Abstract
Up-regulation of the folding machinery of the heat-shock protein 90 (Hsp90) chaperone protein is crucial for cancer progression. The two Hsp90 isoforms (α and β) play different roles in response to chemotherapy. To identify isoform-selective inhibitors of Hsp90(α/β)/cochaperone p23 interactions, we developed a dual-luciferase (Renilla and Firefly) reporter system for high-throughput screening (HTS) and monitoring the efficacy of Hsp90 inhibitors in cell culture and live mice. HTS of a 30,176 small-molecule chemical library in cell culture identified a compound, N-(5-methylisoxazol-3-yl)-2-[4-(thiophen-2-yl)-6-(trifluoromethyl)pyrimidin-2-ylthio]acetamide (CP9), that binds to Hsp90(α/β) and displays characteristics of Hsp90 inhibitors, i.e., degradation of Hsp90 client proteins and inhibition of cell proliferation, glucose metabolism, and thymidine kinase activity, in multiple cancer cell lines. The efficacy of CP9 in disrupting Hsp90(α/β)/p23 interactions and cell proliferation in tumor xenografts was evaluated by non-invasive, repetitive Renilla luciferase and Firefly luciferase imaging, respectively. At 38 h posttreatment (80 mg/kg × 3, i.p.), CP9 led to selective disruption of Hsp90α/p23 as compared with Hsp90β/p23 interactions. Small-animal PET/CT in the same cohort of mice showed that CP9 treatment (43 h) led to a 40% decrease in 18F-fluorodeoxyglucose uptake in tumors relative to carrier control-treated mice. However, CP9 did not lead to significant degradation of Hsp90 client proteins in tumors. We performed a structural activity relationship study with 62 analogs of CP9 and identified A17 as the lead compound that outperformed CP9 in inhibiting Hsp90(α/β)/p23 interactions in cell culture. Our efforts demonstrated the power of coupling of HTS with multimodality molecular imaging and led to identification of Hsp90 inhibitors.
Keywords: drug development, small-molecule inhibitors, co-chaperone p23, bioluminescence imaging, PET/computed tomography imaging
All proteins must be properly folded to perform their biological functions. In mammalian cells, the heat shock protein 90 (Hsp90) chaperone system plays a key role (1, 2). The most important interactions within the Hsp90 chaperone system are between Hsp90 isoforms (α and β) and cochaperone p23, which occur only when Hsp90 is bound to ATP. These interactions are crucial for maturation of Hsp90/client protein complexes and the release of folded proteins (1, 2). Overexpression of Hsp90 in human cancer cells correlates with poor prognosis (3, 4). Small-molecule Hsp90 inhibitors with antitumor effects have been developed by targeting the ATP-binding pocket of Hsp90 (1, 5, 6). They have higher binding affinities to Hsp90 in cancer cells than in normal cells (7) and competitively block ATP binding to Hsp90 and subsequent binding of p23. The prevention of proper protein folding leads to proteasome-mediated degradation of Hsp90 client proteins and inhibition of signaling pathways and growth arrest.
Geldanamycin-based Hsp90 inhibitors (8), purine scaffold– (9–16), pyrazole scaffold– (17), and radicicol-based Hsp90 inhibitors (18, 19), and other compounds are currently in preclinical and/or phase I/II clinical trials (20–22). Despite the diversity of Hsp90 inhibitors reported, inhibitors with better clinical therapeutic efficacy and reduced toxicity are needed. Until now, methods for identifying of Hsp90 inhibitors have been limited to in vitro analyses (13, 23, 24) and phenotypic assays that examine the downstream effects of the Hsp90 inhibition (9, 14, 15, 18). Small-animal PET (25, 26), MRI (27), and ultrasound (28) have been used to monitor the efficacy of Hsp90 inhibitors in mice. However, it was not possible to decipher the contribution of each Hsp90 isoform (α and β) in determining sensitivity to Hsp90 inhibitors, because both isoforms are expressed in cancer cells but play different roles in response to chemotherapy (29).
We previously monitored the efficacy of different Hsp90 inhibitors in disrupting isoform-specific interactions between Hsp90(α/β) and p23 in intact cells in cell culture and in live mice using a genetically encoded split Renilla luciferase (RL) complementation system (Fig. 1A) (30). This system is useful for identifying Hsp90 inhibitors because (i) the potency of the Hsp90 inhibitors in disrupting signals from Hsp90(α/β)/p23 interactions correlates with their binding affinities for cellular Hsp90; (ii) the interaction of each Hsp90 isoform (α or β) with p23 can be monitored individually; (iii) RL does not require ATP for its activity and thus facilitates the screening of inhibitors that target the ATP-binding pocket of Hsp90; (iv) the same reporter cells can be used for high-throughput screening (HTS) in cell culture followed by dynamic monitoring of Hsp90(α/β)/p23 interactions in response to lead compounds in living mice; and (v) the split RL complementation system in combination with other clinical imaging modalities such as PET allows monitoring of the downstream effects of Hsp90 inhibitors.
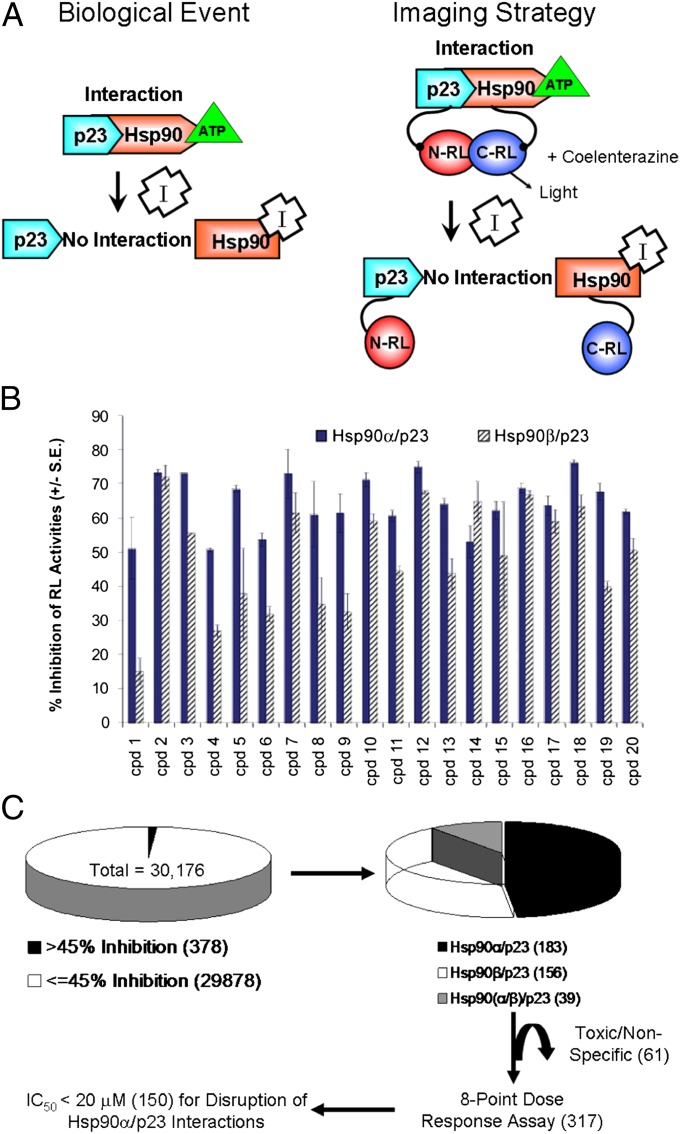
Fig. 1.
HTS of Hsp90 inhibitors using the split RL complementation system and evaluation of lead compounds by BLI. (A) The biological event that we are monitoring (i.e., disruption of Hsp90/p23 interactions by Hsp90 inhibitors) is shown on the left. The schematic diagram for monitoring Hsp90/p23 interaction using the split RL complementation system is shown on the right. The two interacting proteins, p23 and Hsp90, are fused to the inactive N-RL and C-RL portions of RL through a peptide linker (Upper). In the presence of ATP, interactions between p23 and Hsp90 bring N-RL and C-RL into close proximity and lead to complementation of RL enzyme activity and the production of light in the presence of the substrate coelenterazine. (Lower) Binding of Hsp90 inhibitors (I) to Hsp90-CRL leads to a conformation change and prevents ATP from binding, thus diminishing the interaction between N-RL–p23 and Hsp90–C-RL and complementation of RL activity and reduced light output. The effect of Hsp90 inhibitors on cell proliferation is monitored by FL imaging upon introduction of the EGFP-FL fusion reporter. (B) HTS using the 1,280-compound LOPAC for assay standardization. 293T cells stably expressing the Hsp90α/p23 (solid bars) or Hsp90β/p23 (striped bars) split RL reporter constructs were plated in 384-well white plates for 24 h. Baseline RL signals were determined by a luminescence reader upon the addition of the RL substrate EnduRen. Each compound in the LOPAC library (20 μM) or carrier (control) then was added to a well, and RL signals at 24 h after treatment were normalized to those of time 0 h and carrier control-treated. The experiment was repeated twice. The percent of inhibition of RL activity by the top 20 compounds is shown as mean ± SEM. (C) Flow diagram for HTS of a 30,000 small-molecule compound library. Initial screening was performed as in A, but only one well was used for each compound at 8.3 μM. IC50 values for the compounds that resulted in >45% inhibition of Hsp90α/p23 or Hsp90β/p23 interactions were determined by an eight-point dose–response experiment (0.3–20 μM) (Table 1). Toxic compounds were eliminated. Numbers within parentheses denote the number of compounds at each stage during the screening.
To accelerate the development of small-molecule Hps90 inhibitors, we incorporated multimodality molecular imaging into HTS of an uncharacterized chemical library. We successfully identified and monitored the efficacy of a class of 2-{[6-(trifluoromethyl)pyrimidin-2-yl]thio}acetamide-based Hsp90 inhibitors through molecular imaging of Hsp90(α/β)/p23 interactions in living mice.
Results
HTS of Modulators of Hsp90(α/β)/p23 Interactions in Cell Culture.
The initial assay development was performed using the Library of Pharmacologically Active Compounds (LOPAC) (31) with 293T human kidney cancer cells expressing the Hsp90α/p23 or Hsp90β/p23 split RL reporters (30). Baseline bioluminescence imaging (BLI) signals were determined before the addition of LOPAC compounds. The known Hsp90 inhibitor PU-H71 (11) was used a positive control. Twenty compounds were identified with greater than 50% inhibition of BLI signals relative to carrier control-treated cells at 24 h (Fig. 1B). Our initial screen demonstrated that our assay conditions were optimal for discovery of isoform-selective Hsp90(α/β)/p23 modulators in intact cells.
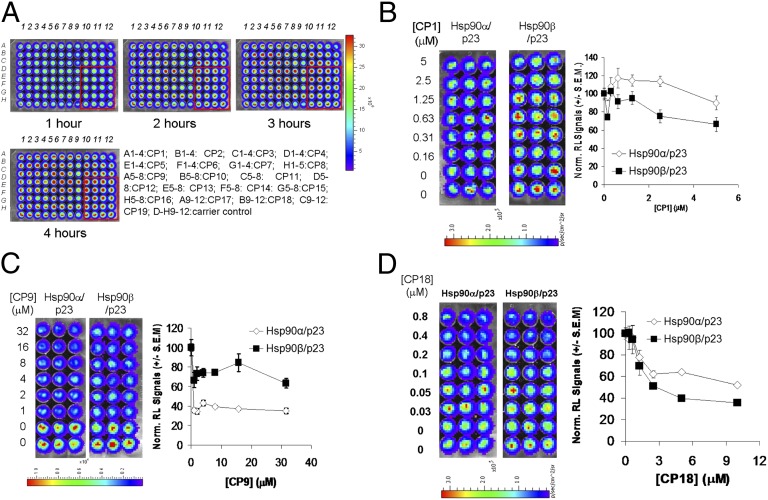
We proceeded to identify Hsp90 inhibitors using a commercially available small-molecule chemical compound library with 30,176 compounds (8.3 μM each) with unknown targets and molecular mechanisms (Fig. 1C). IC50 values were determined for the 317 compounds that led to greater than 45% inhibition of BLI signals relative to carrier control-treated cells. Inhibition of cell proliferation at 20 μM was determined, and toxic compounds were eliminated. The IC50 values for inhibition of BLI signals and cell proliferation by the top 19 compounds with the lowest IC50 values for inhibition of Hsp90α/p23 interactions are shown in Table 1, and their structures are given in Table S1. To determine if any of the top compounds are nonspecific RL inhibitors, 293T cells stably expressing full-length RL were treated with the compounds for different periods of time at levels 10-fold higher than their respective IC50 for inhibition of Hsp90α/p23 interactions (Fig. 2A). Compounds that led to greater than 20% inhibition of RL signals relative to carrier control-treated cells were eliminated (Fig. S1).
Table 1.
Effect of the lead compounds CP1–CP19 on inhibition of Hsp90α/p23 and Hsp90β/p23 interactions and cell proliferation
| Compound | IC50 (Hsp90α/p23) (μM)* | IC50 (Hsp90β/p23) (μM)† | % inhibition cell proliferation (20 μM)‡ |
| CP1 | 0.2 | 0.5 | 44 |
| CP2 | 0.8 | 2.4 | 45 |
| CP3 | 1.2 | 1.6 | 33 |
| CP4 | 1.4 | 1.7 | 46 |
| CP5 | 1.4 | 3.1 | 35 |
| CP6 | 1.8 | 2.8 | 43 |
| CP7 | 1.9 | 2.9 | 45 |
| CP8 | 2.7 | 6.0 | 32 |
| CP9 | 3.2 | 15.3 | 29 |
| CP10 | 1.2 | 2.4 | 11 |
| CP11 | 1.4 | 2.3 | 10 |
| CP12 | 1.4 | 2.4 | 21 |
| CP13 | 2.1 | 3.3 | 27 |
| CP14 | 2.1 | 8.1 | 10 |
| CP15 | 2.3 | 5.7 | 27 |
| CP16 | 2.5 | 4.0 | 2 |
| CP17 | 3.9 | 17.5 | 13 |
| CP18 | 0.07 | 0.04 | 55 |
| CP19 | 0.3 | 0.8 | 50 |
*IC50 was defined as the concentration of the compound required to inhibit bioluminescence signals by 50%, relative to carrier control-treated 293T cells stably expressing Hsp90α/p23 split RL reporters.
†IC50 was defined as the concentration of the compound required to inhibit bioluminescence signals by 50%, relative to carrier control-treated 293T cells stably expressing Hsp90α/p23 split RL reporters.
‡% Inhibition of 293T cells stably expressing Hsp90α/p23 split RL reporters by each compound at 20 μM was determined by Cell Titer-Blue assay as described in Materials and Methods.
Fig. 2.
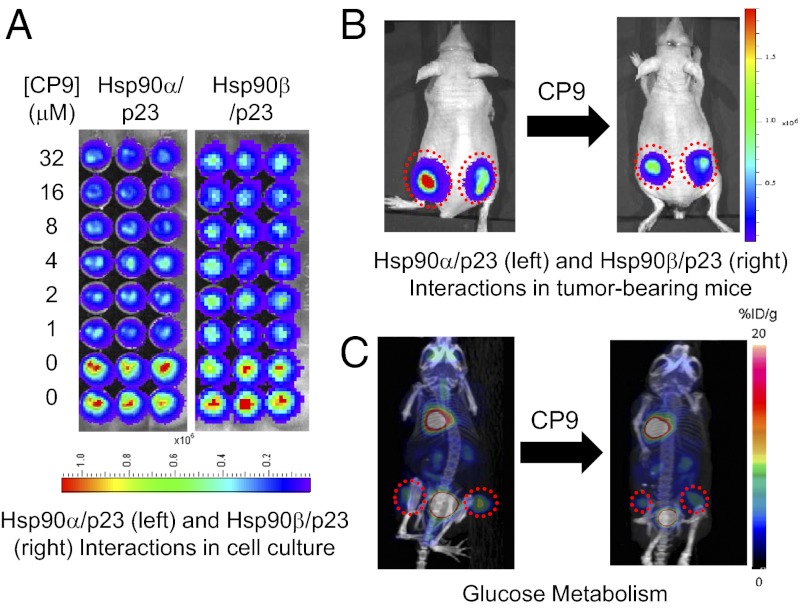
Elimination of nonspecific RL inhibitors and determination of the efficacy of lead compounds from HTS in disruption of Hsp90(α/β)/p23 interactions. (A) Elimination of nonspecific RL inhibitors and validation of the lead compounds from HTS. The top 19 compounds with the lowest IC50 values for inhibition of Hsp90α/p23 interactions from Fig. 1A were tested in cell culture in quadruplicate wells to eliminate compounds that nonspecifically inhibit RL activities. 293T cells stably expressing full-length RL (293T-RL) were treated with carrier (control) (Rows D–H, columns 9–12) or 10-fold higher concentrations of the compounds (CP1–19) at their respective IC50 values for inhibition of Hsp90α/p23 interactions. RL activities in intact 293T-RL cells were determined by BLI at different time points posttreatment upon the addition of EnduRen. (B–D) (Left) Dose-dependent decrease in bioluminescence signals in 293T cells stably expressing Hsp90α/p23 and Hsp90β/p23 split RL reporters treated with CP1 (B), CP9 (C), or CP18 (D) at the indicated doses for 24 h. (Right) Bioluminescence signals were quantified and normalized for cell number and signals of carrier control-treated cells. Results are expressed as mean ± SEM.
Efficacy of Lead Compounds in Inhibiting Hsp90 Chaperone Activities.
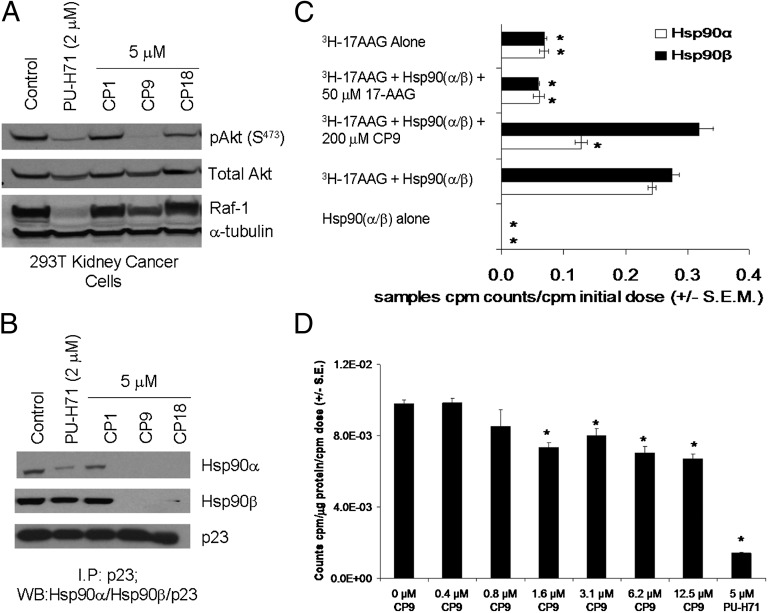
Hsp90 inhibitors lead to degradation of Hsp90 client proteins in cancer cells (20, 32, 33). We initially focused on the two compounds with the greatest inhibition of Hsp90α/p23 BLI signals, CP1 and CP18, and on CP9, N-(5-methylisoxazol-3-yl)-2-[4-(thiophen-2-yl)-6-(trifluoromethyl)pyrimidin-2-ylthio]acetamide, which was more selective for inhibition of Hsp90α/p23 BLI signals than for inhibition of Hsp90β/p23 interactions (Table 1 and Fig. 2 B–D). To determine if these compounds led to degradation of Hsp90 client proteins, 293T cells were treated with CP1, CP9, and CP18, and the expression of phosphorylated Akt relative to total Akt (pAkt/total Akt) and Raf-1 were determined by Western blotting (Fig. 3A). Cells treated with PU-H71 served as a positive control. CP1 did not decrease the levels of any of the three Hsp90 client proteins significantly; CP9 was more effective than CP18 in degrading the pAkt, total Akt, and Raf-1 (Fig. 3A). PU-H71 decreased levels of Raf-1, pAkt, and total Akt, as expected. The disruption of endogenous Hsp90(α/β)/p23 interactions by CP9 and CP18 was confirmed by coimmunoprecipitation (Fig. 3B). Based on our results, we focused our efforts on further characterization of CP9 because of its efficacy as an Hsp90 inhibitor.
Fig. 3.
Validation of the mechanisms of lead compounds as Hsp90 inhibitors. (A) Effect of lead compounds on expression of Hsp90 client proteins. 293T cells stably expressing Hsp90(α/β)/p23 split reporters were treated with carrier (control) or CP1, CP9, or CP18 (5 μM) for 24 h. The known Hsp90 inhibitor PU-H71 (2 μM) was used as a positive control. The expression of Hsp90 client proteins (Raf-1, total Akt, and pAkt) was determined by Western blotting. α-Tubulin was used a loading control. (B) The effects of CP1, CP9, and CP18 on disruption of Hsp90α/p23 and Hsp90β/p23 interactions were determined by co-immunoprecipitation using antibodies against p23 and probing with Hsp90α- and Hsp90β-specific antibodies. The blot was probed with antibodies against p23 to ensure similar immunoprecipitation efficiency. Cells treated with 2 μM of PU-H71 served as a positive control. (C) Competitive in vitro binding of CP9 with 3H-17-AAG to Hsp90(α/β). Purified Hsp90(α/β) was preincubated with CP9 (200 μM) or cold 17-AAG (50 μM) before incubation with 3H-17-AAG. The reaction complexes then were eluted through a desalting column to remove any unbound 3H-17-AAG before scintillation counting to determine the amount of 3H-17-AAG bound to Hsp90(α/β). Columns incubated with 3H-17-AAG alone served as negative controls. Results are expressed as means of triplicates ± SEM. *P < 7.7 × 10−5 vs. Hsp90α + 3H-17AAG. (D) Inhibition of 3H-17-AAG uptake by CP9 in HT29 cells. HT29 cells were preincubated with different concentrations of CP9 along with 3H-17-AAG (0.5 μM) for 1 h followed by 1 h of washout of unbound 3H-17-AAG in fresh Hank's Balanced Salt Solution (HBSS). The amount of 3H-17-AAG incorporated intracellularly was determined by scintillation counting of cell lysates, followed by normalization for protein content and to carrier control-treated cells. Cells incubated with 5 μM of PU-H71 served as a positive control. Results are expressed as means of triplicates ± SEM. *P < 0.05 vs. carrier control-treated cells.
CP9 led to various levels of degradation of pAkt/total Akt and Raf-1 in multiple cancer cell lines, including lung (1975), liver (HUH-7), and breast (BT474) cancer cells, but had no effect on the expression of the Hsp90 client proteins in normal mouse embryonic fibroblasts (MEFs) (Fig. S2A). To confirm the direct binding of CP9 to Hsp90(α/β), an in vitro competitive binding assay was performed using purified Hsp90α/β and the radiolabeled Hsp90 inhibitor [3H]17-allylamino-17-demethoxygeldanamycin (3H-17-AAG). Hsp90 proteins were prebound with CP9 or cold 17-AAG as a control before incubation with 3H-17-AAG. Unbound 3H-17-AAG was removed, and the amount of 3H-17-AAG bound to the Hsp90 proteins was determined. If CP9 binds to Hsp90 (either in the same N-terminal ATP pocket or in other portions of Hsp90 that are involved in binding 17-AAG), that binding will reduce the amount of 3H-17-AAG bound to Hsp90. Fig. 3C shows that CP9 reduced the binding of 3H-17-AAG to purified Hsp90α by about 50% (P < 0.05) but did not affect the binding to Hsp90β significantly (P > 0.05). To confirm cellular Hsp90 as the target of CP9, we performed uptake studies in HT29 cells using 3H-17-AAG. PU-H71 was used as a positive control. CP9 led to a dose-dependent decrease in the uptake of 3H-17-AAG, with a maximum reduction of 30% relative to carrier control-treated cells (P < 0.05) (Fig. 3D).
CP9 Inhibits Cell Proliferation, Glucose Metabolism, and Mammalian Thymidine Kinase Activities in Multiple Cancer Cell Lines.
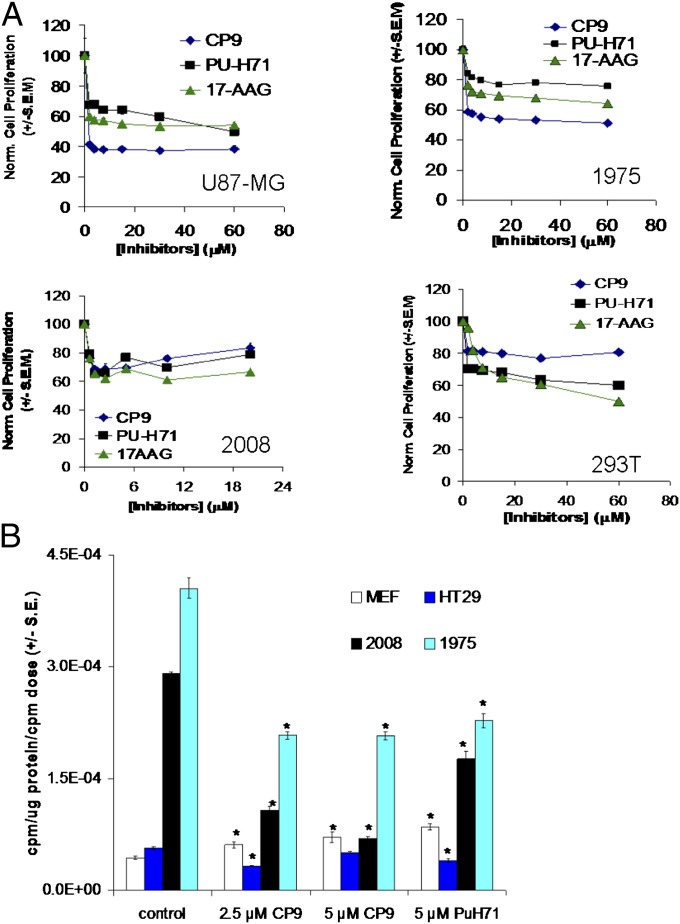
To determine if CP9 also inhibits cell proliferation, 2008 ovarian cancer cells, 293T cells, U87-MG glioblastoma cells, and 1975 cells were treated with different concentrations of CP9, and cell proliferation was monitored. CP9 was more effective than PU-H71 and 17-AAG in inhibiting proliferation of U87-MG and 1975 cells but not of 293T and 2008 cells (Fig. 4A). Because CP9 led to a decrease in pAkt (Fig. 3A and Fig. S2A), we expected it to inhibit glucose metabolism (34, 35), as monitored by cell-uptake studies with [3H]fluorodeoxyglucose (3H-FDG) (Fig. 4B) (36). PU-H71 was used as a positive control. To determine the specificity of CP9 in inhibiting glucose metabolism, normal MEF cells were used as a control. CP9 decreased 3H-FDG uptake in the cancer cells without significantly affecting uptake in MEFs. Likewise, CP9 inhibits uptake of [3H]3-fluorodeoxy-thymidine (3H-FLT) (a surrogate for thymidine kinase activity in mammalian cells) in cancer cell lines but not in normal MEFs (Fig. S2B). Collectively, our data show that CP9 binds to Hsp90 and leads specifically to degradation of Hsp90 client proteins and inhibition of glucose metabolism, thymidine kinase activity, and cell proliferation in cancer cells.
Fig. 4.
Effect of CP9 on glucose metabolism and cell proliferation in different cancer cell lines. (A) The effect of CP9 on cell proliferation was determined by Alamar Blue assay. The indicated cancer cell lines were treated with different concentrations of CP9 for 24 h before incubation with Alamar Blue for 4 h. The Hsp90 inhibitors PU-H71 and 17-AAG were used as positive controls. Emission at 570 nm in CP9-treated cells was normalized to that of carrier control-treated cells. Results are expressed as normalized cell proliferation ± SEM. (B) The effect of CP9 on glucose metabolism was determined in 1975 (lung cancer), 2008 (ovarian cancer), and HT29 (colon cancer) cells by 3H-FDG–uptake studies 24 h after treatment. Normal MEFs were used as a control. Results are expressed as mean counts⋅microgram protein−1⋅initial dose−1 ± SEM. *P < 0.05 vs. carrier control-treated cells.
Noninvasive Monitoring of Disruption of Hsp90(α/β)/p23 Interactions in Live Mice by CP9.
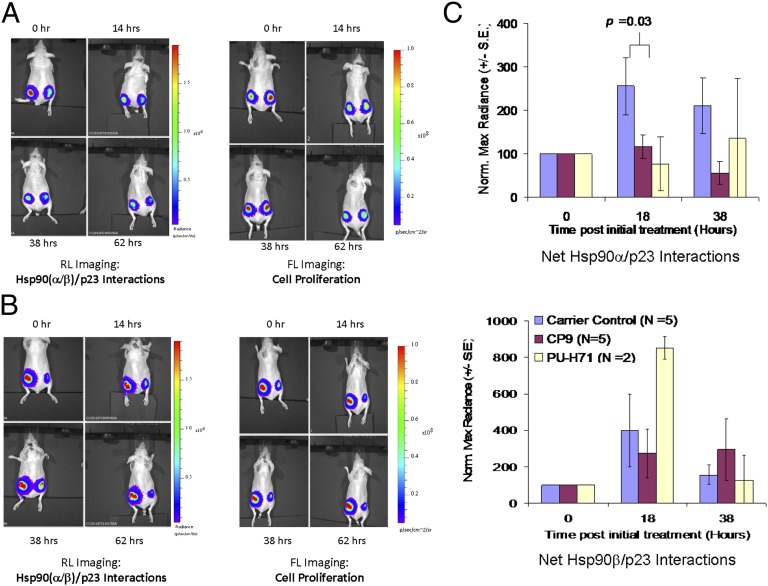
To monitor the inhibition of Hsp90(α/β)/p23 interactions by CP9 in live mice by BLI, we introduced a second reporter, FL-EGFP, into the 293T cells stably expressing Hsp90(α/β)/p23 split RL reporters (hereafter, 293T(α/β)-FG cells). We chose FL imaging because of its high sensitivity and the ease of sequentially performing FL and RL imaging (37, 38). Baseline RL and FL signals in each implanted tumor were determined. Based on our experience with Hsp90 inhibitors (30), we initially tested CP9 (80 mg/kg) in mice (n = 5) by i.p. injection, with four doses delivered immediately after and 16, 24, and 49 h after baseline imaging (Fig. 5A). An equal volume of DMSO was injected into the carrier control-treated group (n = 5) (Fig. 5B). Mice treated with 75 mg/kg PU-H71 (n = 2) served as positive controls. Mice were reimaged for Hsp90(α/β)/p23 interactions and cell proliferation via RL (Fig. 5B, Left) and FL (Fig. 5B, Right) imaging at the time points indicated. To account for the effect of cell number on Hsp90(α/β)/p23 interactions, RL signals were normalized to FL signals for each tumor at each time point, before normalization to signal at time 0 h.
Fig. 5.
BLI of the efficacy of CP9 in disruption of Hsp90(α/β)/p23 interactions and cell proliferation. To monitor the disruption of Hsp90(α/β)/p23 interactions by CP9 noninvasively and repetitively in living subjects, nude mice bearing 293T xenografts expressing the Hsp90(α/β)/p23 split RL reporters and EGFP-FL reporters [293T(α/β)/p23-FG] were used. Mice first were imaged for baseline RL signals from Hsp90α/p23 interactions (tumors on left flank) and Hsp90β/p23 interactions (tumors on right flank) using a cooled CCD camera upon i.v. injection of coelenterazine. The RL signals were allowed to decay for 30 min to allow clearance of coelenterazine, before the baseline cell proliferation was monitored by FL imaging upon i.v. injection of d-luciferin. Mice (n = 5 per group) then were randomized based on FL signals for treatment with CP9 (four doses of 80 mg/kg in DMSO) or carrier (DMSO; control) by i.p. injection. Mice (n = 2) treated with PU-H71 (75 mg/kg) served as positive controls. RL and FL signals were determined at different time points after treatment upon reinjection of coelenterazine and d-luciferin. (A and B) Representative RL (Left) and FL (Right) images from mice treated with CP9 (A) or carrier (control) (B). (C) RL and FL signals for each mouse were quantitated by drawing the same size ROI on each site at all time points. Maximum radiance (photons⋅s−1⋅cm2−1⋅steridian−1) from RL signals was normalized to that of FL signals at each site and at each time point to account for the effect of CP9 on cell proliferation. The RL/FL ratios then were normalized to that of time 0 h to monitor the individual change in Hsp90(α/β)/p23 interactions in each mouse over time. Results are expressed as mean ± SEM for the control and treatment groups for Hsp90α/p23 (Upper) and Hsp90β/p23 (Lower) interactions.
CP9 treatment led to the inhibition of RL signals in tumors bearing Hsp90α/p23 (left flank) and Hsp90β/p23 (right flank) xenografts, relative to time 0 h (Fig. 5A). Normalization of RL signals for cell number shows that CP9 inhibits Hsp90α/p23 interactions in tumor xenografts (P < 0.05 at 38 h vs. carrier control-treated mice) (Fig. 5C). CP9 was less effective in inhibiting Hsp90β/p23 interactions (P > 0.05 at both time points vs. carrier control-treated mice) (Fig. 5D). At 62 h after CP9 treatment, the RL/FL ratios in Hsp90(α/β)/p23 xenografts were similar to those in carrier control-treated mice (P > 0.05). Our data are consistent with selectivity of CP9 in binding to Hsp90α and inhibiting Hsp90α/p23 BLI signals in cell culture, relative to Hsp90β/p23.
CP9 Led to Inhibition of Glucose Metabolism in 293T Xenografts as Shown by Small-Animal [18F]Fluorodeoxyglucose PET/CT Imaging.
[18F]Fluorodeoxyglucose (18F-FDG) PET/CT has been used routinely for repetitive and noninvasive monitoring of chemotherapy responses in small animals and in humans (39, 40). Because CP9 inhibits glucose metabolism in cancer cells (Fig. 4B), we monitored its effect in tumor xenografts by small-animal 18F-FDG PET/CT, using the same cohort of mice used for BLI (Fig. 5). Baseline 18F-FDG uptake was determined before treatment with carrier control or CP9 (80 mg/kg) (Fig. 6A). The 18F-FDG uptake in each tumor site relative to the maximum percent injected dose per gram (maximum %ID/g) was determined upon normalization of injected dose (41). In carrier control-treated mice, 18F-FDG uptake in 293T tumors expressing Hsp90(α/β)/p23 RL reporters (n = 8) increased by 37 ± 18% at 43 h (Fig. 6B). On the other hand, 18F-FDG uptake in CP9-treated tumors (n = 10) decreased by 16 ± 9% (P < 0.005 relative to carrier control-treated mice). Therefore, CP9 inhibits glucose metabolism in tumor xenografts in live mice. We also analyzed the 18F-FDG uptake in the brains of mice using CT images to delineate boundaries. Relative to day 0, the maximum %ID/g of 18F-FDG uptake was 114 ± 11% in mice treated with carrier and 99 ± 4% in mice treated with CP9 (Fig. 6C). There was no statistical difference between the two groups (P > 0.05). Furthermore, there were no significant decreases in weight in CP9-treated mice compared with carrier control-treated mice at 43 h (P > 0.05). Thus, our current data do not indicate that CP9 poses significant toxicity in mice.
Fig. 6.
CP9 led to inhibition of glucose metabolism in tumor xenografts by PET/CT imaging but did not lead to significant degradation of Hsp90 client proteins. (A) To determine the downstream effects of CP9 on glucose metabolism in 293T(α/β)/p23-FG xenografts, small-animal PET/CT imaging was performed using 18F-FDG. To obtain anatomical information, mice were imaged using the CT module of the PET/CT machine before PET scanning. Baseline 18F-FDG uptake was determined at time 0 h before treatment with four doses of CP9 (n = 5 per group) or carrier (control; n = 4 per group). Mice were reimaged at 43 h. PET and maximum intensity projection of the PET/CT images of representative mice treated with CP9 (Left) or carrier (control) (Right) are shown. The dotted red circles outline the locations of Hsp90α/p23 xenografts on the left flanks and Hsp90β/p23 xenografts on the right flanks. The color bar represents %ID/g for 18F-FDG uptake in tumors in PET images. (B) Quantitation of the effect of CP9 on inhibition of glucose metabolism. The PET image of each mouse and each site at each time point was reconstructed and quantified using the OSEM 2D algorithm. The increase in max %ID/g at 43 h relative to 0 h is shown here as average ± SEM. CP9 led to significant decrease in glucose metabolism, compared with carrier control-treated mice (P < 0.005). (C) CP9 did not significantly inhibit 18F-FDG uptake in brain (P > 0.05 relative to carrier control-treated mice). (D) Validation of the mechanism of CP9 as an Hsp90 inhibitor in live mice. After the last imaging time point, tumors were excised from the mice, and the expression of Raf-1, pAkt, and total Akt was determined by Western blotting as in Fig. 3A. α-Tubulin was used as a loading control. CP9 did not lead to a significant decrease in the expression of Hsp90 client proteins, relative to carrier control-treated mice.
Ex Vivo Analyses of the Efficacy of CP9 in Degrading Hsp90 Client Proteins.
To determine if the inhibition of Hsp90(α/β)/p23 interactions also leads to degradation of Hsp90 client proteins in mice, tumors were excised after PET/CT imaging to analyze the expression of pAkt/total Akt and Raf-1 in tumor lysates. Fig. 6D shows that CP9 treatment did not lead to significant degradation of Hsp90 client proteins relative to carrier control-treated mice (P > 0.05). This observation is consistent with our imaging results at 62 h after CP9 treatment, which did not show any significant differences in Hsp90(α/β)/p23 interactions in CP9-treated and carrier control-treated mice (Fig. 5 C and D).
Evaluation of Efficacy of Structural Analogs of CP9 in Disrupting Hsp90(α/β)/p23 Interactions and Degrading Hsp90 Client Proteins.
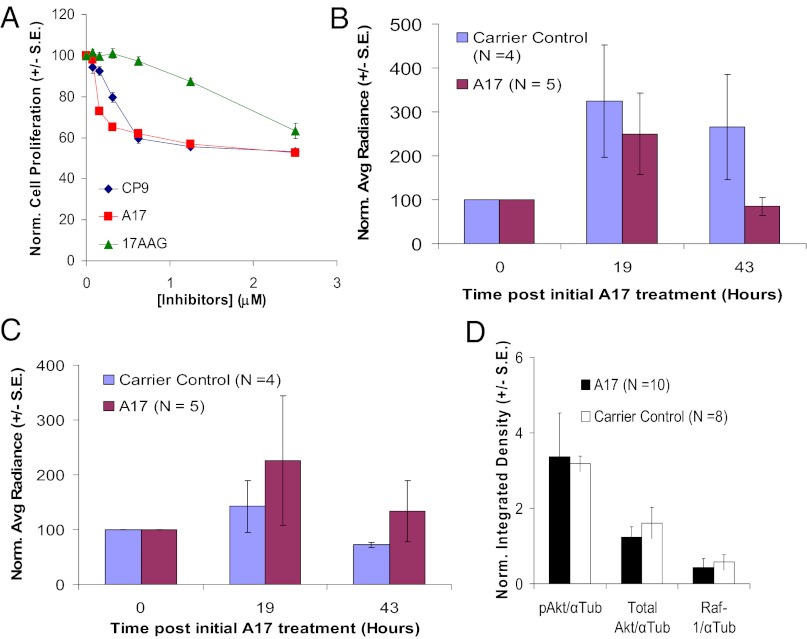
In attempt to improve the efficacy of CP9, we performed a structure–activity relationship (SAR) study using 62 analogs of CP9 with different modifications (Table S2). Their effects on Hsp90(α/β)/p23 interactions and cell proliferation in stable 293T(α/β)-FG cells were monitored by sequential RL (Fig. S3A) and FL imaging. CP9, PU-H71, and 17-AAG were used as positive controls. Compared with the parent compound CP9, the analogs A14 and A17 led to similar inhibition of Hsp90α/p23 and Hsp90β/p23 interactions (Fig. S3B, diamonds and squares, respectively). Time– and dose–response curves were established for the top 13 analogs with the lowest IC50 values for inhibition of Hsp90α/p23 interactions (Fig. S4A).
When we compared derivatives of CP9 with different R3 substitutions (A1–16, A18–31, A35, A37, A39, A42–44, A46, A47, A49, A51, A53, A55, A56, and A59–62), we found that an aromatic moiety is required and that aliphatic substitution led to diminished efficacy in disrupting Hsp90(α/β)/p23 interactions. A single small ortho substitution on the aromatic ring (as in A14 and A23) and methyl substitution in the meta position (A29 and A61) were tolerated. In CP9 analogs with various R1 substitutions (A17, A45, A48, A50, and A57), a five-member aromatic ring seems to be required for binding; analogs containing phenyl rings with various substitutions in the R1 position exhibited lower activity. Only replacement of the thiophen moiety in CP9 by a furanyl substitution (A17) was tolerated and led to higher potency. Methyl substitution in the R2 position was advantageous: A34 and A50 tend to have higher affinity than A40 and A45, respectively.
The best analogue with the lowest IC50 for inhibition of Hsp90α/p23 interactions was A17, which reduced the IC50 for the inhibition of Hsp90α/p23 signals by threefold compared with CP9 (0.15 vs. 0.45 μM) and which had a twofold-lower IC50 for growth inhibition (Fig. 7A). To determine if the reduction in IC50 for Hsp90(α/β)/p23 interactions makes A17 a more efficacious inhibitor of Hsp90, 293T-FG cells were treated with CP9 and A17. PU-H71 and 17-AAG were used as positive controls. A17 was more effective than CP9 in degrading pAkt/total Akt and Raf-1 (Fig, S4B). Because CP9 and its analogs inhibit Hsp90(α/β)/p23 interactions and lead to the degradation of Hsp90 client proteins, we have identified a class of Hsp90 inhibitors. Because A17 has a lower partition coefficient than CP9 (2.8 vs. 4.1), it may be more hydrophilic with less nonspecific binding of serum proteins and better bioavailability in tumors (42). Contrary to cell-culture results, treatment with A17 did not lead to significant decreases in Hsp90(α/β)/p23 interactions in tumor xenografts as compared with carrier control-treated mice using the same dosing regimen (P > 0.05 vs. carrier control-treated mice) (Fig. 7 B and C), perhaps because the bioavailability of A17 led to insufficient intratumoral concentrations for inhibiting Hsp90(α/β)/p23 interactions and subsequently degrading Hsp90 client proteins (Fig. 7D). Despite this outcome, we have established a SAR study platform that allows rapid screening and evaluation of more potent CP9 analogs, first in cell culture and subsequently in live mice.
Fig. 7.
Efficacy of lead CP9 analog A17 in cell culture and in live mice. (A) Effect of A17, CP9, and 17-AAG on proliferation of 293T cells. 293T cells were treated with different concentrations of inhibitors in triplicate for 24 h before cell proliferation was determined by Alamar Blue assay. Emission signals were normalized to those of carrier control-treated cells and are expressed as mean ± SEM. (B) Effect of A17 on Hsp90α/p23 interactions in mice bearing 293T-FG xenografts. Mice were treated with four doses of A17 (80 mg/kg) after the establishment of baseline RL signals (Hsp90α/p23) and FL signals (cell proliferation) and were reimaged at the indicated time points. The net effect of A17 on Hsp90α/p23 interactions was determined as in Fig. 5C. Relative to carrier control-treated mice, A17 led to decreases in Hsp90α/p23 interactions at 43 h, but the differences were not statistically significant (P > 0.05). (C) Effect of A17 on Hsp90β/p23 interactions in mice bearing 293T-FG xenografts was determined as A. A17 did not lead to a significant decrease in Hsp90β/p23 interactions (P > 0.05 relative to carrier control-treated mice). (D) Ex vivo analyses of Hsp90 client proteins in excised tumors from B and C. The expression of pAkt, total Akt, and Raf-1 was determined by Western blotting as in Fig. 6D, and α-tubulin was used a loading control. A17 did not lead to significant decreases in Hsp90 client proteins (P > 0.05 relative to carrier control-treated mice).
Discussion
In the current work we successfully coupled multimodality molecular imaging into HTS and discovered a class of 2-{[6-(trifluoromethyl)pyrimidin-2-yl]thio}acetamide–based Hsp90 inhibitors. We used a dual RL/FL reporter system for monitoring isoform-selective Hsp90(α/β)/p23 interactions and cell proliferation in intact cells. Among the three lead compounds, we focused on CP9 rather than CP1 and CP18 because it was more potent in degrading Hsp90 client proteins including Raf-1, pAkt, and total Akt (Fig. 2A) as well as in disrupting Hsp90(α/β)/p23 interactions (Fig. 2B). Furthermore, CP9 exhibits selectivity in binding to purified Hsp90α and inhibiting Hsp90α/p23 interactions. Therefore we focused our efforts on characterizing CP9 as an Hsp90 inhibitor. We also noninvasively and repetitively monitored the inhibitory effects of CP9 on Hsp90(α/β)/p23 interactions and glucose metabolism in tumor xenografts. Finally, we performed SAR studies and identified a more potent analog of CP9, A17.
CP9 as a 2-{[6-(Trifluoromethyl)Pyrimidin-2-yl]Thio}Acetamide–Based Hsp90 Inhibitor.
CP9 competes with 3H-17-AAG for binding to purified Hsp90(α/β), reduces uptake of 3H-17-AAG in intact HT29 cancer cells, and disrupts Hsp90(α/β)/p23 interactions in intact 293T cells (Fig. 3). Our data indicated that CP9 binds to Hsp90. CP9 treatment leads to the degradation of Hsp90 client proteins and inhibition of glucose metabolism, mammalian thymidine kinase, and cell proliferation in multiple cancer cell lines. The inhibition of Hsp90 chaperone activities is specific for cancer cells, because CP9 had no effect on normal MEFs. The small chemical library that contains CP9 was used by other investigators in bioassays that were not specifically designed for monitoring Hsp90(α/β)/p23 interactions. Targets that were reported in PubChem (http://pubchem.ncbi.nlm.nih.gov/) to be affected by CP9 include β-adrenergic receptor kinase 1, polypyrimidine tract-binding protein 1α, and 5-hydroxytryptamine receptor 1E. It has been reported in PubChem that CP9 inhibits proliferation, protein assembly, and kinase activities in different cancer cell lines. Because Hsp90 is involved in the regulation of kinases, receptors, and protein binding/folding, our data are consistent with others who have reported the downstream effects of CP9 (43).
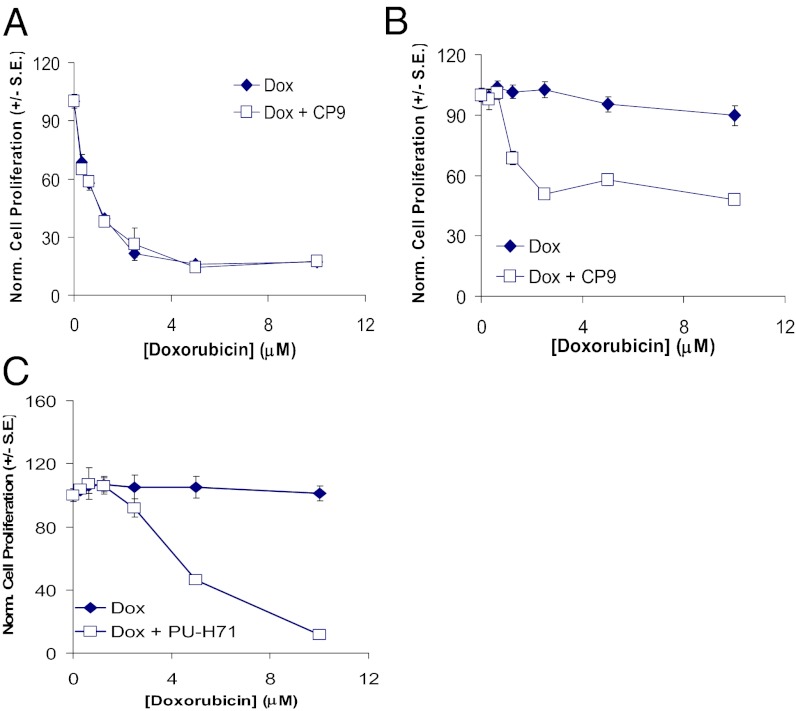
Both Hsp90 isoforms (α and β) are expressed in cancer cells, but Hsp90β is constitutively expressed, whereas the expression of Hsp90α is highly inducible during stress and drug treatment (29, 44). Using our genetically encoded split Renilla reporter system, we were able to decipher the individual contributions of each isoform in determining the sensitivity of Hsp90 inhibitors. It was reported that the transcription of MDR1 is regulated by HSF-1, which in turn is regulated by Hsp90α (45). To determine the biological significance of CP9 selectivity in inhibiting Hsp90α/p23 interactions, we used V79 lung cancer cells and V79/ADR cells that overexpress multidrug-resistant protein and are insensitive to growth inhibition by doxorubicin (DOX). V79 and V79/ADR cells were treated with DOX alone or in the presence of CP9. In V79 cells that do not express MDR-1, the sensitivity to DOX was not affected by CP9 (Fig. 8A). On the other hand, the addition of CP9 sensitized V79/ADR cells to DOX, as shown by the decrease in cell proliferation relative to treatment with DOX alone (Fig. 8B). Similar results were observed with the known Hsp90 inhibitor PU-H71 (Fig. 8C), which is more selective for disruption of Hsp90α/p23 interactions (30). Our results also are consistent with the observation that derivatives of Hsp90 inhibitor peptides sensitize cancer cells with overexpression of MDR to epirubicin (a class of anthracyclines that includes DOX) (46). Further studies will be required to determine the mechanisms of CP9 in reversal of MDR-1–mediated DOX resistance.
Fig. 8.
Inhibition of cell proliferation of DOX-resistant V79/ADR lung cancer cells by CP9 and the Hsp90 inhibitor PU-H71 in cell culture. To determine if CP9 can inhibit proliferation of DOX-resistant lung cancer cells, V79 lung cancer cells and V69/ADR cells that are resistant to DOX were treated with DOX (0.31–10 μM) in the presence or absence of CP9 (8 μM) for 24 h before determination of cell number by Alamar Blue assay. (A) CP9 did not affect the sensitivity of drug-sensitive V79 cells to DOX. (B) CP9 partially restored the sensitivity of drug-resistant V79/ADR cells to DOX. (C) PU-H71 restored the sensitivity of drug-resistant V79/ADR cells to DOX.
We noninvasively and repetitively monitored the inhibitory effects of CP9 in tumor xenografts in live mice by multimodality molecular imaging. BLI shows that CP9 selectively inhibits Hsp90α/p23 interactions relative to Hsp90β/p23 interactions (Fig. 5), and 18F-FDG PET/CT in the same cohort shows that CP9 inhibits glucose metabolism (Fig. 6). The selective inhibition of Hsp90α/p23 interactions and reduction in glucose metabolism in mice were consistent with the cell-culture results. However, CP9 did not decrease the expression of the Hsp90 client proteins significantly. Our results illustrate the challenges of transitioning from cell culture to animal studies in which the efficacy of lead compounds is limited by their tumor bioavailability (47).
Unified System Accelerated Drug Discovery, Mechanism Validation, and Lead Optimization in Living Subjects.
In attempt to improve the potency of CP9, SAR studies were performed on the 62 analogs of CP9. The efficacy in degrading Hsp90 client proteins corresponds to the inhibition of BLI signals (Fig. S4). Furthermore, the most potent analog, A17, was more effective than CP9 in inhibiting Hsp90(α/β)/p23 interactions and degrading Hsp90 client proteins in cell culture (Fig. S4B) but not in live mice (Fig. 6D). Nevertheless, the class of compounds that we have identified is distinct from PU-H71, which was used as a positive control for our experiments. Our data confirm the CP9 family as 2-{[6-(trifluoromethyl)pyrimidin-2-yl]thio}acetamide–based Hsp90 inhibitors. However, further work in medicinal chemistry is necessary to optimize our lead compound in terms of potency, bioavailability, and other parameters important for therapeutic efficacy. Furthermore one could use some of the distinctive components of the chemical structure of CP9 further to derivatize chemically the compounds CP18 and CP19, which exhibited the lowest IC50 values. Our strategy allows rapid evaluation of such structural analogs using small quantities and will reduce the costs of compound syntheses.
In summary, we have identified and validated a class of Hsp90 inhibitors by coupling molecular imaging with HTS. Our work-flow allows rapid identification of cell-permeable lead compounds in cell culture, followed by monitoring of efficacy in live mice. This approach will accelerate significantly the development of the next generation of therapeutics aimed at inhibiting specific chaperone–protein interactions.
Materials and Methods
Chemicals, Enzymes, and Reagents.
Coelenterazine was purchased from Nanolight Technology. Cell culture media, FBS, penicillin/streptomycin (P/S), and 4–12% (wt/vol) gradient SDS/PAGE gels were purchased from Invitrogen. Puromycin hydrochloride and 17-AAG were purchased from Invivogen. Purine-scaffold Hsp90 inhibitor PU-H71 (13, 14, 48) was dissolved as 7.65 mM PBS stock and stored at −20 °C. The slow-kinetic RL substrate EnduRen (Promega) was dissolved in DMSO as 34 mg/mL stock and stored at −20 °C. Doxorubicin hydrochloride was purchased from Sigma.
Cell Culture.
All cell lines used in this study were purchased from American Type Culture Collection and were cultured with their respective medium supplemented with 10% FBS and 1% P/S. 293T HEK cancer cells stably expressing Hsp90(α/β)/p23 split RL reporters (30) were maintained in Eagle's Minimal Essential Medium medium and 1.5 μg/mL of puromycin. MCF-7 human breast adenocarcinoma cells and 2008 human ovarian cancer cells were maintained in RMPI medium. U87MG human glioblastoma cells, SKBr3 human breast carcinoma cells, 1975 lung cancer cells, HUH-7 and 4–4 liver cancer cells (received from Dean Felsher, Stanford University, Stanford, CA), PC-3 human prostate cancer cells, and V79 and V79/ADR hamster lung cancer cells (resistant to doxorubicin) were cultured with DMEM.
HTS of Small-Molecule Chemical Libraries Using 293T Cells Stably Expressing Hsp90(α/β)/p23 Split RL Reporters.
To identify Hsp90 inhibitors the LOPAC from Sigma was used. HTS was performed at the Stanford High-Throughput Bioscience Center. 293T HEK cells (8 × 103) from columns 1–22 (E & K Scientific) expressing Hsp90α/p23 or Hsp90β/p23 split RL reporters were plated in each well of the 384-well white-bottomed plate in 60 μL of medium using the Wellmate matrix (Thermo Scientific) and were allowed to attach for 24 h. Sixty microliters of medium (without cells) was added to columns 23 and 24 to control for background signals caused by substrate alone. Then approximately 100 nL of each LOPAC compound (10 mM stock in DMSO) was added to each well in columns 3–22 (final concentration ∼17 μM) using a PinTool (V&P Scientific) on a Sciclone ALH3000 (Caliper Sciences). Cells in columns 1 and 2 that were not treated with the compounds were used to determine the baseline signals from Hsp90(α/β)/p23 interactions.
Baseline-complemented RL activities were determined 90 min after the addition of 20 μL of EnduRen (10 μM final concentration) (Promega) using the Analyst GT Multimode Reader (Molecular Devices), with an acquisition time of 0.2 s per well. Cells were incubated with compounds for 24 h before redetermination of complemented RL activities and were normalized to the mean RL activities of untreated cells in columns 1 and 2. Another small-molecule chemical library with 30,176 compounds (SPECS) also was used in screening modulators of Hsp90(α/β)/p23 inhibitors (at 8.3 μM, one drug per well for initial screening). For the dose–response confirmation assay the compounds were tested in duplicate in an eight-point (0.156–20 μM) dose–response assay for RL activities for both Hps90(α)/p23 and Hps90(β)/p23 interactions. The effect of each compound on cell proliferation at 20 μM was determined using the Cell Titer-Blue assay (Promega) in duplicate wells.
Elimination of Nonspecific Compounds.
To determine if the 19 lead hit compounds were nonspecific inhibitors of RL activities, 3 × 104 293T cells stably expressing full-length RL were plated in each 96-well black-walled plate for 24 h. They were treated for 4–24 h with different compounds at a 10-fold higher concentration than their respective IC50 for inhibition of Hsp90α/p23 BLI signals. Carrier-treated (0.1% DMSO) cells were used as the positive control. RL signals were determined using a cooled CCD camera upon the addition of EnduRen (90 min, final concentration of 10 μM) and were normalized to that of carrier control-treated cells. Compounds that inhibited RL signals by more than 20% were excluded from further analyses. Other toxic and nonspecific compounds were eliminated based on database analyses of previous screening assays performed at the Stanford High-Throughput Bioscience Center.
Determination of Efficacy of Lead Compounds in Disrupting Hsp90(α/β)/p23 Interactions.
To determine the effect of lead compounds on disruption of Hsp90(α/β)/p23 interactions in intact cells, 3.5 × 104 293T cells stably transfected with split RL reporters (30) were plated in each well in the 96-well black-walled plate (Costar, Corning) and were allowed to attach for 24 h before treatment with different concentrations of the lead compounds for 24 h. Then 10 μg/mL of EnduRen (in 50 μL of cell-culture medium) was added to each well for 1.5 h, and RL activities were determined. BLI signals were normalized to that of the cell number for each well [determined by AlamarBlue assay (Invitrogen)] before normalization to carrier control-treated cells.
Western Blotting and Coimmunoprecipitation.
To validate the mechanisms of the lead compounds and their analogs for inhibiting Hsp90(α/β)/p23 interactions, 293T cells stably expressing the Hsp90(α/β)/p23 split reporters were treated with 5 μM of CP1, CP9, CP18, or carrier (control) for 24 h. Cells treated with 2 μM of PU-H71 served as a positive control. Western blotting of Hsp70, pAkt, and total Akt, and co-immunoprecipitation of Hsp90(α/β)/p23 interactions were performed as described previously (30). The expression of Raf-1 was determined using rabbit polyclonal antibodies against Raf-1 (0.5 μg/mL) (Abcam).
3H-FDG and 3H-FLT Cell-Uptake Studies.
To determine the downstream effects of lead compounds on glucose metabolism, 1.5 × 105 293T, 1975, and 2008 cells and MEFs were plated in a 24-well plate for 24 h before treatment with different concentrations of CP9 for 24 or 48 h and subsequent incubation with 1 μCi of 3H-FDG in 500 μL of medium for 1 h at 37 °C, as described previously (49). Cells treated with 2.5 μM of PU-H71 served as a positive control. The total counts in each sample were normalized to that of the dose added to each well and to the amount of protein and were expressed as mean counts⋅dose−1⋅microgram protein−1 ± SEM. Likewise, the effect of CP9 on cell proliferation was determined using 1 μCi of 3H-FLT per well after 2 h of incubation at 37 °C (50).
Purified Hsp90(α/β) Binding Assay.
To verify the binding of CP9 to Hsp90α and Hsp90β in vitro, a displacement assay using 3H-17-AAG was performed. One microgram of purified Hsp90α or Hsp90β was incubated with different concentrations of CP9 or DMSO carrier (control) in 50 μL of HBS-P+ binding buffer (Biacore) for 1 h on ice on a shaking platform. 17-AAG (final concentration of 200 μM) was used as a positive control. Duplicate samples were used for each condition. One microliter of 3H-17-AAG (final concentration of 0.5 μM) was added, and the mixture was incubated for another 30 min at room temperature. Unbound 3H-17-AAG and inhibitors were removed using the 7-kDa Zeba desalt column (Thermo Scientific). The amount of 3H-17-AAG that remained bound to Hsp90α or Hsp90β was determined by scintillation counting.
Displacement of 3H-17-AAG Uptake by CP9 in HT29 Cells.
To determine if CP9 binds to cellular Hsp90, an uptake study of 3H-17-AAG (Moravek Biochemicals) was performed. HT29 cells (1 × 105 in 500 μL of medium) were plated in each well of a 24-well plate and were allowed to attach for 24 h. Cells were incubated with 0.5 μM 3H-17-AAG in the presence of 0.4–12.5 μM CP9 or carrier (control) for 1 h at 37 °C. PU-H71 (5 μM) was used as a positive control. This incubation was followed by two washes with 500 μL of PBS and 1 h of washout in 500 μL Hank's Balanced Salt Solution (HBSS) to remove unbound 3H-17-AAG. Cells were lysed on ice for 15 min in 300 μL of T-PER tissue extraction buffer in the presence of protease and phosphatase inhibitors (Thermo Scientific), and cell lysates were prepared for scintillation counting and protein determination. The total counts in each sample were normalized to that of the dose added to each well and to the amount of protein and are expressed as mean counts⋅dose−1⋅microgram protein−1 ± SEM.
Optical CCD Imaging in Live Mice.
Animal care and handling were performed in accordance with Stanford University Animal Research Committee guidelines. Mice were gas anesthetized using isofluorane (2% in 100% oxygen, 1 L/min) during all injection and imaging procedures and were kept at 37 °C. Mice were imaged using a cooled CCD camera (IVIS 200; Caliper Life Sciences). Tumor establishment and BLI of 293T cells stably expressing Hsp90(α/β)/p23 split RL reporters and FL-EGFP in 7-wk-old female nude mice (nu/nu; Charles River) were performed as described previously (30). Baseline RL activity in the implanted tumors in live mice was determined by i.v. injection of 30 μg coelenterazine (in 150 μL of 5% ethanol/95% PBS) and image acquisitions of 3 min. After a 30-min wait for RL signals to decay, FL activities were determined by i.v. injection of 163 μg of d-luciferin (Biosynth, Itasca, IL) in 100 μL PBS with image acquisition of 10 sequences (15 s each) to obtain the peak maximum radiance. One set of mice (n = 5 per group) was injected i.p. with 80 mg/kg CP9 dissolved in 100% DMSO in a final volume of 60 μL. Another set of mice (n = 5) was treated with an equal volume of DMSO as control. At different time points after treatment, follow-up RL and FL imaging was performed to monitor the effects of CP9 on complemented Hsp90(α/β)/p23 interactions and cell proliferation. The maximum radiance of RL was divided by that of FL signals at each time point, before normalization to that of time 0 h for each individual mouse, and was expressed as average radiance ± SEM for each treatment group.
Mice were euthanized after the last imaging time points, and tumors were excised and homogenized in tissue extraction buffer in the presence of Halt Complete protease and phosphatase inhibitors (all from Pierce). Protein concentrations were determined by the Bio-Rad Protein DC assay. Expression of pAkt/total Akt, Raf-1, and α-tubulin was determined by Western blotting (30). Western blot images were quantitated using Image J (National Institutes of Health) and were expressed as the ratio of target protein to α-tubulin for each treatment group.
PET/CT Imaging of Glucose Metabolism in Live Mice.
To determine the effects of CP9 on glucose metabolism in 293T xenografts stably expressing Hsp90(α/β)/p23 split RL reporters and FL-EGFP, baseline 18F-FDG uptake in each tumor site for each mouse was determined by small-animal PET imaging using the Inveon PET/CT scanner (Siemens). Mice were placed on a custom-built four-mice holder first for CT image acquisition (632 slices at 206 μm) that was used both for photon attenuation correction and image coregistration with PET image data for anatomical information. A static 5-min PET scan then was performed for [18F]-FDG activity and was reconstructed using the Ordered Subsets Expectation Maximization (OSEM) 2D algorithm (159 slices with 1.5-mm resolution). Region of interest (ROI) analysis was performed using the Inveon Research Workspace software. The maximum %ID/g upon normalization to injected dose was determined before and 43 h after CP9 treatment.
Data Analysis.
Each experiment was repeated at least three times, and results are expressed as mean ± SEM. Statistical differences were determined by Student’s t test using P < 0.05 as the cutoff point. HTS data were analyzed using Accelrys Assay Explorer (Accelrys Inc., San Diego, CA).
Supplementary Material
Acknowledgments
We thank Drs. Tim Doyle and Frezghi Habte for scientific guidance and assistance with optical and PET/CT imaging and image analyses and for the use of the SCi3 Small Animal Imaging Service Center to generate data shown. This research was supported by National Institutes of Health Grant R01 CA082214 and National Cancer Institute In Vivo Cellular and Molecular Imaging Center P50 Grant at Stanford University (to S.S.G.), C.T.C. is the recipient of a Susan G. Komen Postdoctoral Fellowship.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Author Summary on page 14740 (volume 109, number 37).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205459109/-/DCSupplemental.
References
- 1.Zhang H, Burrows F. Targeting multiple signal transduction pathways through inhibition of Hsp90. J Mol Med (Berl) 2004;82:488–499. doi: 10.1007/s00109-004-0549-9. [DOI] [PubMed] [Google Scholar]
- 2.Neckers L, Ivy SP. Heat shock protein 90. Curr Opin Oncol. 2003;15:419–424. doi: 10.1097/00001622-200311000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Yano M, et al. Expression of hsp90 and cyclin D1 in human breast cancer. Cancer Lett. 1999;137:45–51. doi: 10.1016/s0304-3835(98)00338-3. [DOI] [PubMed] [Google Scholar]
- 4.Myung J-K, Afjehi-Sadat L, Felizardo-Cabatic M, Slavc I, Lubec G. Expressional patterns of chaperones in ten human tumor cell lines. Proteome Sci. 2004;2:8. doi: 10.1186/1477-5956-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Brazidec JY, et al. Synthesis and biological evaluation of a new class of geldanamycin derivatives as potent inhibitors of Hsp90. J Med Chem. 2004;47:3865–3873. doi: 10.1021/jm0306125. [DOI] [PubMed] [Google Scholar]
- 6.Chiosis G. Discovery and development of purine-scaffold Hsp90 inhibitors. Curr Top Med Chem. 2006;6:1183–1191. doi: 10.2174/156802606777812013. [DOI] [PubMed] [Google Scholar]
- 7.Kamal A, et al. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 2003;425:407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- 8.Vetcher L, et al. Rapid engineering of the geldanamycin biosynthesis pathway by Red/ET recombination and gene complementation. Appl Environ Microbiol. 2005;71(4):1829–1835. doi: 10.1128/AEM.71.4.1829-1835.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vilenchik M, et al. Targeting wide-range oncogenic transformation via PU24FCl, a specific inhibitor of tumor Hsp90. Chem Biol. 2004;11:787–797. doi: 10.1016/j.chembiol.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 10.Moulick K, et al. Synthesis of a red-shifted fluorescence polarization probe for Hsp90. Bioorg Med Chem Lett. 2006;16:4515–4518. doi: 10.1016/j.bmcl.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 11.Llauger L, et al. Evaluation of 8-arylsulfanyl, 8-arylsulfoxyl, and 8-arylsulfonyl adenine derivatives as inhibitors of the heat shock protein 90. J Med Chem. 2005;48:2892–2905. doi: 10.1021/jm049012b. [DOI] [PubMed] [Google Scholar]
- 12.Immormino RM, Kang Y, Chiosis G, Gewirth DT. Structural and quantum chemical studies of 8-aryl-sulfanyl adenine class Hsp90 inhibitors. J Med Chem. 2006;49:4953–4960. doi: 10.1021/jm060297x. [DOI] [PubMed] [Google Scholar]
- 13.He H, et al. 2006. Identification of potent water soluble purine-scaffold inhibitors of the heat shock protein 90. J Med Chem 49(1):381–390.
- 14.Chiosis G, Lucas B, Shtil A, Huezo H, Rosen N. Development of a purine-scaffold novel class of Hsp90 binders that inhibit the proliferation of cancer cells and induce the degradation of Her2 tyrosine kinase. Bioorg Med Chem. 2002;10:3555–3564. doi: 10.1016/s0968-0896(02)00253-5. [DOI] [PubMed] [Google Scholar]
- 15.Chiosis G, et al. Development of purine-scaffold small molecule inhibitors of Hsp90. Curr Cancer Drug Targets. 2003;3:371–376. doi: 10.2174/1568009033481778. [DOI] [PubMed] [Google Scholar]
- 16.Chiosis G. Targeting chaperones in transformed systems—a focus on Hsp90 and cancer. Expert Opin Ther Targets. 2006;10:37–50. doi: 10.1517/14728222.10.1.37. [DOI] [PubMed] [Google Scholar]
- 17.McDonald E, Jones K, Brough PA, Drysdale MJ, Workman P. Discovery and development of pyrazole-scaffold Hsp90 inhibitors. Curr Top Med Chem. 2006;6:1193–1203. doi: 10.2174/156802606777812086. [DOI] [PubMed] [Google Scholar]
- 18.Soga S, Shiotsu Y, Akinaga S, Sharma SV. Development of radicicol analogues. Curr Cancer Drug Targets. 2003;3:359–369. doi: 10.2174/1568009033481859. [DOI] [PubMed] [Google Scholar]
- 19.Proisy N, et al. Inhibition of Hsp90 with synthetic macrolactones: Synthesis and structural and biological evaluation of ring and conformational analogs of radicicol. Chem Biol. 2006;13:1203–1215. doi: 10.1016/j.chembiol.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 20.Solit DB, Chiosis G. Development and application of Hsp90 inhibitors. Drug Discov Today. 2008;13:38–43. doi: 10.1016/j.drudis.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Biamonte MA, et al. Heat shock protein 90: Inhibitors in clinical trials. J Med Chem. 2010;53:3–17. doi: 10.1021/jm9004708. [DOI] [PubMed] [Google Scholar]
- 22.Kim YS, et al. Update on Hsp90 inhibitors in clinical trial. Curr Top Med Chem. 2009;9:1479–1492. doi: 10.2174/156802609789895728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rowlands MG, et al. High-throughput screening assay for inhibitors of heat-shock protein 90 ATPase activity. Anal Biochem. 2004;327:176–183. doi: 10.1016/j.ab.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 24.Soti C, Vermes A, Haystead TA, Csermely P. Comparative analysis of the ATP-binding sites of Hsp90 by nucleotide affinity cleavage: A distinct nucleotide specificity of the C-terminal ATP-binding site. Eur J Biochem. 2003;270:2421–2428. doi: 10.1046/j.1432-1033.2003.03610.x. [DOI] [PubMed] [Google Scholar]
- 25.Smith-Jones PM, et al. 2004. Imaging the pharmacodynamics of HER2 degradation in response to Hsp90 inhibitors. Nat Biotechnol 22(6):701–706.
- 26.Oude Munnink TH, et al. (89)Zr-trastuzumab PET visualises HER2 downregulation by the HSP90 inhibitor NVP-AUY922 in a human tumour xenograft. Eur J Cancer. 2010;46:678–684. doi: 10.1016/j.ejca.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 27.Ramírez V, Mejía-Vilet JM, Hernández D, Gamba G, Bobadilla NA. Radicicol, a heat shock protein 90 inhibitor, reduces glomerular filtration rate. Am J Physiol Renal Physiol. 2008;295:F1044–F1051. doi: 10.1152/ajprenal.90278.2008. [DOI] [PubMed] [Google Scholar]
- 28.Cao X, et al. Non-invasive MRI tumor imaging and synergistic anticancer effect of HSP90 inhibitor and glycolysis inhibitor in RIP1-Tag2 transgenic pancreatic tumor model. Cancer Chemother Pharmacol. 2008;62:985–994. doi: 10.1007/s00280-008-0688-8. [DOI] [PubMed] [Google Scholar]
- 29.Chang YS, Lo CW, Sun FC, Chang MD, Lai YK. Differential expression of Hsp90 isoforms in geldanamycin-treated 9L cells. Biochem Biophys Res Commun. 2006;344:37–44. doi: 10.1016/j.bbrc.2006.03.157. [DOI] [PubMed] [Google Scholar]
- 30.Chan CT, et al. Molecular imaging of the efficacy of heat shock protein 90 inhibitors in living subjects. Cancer Res. 2008;68:216–226. doi: 10.1158/0008-5472.CAN-07-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inglese J, et al. Quantitative high-throughput screening: A titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci USA. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solit DB, Rosen N. Hsp90: A novel target for cancer therapy. Curr Top Med Chem. 2006;6:1205–1214. doi: 10.2174/156802606777812068. [DOI] [PubMed] [Google Scholar]
- 33.Workman P, Powers MV. Chaperoning cell death: A critical dual role for Hsp90 in small-cell lung cancer. Nat Chem Biol. 2007;3:455–457. doi: 10.1038/nchembio0807-455. [DOI] [PubMed] [Google Scholar]
- 34.Kotliarova S, et al. Glycogen synthase kinase-3 inhibition induces glioma cell death through c-MYC, nuclear factor-kappaB, and glucose regulation. Cancer Res. 2008;68:6643–6651. doi: 10.1158/0008-5472.CAN-08-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nair VD, Olanow CW. Differential modulation of Akt/GSK-3beta pathway regulates apoptotic and cytoprotective signaling responses. J Biol Chem. 2008;283(22):15469–15478. doi: 10.1074/jbc.M707238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dandekar M, Tseng JR, Gambhir SS. Reproducibility of 18F-FDG microPET studies in mouse tumor xenografts. J Nucl Med. 2007;48:602–607. doi: 10.2967/jnumed.106.036608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhaumik S, Lewis XZ, Gambhir SS. Optical imaging of Renilla luciferase, synthetic Renilla luciferase, and firefly luciferase reporter gene expression in living mice. J Biomed Opt. 2004;9:578–586. doi: 10.1117/1.1647546. [DOI] [PubMed] [Google Scholar]
- 38.Bhaumik S, Gambhir SS. Optical imaging of Renilla luciferase reporter gene expression in living mice. Proc Natl Acad Sci USA. 2002;99:377–382. doi: 10.1073/pnas.012611099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aliaga A, et al. A small animal positron emission tomography study of the effect of chemotherapy and hormonal therapy on the uptake of 2-deoxy-2-[F-18]fluoro-D-glucose in murine models of breast cancer. Mol Imaging Biol. 2007;9:144–150. doi: 10.1007/s11307-007-0091-6. [DOI] [PubMed] [Google Scholar]
- 40.de Geus-Oei L-F, Vriens D, van Laarhoven HWM, van der Graaf WTA, Oyen WJG. Monitoring and predicting response to therapy with 18F-FDG PET in colorectal cancer: A systematic review. J Nucl Med. 2009;50(Suppl 1):43S–54S. doi: 10.2967/jnumed.108.057224. [DOI] [PubMed] [Google Scholar]
- 41.Wiant D, Gersh JA, Bennett M, Bourland JD. Evaluation of the spatial dependence of the point spread function in 2D PET image reconstruction using LOR-OSEM. Med Phys. 2010;37:1169–1182. doi: 10.1118/1.3310381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma Q, Lu AYH. Pharmacogenetics, pharmacogenomics, and individualized medicine. Pharmacol Rev. 2011;63:437–459. doi: 10.1124/pr.110.003533. [DOI] [PubMed] [Google Scholar]
- 43.Zuehlke A, Johnson JL. Hsp90 and co-chaperones twist the functions of diverse client proteins. Biopolymers. 2010;93:211–217. doi: 10.1002/bip.21292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu YP, Kita K, Suzuki N. Yu-ping Wu KKNS Involvement of human heat shock protein 90 alpha in nicotine-induced apoptosis. Int J Cancer. 2002;100:37–42. doi: 10.1002/ijc.10449. [DOI] [PubMed] [Google Scholar]
- 45.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10:537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molnár J, et al. 2007. Effects of nontoxic heat shock protein 90 inhibitor peptide derivatives on reversal of MDR of tumor cells. In Vivo 21(2):429–433.
- 47.James ML, Gambhir SS. A molecular imaging primer: Modalities, imaging agents, and applications. Physiol Rev. 2012;92(2):897–965. doi: 10.1152/physrev.00049.2010. [DOI] [PubMed] [Google Scholar]
- 48.Zhou V, et al. A time-resolved fluorescence resonance energy transfer-based HTS assay and a surface plasmon resonance-based binding assay for heat shock protein 90 inhibitors. Anal Biochem. 2004;331:349–357. doi: 10.1016/j.ab.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 49.Tseng JR, et al. Preclinical efficacy of the c-Met inhibitor CE-355621 in a U87 MG mouse xenograft model evaluated by 18F-FDG small-animal PET. J Nucl Med. 2008;49:129–134. doi: 10.2967/jnumed.106.038836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tseng JR, et al. Reproducibility of 3′-deoxy-3′-(18)F-fluorothymidine microPET studies in tumor xenografts in mice. J Nucl Med. 2005;46:1851–1857. [PMC free article] [PubMed] [Google Scholar]