Abstract
Nectins are members of the Ig superfamily that mediate cell–cell adhesion through homophilic and heterophilic interactions. We have determined the crystal structure of the nectin-2 homodimer at 1.3 Å resolution. Structural analysis and complementary mutagenesis studies reveal the basis for recognition and selectivity among the nectin family members. Notably, the close proximity of charged residues at the dimer interface is a major determinant of the binding affinities associated with homophilic and heterophilic interactions within the nectin family. Our structural and biochemical data provide a mechanistic basis to explain stronger heterophilic versus weaker homophilic interactions among these family members and also offer insights into nectin-mediated transinteractions between engaging cells.
Keywords: nectin-2 dimer, cell adhesion molecule, immunoglobulin superfamily, X-ray crystallography
Nectins are members of the Ig superfamily (IgSF) that engage in calcium-independent adhesive interactions critical for a wide range of biological processes (1, 2). The nectin family is composed of four single-pass type I membrane glycoproteins (nectin-1 to nectin-4), whose extracellular domains are composed of an N-terminal variable-type Ig (IgV) domain, followed by two constant-type Ig (IgC) domains (1–3). These ectodomains participate in a complex network of interactions that includes both homophilic and heterophilic adhesive associations between interacting cells (Fig. S1). The cytoplasmic tails of the nectins bind the F-actin–binding protein afadin and support connections with the actin cytoskeleton similar to those observed in the cadherin and integrin systems (4). Nectin-based cell–cell adhesions are prominent in adherens junctions in fibroblast and epithelial cells and in synaptic junctions in neurons, either autonomously or in concert with other cell adhesion molecules like cadherins (1, 2, 5).
Although homophilic and heterophilic transinteractions of nectins are implicated in cell–cell adhesion, several studies suggest that heterophilic interactions are stronger than homophilic interactions (6, 7). Furthermore, it has been shown that heterophilic interactions between nectins contribute to several critical cellular functions (8, 9). For example, nectin-2 and nectin-3 are expressed in Sertoli cells and spermatids, respectively, and their heterophilic transinteraction regulates the organization of the Sertoli cell-spermatid junctions (8). It has also been shown that the homozygous deletion of either nectin-2 or nectin-3 leads to male-specific infertility in mice (10, 11). Another remarkable example is provided by the interaction of nectin-1 and nectin-3. These proteins are expressed in commissural axons and floor plate cells, respectively, during the early development of the vertebrate central nervous system, and this heterophilic transinteraction is critically involved in the control of axonal guidance (12). In addition to cell–cell adhesion, the heterophilic interactions of nectins with a member of other protein families lead to different biological outcomes such as immune modulation and host–pathogen interactions (13–15).
To define the molecular and structural determinants underlying the homophilic and heterophilic interactions between nectins, we examined the biochemical, biophysical, and structural properties of human nectin-2, a representative member of the family. The 1.3 Å structure of the nectin-2 IgV homodimer defined the chemical and physical determinants contributing to the homophilic interface. In combination with complementary biochemical data, these studies provide a mechanistic basis for the observation that heterophilic interactions are stronger than homophilic interactions among these family members and suggest a model for nectin-associated transinteractions.
Results and Discussion
IgV Domain of Nectin-2 Exists as a Dimer in Solution.
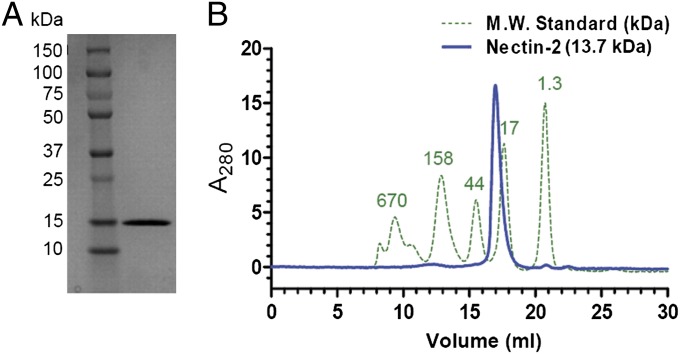
The human nectin-2 IgV domain (molecular weight of 13.68 kDa) was expressed as inclusion bodies in Escherichia coli, refolded, and purified by size-exclusion chromatography (SEC) (Fig. 1A). The purified protein eluted as a monodisperse peak on a calibrated gel-filtration column with apparent molecular mass consistent with a dimeric species (Fig. 1B). Analytical sedimentation equilibrium results are consistent with a dimer (molecular weight of 24,825 Da) characterized by an equilibrium dissociation constant (Kd) of <10 μM. Together, SEC and sedimentation equilibrium analysis demonstrate that the IgV domain of human nectin-2 exists as a dimer in solution.
Fig. 1.
(A) SDS/PAGE showing refolded and purified nectin-2 (13.7 kDa) in the right lane. Molecular weight standards are in the left lane. (B) Nectin-2 (blue) and molecular weight standards (green dotted) were analyzed by size-exclusion chromatography on a Superdex 200 column. The standards are labeled with their molecular weights (in kDa) on the top of each peak. Nectin-2 eluted after the 44-kDa peak and just before the 17-kDa peak, which indicates a dimer in solution.
Crystal Structure of Nectin-2 IgV Exhibits a Dimeric Assembly.
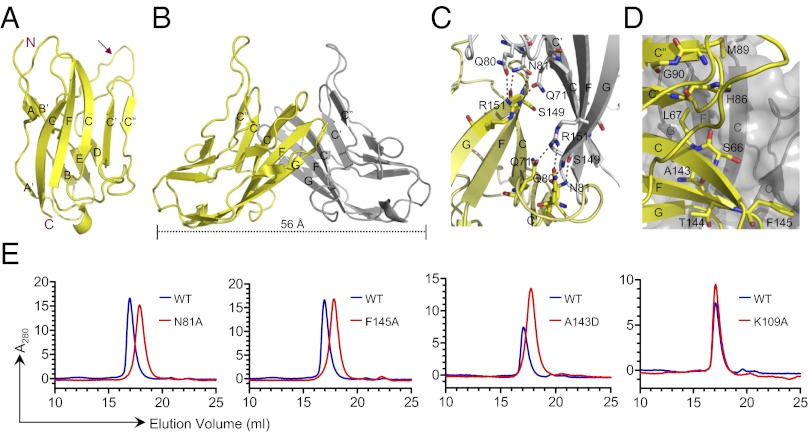
The structure of nectin-2 was determined and refined at 1.3 Å resolution with Rwork and Rfree of 15.2 and 17.3%, respectively (Table S1). The asymmetric unit contains one copy of the nectin-2 IgV domain, which exhibits the classic two-layer β-sandwich topology observed in other IgV structures, with front and back sheets composed of the GFCC′C′′ and ABED strands, respectively (Fig. 2A). Similar to other conventional IgSF members, the nectin-2 IgV domain possesses the hallmark disulfide bond that links the B and F strands. A distinctive structural feature of nectin-2 is the insertion of a long loop between the D and E strands (Fig. 2A), which is absent in other nectins (Fig. 3). Although this insertion is present in all nectin-2 orthologs, the sequences are not conserved in this region. Molecules related by twofold crystallographic symmetry generate a dimer, with the interface formed by the nearly orthogonal association of C, C′, C′′, and F strands from the front sheets of two engaging IgV domains (Fig. 2B). This organization results in a “kinked” dimer with an end-to-end distance of 56 Å (Fig. 2B), which is similar to the quaternary structure observed in numerous other dimers involving IgSF members (Fig. S2).
Fig. 2.
(A) Structure of the IgV domain of human nectin-2 showing a classical two-layer β-sandwich topology. The front and back sheets of the domain are composed of the GFCC′C′′ and ABED strands, respectively. The unusual long loop between the D and E strands is identified by an arrow. (B) Two monomers (yellow and gray) interact in a nearly orthogonal fashion to form the dimer, resulting in an end-to-end distance of ∼56 Å. The dimer interface is formed by the orthogonal association of the front β-sheets with predominant contribution from the C, C′, C′′, and F strands of each monomer. (C) Residues participating in hydrogen bonds at the dimer interface are shown in ball-and-stick representation (dashed lines represent hydrogen bonds). (D) Residues participating in van der Waals contacts at the dimer interface are shown by ball-and-stick representation from one molecule only. (E) Confirmation of dimer interface by size-exclusion chromatography. The elution profile of wild-type nectin-2 is shown by the blue lines, and the mutants N81A, F145A, A143D, and K109A are represented by the red lines. In all of the cases except K109A (which is located outside of the dimer interface), the elution volume of the mutants increases significantly.
Fig. 3.
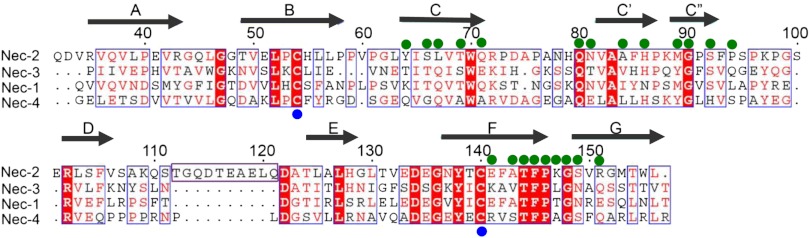
Structure-based sequence alignment of human nectins. The secondary structure of nectin-2 IgV domain is labeled on the top of the alignment. Residues with similar properties are marked in red, whereas identical residues are in white with red shading. Interfacial residues (green circle) and the cysteine residues involved in the formation of disulfide bond between B and F strands (blue circle) are marked. The long loop between the D and E strands of nectin-2, which is absent in other nectins, is also shown.
Homophilic Interface of Nectin-2.
The nectin-2 homophilic interface buries a total surface area of ∼1,823 Å2 , which is significantly larger than most IgV domain interfaces (Table S2). Ten residues (Gln-71, Gln-80, Asn-81, Ser-149, Arg-151, and their symmetry mates) are involved in eight potential hydrogen bonds (Fig. 2C). van der Waals contacts also make an important contribution to this homophilic interface, including Ser-66, Leu-67, His-86, Met-89, Gly-90, Ala-143, Thr-144, and Phe-145, which are partially or completely buried at the dimer interface (Fig. 2D). A complete list of interfacial residues is provided in Table S3.
To confirm the homophilic interface observed in the crystalline state, we examined a series of mutants. Point mutations (N81A, M89A, A143D, and F145A) in the putative interface severely affected the homophilic association and resulted in a strictly monomeric population in SEC, whereas K109A, which is outside the putative interface, had no effect (Fig. 2E). Despite the large extent of this interface, these data suggest that the homophilic interactions in the nectin family are highly sensitive to perturbation, as the change of a single residue significantly alters the monomer–dimer equilibrium. This sensitivity may be related to the twofold symmetric organization of the homophilic dimer, as a single mutation in the primary sequence results in the disruption of two sets of contacts at the binding interface.
Close Proximity of Two Charged Residues at the Dimer Interface: A Major Determinant of Homophilic and Heterophilic Binding Affinities.
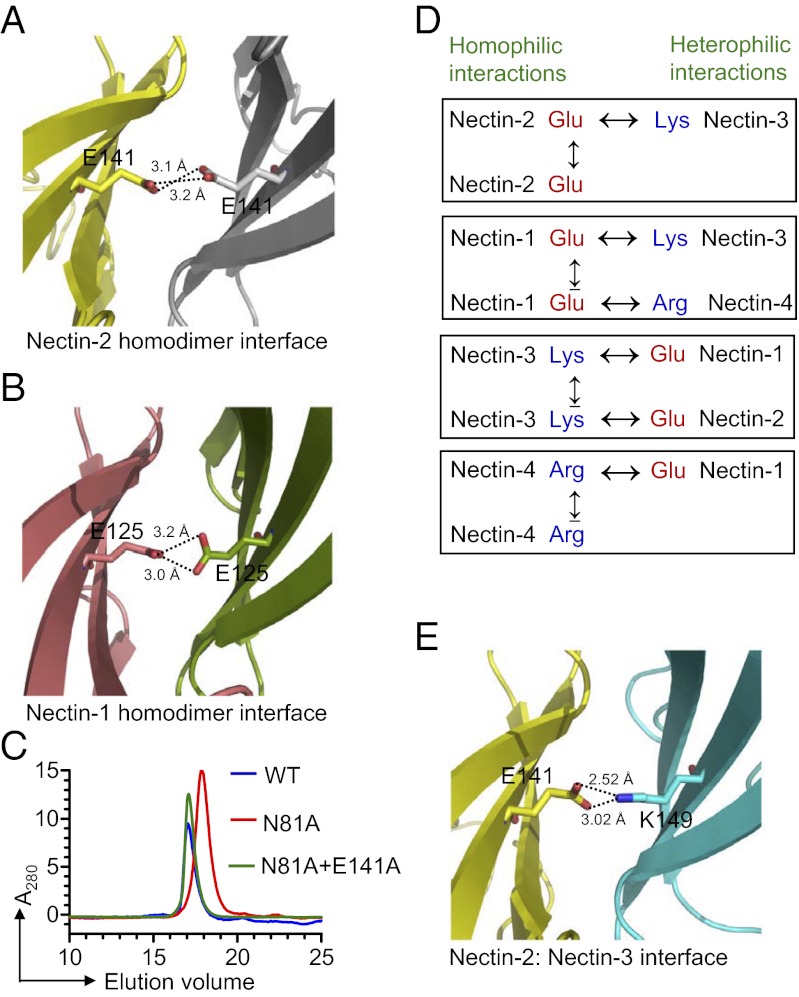
One notable feature is the presence of an apparently unfavorable electrostatic interaction resulting from the close proximity of two glutamic-acid side chains (E141 from F strand) in the center of the nectin-2 homophilic interface (Fig. 4A). A similar unfavorable electrostatic interaction is observed in the recently reported nectin-1 homophilic interface [Protein Data Bank (PDB) ID code 3ALP] (Fig. 4B) (16). Structure-based sequence alignment predicts that the glutamic-acid residue is replaced by lysine and arginine in nectin-3 and nectin-4, respectively, again suggestive of unfavorable electrostatics at these homodimer interfaces (Fig. 3 and Fig. S3). Notably, these charged residues are strictly conserved across species (see sequence alignment in Fig. S4). To further investigate the impact of these putative unfavorable interactions at the dimer interface, we mutated Glu-141 to alanine in nectin-2. Because wild-type nectin-2 forms a tight dimer, with a dissociation that is below the detection limit of analytical ultracentrifugation (AUC), the effect of E141A mutation on the dimerization would be difficult to assess. To circumvent this problem, the E141A mutation was generated in the background of the N81A mutant, which on its own behaves as a monomer by size-exclusion chromatography (Fig. 2E). Notably, in this background, the E141A mutation causes a dramatic shift of the monomer to a predominately dimeric population (Fig. 4C), consistent with the notion that the presence of Glu-141 at the dimer interface results in a reduction of the homophilic affinity.
Fig. 4.
(A) Dimer interface of human nectin-2 showing the close proximity of two negatively charged side chains; Glu-141 is contributed from the F strand of each monomer (yellow and gray). (B) Dimeric interface of nectin-1 (PDB ID code 3ALP) showing the similar unfavorable repulsive electrostatics as depicted in the case of nectin-2. (C) Size-exclusion chromatography showing that the E141A mutation in the background of the N81A mutation causes the shift of monomer (an effect of N81A mutation) to the dimer population. (D) The presence of the same charges at the homodimer interface causes unfavorable electrostatics during homophilic interactions, whereas the presence of two different charges at the heterodimer interface can eliminate the unfavorable electrostatics as seen at the homodimer interface. (E) Molecular model showing the heterophilic interaction between nectin-2 (yellow) and nectin-3 (cyan). The modeling suggests that E141 of nectin-2 contacts K149 of nectin-3, forming a putative H-bond at the center of the interface, which favors a strong heterophilic interaction.
In contrast to the homophilic dimers, the heterophilic interactions in the nectin family involve opposite charges at the position analogous to E141 (Fig. 4D). This feature is illustrated in a model of the nectin-2:nectin-3 heterodimer, where Glu-141 from nectin-2 and Lys-149 from nectin-3 may participate in favorable polar interaction that promotes heterophilic association (Fig. 4E). Notably, this behavior is consistent with the higher affinities reported for heterophilic interactions compared with homophilic interactions in the nectin family (6, 7). These observations also highlight the critical concept that receptor–ligand interactions do not evolve to produce the highest possible affinity, but instead evolve to select the affinity that allows for the specificity, kinetics, and associated signaling properties that are biologically optimal. It is this range of affinities, and the quantitative differences, exhibited by the homophilic and heterophilic interactions of the nectin family that support the intersection of diverse adhesive processes required for a wide range of biological functions.
Structural Comparison of Nectin-2 and Nectin-1.
A structural alignment of the IgV domains of nectin-1 and nectin-2 demonstrates high sequence identity of about 40% and high structural similarity with an rmsd of 1.7 Å for 111 equivalent Cα atoms. Furthermore, the dimer structure and dimer interface of nectin-2 are comparable to the dimer structure and interface of nectin-1 exhibiting an rmsd of 2.04 Å for 212 equivalent Cα atoms, with 13 of the 22 (∼59%) nectin-2 interface residues being identical between the two proteins. A structure-based sequence alignment revealed that many of the nectin-2 homophilic interface residues are not conserved in the family (Fig. 3). However, the sequence motif TFPXG, corresponding to residues 144–148, contributed by the FG loop of nectin-2 is well conserved (Fig. 3).
Nectin-2 Dimer Represents a Result of Transinteraction.
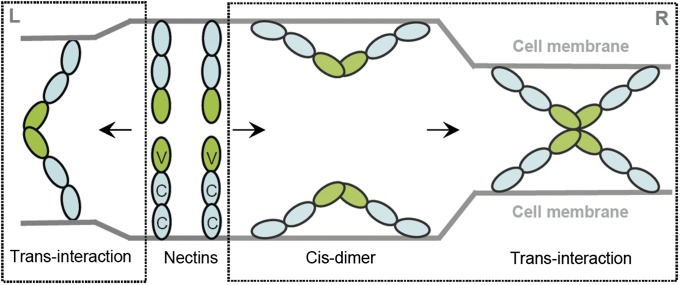
Both cis- and transhomophilic interactions may contribute to nectin function. Based on the behavior of cells expressing a series of deletion mutants, the first IgC domain was initially proposed to be responsible for cis-interactions (1, 2, 17). More recently, the crystal structure of human nectin-1 (PDB ID code 3ALP) was interpreted to suggest that nectin-1 forms a cis-dimer via interactions involving the orthogonal association of IgV domains similar to that observed in the current structure of the nectin-2 IgV domain (16). It was further proposed that lateral clustering of these cis-dimers was driven by interactions involving the first IgC domains and that these clusters of cis-dimers formed transinteractions via “head-to-head” contacts between the IgV domains of molecules on engaging cells (16).
The different mechanisms offered for nectin assembly reflect the complexity associated with systems possessing multiple interaction domains (e.g., IgV and IgC) and multiple geometric modes of association (i.e., cis and trans). Notably, the interface and overall architecture of the homophilic dimers formed by the IgV from nectin-1 and nectin-2 are similar to each other, as well as to a wide range of dimeric complexes formed by other members of the IgSF that are known to function via biologically competent transinteractions (Fig. S2). In contrast, the proposed head-to-head transinteraction of cis-dimers is likely to be sterically precluded by the prior participation of the IgV domains in the putative cis-interaction (Fig. 5). Our solution studies (SEC and AUC) also indicate that the IgV domain of nectin-2 exists only as a dimer in solution, and higher oligomeric states were not observed (Fig. 1A) as would be required by the proposed head-to-head transinteraction of cis-dimers (Fig. 5). Furthermore, in murine nectin-2α, Phe-136 was previously reported to be crucial for its transinteraction but not for the cis-dimerization (18). In human nectin-2, Phe-145 is the equivalent residue and buried at the dimer interface (82.8% buried) (Fig. S5). Introduction of a single mutant (F145A) disrupts the dimer in solution, consistent with our interpretation of the current structure as a physiologically relevant model of the productive transinteraction (Fig. 2E). Thus, the preponderance of the structural, biochemical, and comparative data support the crystallographically observed association of IgV domains as an appropriate model for the transinteraction that underlies the biological function of the nectins. However, it remains a possibility that the cis-interaction between IgV domains proposed on the basis of the nectin-1 structure could act as a regulatory mechanism to control the extent of transinteractions.
Fig. 5.
Model for the transinteraction of nectins. (Left) The transinteraction of two nectin molecules from opposing cells through their IgV domains, which represents a physiologically relevant model of the productive transinteraction. (Right) Models in which two nectin molecules first form a cis-dimer on the cell surface via their IgV domains; these cis-dimers subsequently form transinteractions in a head-to-head manner as proposed by Narita et al. (16).
In summary, on the basis of our structural and biochemical data, as well as previously reported data (16–19), we believe that our nectin-2 dimer represents the biologically relevant homophilic transinteraction shown in Fig. 5. Moreover, the present study provides structural and biochemical insights into the chemical and physical determinants responsible for the spectrum of affinities and interactions associated with the homophilic and heterophilic dimers within the nectin family. These insights will drive the design, execution, and interpretation of experiments to further dissect the mechanistic basis of cell–cell adhesion.
Materials and Methods
Cloning, Expression, and Purification of Nectin-2.
The IgV domain of human nectin-2 (residues 32–158) was cloned into pET3a (Novagen). Protein was expressed in E. coli strain BL21 (DE3) pLysS as insoluble inclusion bodies. Protein expression was induced at an OD600 of 0.5 with 1.0 mM isopropyl 1-thio-d-galactopyranoside. Cells were harvested and suspended in buffer containing 50 mM Tris⋅HCl (pH 8.0), 100 mM NaCl, 20% (wt/vol) sucrose, 1 mM EDTA, and 10 mM DTT. DNase I (10 μg/mL) was added to the suspension, the cells were lysed, and insoluble protein was pelleted by centrifugation. The inclusion bodies were washed three times with buffer containing 10 mM Tris⋅HCl (pH 8.0), 100 mM NaCl, 0.5% Triton X-100, 1 mM EDTA, and 10 mM DTT. The detergent was removed by washing the inclusion bodies twice with this buffer but omitting the Triton X-100. Protein purity was confirmed by SDS/PAGE.
The purified, detergent-free inclusion bodies were solubilized in buffer containing 6 M guanidine hydrochloride, 10 mM Na-acetate (pH 4.5), 5 mM EDTA, and 1 mM DTT. The solubilized material was refolded by rapid dilution (20, 21) in buffer containing 400 mM arginine–HCl, 100 mM Tris⋅HCl (pH 8.0), 1 mM EDTA, 5 mM reduced glutathione, and 0.5 mM oxidized glutathione. Finally, the refolded material was purified by size-exclusion chromatography with a buffer composed of 20 mM Hepes (pH 7.0), 150 mM NaCl, and 1 mM EDTA.
Analytical Ultracentrifugation Sedimentation Equilibrium Analysis.
Sedimentation equilibrium experiments were performed at 20 °C using a Beckman XL-I analytical ultracentrifuge, six-sector cells, and an AN-60Ti rotor. The 280-nm absorption scans of Nectin-2 present in 20 mM Hepes (pH 7.0), 150 mM NaCl, and 1 mM EDTA was globally analyzed at 10, 23, and 53 μM protein and 20,000 and 25,000 × g using HeteroAnalysis ver 1.1.44 (22). Buffer density and the partial specific volume were calculated using SEDNTREP version 1.01 (23). AUC data and fits are shown in Dataset S1.
Crystallization and Structure Determination.
The IgV domain of nectin-2 [10 mg/mL in 20 mM Hepes (pH 7.0), 150 mM NaCl, and 1 mM EDTA] was crystallized using the sitting drop vapor diffusion method at room temperature by mixing 0.5 μL of protein with 0.5 μL of precipitant composed of 0.2 M MgCl2·6H2O, 0.1 M Tris⋅HCl (pH 8.5), and 30% PEG 4000 and by equilibrating over 70 μL of precipitant. A number of tetragonal crystals were obtained within 3–4 d. Crystals were cryoprotected in mother liquor supplemented with 15% ethylene glycol before flash-cooling in liquid nitrogen. Diffraction was consistent with the space group P41212 (a = b = 57.96 Å; c = 67.54 Å; with one molecule per asymmetric unit). Data were collected at the X29A beam line (National Synchrotron Light Source) and integrated and scaled with HKL2000 (24). The structure was determined by molecular replacement with the program MOLREP (CCP4) using the model 1NEU (Myelin P0 protein) (25). Initial placement and rigid body refinement with REFMAC5 (26) resulted in clear density for the whole IgV domain. The model was further improved by alternative cycles of manual revision with COOT and refinement with REFMAC5. The final model was refined to 1.3 Å resolution, with Rwork and Rfree of 15.23% and 17.33%, respectively. Analysis shows that 99.2% of the residues are in most favored regions and 0.8% in additionally allowed regions of the Ramachandran plot (Table S1). The atomic coordinates of this structure were submitted to Protein Data Bank (PDB ID code 3R0N).
Molecular Modeling.
The structure models of nectin-2:nectin-3, nectin-3:nectin-3, and nectin-4:nectin-4 complexes were constructed with the MODELER program (27) using a combination of nectin-2 and nectin-1 (PDB ID codes 3R0N and 3ALP) as structural templates.
Supplementary Material
Acknowledgments
We thank the staff of the X29A beam line at the National Synchrotron Light Source; Michael Brenowitz for help with analytical ultracentrifugation; and Teresa P. DiLorenzo, Rajesh K. Prakash, and Gayatri Mukherjee for their critical reading of the manuscript. This work was supported by National Institutes of Health Grants AI007289 and U01GM094665 (to S.G.N. and S.C.A.) and U54GM093342 (to S.C.A.) and by the Albert Einstein Cancer Center Grant 3P30CA013330.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3R0N).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212912109/-/DCSupplemental.
References
- 1.Takai Y, Ikeda W, Ogita H, Rikitake Y. The immunoglobulin-like cell adhesion molecule nectin and its associated protein afadin. Annu Rev Cell Dev Biol. 2008;24:309–342. doi: 10.1146/annurev.cellbio.24.110707.175339. [DOI] [PubMed] [Google Scholar]
- 2.Takai Y, Miyoshi J, Ikeda W, Ogita H. Nectins and nectin-like molecules: Roles in contact inhibition of cell movement and proliferation. Nat Rev Mol Cell Biol. 2008;9:603–615. doi: 10.1038/nrm2457. [DOI] [PubMed] [Google Scholar]
- 3.Fuchs A, Colonna M. The role of NK cell recognition of nectin and nectin-like proteins in tumor immunosurveillance. Semin Cancer Biol. 2006;16:359–366. doi: 10.1016/j.semcancer.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, et al. Nectin/PRR: An immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with Afadin, a PDZ domain-containing protein. J Cell Biol. 1999;145:539–549. doi: 10.1083/jcb.145.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizoguchi A, et al. Nectin: An adhesion molecule involved in formation of synapses. J Cell Biol. 2002;156:555–565. doi: 10.1083/jcb.200103113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satoh-Horikawa K, et al. Nectin-3, a new member of immunoglobulin-like cell adhesion molecules that shows homophilic and heterophilic cell-cell adhesion activities. J Biol Chem. 2000;275:10291–10299. doi: 10.1074/jbc.275.14.10291. [DOI] [PubMed] [Google Scholar]
- 7.Reymond N, et al. Nectin4/PRR4, a new afadin-associated member of the nectin family that trans-interacts with nectin1/PRR1 through V domain interaction. J Biol Chem. 2001;276:43205–43215. doi: 10.1074/jbc.M103810200. [DOI] [PubMed] [Google Scholar]
- 8.Ozaki-Kuroda K, et al. Nectin couples cell-cell adhesion and the actin scaffold at heterotypic testicular junctions. Curr Biol. 2002;12:1145–1150. doi: 10.1016/s0960-9822(02)00922-3. [DOI] [PubMed] [Google Scholar]
- 9.Togashi H, et al. Nectins establish a checkerboard-like cellular pattern in the auditory epithelium. Science. 2011;333:1144–1147. doi: 10.1126/science.1208467. [DOI] [PubMed] [Google Scholar]
- 10.Bouchard MJ, et al. Defects in nuclear and cytoskeletal morphology and mitochondrial localization in spermatozoa of mice lacking nectin-2, a component of cell-cell adherens junctions. Mol Cell Biol. 2000;20:2865–2873. doi: 10.1128/mcb.20.8.2865-2873.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mueller S, Rosenquist TA, Takai Y, Bronson RA, Wimmer E. Loss of nectin-2 at Sertoli-spermatid junctions leads to male infertility and correlates with severe spermatozoan head and midpiece malformation, impaired binding to the zona pellucida, and oocyte penetration. Biol Reprod. 2003;69:1330–1340. doi: 10.1095/biolreprod.102.014670. [DOI] [PubMed] [Google Scholar]
- 12.Okabe N, et al. Contacts between the commissural axons and the floor plate cells are mediated by nectins. Dev Biol. 2004;273:244–256. doi: 10.1016/j.ydbio.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 13.Stanietsky N, et al. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc Natl Acad Sci USA. 2009;106:17858–17863. doi: 10.1073/pnas.0903474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 15.Zhang N, et al. Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion. Nat Commun. 2011;2:577. doi: 10.1038/ncomms1571. [DOI] [PubMed] [Google Scholar]
- 16.Narita H, et al. Crystal structure of the cis-dimer of Nectin-1: Implications for the architecture of cell-cell junctions. J Biol Chem. 2011;286:12659–12669. doi: 10.1074/jbc.M110.197368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Momose Y, et al. Role of the second immunoglobulin-like loop of nectin in cell-cell adhesion. Biochem Biophys Res Commun. 2002;293(1):45–49. doi: 10.1016/S0006-291X(02)00183-3. [DOI] [PubMed] [Google Scholar]
- 18.Miyahara M, et al. Interaction of nectin with afadin is necessary for its clustering at cell-cell contact sites but not for its cis dimerization or trans interaction. J Biol Chem. 2000;275:613–618. doi: 10.1074/jbc.275.1.613. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, et al. Crystal structure of cell adhesion molecule nectin-2/CD112 and its binding to immune receptor DNAM-1/CD226. J Immunol. 2012;188:5511–5520. doi: 10.4049/jimmunol.1200324. [DOI] [PubMed] [Google Scholar]
- 20.Garboczi DN, et al. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. [PubMed] [Google Scholar]
- 21.Zhang X, Schwartz JC, Almo SC, Nathenson SG. Expression, refolding, purification, molecular characterization, crystallization, and preliminary X-ray analysis of the receptor binding domain of human B7-2. Protein Expr Purif. 2002;25(1):105–113. doi: 10.1006/prep.2002.1616. [DOI] [PubMed] [Google Scholar]
- 22.Cole JL. Analysis of heterogeneous interactions. Methods Enzymol. 2004;384:212–232. doi: 10.1016/S0076-6879(04)84013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glasel JA. Introduction to Biophysical Methods for Protein and Nucleic Acid Research. San Diego: Academic Press; 1995. [Google Scholar]
- 24.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.Shapiro L, Doyle JP, Hensley P, Colman DR, Hendrickson WA. Crystal structure of the extracellular domain from P0, the major structural protein of peripheral nerve myelin. Neuron. 1996;17:435–449. doi: 10.1016/s0896-6273(00)80176-2. [DOI] [PubMed] [Google Scholar]
- 26.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 27.Eswar N, et al. Comparative Protein Structure Modeling with MODELLER. Current Protocols in Bioinformatics. New York: John Wiley & Sons, Inc.; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.