Abstract
A conserved injury-defense mechanism is present in plants and animals, in which the production of reactive oxygen species (ROS) and lipid metabolism are essential to the response. Here, we describe that in the filamentous fungus Trichoderma atroviride, injury results in the formation of asexual reproduction structures restricted to regenerating cells. High-throughput RNA-seq analyses of the response to injury in T. atroviride suggested an oxidative response and activation of calcium-signaling pathways, as well as the participation of lipid metabolism, in this phenomenon. Gene-replacement experiments demonstrated that injury triggers NADPH oxidase (Nox)–dependent ROS production and that Nox1 and NoxR are essential for asexual development in response to damage. We further provide evidence of H2O2 and oxylipin production that, as in plants and animals, may act as signal molecules in response to injury in fungi, suggesting that the three kingdoms share a conserved defense-response mechanism.
Keywords: conidiation, stress, transcriptome
The study of responses to mechanical damage has implications for medical applications related to tissue regeneration and repair (1). A conserved defense mechanism is present in plants and animals, in which the production of reactive oxygen species (ROS) and lipid metabolism are essential to the response (2). In contrast, our current knowledge of how filamentous fungi respond to injury is limited to the sealing of septal pores by Woronin bodies, thereby preventing excessive loss of cytoplasmic content and cell death (3). After septal plugging, one or more hyphal tips are formed at the plugged septum, resulting in reinitiation of growth and hyphal reconnection (3). Additionally, a few reports suggest that mechanical damage may induce oxidative stress in fungal cells (4, 5). However, accumulation of ROS in response to mechanical damage has been demonstrated only for hyphae of Glomus intraradices (6).
Leonard and Dick (7) observed that the fungus Schizophyllum commune produced fruiting bodies in response to mechanical damage and suggested that wounding could generate oxidative stress, responsible for fruiting body formation. Various studies have shown that antioxidants inhibit conidiation in Neurospora crassa, and the lack of antioxidant enzymes results in higher levels of ROS and enhanced cell differentiation, whereas elimination of prooxidant enzymes results in arrested cell differentiation (8, 9).
All aerobic organisms generate ROS as an inevitable product of metabolism, primarily through respiration (8). NADPH oxidases (Noxs) are the most important enzymes capable of producing ROS (9). The damaging effects of ROS on cell components and their role in aging are well established. However, there is increasing evidence supporting an alternative view, in which the production of ROS by Noxs regulates cell differentiation, among other functions (9).
The Nox enzyme family was originally related to responses to exposure to pathogens in both plants and animals, resulting in the generation of high levels of ROS (2, 9). In animals, Noxs are activated upon mechanical injury, producing high levels of ROS, which may aid in sterilization of the wound and serve as a chemotactic signal to recruit neutrophils (9). In plants, Noxs have a variety of functions, including response to wounding, herbivory and programed cell death (PCD), where ROS produced by Noxs forms a gradient of H2O2, indicating that it functions as propagation signal (2, 9). Fungi contain from one to three Nox genes (noxA, noxB, noxC), depending on the species (8), and a NoxR protein that regulates NoxA/NoxB (10). Fungal Noxs play key roles in plant host interactions, aging, and differentiation (9). Lara-Ortíz et al. (11) first reported a direct link between Nox-derived ROS and sexual differentiation. Accordingly, in N. crassa, nox1 elimination results in complete female sterility, slightly decreased asexual development, and reduction of hyphal growth (12).
Trichoderma atroviride are common soil fungi applied in the field to aid disease control in the form of commercial formulations based on conidia (13). Interestingly, in T. atroviride, conidiation is triggered by mechanical injury (14). However, the mechanism of response to mechanical injury is unknown.
Results
Initiation of Conidiophore Formation Following Injury Is Restricted to Cells Adjacent to the Dead Cell.
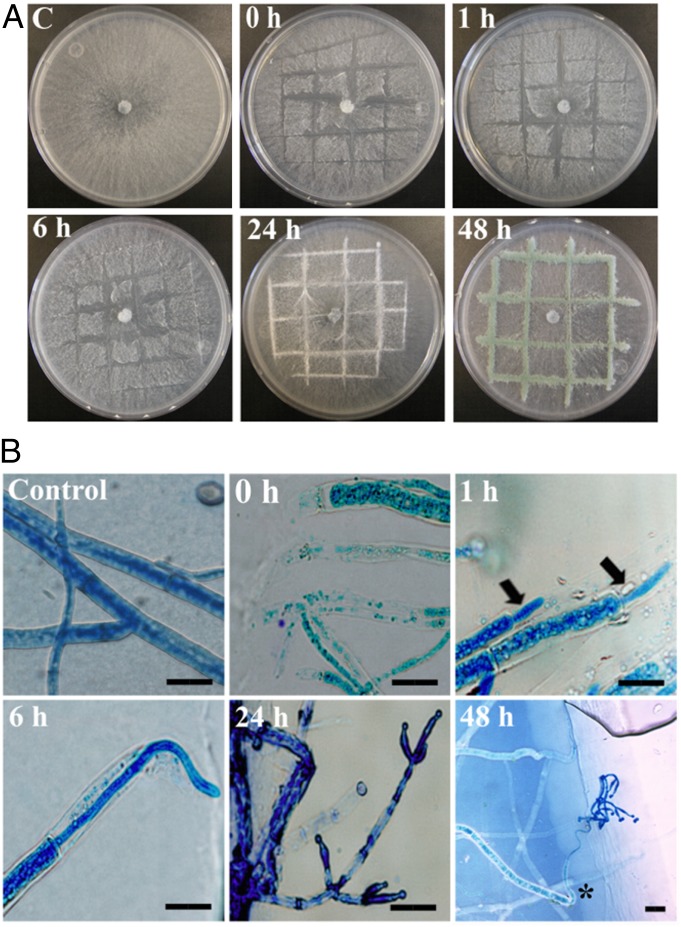
T. atroviride colonies were injured with a scalpel and observed macro- and microscopically. Macroscopically, the first response is the formation of aerial mycelium, observed 24 h after injury. Forty-eight hours after injury, the presence of mature green conidia is observed at the wound site (Fig. 1A). The first morphological change observed microscopically is the regeneration of the damaged hyphae with the formation of “new,” significantly thinner, hyphal tips 1 h after injury. Interestingly, after 24 h, the formation of phialides is observed, and 48 h later, conidiophores with mature spores have formed exclusively from the “new” hyphae (Fig. 1B).
Fig. 1.
Conidiophores are formed from regenerating hyphae. (A) Photographs of the WT strain growing on MMV-CN were taken at the indicated times after injury. (B) Light microscopic photographs taken at the indicated times after injury. Arrows indicate “new” hyphae. An uninjured control (C) is shown in both A and B. Hyphae were stained with lactophenol cotton blue. An asterisk indicates the generation of conidiophores from the regenerated hyphae. (Scale bar: 20 μm.)
In-Depth Transcriptome Analysis Indicates That T. atroviride Responds to Injury Using Conserved Mechanisms.
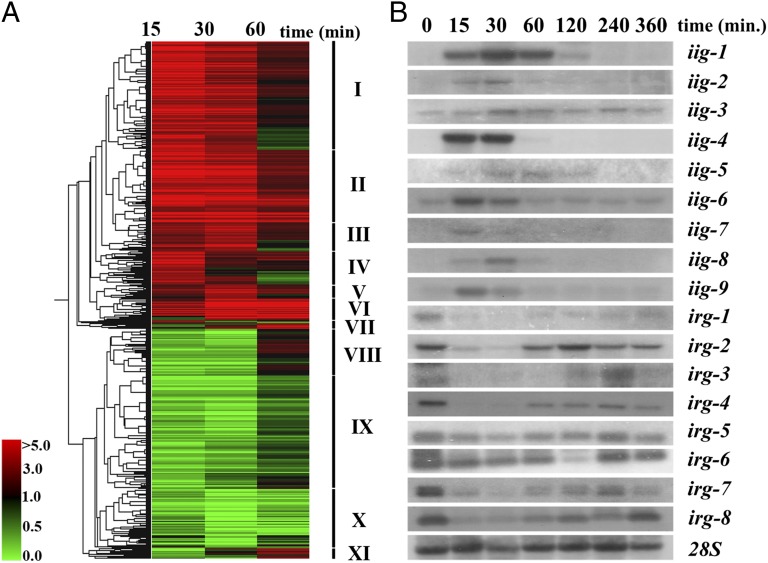
T. atroviride colonies grown in the dark were wounded and allowed to grow for a further 0, 4, 8, and 12 h in the dark and then treated with the transcription inhibitor 5-fluorouracyl (5-FU). Treatment with 5-FU indicates that 12 h are required for the synthesis of all RNA necessary for injury-induced conidiation (Fig. S1A). RNA-seq was carried out on RNA samples obtained 15, 30, and 60 min after mycelial injury, and an RNA sample from uninjured mycelia was included as control (Dataset S1). All genes that fulfilled the statistical criteria established and showed a change in expression of twofold or higher with respect to the uninjured control, in at least one of the three sequenced samples, were considered to be differentially expressed. A total of 933 differentially expressed genes (522 induced and 411 repressed) were detected (Fig. 2A and Dataset S2). The corresponding proteins were compared against the Functional Catalogue Database (FunCatDB) (15), providing us with a global view of the biological processes associated with the transcriptional response to mycelial injury (Fig. S1 B and C and Dataset S2). The 933 proteins were compared with the protein families database (Pfam). A domain clustering based on their possible cellular functions indicated that the most represented functions are: redox reactions (140), transport (80), and transcription factors (TF) (34) (Dataset S2). Comparison of all proteins to the National Center for Biotechnology Information (NCBI) nonredundant database using BLASTP showed that 91% had a significant hit (Dataset S2).
Fig. 2.
Transcriptome analysis of the early response to injury. (A) Hierarchical clustering of temporal expression of 933 injury-responsive genes was performed using smooth correlation and average linkage clustering in GeneSpring GX 7.3.1 software (Agilent Technologies). Roman numbers indicate independent clusters. (B) Time-course analysis of the expression of injury-induced (iig) and -repressed genes (irg) by Northern blot. iig-1 (Id: 127833; NADP-dependent leukotriene B4 12-hydroxydehydrogenase), iig-2 [Id: 297699; common in several fungal extracellular membrane proteins (CFEM) domain protein], iig-3 (Id: 297381; proliferating-cell nuclear antigen), iig-4 (Id: 33350; lipoxygenase), iig-5 (Id: 36070; hypothetical protein), iig-6 [Id: 301592; Ca+2/calmodulin-dependent kinase 1 (CAMK-1)], iig-7 (Id: 126859; cytochrome b5), iig-8 [Id: 314604; basic leucine zipper (bZIP) transcription factor], iig-9 (Id: 219770; metacaspase-1A), irg-1 (Id: 297389; hypothetical protein), irg-2 (Id: 299661), irg-3 (Id: 297668; peroxisomal catalase), irg-4 (Id:300386; hypothetical protein), irg-5 (Id:297186; hypothetical protein), irg-6 (Id:300960; hypothetical protein), irg-7 (Id:253020; hypothetical protein), and irg-8 (Id: 155960; peroxidase/catalase). Time 0 refers to control without injury. The 28S ribosomal RNA was used as loading control.
Hierarchical clustering of the expression pattern of the 933 genes grouped them in 11 clusters (Fig. 2A). Among the early-induced genes (clusters I, II, and IV), the presence of genes involved in calcium signaling and transport, redox balance, stress responses, cell cycle, and cell death, as well as TF, among others should be highlighted (Dataset S2). Late-induced genes (clusters III, V, VI, VII, and XI) mainly encode proteins involved in DNA damage and cell cycle and at least 11 proteins with oxide-reductase activity (Dataset S2). In contrast, early-repressed genes (clusters VIII and IX) include mainly genes involved in metabolism, transcription factors, and ROS-scavenging proteins. The set of genes involved in ROS scavenging is strongly repressed at early stages of the response, whereas their expression tends to increase at later stages and returning to levels found in the control (Dataset S2). Finally, cluster X contains late repressed genes, affecting mainly metabolism. Nevertheless, seven genes encoding ROS scavenging proteins also belong to this cluster (Dataset S2).
The expression of 10 injury-induced genes (iig) and 10 injury-repressed genes (irg), selected based on their level of expression and putative function, was verified by Northern blot analysis of samples collected 15, 30, 60, 120, 240, and 360 min after injury. Seven genes cataloged as induced show no detectable levels of transcript in the control sample (iig-1, iig-2, iig-4, iig5, iig-7, iig-8, and iig-9) and two more have a basal level (iig-3 and iig-6). The nine transcripts accumulate rapidly after 15 min, and by 120 min, their transcripts return to the basal level observed. Six of the repressed genes display a transitory decrease in transcription at 15 and 30 min, and their expression starts increasing until 60 min following wounding (irg-1, irg-2, irg-3, irg-4, irg-7, and irg-8). Two genes show only a subtle but detectable decrease in transcription (irg-5 and irg-6) (Fig. 2B). No transcript could be detected for three of the genes tested.
Antioxidants and a Flavin Oxidase Inhibitor Block Injury-Induced Conidiation.
Given the transcriptional response of a significant number of genes involved in oxidative stress, we tested the effect of the antioxidants ascorbic acid (AA) and N-acetyl l-cysteine (NAC) in injury-induced conidiation. Our results show that both AA and NAC cause a clear decrease in conidiation (Fig. S2B) and slightly affected radial growth. In contrast, the control with N-acetyl glycine (NAG) (structurally similar to NAC but not an antioxidant) conidiated as efficiently as the untreated control (Fig. S2B). Furthermore, preliminary experiments using nitroblue tetrazolium chloride (NBT) allowed us to detect the formation of superoxide along the injured area (Fig. S2A). Therefore, we decided to use diphenyleneiodonium chloride (DPI), a flavin oxidase inhibitor (11), to test the possible involvement of Noxs in injury-induced conidiation. Our results clearly demonstrate that injury-induced conidiation is completely blocked upon treatment with DPI, whereas application of the DPI solvent [dimethyl sulfoxide (DMSO)] alone allowed conidia formation (Fig. S2C).
Nox1 Is Essential for Injury-Induced Conidiation.
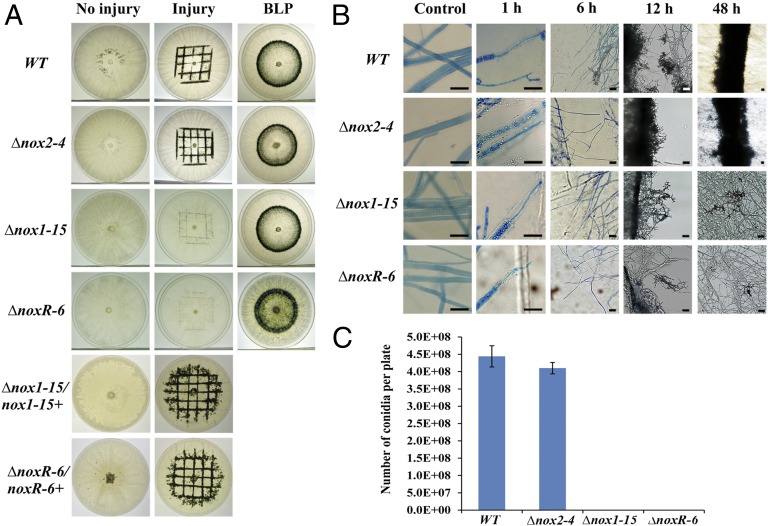
Transcriptome analysis suggested the involvement of oxidative stress, and pharmacological evidence indicated a potential role for Noxs in the injury response. Accordingly, we decided to generate gene-replacement mutants of the nox genes in T. atroviride. We identified two genes encoding Nox catalytic subunits, nox1 with protein identifier (Id): 302802 and nox2 (Id: 300495), and one encoding a regulatory subunit noxR (Id: 315943). Gene-replacement mutants of nox1, nox2, and noxR were obtained. To determine the role of Noxs in the response to injury, mutants were subjected to injury. The Δnox1 and ΔnoxR mutants did not conidiate in response to injury, although they showed a slightly slower radial growth than the wild type (WT), whereas the Δnox2 mutants conidiated as efficiently as the WT (Fig. 3A). The Δnox1 and ΔnoxR mutants were retransformed with the corresponding WT gene, and, as expected, the retransformed strains (Δnox1/nox1+ and ΔnoxR/noxR+) responded normally to injury (Fig. 3A). Bright-field microscopy clearly showed that the Δnox1 and ΔnoxR mutants are severely affected in their capacity to form conidiophores in response to injury. Interestingly, hyphal regeneration was not compromised. In contrast, conidiation of the Δnox2 mutants was indistinguishable from that observed in the WT (Fig. 3B). Quantification of conidia showed a 99.9% decrease in the production of conidia by the Δnox1 and ΔnoxR compared with the WT (Fig. 3C). Nox mutants were also exposed to a pulse of blue light (1200 μmol/m2), which responded to this cue by forming a ring of conidia indistinguishable from that observed in WT (Fig. 3A).
Fig. 3.
Nox1 and NoxR are essential for injury-induced conidiation. (A) Light- and injury-induced conidiation assay. The WT and indicated mutants were induced to conidiate with a pulse of blue light or by injury, and retransformed strains (Δnox1-15/nox1+ and ΔnoxR-6/noxR+) were induced to conidiate by injury and photographed 48 h later. (B) Microscopic changes observed upon injury. Mycelial samples of the WT and mutants were stained with lactophenol cotton blue and examined under the light microscope at the indicated times after injury. Samples of uninjured mycelium were used as controls. (Scale bar: 20 μm.) (C) Quantification of conidia. The number of conidia indicated is the result of subtracting conidia produced in the uninjured control from the number of conidia produced after injury for each strain. Errors bars represent the means ± SEM of two biological replicas per duplicate.
Injury Stimulates ROS Production Through Nox1.
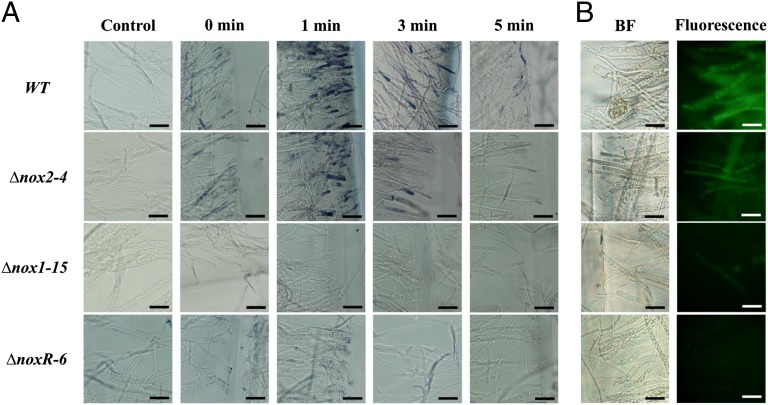
To determine whether Noxs are necessary for ROS production in response to injury, WT, Δnox2, Δnox1, and ΔnoxR strains were subjected to injury and then incubated in the dark for 0, 1, 3, and 5 min. Samples of each injured colony were then stained using NBT to determine the production of superoxide in the damaged hyphae. As shown in Fig. 4A, in the WT strain, high levels of formazan were observed 1 min after injury, as judged by the intensity of the staining, which was restricted to the cells adjacent to those wounded. Similar staining was observed in the Δnox2 mutant 1 min after injury, although not as intense as that observed for the WT. In both cases the intensity of staining decreased by 3 min, and no staining was observed 5 min after injury. In contrast, no superoxide production was detected in the Δnox1 and ΔnoxR mutants. To determine whether other types of ROS were produced, 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) was used as indicator of ROS production (11). As described for the assay using NBT, samples of mycelia collected 1 min after injury were incubated in H2DCFDA and examined using a fluorescence microscope. The WT strain, as well as the Δnox2 mutant, showed fluorescence along the injured sites, indicating ROS production, most likely H2O2, although fluorescence was less intense in the case of the mutant. Interestingly, in contrast with what was observed using NBT, production of H2O2 is not restricted to the cell adjacent to the damaged one but extends to other cells in the vicinity. No fluorescence was detected in the assays using the Δnox1 and ΔnoxR mutants, indicating the lack of production of H2O2 (Fig. 4B).
Fig. 4.
Nox1-dependent ROS production. (A) Production of superoxide in response to injury. Samples of mycelium were incubated in a 0.3 mM NBT solution and examined by bright-field microscopy (BF). A blue/purple coloration indicates the production of superoxide (Formazan generation). (B) Production of H2O2 in response to injury. H2O2 production was visualized using 40 mM H2DCFDA and fluorescence microscopy. (Scale bar: 20 μm.)
Effect of noxR Mutation in Injury Late-Responsive Genes.
Because Nox1 and NoxR participate directly in ROS formation and conidiation in response to injury, we decided to analyze the transcriptome of the ΔnoxR mutant in response to injury. Data obtained from an RNA-seq experiment comparing the ΔnoxR mutant and the WT strain at late stages after injury indicate that there is a clear difference in the transcriptional response between the two strains by 1–2 h after injury (Fig. S3A and Dataset S3). A total of 189 genes are induced in response to injury only in the WT strain and are repressed or not responsive in the ΔnoxR mutant (Fig. S3 A and B and Dataset S3). This set mainly involves genes participating in calcium signaling and oxylipin biosynthesis [including an α-dioxygenase homolog (Id: 293464)] (Fig. S3C and Dataset S3). In contrast, 300 genes are repressed in the WT strain but induced in the ΔnoxR mutant (Fig. S3 A and B and Dataset S3). Interestingly, this group includes mainly genes encoding ROS-scavenging proteins, including a superoxide dismutase-2 (Id: 224288) and a catalase 2 (Id: 88135), transcription factors or proteins involved in secondary metabolism (Fig. S3C and Dataset S3).
Discussion
The few existing reports on the responses of filamentous fungi to mechanical damage are limited to S. commune (7), Sclerotium rolfsii (16), and G. intraradices (6) and do not provide mechanistic insights. Here, we report that mechanical damage in T. atroviride results in the production of conidiophores with mature spores formed exclusively from the “new” hyphae that result from regeneration of damaged cells. Furthermore, de novo synthesis of mRNA and proteins is required for conidiation during 12 h following the injury. Our results show the regulation at least 25 genes related to cytoskeletal organization, DNA replication, and cell cycle (Dataset S2), including a gene with homology to the severe depolymerization of actin 1 (sda1) gene of Saccharomyces cerevisiae (Id: 43203), which plays a critical role in the passage of cells arrested in G0 phase into G1, reinitiation of the cell cycle (17), suggesting that these genes might play an important role in the regeneration of the damaged hyphae.
Fester and Hause (6) showed the cytoplasmic accumulation of ROS in damaged hyphae of G. intraradices. In agreement with their report, our transcriptome analyses indicated that transcriptional responses are consistent with generation of oxidative stress. At least 60 genes encoding proteins related to redox reactions are regulated by injury, including one gene encoding a cytochrome b5 reductase that is strongly induced, which has been related to oxidative stress in fungi through the reduction of the free radical d-erythroascorbyl and the generation of high reducing power (18). In this sense, Gessler et al. (5) suggested that impairment of the intracellular redox status, as a result of an increase in generation of ROS, can generate oxidative stress. Accordingly, our results show that in the WT strain, 16 genes encoding ROS scavenging proteins are strongly repressed at short times after injury, and later, their levels begin increasing to finally recover the level found in the noninjured control, or reach even higher levels, indicating that the cell initially allows the generation of oxidative stress upon injury. In contrast, in the ΔnoxR strain ROS-scavenging proteins are induced upon injury, and those involved in ROS production are repressed.
In plants and animals, the response to injury involves conserved mechanisms, including an increase in intracellular calcium and the activation of the calcium signaling machinery (19). In fungi, Nelson et al. (20) showed that mechanical perturbation provokes a transient increase in intracellular calcium. In full agreement with these reports, our results show a series of induced genes involved in calcium signaling and transport. These include calcium transporters, phospholipase C, and a Ca2+/calmodulin-dependent kinase-1 (CAMK-1), all considered key components in calcium signaling in response to different types of stress (21). Another induced gene is a homolog of the Calcineurin-responsive zinc finger transcription factor (crz1), which regulates responses to calcium and is essential for membrane integrity and oxidative stress responses (22). These data suggest that calcium signaling might play an important role in the injury response of T. atroviride (Fig. S4A).
Lipid metabolism also plays an important role in the response to injury of plants and animals, where activation of phospholipase A2 is important for the generation of oxylipins, which mediate wound responses (23). In this sense, our results show the induction of a gene encoding a protein with a Patatin domain closely related to phospholipase A2, which in plants is involved in responses to oxidative stress and pathogen attack that possesses the same catalytic activity (23). Thus, it could be suggested that in T. atroviride, this protein has phospholipase A2 activity and participates in oxylipin generation. Moreover, production of lipoperoxides and oxylipins has been reported during cell differentiation in fungi (24). Interestingly, our data indicate that genes encoding a lipoxygenase, a cytochrome P450, and a 12-oxophytodienoate reductase are induced. These proteins in plants are also important in the response to wounding and herbivory, and they play a relevant role in plant-pathogen interactions and oxidative burst. In addition, they are involved in the synthesis of the oxylipin jasmonates and the formation of lipoperoxides during the hypersensitive response (25). Conversely, a COP9 signalosome (CSN) subunit, reported as a negative regulator of the biosynthesis of oxylipins (24), is repressed. Moreover, in our transcriptome analysis, at later times, a gene homologous to Velvet A (veA), is repressed in the ΔnoxR mutant, which in Aspergillus spp. is reported as a regulator of sexual and asexual development and of oxylipin biosynthesis (26). Furthermore, a gene encoding a protein with homology to the α-dioxygenase (α-Dox) of Arabidopsis thaliana is induced in the WT strain and does not respond in the ΔnoxR mutant. Interestingly, in A. thaliana, expression of α-Dox is induced by pathogens, salicylic acid, and intracellular ROS (27). Our transcriptome analysis in the ΔnoxR mutant shows that all genes that are related to oxylipin biosynthesis are repressed or not responsive to injury (Datasets S2 and S3), suggesting that oxylipin biosynthesis is compromised in the ΔnoxR mutant. Tsitsigiannis and Keller (24) proposed that oxylipins could bind to and sensitize self G protein–coupled receptors, activating downstream signaling cascades. Furthermore, oxylipins regulate conidiation of A. nidulans and Penicillium chrysogenum through the transcription factor Bristle A (BrlA) (28), leading us to suggest that, as in plants and animals, oxylipins might play an important role in the response to injury of T. atroviride through the formation of lipoperoxides along the injured sites and oxylipins could serve as signaling molecules (Fig. S4).
Another response to injury in plants and animals involves an increase of ROS, which are generated by Nox enzymes, provoking an oxidative burst and later induction of cell death (10). Our results demonstrate the participation of Nox1 and NoxR in the response to injury through the production of ROS; in addition, like in plants and animals, formation of H2O2 is not restricted to damaged cells, suggesting that H2O2 could serve as a propagation signal (Fig. S4). Moreover, in plants and animals, high levels of ROS and intracellular calcium provoke the death of damaged cells (2, 29). In fungi, it has also been reported that elevated levels of ROS induce PCD (30), and generation of ROS depends on Nox activity (31). In this sense, we observed induction of two genes coding for metacaspases and at least six genes encoding proteins related to heterokaryon incompatibility that have been correlated with cell death (32). Thus, our results also indicate that PCD is induced by injury in T. atroviride (Fig. S4A).
For ascomycetes, it has been reported that oxidative stress can induce differentiation (8, 16). In full agreement, our analyses using antioxidants show an important decrease in conidiation in response to injury, which, together with the production of ROS by Nox1, indicate that mechanical injury generates oxidative stress, leading to conidiation in T. atroviride. In this regard, various reports in fungi demonstrated that NoxA is involved in sexual development (11, 12), whereas NoxB does not affect this process, and mutants in noxR present the phenotypes observed in both noxA and noxB deletion mutants (12). Interestingly, our results indicate that conidiation in response to injury is mainly attributable to Nox1, and ROS production is an early, transient event. Furthermore, regeneration of injured hyphae is not affected by the lack of either nox1 or noxR, whereas conidiophore development is severely affected, suggesting that at least two signaling pathways are activated by mechanical injury in T. atroviride. Thus, the transient stimulation of ROS production through Nox1 is essential for conidiation but dispensable for hyphal regeneration in T. atroviride.
In summary, our transcriptome analyses suggest that injury would provoke an increase in the concentration of intracellular calcium, leading to changes in membrane potential, resulting in optimal conditions for Nox activity and Nox1-/NoxR-dependent ROS production. Moreover, injury activates genes involved in oxylipin synthesis (Fig. S4). Thus, the mycelial damage response of Trichoderma could use oxylipins and H2O2 as signaling molecules (Fig. S4), suggesting that the injury response of T. atroviride involves signaling pathways sharing common features with the response to injury of animals and plants.
Materials and Methods
Strains and Culture Conditions.
T. atroviride IMI 206040 was used as WT strain and propagated on potato dextrose agar (PDA) (Difco) at 28 °C.
Mechanical Injury and Blue Light–Induced Conidiation Assays.
For mechanical injury, fungal colonies were grown in total darkness on PDA or Vogel's minimal media supplemented with 2% glucose and 10 mM NH4NO3 (MMV-CN) at 28 °C for 40 h, cut in a grid pattern with a scalpel under a red safety light, and incubated for an additional 48 h in the dark at 28 °C. Conidiation was induced by blue light as described by Casas-Flores et al. (14) but using a light emitting diode (LED)-equipped growth chamber with a fluency of 1200 μmol/m2.
Morphological Analysis.
To record the morphological changes, which occur during the injury response, the WT strain grown on plates containing MMV-CN was wounded and photographed at different times after injury (0, 1, 6, 24, and 48 h). For microscopic changes, plugs of mycelium of the WT strain were used to inoculate microcultures in MMV-CN on microscope slides. Following incubation in the dark at 28 °C for 24 h, the colonies were wounded and stained with lactophenol cotton blue (0.5%). Samples were observed on an Olympus U-LH100HG microscope and photographed at the indicated times after injury.
ROS Detection Assays.
Superoxide and hydrogen peroxide detection was performed as described by Lara-Ortíz et al. (11) with slight modifications (see SI Materials and Methods for details).
Generation and Analysis of Transcriptome Data.
For early injury responses mycelia of the WT strain was collected at 15, 30, and 60 min after damage and frozen immediately. For late times, mycelia from the WT and ΔnoxR strains was collected at 1, 2, and 6 h after injury and frozen immediately. In all cases, a control without injury was included. Total RNA was extracted with TRIzol (Invitrogen), converted to cDNA, and sequenced as described previously (33); 50-bp-long reads were obtained using SOLiD’s standard preprocessing pipeline. Reads were mapped to the genome using GeneSifter net software (http://www.geospiza.com). Read counts were normalized in the R environment (R Development Core Team 2004, version 2.1.1) using a quantile normalization procedure as described previously by Bullard et al. (34). To assess the relative abundance of gene transcripts between cDNA samples, we applied the statistical R test (35). Early-responses genes with an R value of ≥9, and late-responses genes with an R value of ≥15 for the WT strain and ≥12 for the ΔnoxR mutant, were considered positive (true positive rate of ∼98%). Northern blot analysis of genes was carried out using 15 μg of total RNA isolated from mycelium samples obtained at different times after injury following standard procedures. The indicated probes were obtained by PCR; the set of primers used is indicated in Table S1.
Generation of Δnox1, Δnox2, and ΔnoxR Mutants.
The open reading frames (ORFs) of genes nox1, nox2, and noxR were replaced by a hygromycin-resistance cassette as described previously (36). The set of primers used is indicated in Table S1. Gene-replacement constructs were directly used for polyethylene glycol (PEG)-mediated protoplast transformation of the WT strain as described previously (36), and transformants were subjected to three rounds of single-spore isolation.
Complementation of Δnox1 and ΔnoxR Mutants with the WT Nox1 and NoxR genes.
The complete nox1 and noxR genes were amplified by PCR, sequenced, cloned in the pcR2.1 vector, and cotransformed with plasmid pII99, which carries a Geneticin-resistance cassette into the Δnox1-15 and ΔnoxR-6 strains. The set of primers used is indicated in Table S1. The presence of the nox1 and noxR genes was confirmed by PCR.
Supplementary Material
Acknowledgments
We thank J. Simpson, A. Sanchez, N. Carreras, and L. D. Alcaraz for critical reading of the manuscript and E. Ibarra and L. D. Alcaraz for help with the bioinformatic and statistical analyses. M.A.H.-O. was supported by CONACYT for a doctoral fellowship. This work was supported by Grant FOINS-CONACYT (I0110/193/10FON.INST. -30-10) and by the Terciary Education Commission of New Zealand (to A.H.H.-E., A.M.-M., and A.S., respectively).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the NCBI GenBank as sequence read archives accession numbers SRS356201 and SRS356202.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209396109/-/DCSupplemental.
References
- 1.Chang C, et al. Reptile scale paradigm: Evo-Devo, pattern formation and regeneration. Int J Dev Biol. 2009;53:813–826. doi: 10.1387/ijdb.072556cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mittler R, et al. ROS signaling: The new wave? Trends Plant Sci. 2011;16:300–309. doi: 10.1016/j.tplants.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Jedd G. Fungal evo-devo: Organelles and multicellular complexity. Trends Cell Biol. 2011;21:12–19. doi: 10.1016/j.tcb.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Belozerskaya TA, Gessler NN. Reactive oxygen species and the strategy of antioxidant defense in fungi: a review. Prikl Biokhim Mikrobiol. 2007;43:565–575. (in Russian) [PubMed] [Google Scholar]
- 5.Gessler NN, Aver’yanov AA, Belozerskaya TA. Reactive oxygen species in regulation of fungal development. Biochemistry (Mosc) 2007;72:1091–1109. doi: 10.1134/s0006297907100070. [DOI] [PubMed] [Google Scholar]
- 6.Fester T, Hause G. Accumulation of reactive oxygen species in arbuscular mycorrhizal roots. Mycorrhiza. 2005;15:373–379. doi: 10.1007/s00572-005-0363-4. [DOI] [PubMed] [Google Scholar]
- 7.Leonard TJ, Dick S. Induction of haploid fruiting by mechanical injury in Schizophyllum commune. Mycology. 1973;65:809–821. [Google Scholar]
- 8.Aguirre J, Ríos-Momberg M, Hewitt D, Hansberg W. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 2005;13:111–118. doi: 10.1016/j.tim.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Aguirre J, Lambeth JD. Nox enzymes from fungus to fly to fish and what they tell us about Nox function in mammals. Free Radic Biol Med. 2010;49:1342–1353. doi: 10.1016/j.freeradbiomed.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takemoto D, Tanaka A, Scott B. NADPH oxidases in fungi: Diverse roles of reactive oxygen species in fungal cellular differentiation. Fungal Genet Biol. 2007;44:1065–1076. doi: 10.1016/j.fgb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Lara-Ortíz T, Riveros-Rosas H, Aguirre J. Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans. Mol Microbiol. 2003;50:1241–1255. doi: 10.1046/j.1365-2958.2003.03800.x. [DOI] [PubMed] [Google Scholar]
- 12.Cano-Domínguez N, Alvarez-Delfín K, Hansberg W, Aguirre J. NADPH oxidases NOX-1 and NOX-2 require the regulatory subunit NOR-1 to control cell differentiation and growth in Neurospora crassa. Eukaryot Cell. 2008;7:1352–1361. doi: 10.1128/EC.00137-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shoresh M, Harman GE, Mastouri F. Induced systemic resistance and plant responses to fungal biocontrol agents. Annu Rev Phytopathol. 2010;48:21–43. doi: 10.1146/annurev-phyto-073009-114450. [DOI] [PubMed] [Google Scholar]
- 14.Casas-Flores S, Rios-Momberg M, Bibbins M, Ponce-Noyola P, Herrera-Estrella A. BLR-1 and BLR-2, key regulatory elements of photoconidiation and mycelial growth in Trichoderma atroviride. Microbiology. 2004;150:3561–3569. doi: 10.1099/mic.0.27346-0. [DOI] [PubMed] [Google Scholar]
- 15.Ruepp A, et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgiou CD, Patsoukis N, Papapostolou I, Zervoudakis G. Sclerotial metamorphosis in filamentous fungi is induced by oxidative stress. Integr Comp Biol. 2006;46:691–712. doi: 10.1093/icb/icj034. [DOI] [PubMed] [Google Scholar]
- 17.Zimmerman ZA, Kellogg DR. The Sda1 protein is required for passage through start. Mol Biol Cell. 2001;12:201–219. doi: 10.1091/mbc.12.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Syed K, Kattamuri C, Thompson TB, Yadav JS. Cytochrome b5 reductase-cytochrome b5 as an active P450 redox enzyme system in Phanerochaete chrysosporium: Atypical properties and in vivo evidence of electron transfer capability to CYP63A2. Arch Biochem Biophys. 2011;509:26–32. doi: 10.1016/j.abb.2011.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arimura G-I, Maffei ME. Calcium and secondary CPK signaling in plants in response to herbivore attack. Biochem Biophys Res Commun. 2010;400:455–460. doi: 10.1016/j.bbrc.2010.08.134. [DOI] [PubMed] [Google Scholar]
- 20.Nelson G, et al. Calcium measurement in living filamentous fungi expressing codon-optimized aequorin. Mol Microbiol. 2004;52:1437–1450. doi: 10.1111/j.1365-2958.2004.04066.x. [DOI] [PubMed] [Google Scholar]
- 21.Zelter A, Bencina M, Bowman BJ, Yarden O, Read ND. A comparative genomic analysis of the calcium signaling machinery in Neurospora crassa, Magnaporthe grisea, and Saccharomyces cerevisiae. Fungal Genet Biol. 2004;41:827–841. doi: 10.1016/j.fgb.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Schumacher J, de Larrinoa IF, Tudzynski B. Calcineurin-responsive zinc finger transcription factor CRZ1 of Botrytis cinerea is required for growth, development, and full virulence on bean plants. Eukaryot Cell. 2008;7:584–601. doi: 10.1128/EC.00426-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dhondt S, Geoffroy P, Stelmach BA, Legrand M, Heitz T. Soluble phospholipase A2 activity is induced before oxylipin accumulation in tobacco mosaic virus-infected tobacco leaves and is contributed by patatin-like enzymes. Plant J. 2000;23:431–440. doi: 10.1046/j.1365-313x.2000.00802.x. [DOI] [PubMed] [Google Scholar]
- 24.Tsitsigiannis DI, Keller NP. Oxylipins as developmental and host-fungal communication signals. Trends Microbiol. 2007;15:109–118. doi: 10.1016/j.tim.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 25.Andreou A, Brodhun F, Feussner I. Biosynthesis of oxylipins in non-mammals. Prog Lipid Res. 2009;48:148–170. doi: 10.1016/j.plipres.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 26.Tsitsigiannis DI, Zarnowski R, Keller NP. The lipid body protein, PpoA, coordinates sexual and asexual sporulation in Aspergillus nidulans. J Biol Chem. 2004;279:11344–11353. doi: 10.1074/jbc.M310840200. [DOI] [PubMed] [Google Scholar]
- 27.Mosblech A, Feussner I, Heilmann I. Oxylipin signaling and plant growth. In: Munnik T, editor. Lipid Signaling in Plants, Series: Plant Cell Monographs. Vol 16. Berlin: Springer; 2010. pp. 277–291. [Google Scholar]
- 28.Hegedüs N, Sigl C, Zadra I, Pócsi I, Marx F. The paf gene product modulates asexual development in Penicillium chrysogenum. J Basic Microbiol. 2011;51:253–262. doi: 10.1002/jobm.201000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das DK, Maulik N. Conversion of death signal into survival signal by redox signaling. Biochemistry (Mosc) 2004;69:10–17. doi: 10.1023/b:biry.0000016345.19027.54. [DOI] [PubMed] [Google Scholar]
- 30.Leiter E, Marx F, Pusztahelyi T, Haas H, Pócsi I. Penicillium chrysogenum glucose oxidase — a study on its antifungal effects. J Appl Microbiol. 2004;97:1201–1209. doi: 10.1111/j.1365-2672.2004.02423.x. [DOI] [PubMed] [Google Scholar]
- 31.Silar P. Peroxide accumulation and cell death in filamentous fungi induced by contact with a contestant. Mycol Res. 2005;109:137–149. doi: 10.1017/s0953756204002230. [DOI] [PubMed] [Google Scholar]
- 32.Glass NL, Jacobson DJ, Shiu PKT. The genetics of hyphal fusion and vegetative incompatibility in filamentous ascomycete fungi. Annu Rev Genet. 2000;34:165–186. doi: 10.1146/annurev.genet.34.1.165. [DOI] [PubMed] [Google Scholar]
- 33.Vega-Arreguín JC, et al. Deep sampling of the Palomero maize transcriptome by a high throughput strategy of pyrosequencing. BMC Genomics. 2009;10:299–308. doi: 10.1186/1471-2164-10-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94–107. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stekel DJ, Git Y, Falciani F. The comparison of gene expression from multiple cDNA libraries. Genome Res. 2000;10:2055–2061. doi: 10.1101/gr.gr-1325rr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castellanos F, et al. Crucial factors of the light perception machinery and their impact on growth and cellulase gene transcription in Trichoderma reesei. Fungal Genet Biol. 2010;47:468–476. doi: 10.1016/j.fgb.2010.02.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.