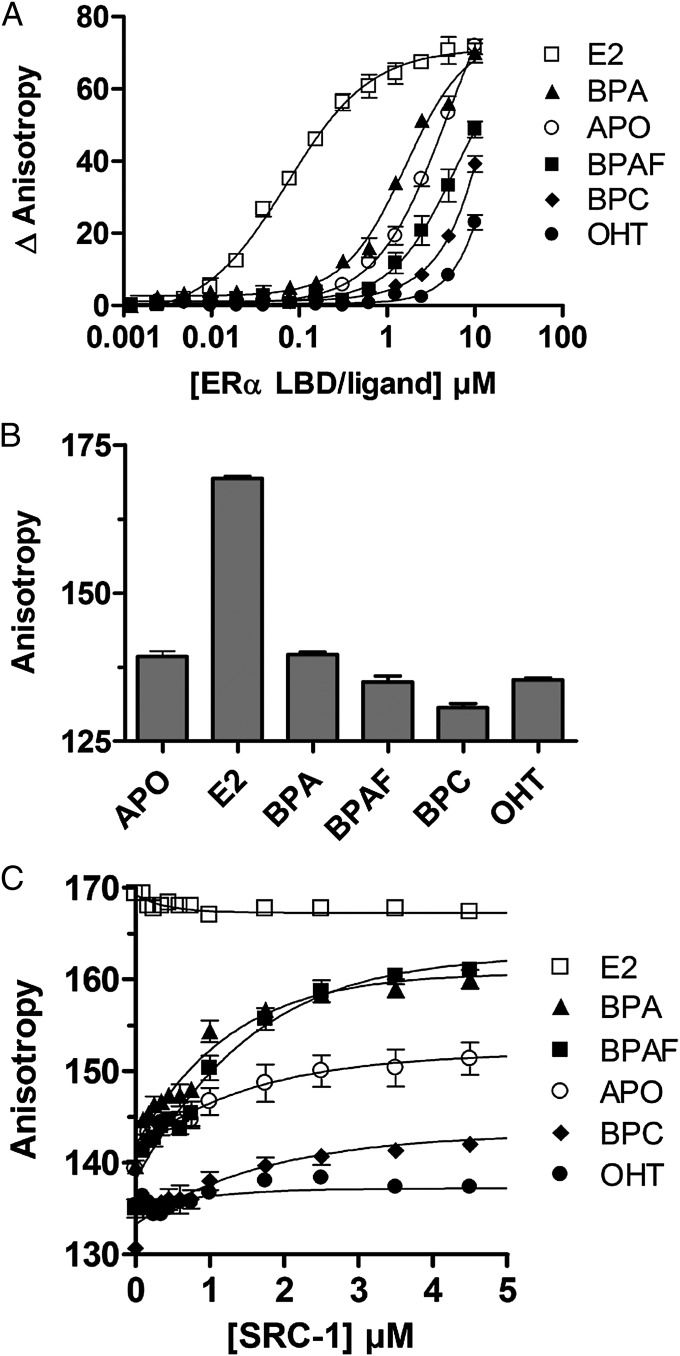
To further characterize the capacity of bisphenols to induce the recruitment of coactivators to the AF-2 surface of ERα, we studied their effects on the interaction of the fluorescein-labeled NR box2 peptide of SRC-1 (SRC-1 NR2) with ERα LBD by fluorescence anisotropy (Fig. 2A). In keeping with their respective agonistic or antagonistic activities, E2 and 4-hydroxytamoxifen (OHT), respectively, strongly enhance and decrease the binding affinity of SRC-1 NR2 (apparent Kd values are 0.07 ± 0.01 μM, 4.95 ± 1.12 μM, and >10 μM for E2-, apo-, and OHT-ERα LBDs, respectively). In contrast, we observed a weaker impact of bisphenols on the interaction with the coactivator-derived peptide and a progressive transition from weak agonist (BPA; Kd = 1.61 ± 0.57 μM) to antagonist (BPC; Kd > 8 μM).
Fig. 2.
Bisphenol-induced coactivator recruitment and structural dynamics. (A) Titration of fluorescein-labeled SRC-1 NR2 peptide by ERα LBD in the absence of ligand or in the presence of E2 (agonist), OHT (antagonist), BPA, BPAF, or BPC. (B) Anisotropy measurements of fluorescein-labeled ERα LBD in the presence of saturating concentrations of bisphenols, E2, or OHT. (C) Similar experiments performed in the presence of increasing concentrations of the coactivator-derived peptide SRC-1 NR2.
As previously reported with RXR (33), we used fluorescence anisotropy measurements of a fluorescein moiety attached to the C terminus of ERα LBD to monitor the effect of bisphenols on H12 dynamics. We showed that anisotropy is strongly enhanced upon addition of E2, reflecting the stabilization of ERα H12 in the active conformation (Fig. 2B). In contrast, binding of bisphenols or OHT slightly (BPA and BPAF) or markedly (BPC and OHT) decreases anisotropy, revealing a higher mobility of H12 in the presence of these compounds. These data fully support the aforementioned results and suggest that bisphenols fail to efficiently stabilize the active receptor conformation, implying that they may act as weak AF-2 agonists or antagonists. To unambiguously characterize the functional profile of bisphenols, we monitored H12 dynamics in the various bisphenol-bound ERα complexes and in the presence of increasing concentrations of unlabeled SRC-1 NR2. Interestingly, addition of SRC-1 NR2 caused a clear dose-dependent anisotropy increase of the ERα LBD bound to BPA and BPAF, indicating that peptide binding helps reducing H12 mobility by shifting the equilibrium toward the active conformation (Fig. 2C). In contrast, even high doses of SRC-1 NR2 failed to stabilize H12 in the presence of BPC or OHT, supporting the notion that, like OHT, BPC acts as an AF-2 antagonist, preventing coactivator binding to ERα LBD. Taken as a whole, these data support the aforementioned cell-based assays and reveal that bisphenols fail to efficiently stabilize the proper LBD interaction surface with coactivators. However, the graded effect of these compounds on H12 dynamics accounts for their differential impact on coactivator recruitment. BPA and BPAF allow some interaction, provided that coactivators are present in sufficient amount in the cellular environment, whereas BPC permanently prevents any interaction of ERα LBD with coactivators.
Bisphenols Interact with ER via Two Binding Modes.
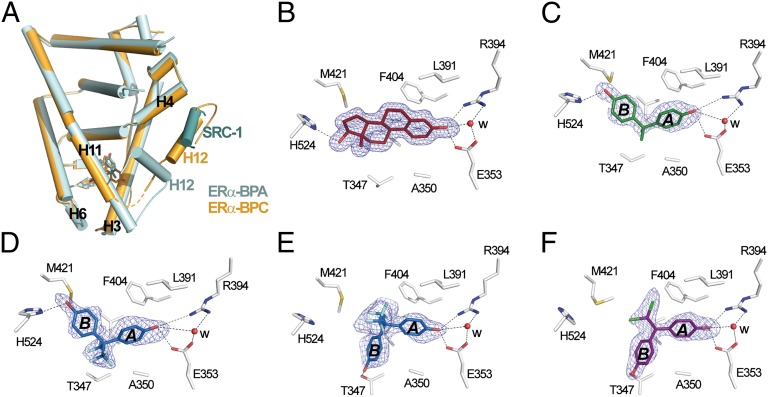
In an attempt to gain structural insight into the binding mode of BPA to ERs, we subjected the WT ERα LBD complexed with BPA to crystallization assays. After several rounds of unsuccessful trials, we used the recently reported ERα LBD mutant (Y537S), which has been shown to stabilize the agonist-bound conformation of ERα, and, in turn, facilitate crystallization of weak agonists (34). To ensure that this mutation at the surface of the protein will not compromise the accuracy of our structural analysis, we cocrystallized ERα-Y537S LBD with E2 and SRC-1 NR2. Comparison of the obtained structure with that of the corresponding WT receptor [Protein Data Bank (PDB) code 1GWR] indicated a very high degree of similarity in the overall structure (rmsd of 0.48 Å for 230 backbone atoms) and in the details of the protein–ligand interactions (SI Appendix, Fig. S8 A and B). These data complement other comparisons that were made earlier with a nonsteroidal ligand (34, 35). Additional characterization of the Y537S mutant via transient transfection of HeLa cells, Thermofluor and fluorescence anisotropy indicated that the mutation stabilizes the active conformation of the receptor without modifying the relative potencies of compounds (SI Appendix, Figs. S9 and S10). Earlier comparative studies on the ERα-Y537S mutant established that it had somewhat elevated affinity for E2 (36).
Subsequently, we crystallized ERα-Y537S LBD in complex with BPA or BPAF. Most likely because of the stronger antagonistic character of BPC, crystals with this compound could be obtained by using the WT construct but not with the H12-stabilized ERα mutant. Details of structure determination and refinement are summarized in SI Appendix, Table S1. The structures with BPA and BPAF display the canonical active conformation, with H12 capping the ligand binding pocket (LBP) and the SRC-1 peptide bound to the AF-2 surface (Fig. 3A). In agreement with the aforementioned functional data, the structure with BPC displays an antagonist conformation similar to that observed in the OHT-bound structure (PDB code 3ERT), with H12 occupying the coactivator binding groove (Fig. 3A). All compounds could be precisely placed in their respective electron density (Fig. 3 B–F), revealing two discrete orientations of the bisphenols in the LBP. As shown in Fig. 3C, BPA adopts a binding mode reminiscent of that used by E2 (Fig. 3B) with the two phenol groups hydrogen-bonded to three polar residues located at the two ends of the pocket, namely H524 (H11) on one side and E353 (H3) and R394 (H5) on the other side. The remaining contacts involve 51 van der Waals interactions (4.2 Å cutoff) in the E2 complex but only 42 in the complex with BPA, with this difference accounting, at least in part, for the weaker affinity of the bisphenol for ERs. In the complex with BPC, the ligand is positioned in the pocket so as to draw the phenol ring B into an alternate position compared with that of the corresponding ring in BPA. A rotation by 180° around the main axis of phenol ring A, which remains anchored to E353 and R394, orients ring B toward H12 (Fig. 3 C and F). A molecular modeling approach reveals that the “BPA-like” mode of binding would position one of the two chlorine atoms of BPC in very close proximity of A350 in helix H3 (SI Appendix, Fig. S11), thus explaining the “antagonist orientation” adopted by BPC. Finally, BPAF displays an intermediate situation with each subunit of the ERα homodimer containing one BPAF molecule with the “agonist, BPA-like” or “antagonist, BPC-like” positioning (Fig. 3 D and E). The observation that two distinct orientations of BPAF are found in each monomer rather than occurring randomly is intriguing and suggests the existence of a regulatory crosstalk between the two subunits whereby, as recently reported, ligand and/or coregulator binding to one monomer can affect ligand and coregulator binding to the second monomer of a dimer (37). However, such a situation is not observed in the E2 or BPA complexes, which yet crystallize in the same crystal form as with BPAF and are therefore engaged in similar packing contacts. This apparent discrepancy could indicate that there are some chemical requirements for a ligand to promote such allosteric regulation, thereby providing new perspectives for drug design.
Fig. 3.
Two different binding modes of bisphenols. (A) The whole structure of the ERα Y537S LBD in complex with SRC-1 NR2 and BPA (cyan) superimposed on that of WT ERα LBD bound to BPC (orange). The orange dashed line denotes residues not visible in the electron density map. (B–F) Interaction networks of E2 (B), BPA (C), BPAF (D and E), and BPC (F) with LBP residues in ERα. Oxygen, nitrogen, sulfur, fluorine, and chlorine atoms are colored in red, blue, yellow, cyan, and green, respectively. Hydrogen bonds are indicated by black dashed lines. For clarity, not all protein–ligand interactions are depicted. The blue electron density represents a Fo-Fc simulated annealing omit map contoured at 3σ.
Key Contacts Are Missing in ER–Bisphenol Complexes.
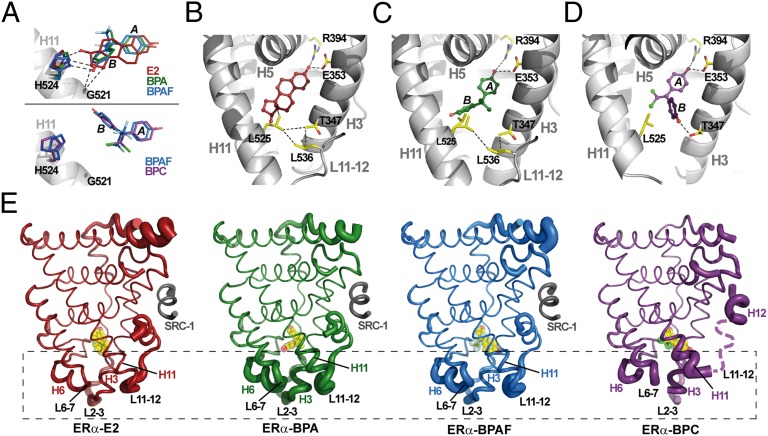
We next considered how the two binding modes of bisphenols may contribute to the destabilization of the AF-2 surface. By comparing our four structures, we observed that previously recognized ligand–H11 stabilizing interactions observed in the E2-bound structure are altered in the bisphenol-containing complexes (34, 35). For example, E2 makes an important stabilizing interaction with G521 (3.95 Å), which is severely weakened in the structures with bisphenols in the “BPA-like” conformation (4.65 Å) or completely abolished in the structures with bisphenols in the “BPC-like” conformation (>5.20 Å; Fig. 4A). We also noticed significant differences in the geometry of the interaction between H524 and the hydroxyl moieties of E2 or bisphenols in the BPA-like orientation (Fig. 4A) and an absence of interaction of this residue with bisphenols in the BPC-like orientation (Fig. 4A). These suboptimal or complete lack of interactions induce a substantial reorientation of the H524 imidazole ring, which, as previously reported by Nettles et al. (34), perturbs a key hydrogen bond network involving residues from loop L6-7, H3, and H11. Last but not least, we observed that, in the E2-bound structure, the 18-methyl group of E2 is in contact with L525 and imposes a conformation that strengthens a network of van der Waals interactions involving T347, L525, and L536 (Fig. 4B). This cluster of interactions is of utmost importance because it holds together helices H3 and H11 and the loop preceding H12, thereby stabilizing the AF-2 surface. In the structure with BPA, the stabilizing interaction between the bisphenol and L525 is absent. This renders L525 more dynamic, so that its side chain adopts different conformations in the two subunits of the homodimer (Fig. 4C). The situation is even worse in the structure with BPC, in which the side chain of T347 rotates by 180° to form a hydrogen bond with the hydroxyl moiety from the phenol ring B of BPC (Fig. 4D). The complete disruption of the hydrophobic cluster by BPC probably accounts for the marked antagonistic character of this bisphenol. This massive loss of stabilizing contacts provoked by bisphenols renders the lower part of the ERα LBP more dynamic (Fig. 4E). Indeed, this portion of LBDs has been previously shown to have some of the characteristics of a molten globule and a large part of the agonistic properties of a ligand relies in its capacity to stabilize this region encompassing the H3/H11 docking surface for H12 in the active conformation (38, 39). These findings reveal the mechanisms by which bisphenols interact with ERα and highlight how key secondary structural elements sense and allosterically convey ligand activities to the AF-2 surface through modifications of H12 positioning and/or dynamics. In this respect, it is noteworthy that the structural effects observed in the background of a H12-stabilized ERα mutant and in a crystalline context would be more pronounced with the WT receptor in solution as indicated by fluorescence anisotropy data (Fig. 2).
Fig. 4.
Bisphenol binding promotes ERα structural dynamics. (A) Differential interactions of bisphenols and E2 with G521 and H524. (B) The interaction of E2 with L525 strengthens a van der Waals interactions network involving T347 (H3), L525 (H11), and L536 (L11–L12). (C) Because of a lack of contact with BPA, L525 is not stabilized and adopts two different conformations. (D) In the BPC-bound ERα structure, T347 rotates by 180° to form a hydrogen bond with the bisphenol, resulting in the disruption of the hydrophobic network. (E) Ribbon representation of ERα LBD in complex with E2 (red), BPA (green), BPAF (blue), and BPC (purple; dashed line denotes missing residues). Ligands are shown in yellow. The diameter of the ribbon is directly proportional to the temperature factor B and highlights the dynamics all along the polypeptide chain.
Focused Virtual Screening of Bisphenols on NRs.
Having characterized the interaction of three bisphenols with ERs at the functional and structural levels, we reasoned that this information could aid in the development of a computational tool to predict binding of any bisphenol to this receptor and the induced functional outcome. We took advantage of our server @TOME-2 (40) to select optimal ERα conformations for virtual screening. First, the various ERα crystal structures available in PDB were partitioned into two groups according to their agonist or antagonist conformation. Within each group, we performed so-called comparative docking, by which each ligand contained in a particular structure is transferred into the other structures of this group through protein–protein superimposition. This cross-docking allows the exploration of a wide range of binding site conformations and ligand orientations, and builds up an array of optimal shape restraints to focus virtual screening. Implementation of an interface between the server @TOME-2 and the docking program PLANTS (41) allowed virtual screening of bisphenols with the complexes described earlier used as anchoring models. Binding affinities were evaluated by using several scoring functions, including the recently developed DSX (42).
In our test case, we observed that BPAF adopts the two alternative BPA-like and BPC-like orientations in the agonist and antagonist groups of ERα conformations, whereas BPC docks mostly in the BPC-like orientation whatever the conformation screened (results available at http://atome.cbs.cnrs.fr/EDCNR.html). These results are in full agreement with crystallographic data (SI Appendix, Fig. S12A). In the case of BPA, the server predicted that it could adopt the two orientations (with a majority of poses in the BPA-like conformation), whereas the corresponding crystal structure shows only one orientation of the ligand (Fig. 3C). Note that these results were obtained before deposition of the various bisphenol-bound ERα structures to PDB. The rough affinity predictions of BPA, BPAF, and BPC for ERα nicely matched the experimental ones (http://atome.cbs.cnrs.fr/EDCNR.html and Fig. 3). Subsequently, we applied this screening approach to other bisphenols and found that the ranking order of affinity with BPAF, BPC, BPB better than BPA, BPE, BPF better than BPS agrees well with that obtained experimentally (SI Appendix, Fig. S13).
Then, we extended further this in silico approach to ERβ, AR, and ERRγ, which are also known targets of bisphenols. Predictions indicated binding modes similar to those found in ERα (http://atome.cbs.cnrs.fr/EDCNR.html and SI Appendix, Fig. S12 B–D). Interestingly, all studied bisphenols appeared to bind to AR exclusively in the antagonist BPC-like orientation, in agreement with the observation that these compounds act as AR antagonists (20) (SI Appendix, Fig. S14). This orientation appears to be stabilized by formation of a hydrogen bond between one hydroxyl group of the ligand and N705, a polar residue specific of AR (SI Appendix, Fig. S12C). This situation is mirrored with ERRγ, in which all bisphenols studied adopt the agonist BPA-like position as a result of a hydrogen bond with N346 from helix H7 (SI Appendix, Fig. S12D). This in silico result correlates with functional data showing that bisphenols are ERRγ activators (SI Appendix, Fig. S15) and the crystal structure of ERRγ in complex with BPA (SI Appendix, Fig. S8C) (43, 44). Therefore, it appears that most bisphenols are rather weak binders (i.e., micromolar range) of several NRs and that their binding mode varies according to their chemical structure as well as the receptor under scrutiny.
Concluding Remarks
Deregulation of NR-mediated transcription accounts for the deleterious effects of many EDCs. Thus, characterization of the harmful interaction between receptors and environmental compounds at the structural and functional levels, as well as the development of robust in vivo, in vitro, and in silico screening methods, are important for assessment of the toxic potential of large numbers of chemicals. In addition, because of mounting restrictions on the use of many synthetic chemicals used in consumer products (e.g., BPA), especially in the European Union, Canada, and the United States, there is a huge demand for alternative safer substitutes for industrial applications.
In this context, by using complementary approaches, we have dissected the mechanisms by which an important class of environmental endocrine disruptors interferes with ER signaling. We have found that bisphenols are SERMs that function in a cell- and tissue-selective manner (24). As a consequence, bisphenols might exert E2-like activities in some tissues but not in others, implying that cell, tissue, or animal models used in which to assess the risk to human health should be cautiously designed and the results carefully interpreted. Most of the methods used in this study, including fluorescence anisotropy, thermal denaturation shift, and cell-based assays have been implemented in a medium-throughput setting, allowing for rapid assessment of the endocrine-disruptive potential of large numbers of EDCs. Moreover, use of the previously described H12-stabilized ERα mutant (34) facilitating EDC-bound ER LBD crystallization will permit a rapid increase of our knowledge of the structural mechanisms and molecular interactions used by ERs and a wide range of structurally and chemically diverse compounds. To add to the tool box, we have developed a 3D structure-based computational method the aim of which is to help evaluation of the interference of EDCs with hormonal signaling. By using this tool, we have been able to discern with a high level of accuracy the docking modes of bisphenols in four different NRs, thereby allowing for the prediction of their activity profiles, and the ranges of binding affinities of these compounds. Although currently restricted to virtual screening of bisphenols on four human NRs, future developments of the server will allow examination of (i) other EDC families, (ii) an extended set of NRs, and (iii) other species, including mouse, zebrafish, and xenope. We believe the structural insights gained at a near-atomic resolution, together with the experimental and computational tools developed in this study, could facilitate evaluation of the EDC activity of chemicals and aid in the design of novel compounds with the promise to separate their industrial characteristics from their unwanted toxic effects.
Materials and Methods
Reporter Cell Lines and Culture Conditions.
Luciferase, cell proliferation, and whole-cell ER competitive binding assays have been performed by using the stably transfected luciferase reporter MELN, HELN-ERα, -ERβ, -ΔAB-ERα, and -ΔAB-ERβ cell lines as described previously (32).
Structure Determination.
The ERα LBD and ERα-Y537S LBD mutant were cloned into the pET-32a vector and expressed in BL21(DE3) cells. Protein domains were purified by using a nickel affinity column and size exclusion chromatography. The purified ERα-Y537S LBD was mixed with E2, BPA or BPAF and SRC-1 NR2, and ERα LBD was mixed with BPC. All complexes were crystallized by using the vapor diffusion method. Data were collected on the ID14-1, ID23-2, or ID29 beam lines at the European Synchrotron Radiation Facility (Grenoble, France). Data were processed as described in SI Appendix, Materials and Methods.
Fluorescence Anisotropy Measurements.
H12 dynamics were monitored by using fluorescein-labeled ERα LBD prepared following the previously described protocol (45). Assays were performed by using a Safire2 microplate reader (TECAN) at a protein concentration of 0.140 μM. The excitation wavelength was set at 470 nm, with emission measured at 530 nm. SRC-1 NR2 was added to protein samples containing 5 μM of ligand to a final concentration of 10 μM. Then, samples were diluted successively with 20 mM Tris, pH 8.0, 180 mM NaCl, 5 mM DTT, and 10% glycerol supplemented with 0.140 μM of protein and 5 μM of ligand. Details of the experimental procedures and associated references are given in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
The authors acknowledge the experimental assistance from the staff of the European Synchrotron Radiation Facility during data collection. This work was supported by Programme National de Recherche sur les Perturbateurs Endocriniens (P.B.) and Agence Nationale de Recherches Contaminants, Ecosystèmes, Santé Project “BISCOT” 2010 Contaminants et Environnements: Métrologie, Santé, Adaptabilité, Comportements et Usages 004 02 (to P.B. and W.B.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3UU7, 3UUA, 3UUC, and 3UUD).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203574109/-/DCSupplemental.
References
- 1.Vandenberg LN, et al. Exposure to environmentally relevant doses of the xenoestrogen bisphenol-A alters development of the fetal mouse mammary gland. Endocrinology. 2007;148:116–127. doi: 10.1210/en.2006-0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dekant W, Völkel W. Human exposure to bisphenol A by biomonitoring: Methods, results and assessment of environmental exposures. Toxicol Appl Pharmacol. 2008;228:114–134. doi: 10.1016/j.taap.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Vandenberg LN, et al. Urinary, circulating, and tissue biomonitoring studies indicate widespread exposure to bisphenol A. Environ Health Perspect. 2010;118:1055–1070. doi: 10.1289/ehp.0901716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zalko D, Jacques C, Duplan H, Bruel S, Perdu E. Viable skin efficiently absorbs and metabolizes bisphenol A. Chemosphere. 2011;82:424–430. doi: 10.1016/j.chemosphere.2010.09.058. [DOI] [PubMed] [Google Scholar]
- 5.Crain DA, et al. An ecological assessment of bisphenol-A: Evidence from comparative biology. Reprod Toxicol. 2007;24:225–239. doi: 10.1016/j.reprotox.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Richter CA, et al. In vivo effects of bisphenol A in laboratory rodent studies. Reprod Toxicol. 2007;24:199–224. doi: 10.1016/j.reprotox.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung E, Genco MC, Megrelis L, Ruderman JV. Effects of bisphenol A and triclocarban on brain-specific expression of aromatase in early zebrafish embryos. Proc Natl Acad Sci USA. 2011;108:17732–17737. doi: 10.1073/pnas.1115187108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tyl RW, et al. Two-generation reproductive toxicity study of dietary bisphenol A in CD-1 (Swiss) mice. Toxicol Sci. 2008;104:362–384. doi: 10.1093/toxsci/kfn084. [DOI] [PubMed] [Google Scholar]
- 9.Rubin BS, Soto AM. Bisphenol A: Perinatal exposure and body weight. Mol Cell Endocrinol. 2009;304:55–62. doi: 10.1016/j.mce.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newbold RR, Jefferson WN, Padilla-Banks E. Prenatal exposure to bisphenol a at environmentally relevant doses adversely affects the murine female reproductive tract later in life. Environ Health Perspect. 2009;117:879–885. doi: 10.1289/ehp.0800045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.vom Saal FS, Hughes C. An extensive new literature concerning low-dose effects of bisphenol A shows the need for a new risk assessment. Environ Health Perspect. 2005;113:926–933. doi: 10.1289/ehp.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vandenberg LN, Maffini MV, Sonnenschein C, Rubin BS, Soto AM. Bisphenol-A and the great divide: a review of controversies in the field of endocrine disruption. Endocr Rev. 2009;30:75–95. doi: 10.1210/er.2008-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuiper GG, et al. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinology. 1997;138:863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- 14.Soriano S, et al. Rapid insulinotropic action of low doses of bisphenol-A on mouse and human islets of Langerhans: Role of estrogen receptor β. PLoS ONE. 2012;7:e31109. doi: 10.1371/journal.pone.0031109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mnif W, et al. Estrogens and antiestrogens activate hPXR. Toxicol Lett. 2007;170:19–29. doi: 10.1016/j.toxlet.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 16.Sui Y, et al. Bisphenol A and its analogues activate human pregnane X receptor. Environ Health Perspect. 2012;120:399–405. doi: 10.1289/ehp.1104426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okada H, et al. Direct evidence revealing structural elements essential for the high binding ability of bisphenol A to human estrogen-related receptor-gamma. Environ Health Perspect. 2008;116:32–38. doi: 10.1289/ehp.10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riu A, et al. Peroxisome proliferator-activated receptor γ is a target for halogenated analogs of bisphenol A. Environ Health Perspect. 2011;119:1227–1232. doi: 10.1289/ehp.1003328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riu A, et al. Characterization of novel ligands of ERα, Erβ, and PPARγ: The case of halogenated bisphenol A and their conjugated metabolites. Toxicol Sci. 2011;122:372–382. doi: 10.1093/toxsci/kfr132. [DOI] [PubMed] [Google Scholar]
- 20.Paris F, et al. Phenylphenols, biphenols, bisphenol-A and 4-tert-octylphenol exhibit alpha and beta estrogen activities and antiandrogen activity in reporter cell lines. Mol Cell Endocrinol. 2002;193:43–49. doi: 10.1016/s0303-7207(02)00094-1. [DOI] [PubMed] [Google Scholar]
- 21.Fini JB, et al. Parallel biotransformation of tetrabromobisphenol A in Xenopus laevis and mammals: Xenopus as a model for endocrine perturbation studies. Toxicol Sci. 2012;125:359–367. doi: 10.1093/toxsci/kfr312. [DOI] [PubMed] [Google Scholar]
- 22.Moriyama K, et al. Thyroid hormone action is disrupted by bisphenol A as an antagonist. J Clin Endocrinol Metab. 2002;87:5185–5190. doi: 10.1210/jc.2002-020209. [DOI] [PubMed] [Google Scholar]
- 23.Levin ER. Minireview: Extranuclear steroid receptors: Roles in modulation of cell functions. Mol Endocrinol. 2011;25:377–384. doi: 10.1210/me.2010-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gronemeyer H, Gustafsson JA, Laudet V. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov. 2004;3:950–964. doi: 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- 25.Heldring N, et al. Estrogen receptors: How do they signal and what are their targets. Physiol Rev. 2007;87:905–931. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- 26.Perissi V, Rosenfeld MG. Controlling nuclear receptors: The circular logic of cofactor cycles. Nat Rev Mol Cell Biol. 2005;6:542–554. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- 27.Benecke A, Chambon P, Gronemeyer H. Synergy between estrogen receptor alpha activation functions AF1 and AF2 mediated by transcription intermediary factor TIF2. EMBO Rep. 2000;1:151–157. doi: 10.1093/embo-reports/kvd028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Métivier R, Penot G, Flouriot G, Pakdel F. Synergism between ERalpha transactivation function 1 (AF-1) and AF-2 mediated by steroid receptor coactivator protein-1: requirement for the AF-1 alpha-helical core and for a direct interaction between the N- and C-terminal domains. Mol Endocrinol. 2001;15:1953–1970. doi: 10.1210/mend.15.11.0727. [DOI] [PubMed] [Google Scholar]
- 29.Nahoum V, Bourguet W. Androgen and estrogen receptors: Potential of crystallography in the fight against cancer. Int J Biochem Cell Biol. 2007;39:1280–1287. doi: 10.1016/j.biocel.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Pike AC. Lessons learnt from structural studies of the oestrogen receptor. Best Pract Res Clin Endocrinol Metab. 2006;20:1–14. doi: 10.1016/j.beem.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Huang P, Chandra V, Rastinejad F. Structural overview of the nuclear receptor superfamily: Insights into physiology and therapeutics. Annu Rev Physiol. 2010;72:247–272. doi: 10.1146/annurev-physiol-021909-135917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molina-Molina JM, et al. Profiling of benzophenone derivatives using fish and human estrogen receptor-specific in vitro bioassays. Toxicol Appl Pharmacol. 2008;232:384–395. doi: 10.1016/j.taap.2008.07.017. [DOI] [PubMed] [Google Scholar]
- 33.Nahoum V, et al. Modulators of the structural dynamics of the retinoid X receptor to reveal receptor function. Proc Natl Acad Sci USA. 2007;104:17323–17328. doi: 10.1073/pnas.0705356104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nettles KW, et al. NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses. Nat Chem Biol. 2008;4:241–247. doi: 10.1038/nchembio.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruning JB, et al. Coupling of receptor conformation and ligand orientation determine graded activity. Nat Chem Biol. 2010;6:837–843. doi: 10.1038/nchembio.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carlson KE, Choi I, Gee A, Katzenellenbogen BS, Katzenellenbogen JA. Altered ligand binding properties and enhanced stability of a constitutively active estrogen receptor: Evidence that an open pocket conformation is required for ligand interaction. Biochemistry. 1997;36:14897–14905. doi: 10.1021/bi971746l. [DOI] [PubMed] [Google Scholar]
- 37.Osz J, et al. Structural basis for a molecular allosteric control mechanism of cofactor binding to nuclear receptors. Proc Natl Acad Sci USA. 2012;109:E588–E594. doi: 10.1073/pnas.1118192109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagy L, Schwabe JW. Mechanism of the nuclear receptor molecular switch. Trends Biochem Sci. 2004;29:317–324. doi: 10.1016/j.tibs.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 39.Gee AC, Katzenellenbogen JA. Probing conformational changes in the estrogen receptor: Evidence for a partially unfolded intermediate facilitating ligand binding and release. Mol Endocrinol. 2001;15:421–428. doi: 10.1210/mend.15.3.0602. [DOI] [PubMed] [Google Scholar]
- 40.Pons JL, Labesse G. @TOME-2: A new pipeline for comparative modeling of protein-ligand complexes. Nucleic Acids Res. 2009;37(web server issue):W485–W491. doi: 10.1093/nar/gkp368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korb O, Stützle T, Exner TE. Empirical scoring functions for advanced protein-ligand docking with PLANTS. J Chem Inf Model. 2009;49:84–96. doi: 10.1021/ci800298z. [DOI] [PubMed] [Google Scholar]
- 42.Neudert G, Klebe G. DSX: A knowledge-based scoring function for the assessment of protein-ligand complexes. J Chem Inf Model. 2011;51:2731–2745. doi: 10.1021/ci200274q. [DOI] [PubMed] [Google Scholar]
- 43.Abad MC, et al. Structural determination of estrogen-related receptor gamma in the presence of phenol derivative compounds. J Steroid Biochem Mol Biol. 2008;108:44–54. doi: 10.1016/j.jsbmb.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 44.Matsushima A, et al. Structural evidence for endocrine disruptor bisphenol A binding to human nuclear receptor ERR gamma. J Biochem. 2007;142:517–524. doi: 10.1093/jb/mvm158. [DOI] [PubMed] [Google Scholar]
- 45.Pérez Santín E, et al. Modulating retinoid X receptor with a series of (E)-3-[4-hydroxy-3-(3-alkoxy-5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)phenyl]acrylic acids and their 4-alkoxy isomers. J Med Chem. 2009;52:3150–3158. doi: 10.1021/jm900096q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.