Abstract
Water is critical for the structure, stability, and functions of macromolecules. Diffraction and NMR studies have revealed structure and dynamics of bound waters at atomic resolution. However, localizing the sites and measuring the dynamics of bound waters, particularly on timescales relevant to catalysis and macromolecular assembly, is quite challenging. Here we demonstrate two techniques: first, temperature-dependent radiolytic hydroxyl radical labeling with a mass spectrometry (MS)-based readout to identify sites of bulk and bound water interactions with surface and internal residue side chains, and second, H218O radiolytic exchange coupled MS to measure the millisecond dynamics of bound water interactions with various internal residue side chains. Through an application of the methods to cytochrome c and ubiquitin, we identify sites of water binding and measure the millisecond dynamics of bound waters in protein crevices. As these MS-based techniques are very sensitive and not protein size limited, they promise to provide unique insights into protein–water interactions and water dynamics for both small and large proteins and their complexes.
Keywords: protein structure, footprinting, time resolved, covalent labeling
Water plays an important role in protein structure, folding, and stability (1–9). For proteins in solution, we broadly categorize water into three different types: bulk or free water, water that interacts with the protein surface, and internal water molecules (10, 11). Bulk water has no interactions with the protein. As bulk waters approach the protein surface, water diffusion rates are slowed and water concentration is elevated due to interactions with protein surface residues (12–14). Waters can also provide important elements of the internal protein structure. Atomic resolution details of water structure and dynamics are explored by X-ray and neutron diffraction and NMR studies and reveal water molecules co-localized with strategically placed polar or charged amino acid groups that are highly conserved (1–5). Water dynamics explored using high-field NMR, such as nuclear Overhauser effect (NOE) and magnetic relaxation dispersion (MRD) methodologies, can indirectly calculate residence times of internal waters in proteins; these residence times range from subnanoseconds to milliseconds depending on the interaction of the water molecules in question (15–21). However, experimental approaches that can reveal specific sites of water binding to the protein surface and the interior as well as the dynamics of surface and internal waters are limited. Direct measures of both sites of water occupancy and water dynamics in solution could address a wide range of questions related to the role of water in mediating protein interactions with ligands and in assembly of protein complexes.
Structural mass spectrometry (MS) approaches can probe the interactions of bulk water and the hydration layer with protein structure and provide specific information on backbone and side-chain interactions with water (22–24). Hydroxyl radical-mediated approaches are effective probes of water interactions with the protein surface; recently, we have shown that radiolysis-mediated hydroxyl radical labeling (footprinting) coupled to MS can probe the structure of internal waters in membrane proteins (25, 26). Given this potential for utilizing water as a reagent to probe protein structure, we explored applications of the footprinting technique using a combination of temperature and radiolytic 18O labeling. In particular, we were interested to expand the approach to soluble proteins and in identifying waters bound to the protein surface. For the soluble proteins cytochrome c (cyt c) and ubiquitin, we differentiated radiolytic labeling arising from bulk vs. surface-bound waters and measured the exchange rates of internal water interactions with specific side chains directly. As these approaches are coupled to sensitive MS-based detection methods, they can be widely applied to understand the locations and dynamics of bound waters in complex macromolecular systems in vitro and provide a useful complement to evolving NMR-based studies of water structure and dynamics.
Results
Radiolytic Labeling of cyt c and Ubiquitin in Liquid vs. Frozen State Distinguishes Bound vs. Bulk Water.
Radiolytic labeling of the protein surface relies on the activation of bulk and surface waters forming hydroxyl radicals, followed by the diffusion of radicals to reactive and accessible sites on the protein (23, 24). In order to explore the effect of diffusion on radiolytic labeling of proteins, we subjected cyt c and ubiquitin to 0—to 40-ms pulses of focused synchrotron X-rays followed by rapid quenching, protease digestion, and analysis by liquid-chromatography electrospray-ionization mass spectrometry (LC-ESI-MS) and MS/MS (Figs. S1 and S2) while comparing the labeling seen at room temperature (RT) to that for samples rapidly frozen and maintained at -35 °C (Figs. S3 and S4A). The rates of modification were vastly reduced for some residues at -35 °C, while they were unchanged for others (Fig. 1, and Tables S1 and S2). To understand the molecular basis of this effect, we explored radiolysis of the small molecule Alexa 488, which is used as a “target” dosimeter to optimize the radiolysis conditions for X-ray footprinting (24). Upon radiolysis of Alexa 488 solution at RT, its intensity of fluorescence decreases rapidly (k = 176 s-1; Fig. S3), whereas when irradiated at -35 °C the rate of modification is reduced > 200-fold (k = 0.8 s-1). Essentially, ice formation blocks diffusion of hydroxyl radicals and limits their reaction with Alexa 488. Thus, it can be deduced that macromolecule residues that had significant modification at RT but no discernible modification upon freezing are primarily modified based on hydroxyl radicals derived from bulk water.
Fig. 1.
Pictorial representation of modification sites on cyt c (A and B) and ubiquitin (C and D) in two orientations. Colored surface representation based on the X-ray crystal structure of cyt c (1HRC) (27) and ubiquitin (1UBQ) (28) indicate the side-chain residues that are consistently modified after irradiation under RT and/or frozen conditions. Blue indicates the residues that show 13- to 200-fold decreases in modification upon freezing, violet indicates 3- to 10-fold decreases, and red indicates minimal to no change (< 2-fold) in the modification rate when sample is frozen compared to RT. Y67 in cyt c is completely buried inside the heme cavity and not visible in these orientations.
The side-chain residues H33, F36, R38, T47, K53, K60, E61-62, L64, P71, P76, and K86 in cyt c are clearly modified at RT, but we do not see any modifications at -35 °C; these residues have temperature-dependent behavior identical to that of Alexa 488 dye (Fig. S3). The X-ray crystal structure shows that for these amino acid residues both the side-chain and the backbone atoms are located on the surface of the protein (Fig. 1 A and B); presumably, ice formation adjacent to these residues blocks modification at -35 °C (27). On the other hand, residues F46, Y48, W59, M65, Y67 and M80 exhibit minimal changes in their modification rate as a function of temperature (< 2-fold; Fig. S3). This indicates that waters adjacent to these residues are activated by radiolysis while in close proximity and thus can react without significant diffusional motion or that ice formation is inhibited in the immediate vicinity of these residues. The X-ray crystal structure shows that Y67 is fully, and F46, Y48, W59, and M80 are partly, buried inside the heme cavity (Fig. 1 A and B). Y48, Y67, and M80 are in close proximity to internal water molecules detected in the X-ray structure; these waters are likely candidates driving the modification of these residues at -35 °C. Residues K99, C14/17, and 100–104 have detectible modifications at -35 °C but these are approximately 3- to 10-fold reduced compared to RT; thus, they may be modified primarily by bulk water at RT while they are also adjacent to bound surface or internal waters that can oxidize them at -35 °C. In contrast to the behavior for cyt c, ubiquitin, which lacks internal water molecules (28), shows large decreases in the modification rate constants for all the modified residues (Fig. 1 C and D and Fig. S4A). However, residual modifications of residues M1, K48, L67, and L69, and modifications on multiple residues on peptide 5–15 and within peptide 12–27, are seen at low temperature indicating multiple sites of surface water interactions.
Radiolytic Labeling of Proteins with 18O-Water and 18O/16O Exchange.
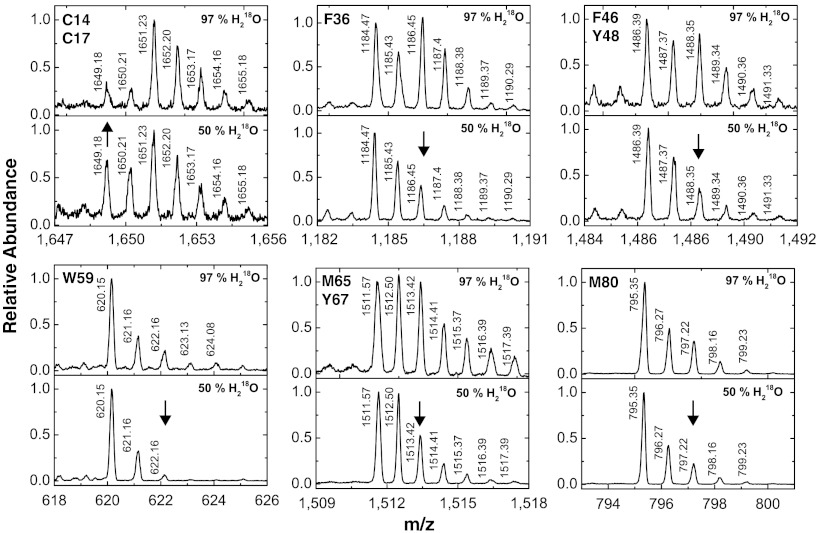
Addition of hydroxyl radical to π bonds in an aromatic ring results in a resonance delocalized radical, which after reacting with O2 will rearomatize through the loss of hydroperoxide radical (24, 29) (Fig. S5). Besides, hydroxyl radical can also attack sulfur-containing side chains, wherein the incorporated oxygen may also be derived either from O2 or OH• (Fig. S5). Thus, irradiation of solutions containing H218O will generate  that will selectively label Met, Cys, Phe, Tyr, and Trp residues (30). In order to establish a baseline for 1∶1 H218O/H216O exchange experiments, we subjected cyt c and ubiquitin at 10–30 μM concentration in 97% H218O containing buffer to X-ray irradiation for 5 ms and compared the results to the proteins irradiated in 50∶50 H216O∶H218O. Fig. 2 illustrates the isotopologues of the radiolabeled singly protonated peptide 9–22, 28–38, 40–53, 61–72, and 80–86 of cyt c (+16 Da or +18 Da) under the two aforementioned conditions. The relative levels of 18O/16O labeling in 97% H218O compared to 50∶50 H216O∶H218O is consistent with the reaction mechanisms of hydroxylation mentioned above (29) and indicates a robust signal for examining the exchange processes (Fig. 2). We characterized nine sites (C14, C17, F36, F46, Y48, W59, M65, Y67, and M80) that show 18O labeling and 16O exchange that are located either close to bound waters (in the heme groove) or are on the exposed surface and are labeled by bulk or surface water. In contrast, ubiquitin shows 18O labeling only for M1 and F45 residues from the peptide fragments 1–6 and 43–48, respectively (Table S2 and Fig. S4B), and exchange can be measured only for these two residues.
that will selectively label Met, Cys, Phe, Tyr, and Trp residues (30). In order to establish a baseline for 1∶1 H218O/H216O exchange experiments, we subjected cyt c and ubiquitin at 10–30 μM concentration in 97% H218O containing buffer to X-ray irradiation for 5 ms and compared the results to the proteins irradiated in 50∶50 H216O∶H218O. Fig. 2 illustrates the isotopologues of the radiolabeled singly protonated peptide 9–22, 28–38, 40–53, 61–72, and 80–86 of cyt c (+16 Da or +18 Da) under the two aforementioned conditions. The relative levels of 18O/16O labeling in 97% H218O compared to 50∶50 H216O∶H218O is consistent with the reaction mechanisms of hydroxylation mentioned above (29) and indicates a robust signal for examining the exchange processes (Fig. 2). We characterized nine sites (C14, C17, F36, F46, Y48, W59, M65, Y67, and M80) that show 18O labeling and 16O exchange that are located either close to bound waters (in the heme groove) or are on the exposed surface and are labeled by bulk or surface water. In contrast, ubiquitin shows 18O labeling only for M1 and F45 residues from the peptide fragments 1–6 and 43–48, respectively (Table S2 and Fig. S4B), and exchange can be measured only for these two residues.
Fig. 2.
Identification and quantification of 18O labeled residues. MS analysis in zoom scan mode of cyt c samples in 97% and 50% H218O provide a quantitative analysis of isotopologues of singly protonated 18O and 16O labeled peptides 9–22, 28–38, 40–53, 56–60, 61–72, and 80–86 of cyt c with side-chain modifications on the residues C14/C17, F36, F46/Y48, W59, M65/Y67, and M80, respectively. The first labeled peak indicates the position of the monoisotopic mass of the 16O-adduct; the peaks shifted by 2 m/z units from this mass contain a mixture of the 18O monoisotopic mass and the (two) C13 containing 16O-isotope. The decreased abundance at this m/z value relative to the monotisotopic mass, for 50% vs. 97% H218O, is indicated by an arrow and represents the potential signal for an exchange experiment for the respective peptide. For peptide 9–22, the largest signal is the increase in the 9–16 monoisotopic peak.
Time-Resolved 18O/16O Exchange on Millisecond and Submillisecond Timescales.
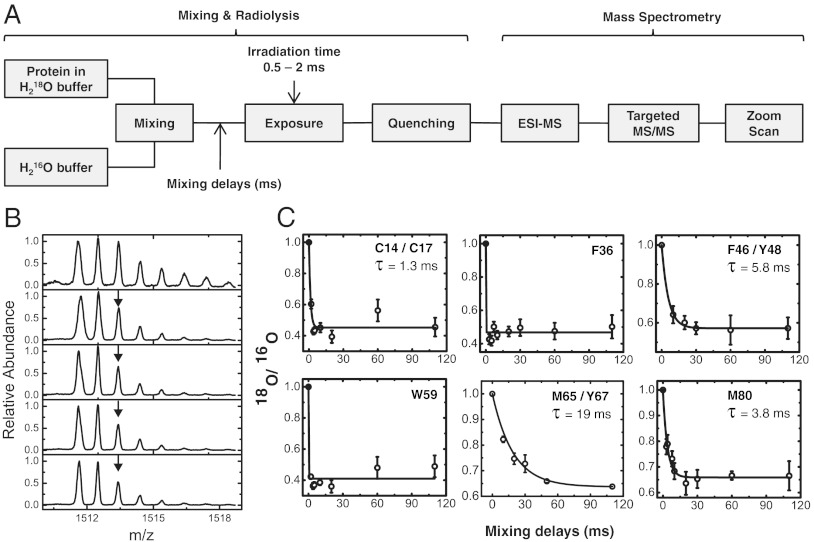
Rapid 1∶1 mixing of proteins at 2 °C in 97% H218O buffer solution with 100% H216O-buffer was carried out followed by synchrotron X-ray exposure and LC-ESI-MS (Fig. 3A). Zoom scans were used to quantify the ratio of 18O vs. 16O labeling for various mixing delays (Fig. 3B and Fig. S6). Progress curves that monitored the exchange of H218O water with added H216O were obtained by plotting the ratio of peak intensity of 18O-isotopologue to that of the 16O-isotopologue vs. the delay time (Fig. 3C). Residues F36 and W59 of cyt c showed exchange that was too fast to measure with the current mixing dead time. F36 is exposed toward the surface of the cyt c molecule (Fig. 1 A and B). The submillisecond exchange and the fact that low temperature abolishes modification support the idea that modification is mediated by bulk water with no evidence of bound waters in the vicinity. The side chain of W59 residue is projected toward the heme cavity (Fig. 4), and the temperature dependence indicates modification by bound water that can be exchanged with bulk water rapidly.
Fig. 3.
Time-resolved radiolytic 18O labeling and water exchange in cyt c. (A) Rapid mixing combined with 18O-mediated hydroxyl radical labeling monitored the time course of exchange of water in cyt c. LC-ESI-MS is used to identify and isolate the modified peptides, targeted MS/MS is used to identify the sites of 18O labeling, and zoom scan is used quantify the ratio of 18O vs. 16O labeling at various mixing delays. (B) Zoom scans for singly protonated peptide 61–72 showing decease in the abundance of the 2 m/z shifted 18O monoisotopic mass (arrow) that corresponds to the water exchange at M65 and Y67 with increase in the mixing delays. (C) Progress curves (circles and error bars) of water exchange for the 18O labeled side-chain residues. The solid line represents the fit to single exponential function. Residues W59 and F36 have exchange that is complete at the first measurement, while the rates of exchange of C14/C17, F46/Y48, M65/Y67, and M80 are discretely measured.
Fig. 4.
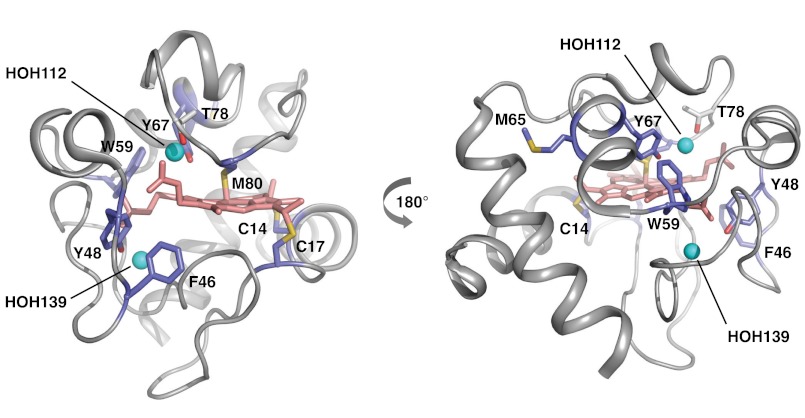
Cyt c 18O-labeling map. The sites of 18O-modifications are visualized from the crystal structure 1HRC (27) using PyMOL. The 18O-labeled residues (light blue) in and around the heme (light pink) crevices, and the position of residue T78 (gray) and conserved waters (cyan spheres) HOH112, HOH139 are shown in two orientations of the cyt c molecule.
The slowest rate of exchange was observed for peptide 61–72, which contains mixed modification products at M65 and Y67 (Fig. S2). The lifetime of 19 ms thus reflects an average exchange rate for M65 and Y67 residues. M65 is surface accessible, but Y67 is fully buried; this slow exchange and the lack of temperature dependence of the modification rate indicates that bound waters around M65 and an internal water close to Y67 (HOH112) are likely responsible for their modifications (31). Explicit exchange lifetimes ranging from 1.3–5.8 ms were observed for resides C14/17, F46/Y48, and M80 of cyt c (Fig. 3C). The observed exchange rate for peptide 40–53 reflects an average exchange rate constant for F46 and Y48. Residue Y48 and part of residue F46 are located deep inside the heme cavity (Fig. 4). The millisecond exchange and the lack of temperature dependence indicate that the modifications arise from interactions with bound waters; HOH139 and HOH112, which are in close proximity to Y48 and Y67, respectively (Fig. 3C). HOH112 makes H-bonding interactions with both Y67 and T78, which are in close proximity to M80. The millisecond exchange for C14 and C17 (Fig. 3C and Fig. S6A), which are covalently connected to the heme, and the approximately 3-fold decrease in temperature dependency purport interactions with bound waters. Molecular dynamics (MD) simulations show rotameric changes in K79 of cyt c that can permit access of water into the heme pocket within 4 ns (Fig. S7). This appears to facilitate rapid exchange adjacent to W59, while for other residues inside the heme pocket water exchange is delayed due to water protein interactions. Our exchange studies of ubiquitin reveal that M1 and F45 exchange too fast to be directly measured by our approaches (Fig. S4B); nevertheless, the residual modifications at lower temperatures provide evidence for bound water interactions with the protein surface, which may experience rapid exchange.
Discussion
Detection of Bound Water at the Protein Surface and in the Protein Interior.
Evidence for the sites of protein-bound water comes from analysis of high-resolution crystal structures having reliable water assignments; these data indicate areas of the protein surface adjacent to and within grooves that bind many more water molecules than other areas of the surface (13), and these bound waters form extensive hydrogen bonding networks with amino acid residues inside protein grooves. In these studies, the temperature dependence of radiolysis is used to infer the presence of bound water both on the surface and inside the protein in cyt c. In the temperature range of -35 °C to -50 °C, NMR line broadening studies suggest the existence of unfrozen water bound to the protein surface (6, 32). Thus, our radiolysis results at -35 °C may reflect reactivity of patches of bound water molecules that remain in a semihydrated state. To gain additional insight into the temperature dependence of labeling and water structure, we carried out the radiolytic labeling of cyt c at -192 °C, where all liquid water is abolished. The residues that were modified at -35 °C undergo additional decreases in modification rates at cryogenic temperatures (Table S1). However, most residues on the surface have no observable modifications remaining, with the exception of E61-62 and P71, while residues projecting toward the heme cavity or inside the heme pocket still have significant (but reduced) modifications. At cryogenic temperatures, water molecules have minimal motions except for localized vibrational motions (33). The continued observation of modification at cryogenic temperatures implies the reaction of activated waters with immediately adjacent residues, and the reductions in surface modifications imply the elimination of the semihydrated state of bound waters that mediates reactivity at -35 °C. Thus, we suspect that modification of surface residues seen at -35 °C for both ubiquitin and cyt c reflects interactions with water in a semihydrated state bound to specific regions of the protein surface.
Rates and Sites of Bound Water Exchange in Proteins.
Water is of increasing interest in the study of protein structure and function (34). “De-wetting” is required prior to oligomerization of some complexes, and reorganization of water in substrate binding cavities is an important process whose dynamics are of considerable interest. Experimental studies of water dynamics, primarily by NMR, have suggested a wide range of exchange rates, from nanoseconds to milliseconds for globular proteins, although water residence times can be even longer. For example, waters residing in the transmembrane regions of membrane proteins in some cases are extremely resistant to exchange (25, 26). An MRD examination of cyt c suggested exchange rates on microsecond timescales contributed from buried water molecules (35), while our studies reveal that some cyt c residues exhibit millisecond exchange. Methodological differences between the MRD experiments and those here likely account for the differences. First, these MRD experiments indirectly measured the exchange for the ensemble of buried waters, not distinguishing the individual water–side-chain interactions. Second, the above MRD experiments were carried out at 27 °C (vs. 2 °C here), and water residence times can be quite temperature dependent (10). Additionally, some of our cyt c exchange rates are submillisecond (F36 and W59) and may be microseconds or even faster, consistent with the MRD results. Millisecond exchange for internal, bound waters is well established as the residence time of the most long-lived buried water in Bovine Pancreatic Trypsin Inhibitor was determined to 3 ms at 4 °C by 17O and 2H MRD (20). Also, 17O and 2H MRD data of medium-sized to large proteins indicate that most integral water molecules have residence times ranging from nanoseconds to 1 ms at 27 °C (10), consistent with the range of exchange rates for cyt c observed here.
Ubiquitin exhibits multiple sites of surface water interaction but shows greater reductions in modification rate as a function of temperature and only submillisecond exchange for M1 and F45, the two side-chain residues for which exchange can be observed. A recent study by Nucci et al. (36) characterized the hydration layer of ubiquitin by encapsulation within a reverse micelle; dozens of hydration waters were observed, and NOE indicated a considerable range of hydration water dynamics present on the protein surface. The encapsulated ubiquitin showed a “very-long lived” water molecule localized to a surface pocket adjacent to Val 69 (human sequence) and Lys 6. The data here, based on residual modification observed at -35 °C, indicates formation of a semihydrated state at -35 °C for surface waters near Leu 67, Leu 69, Met 1, and near peptide 5–15 (including Lys 6), consistent with the above results. The submillisecond exchange for M1 and F45 are also consistent with previous MRD studies of several globular proteins, including ubiquitin, which showed the fastest exchange rate (10, 37). Thus, the MS-based approach here, although quite different from the NMR approaches overall, appears to provide a consistent and complementary picture relative to existing studies, giving confidence in the overall method.
To summarize, the temperature dependence of radiolytic labeling identifies residues that are oxidized by bound vs. bulk solvent, and our studies directly reveal multiple sites of bound water on the surface of cyt c and a more limited set of such sites on ubiquitin. Specific exchange rates of water for selected aromatic and sulfur-containing residues can also be measured using H218O coupled radiolysis. For cyt c, we provide an explicit picture of the dynamics of exchange of water for specific residues within the heme pocket, with exchange rates that vary from submillisecond to tens of milliseconds. Because water is ubiquitous in its interactions with macromolecules, these two technologies will be powerful tools in understanding active site dynamics in enzyme function involving bound water molecules, dynamics of bound water in the activation of receptors, the movement of water across pores and channels associated with gating, as well as ligand-dependent structural assembly of macromolecules.
Methods
Synchrotron X-Ray Radiolysis of Protein or Radiolytic Labeling.
X-ray radiolysis was performed at beamline X28C at National Synchrotron Light Source of Brookhaven National Laboratory, New York. X-ray focusing mirror was used to carry out radiolytic labeling under optimal flux density (38, 39). Solutions of 5 μM Alexa 488 (Invitrogen), 10 μM cyt c (horse heart) and 30 μM ubiquitin (bovine erythrocyte) were prepared in 10 mM sodium phosphate buffer pH 7.2. Samples were loaded into 200-μL PCR tubes in 5-μL volume and irradiated for 0–40 ms using a millisecond shutter in the standard multiple sample irradiation set up at RT (25 °C) (40). The same experiment was performed after snap-freezing of samples in PCR tubes in liquid nitrogen, while maintaining the temperature at -35 °C by Peltier cooling coupled to the multiple sample holder following the protocol for the X-ray footprinting of frozen cell samples (40). For the experiment at cryogenic temperature (-190 °C), a cryo-cooled (liquid N2) sample holder was used to irradiate sample. Exposed samples were treated with methionine-amide to stop unwanted secondary free radicals reactions, and the samples were stored at -80 °C. UV-visible spectrum of cyt c before and after X-ray irradiation showed no protein damage. Protein digestion with trypsin for cyt c, and trypsin and pepsin digestion for ubiquitin, was carried out using standard procedures. For 18O-labeling experiments, proteins were prepared in buffer solution containing 97% and 50% H218O followed by the radiolytic labeling at RT. Irradiated samples were completely evaporated to dryness under vacuum at 60 °C to remove any water from the sample. Samples were reconstituted in the digestion buffer followed by protease digestion.
Time-Resolved Radiolytic Labeling with 18O and Water Exchange.
Time-resolved radiolysis was carried out at 2 °C in a modified KinTek® (KinTek Corporation) apparatus using a two-step (push-pause-push) flow method. In the first step, the 10 μL of 10 μM cyt c in 97% H218O buffer was mixed with equal amount of H216O buffer (1∶1 mixing) by a T-mixer. In the second step after millisecond delay time (0–110 ms), the mixed sample was passed through an irradiation cell. The flow speed was adjusted to achieve irradiation times from 0.5–2 ms. The overall dead time was dependent on the sum of the travel time of the mixed solution right up to the irradiation cell and the sample irradiation time. The “zero” millisecond delay was obtained by mixing the 10 μL of 10 μM cyt c in 97% H218O buffer with equal volume of the same solution followed by irradiation. Exposed samples were treated with methionine-amide to stop unwanted secondary free radicals reactions, and the samples were stored at -80 °C. Protein digestion with trypsin for cyt c and trypsin and pepsin digestion for ubiquitin was carried out using standard procedures.
Mass Spectrometry and Data Analysis.
Protease digest of cyt c and ubiquitin were analyzed by Thermo-Fisher LCQ Classic and LCQ DecaXP plus mass spectrometer interfaced with Waters Alliance 2695 HPLC according to standard LC-ESI-MS procedures (Fig. S1) (41). LC-MS/MS parameters were set for carrying out a full data-dependent scan for 16O modification and also by specifically selecting mass ranges targeting identification of 16O and 18O modified peptides of interest (Fig. S2). MS/MS spectra for the unmodified and modified peptides were manually interpreted with the aid of the ProteinProspector (University of California, San Francisco) algorithm and Bioworks 3.3 software (Fig. S2). Selected ion chromatograms from the LC-ESI-MS results were used to quantify the extent of modification (Fig. S1). The fraction unmodified for each peptide from the chromatogram was calculated as the ratio of integrated area of the unmodified peptide to the sum of integrated areas from the modified and unmodified peptides. The dose-response curve (fraction unmodified vs. X-ray exposure time) was fitted to first-order decay function with Origin® Version 8.5 (OriginLabs) to determine the modification rate constants (Figs. S3 and S4A). Rate constants of modifications were correlated to the solvent-accessible surface area of side-chain residues calculated by the program MODELLER (42) and compared between liquid (RT) and frozen states ( -35 °C and -190 °C) (Tables S1 and S2). MS analysis of 18O-labeling experiments was carried out in zoom scan mode around the monoisotopic m/z (± 5 m/z) of 16O modified peak (Figs. 2 and 3B and Fig. S6). The intensities of the isotopes that correspond to 18O- and 16O-labelled peptides were used to determine relative amount of 18O labeling. Zoom scans were used to quantify the ratio of 18O vs. 16O labeling for various mixing delays and to plot the progress curves for site-specific water exchange processes (Fig. 3 B and C and Figs. S4B and S6). Data were best fitted to a single exponential decay Origin® Version 8.5 (OriginLabs), which resulted in the determination of lifetimes of water exchange processes at the radiolabeled side-chain residues.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Manish Datt for MD simulations of cytochrome c. This work is supported in part by the National Institute for Biomedical Imaging and Bioengineering under Grants P30-EB-09998 and R01-EB-09688. The National Synchrotron Light Source at Brookhaven National Laboratory is supported by the Department of Energy under Contract DE-AC02-98CH10886.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209060109/-/DCSupplemental.
References
- 1.Angel TE, Chance MR, Palczewski K. Conserved waters mediate structural and functional activation of family A (rhodopsin-like) G protein-coupled receptors. Proc Natl Acad Sci USA. 2009;106:8555–8560. doi: 10.1073/pnas.0903545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carugo O, Bordo D. How many water molecules can be detected by protein crystallography? Acta Crystallogr D Biol Crystallogr. 1999;55:479–483. doi: 10.1107/s0907444998012086. [DOI] [PubMed] [Google Scholar]
- 3.Jiang JS, Brunger AT. Protein hydration observed by X-ray diffraction. Solvation properties of penicillopepsin and neuraminidase crystal structures. J Mol Biol. 1994;243:100–115. doi: 10.1006/jmbi.1994.1633. [DOI] [PubMed] [Google Scholar]
- 4.Royer WE, Jr, et al. Ordered water molecules as key allosteric mediators in a cooperative dimeric hemoglobin. Proc Natl Acad Sci USA. 1996;93:14526–14531. doi: 10.1073/pnas.93.25.14526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams MA, Goodfellow JM, Thornton JM. Buried waters and internal cavities in monomeric proteins. Protein Sci. 1994;3:1224–1235. doi: 10.1002/pro.5560030808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuntz ID, Jr, et al. Hydration of macromolecules. Science. 1969;163:1329–1331. doi: 10.1126/science.163.3873.1329. [DOI] [PubMed] [Google Scholar]
- 7.Chaplin M. Do we underestimate the importance of water in cell biology? Nat Rev Mol Cell Biol. 2006;7:861–866. doi: 10.1038/nrm2021. [DOI] [PubMed] [Google Scholar]
- 8.Ebbinghaus S, et al. An extended dynamical hydration shell around proteins. Proc Natl Acad Sci USA. 2007;104:20749–20752. doi: 10.1073/pnas.0709207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ball P. Water as an active constituent in cell biology. Chem Rev. 2008;108:74–108. doi: 10.1021/cr068037a. [DOI] [PubMed] [Google Scholar]
- 10.Denisov VP, Halle B. Protein hydration dynamics in aqueous solution. Faraday Discuss. 1996;103:227–244. doi: 10.1039/fd9960300227. [DOI] [PubMed] [Google Scholar]
- 11.Tarek M, Tobias DJ. The dynamics of protein hydration water: A quantitative comparison of molecular dynamics simulations and neutron-scattering experiments. Biophys J. 2000;79:3244–3257. doi: 10.1016/S0006-3495(00)76557-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellissent-Funel M-C, Zanotti JM, Chenb SH. Slow dynamics of water molecules on the surface of a globular protein. Faraday Discuss. 1996;103:281–294. [Google Scholar]
- 13.Kuhn LA, et al. The interdependence of protein surface topography and bound water molecules revealed by surface accessibility and fractal density measures. J Mol Biol. 1992;228:13–22. doi: 10.1016/0022-2836(92)90487-5. [DOI] [PubMed] [Google Scholar]
- 14.Bagchi B. Water dynamics in the hydration layer around proteins and micelles. Chem Rev. 2005;105:3197–3219. doi: 10.1021/cr020661+. [DOI] [PubMed] [Google Scholar]
- 15.Bertini I, et al. Protein hydration and location of water molecules in oxidized horse heart cytochrome c by (1)H NMR. J Magn Reson. 2000;147:1–8. doi: 10.1006/jmre.2000.2131. [DOI] [PubMed] [Google Scholar]
- 16.Otting G, Liepinsh E, Wuthrich K. Protein hydration in aqueous solution. Science. 1991;254:974–980. doi: 10.1126/science.1948083. [DOI] [PubMed] [Google Scholar]
- 17.Otting G, Liepinsh E, Wuthrich K. Proton exchange with internal water molecules in the protein BPTI in aqueous solution. J Am Chem Soc. 1991;113:4363–4364. [Google Scholar]
- 18.Brunne RM, et al. Hydration of proteins. A comparison of experimental residence times of water molecules solvating the bovine pancreatic trypsin inhibitor with theoretical model calculations. J Mol Biol. 1993;231:1040–1048. doi: 10.1006/jmbi.1993.1350. [DOI] [PubMed] [Google Scholar]
- 19.Persson E, Halle B. Nanosecond to microsecond protein dynamics probed by magnetic relaxation dispersion of buried water molecules. J Am Chem Soc. 2008;130:1774–1787. doi: 10.1021/ja0775873. [DOI] [PubMed] [Google Scholar]
- 20.Denisov VP, et al. Using buried water molecules to explore the energy landscape of proteins. Nat Struct Biol. 1996;3:505–509. doi: 10.1038/nsb0696-505. [DOI] [PubMed] [Google Scholar]
- 21.Nucci NV, Pometun MS, Wand AJ. Site-resolved measurement of water-protein interactions by solution NMR. Nat Struct Mol Biol. 2011;18:245–249. doi: 10.1038/nsmb.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcsisin SR, Engen JR. Hydrogen exchange mass spectrometry: What is it and what can it tell us? Anal Bioanal Chem. 2010;397:967–972. doi: 10.1007/s00216-010-3556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takamoto K, Chance MR. Radiolytic protein footprinting with mass spectrometry to probe the structure of macromolecular complexes. Annu Rev Biophys Biomol Struct. 2006;35:251–276. doi: 10.1146/annurev.biophys.35.040405.102050. [DOI] [PubMed] [Google Scholar]
- 24.Xu G, Chance MR. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem Rev. 2007;107:3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 25.Angel TE, et al. Structural waters define a functional channel mediating activation of the GPCR, rhodopsin. Proc Natl Acad Sci USA. 2009;106:14367–14372. doi: 10.1073/pnas.0901074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orban T, et al. Visualizing water molecules in transmembrane proteins using radiolytic labeling methods. Biochemistry. 2010;49:827–834. doi: 10.1021/bi901889t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bushnell GW, Louie GV, Brayer GD. High-resolution three-dimensional structure of horse heart cytochrome c. J Mol Biol. 1990;214:585–595. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]
- 28.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 A resolution. J Mol Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 29.Nukuna BN, Sun G, Anderson VE. Hydroxyl radical oxidation of cytochrome c by aerobic radiolysis. Free Radic Biol Med. 2004;37:1203–1213. doi: 10.1016/j.freeradbiomed.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 30.Maleknia SD, Brenowitz M, Chance MR. Millisecond radiolytic modification of peptides by synchrotron X-rays identified by mass spectrometry. Anal Chem. 1999;71:3965–3973. doi: 10.1021/ac990500e. [DOI] [PubMed] [Google Scholar]
- 31.Qi PX, et al. Structural water in oxidized and reduced horse heart cytochrome c. Nat Struct Biol. 1994;1:378–382. doi: 10.1038/nsb0694-378. [DOI] [PubMed] [Google Scholar]
- 32.Tompa K, et al. Interfacial water at protein surfaces: Wide-line NMR and DSC characterization of hydration in ubiquitin solutions. Biophys J. 2009;96:2789–2798. doi: 10.1016/j.bpj.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halle B. Biomolecular cryocrystallography: Structural changes during flash-cooling. Proc Natl Acad Sci USA. 2004;101:4793–4798. doi: 10.1073/pnas.0308315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ball P. Biophysics: More than a bystander. Nature. 2011;478:467–468. doi: 10.1038/478467a. [DOI] [PubMed] [Google Scholar]
- 35.Bertini I, et al. Redox-dependent hydration of cytochrome c and cytochrome b(5) studied through (17)O NMRD. J Am Chem Soc. 2001;123:12925–12926. doi: 10.1021/ja0163624. [DOI] [PubMed] [Google Scholar]
- 36.Nucci NV, Pometun MS, Wand AJ. Site-resolved measurement of water-protein interactions by solution NMR. Nat Struct Mol Biol. 2011;18:245–249. doi: 10.1038/nsmb.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Persson E, Halle B. Nanosecond to microsecond protein dynamics probed by magnetic relaxation dispersion of buried water molecules. J Am Chem Soc. 2008;130:1774–1787. doi: 10.1021/ja0775873. [DOI] [PubMed] [Google Scholar]
- 38.Gupta S, et al. The Beamline X28C of the Center for Synchrotron Biosciences: A national resource for biomolecular structure and dynamics experiments using synchrotron footprinting. J Synchrotron Radiat. 2007;14:233–243. doi: 10.1107/S0909049507013118. [DOI] [PubMed] [Google Scholar]
- 39.Sullivan MR, et al. Installation and testing of a focusing mirror at beamline X28C for high flux x-ray radiolysis of biological macromolecules. Rev Sci Instrum. 2008;79:025101. doi: 10.1063/1.2839027. [DOI] [PubMed] [Google Scholar]
- 40.Adilakshmi T, Soper SF, Woodson SA. Structural analysis of RNA in living cells by in vivo synchrotron X-ray footprinting. Methods Enzymol. 2009;468:239–258. doi: 10.1016/S0076-6879(09)68012-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiselar JG, et al. Structural analysis of gelsolin using synchrotron protein footprinting. Mol Cell Proteomics. 2003;2:1120–1132. doi: 10.1074/mcp.M300068-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.