Abstract
A convenient way to estimate internal body time (BT) is essential for chronotherapy and time-restricted feeding, both of which use body-time information to maximize potency and minimize toxicity during drug administration and feeding, respectively. Previously, we proposed a molecular timetable based on circadian-oscillating substances in multiple mouse organs or blood to estimate internal body time from samples taken at only a few time points. Here we applied this molecular-timetable concept to estimate and evaluate internal body time in humans. We constructed a 1.5-d reference timetable of oscillating metabolites in human blood samples with 2-h sampling frequency while simultaneously controlling for the confounding effects of activity level, light, temperature, sleep, and food intake. By using this metabolite timetable as a reference, we accurately determined internal body time within 3 h from just two anti-phase blood samples. Our minimally invasive, molecular-timetable method with human blood enables highly optimized and personalized medicine.
Keywords: metabolomics, circadian rhythm, liquid chromatography mass spectrometry, diagnostic tool
Many organisms possess a molecular time-keeping mechanism, a circadian clock, which has endogenous, self-sustained oscillations with a period of about 24 h. Circadian regulation of cell activity occurs in diverse biological processes such as electrical activity, gene/protein expression, and concentration of ions and substances (1, 2). In mammals, for example, several clock genes regulate circadian gene expression in central and peripheral clock tissues (3–9), as well as metabolites in the blood (10–15). Reflecting circadian regulation of such processes, the potency and toxicity of administered drugs depends on an individual's body time (BT) (16–22). Drug delivery according to body time improves the outcome of pharmacotherapy by maximizing potency and minimizing toxicity (23), and administrating drugs at an inappropriate body time can result in severe side effects (22). For example, rhythm disturbances were induced by administration of IFN-α during the early active phase in mice, although unaffected during the early rest phase (22); and the time of administration of two anticancer drugs, adriamycin (6:00 AM) and cisplatin (6:00 PM), made a lower toxicity effect than its antiphasic administration (24). However, several reports showed that internal body time varies by 5–6 h in healthy humans (25, 26) and as much as 10–12 h in shift workers without forced entrainments (27, 28). Therefore, for efficient application of body-time drug delivery or “chronotherapy” (16–20) in a clinical setting, a simple and robust method for estimating an individual’s internal body time is needed.
Additionally, the timing of food intake may contribute to weight gain (29) and metabolic disease (30) because energy regulation and circadian rhythms are molecularly and physiologically intertwined (31–41). For example, mice fed a high-fat diet during a 12-h light phase gain significantly more weight than mice fed only during a 12-h dark phase (29). These results suggest that food intake at different body times can alleviate or exacerbate diet-induced obesity. Therefore, an accurate and convenient way to detect internal body time may improve time-restricted diet strategies.
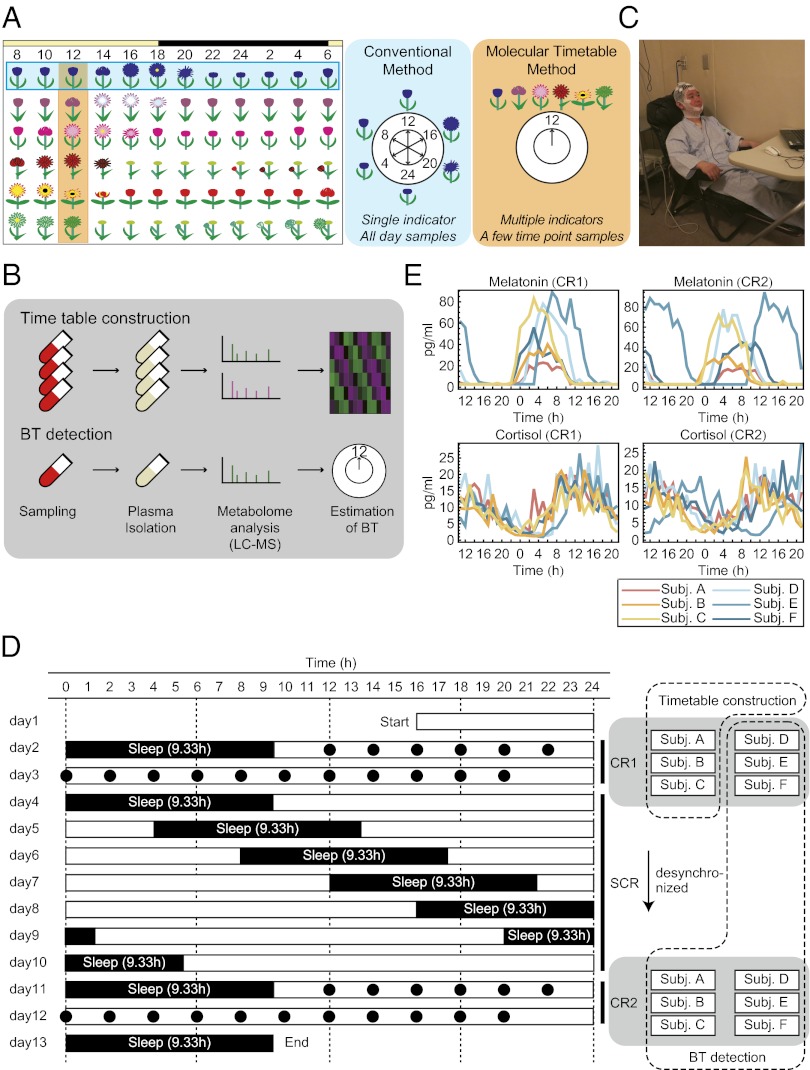
One conventional way to estimate human internal body time is to periodically sample for more than 24 h the level of melatonin or cortisol, which have robust circadian oscillations in the blood (10–12). Although this strategy directly measures internal body time, it requires labor-intensive constant sampling under controlled environmental conditions to reveal metabolite peaks and rhythms. To reduce this burden, we previously developed a molecular-timetable method (12, 42), which was inspired by Linné’s flower clock (Fig. 1A). In Linné’s flower clock, one can estimate the time of day by watching the opening and closing pattern of various flowers. Similarly, by using our molecular timetable method, one can estimate the body time of day by profiling the circadian oscillation patterns of gene expression or metabolites in the body. Compared with the conventional method, the molecular-timetable method requires only one or few time-point samplings. In mice, this method works with the expression activity of clock-controlled genes in different organs (42, 43) and with oscillatory metabolites in blood plasma (12).
Fig. 1.
Experimental condition. (A) The concept of body-time detection with conventional and molecular timetable methods illustrated by Linné’s flower clock. In the conventional method a single indicator monitored over a few days detects internal body time; in the molecular timetable method, multiple metabolic “flowers” are simultaneously measured at a few time points, which reduce efforts in sampling. (B) Metabolite timetable construction. We collected time-course blood samples, isolated plasma, measured with LC–MS, and selected circadian-oscillating metabolites as indicators. In body-time detection, we collected blood samples at a few time points, isolated plasma, measured with LC–MS, and estimated body time. (C) Image of the constant routine (CR) experiments. During CR, subjects stayed in chairs while various measurements were performed. Note: the man in this picture is demonstrating the set-up and is not an actual subject in this study. (D) Sampling schedule for each subject. Black circles indicate the time points when blood samples were taken and when subjects ate during CR. White boxes indicate the time when subjects were awake, and black boxes indicate when they were asleep. The blood samples of three subjects during CR1 were used for timetable construction, and other samples were used for body-time estimation. (E) Measured melatonin (Upper) and cortisol (Lower) rhythms in the collected blood samples during CR1 (Left) and CR2 (Right). The cortisol and melatonin levels show that some subjects (e.g., subject E in CR2) have shifted internal body time against sampled time. CR, constant routine; SCR, semiconstant routine; BT, body time; LC–MS, liquid chromatography mass spectrometry.
In this study, we applied this method to detect internal body time using blood samples in humans (Fig. 1B). First, we used liquid chromatography mass spectrometry (LC–MS) (44–46) to measure the abundance of various metabolites in blood plasma samples from several healthy human subjects over 1.5 d while controlling for activity level, environmental changes, sleep, and food intake. From these data we constructed a molecular timetable of metabolites that oscillate over 24-h, arranged according to their peak abundance during the day. This reference metabolite timetable enabled us to accurately detect an individual’s body time with only two samples of blood drawn 12 h apart. Our study demonstrates that human internal body time can be detected using limited time-point sampling and a reference metabolite timetable.
Results
Construction of Human Blood Molecular Timetable.
First, we aimed to construct a molecular timetable that includes a list of oscillating metabolites in human blood with information on mean, amplitude, and peak time (PT) of metabolite abundance over 24 h. We recruited six healthy volunteers (subjects A–F) who stayed in an environmentally controlled sleep laboratory for about 2 wk and underwent a forced desynchronization (FD) protocol consisting of three activity routines—constant routine one (CR1), then semiconstant routine (SCR), and finally constant routine two (CR2) (Fig. 1 C and D). The FD protocol used in this study was composed of the following three parts: (i) measurement under CR1 (47) followed by (ii) a 28-h sleep–wake schedule for 7 d during SCR (48), and (iii) a second measurement under CR2. The 28-h sleep–wake schedule enforces desynchronization between sleep homeostasis and circadian cycle. Therefore, the circadian clock of each individual runs according to his/her own period length and hence reveals the variability of its natural frequency or phase during CR2 after SCR.
Blood samples were collected every hour for 1.5 d in CR1 and CR2 with a defined caloric amount of food eaten every 2 h. We measured the time-course abundance of melatonin and cortisol in these blood plasma samples by RIA and found a wide variation in melatonin and cortisol rhythms (Fig. 1E). We used blood samples from three subjects (subjects A–C) in CR1 to construct a standard metabolic timetable because their body time most closely matched each other and previously reported melatonin and cortisol rhythms. We note that constructing a timetable with any combination of subjects did not change the results of our study (SI Results and Discussion).
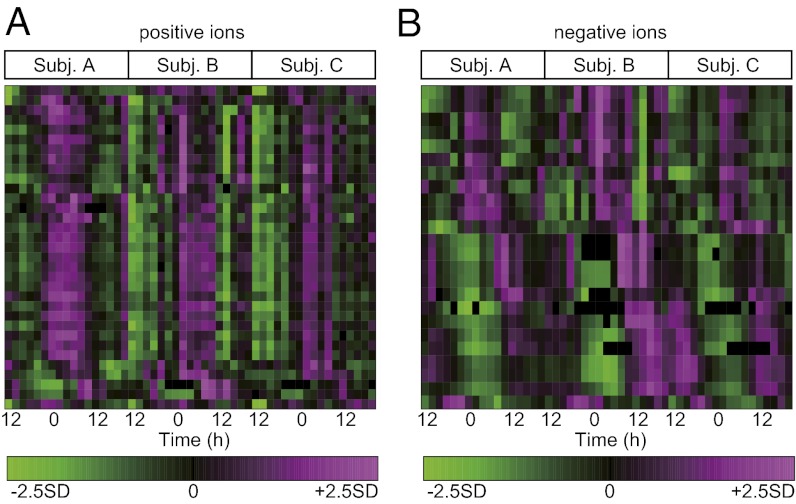
We used an LC–MS system to obtain the abundance of various metabolites for each subject at 17 time points (every 2 h) in each constant routine (CR1 or CR2) for a total of 204 samples. Using the three blood sample sets of CR1 from subjects A–C (51 samples in total) for molecular timetable construction, 2,541 and 1,796 metabolite peaks were detected in the positive and negative ion modes, respectively. To select circadian-oscillating metabolite peaks from these, we performed cosine fitting to the mean peak area of each sample set, which is averaged over three individuals at the same time point, and tested the significance of circadian rhythmicity by permutation tests (i.e., calculating the probability by using randomly shuffled time-course data of mean peak area). With the cutoff false discovery rate (FDR; the expected proportions of false positives in the selected set) of ≤0.1, we obtained 142 and 168 metabolite peaks of positive and negative ions, respectively, as circadian-oscillating metabolite peaks. These peaks include circadian-oscillating metabolite peaks with larger or smaller amplitudes and, as a result, some of them show variability among the three individuals. This is because our initial test only selects for peaks with significant circadian rhythmicity, but not significant amplitude. Thus, we further selected circadian-oscillating metabolite peaks with significant amplitude, whose circadian amplitude was significantly larger than the variations among individuals, with the cutoff FDR of ≤0.1 in a one-way ANOVA analysis. Consequently, the substantial overall cutoff is 0.1 × 0.1 = 0.01 because metabolite peaks were filtered by two independent criteria (FDR ≤ 0.1), and this is equivalent to FDR ≤ 0.01 used in our previous mouse study (12). As a final set of circadian-oscillating metabolites (with significant rhythmicity and amplitude), we obtained 33 and 25 metabolite peaks of positive and negative ions, respectively (Fig. 2 and Dataset S1). The procedure for the molecular timetable construction is illustrated in Fig. S1. The number of metabolite peaks (58 peaks) is adequate for body-time estimation because we previously found that 20 metabolites were sufficient for accurate body-time estimation (12).
Fig. 2.
Heat maps of circadian-oscillating metabolites in the human timetable. Circadian-oscillating metabolite peaks in the plasma samples [positive ions (A); negative ions (B)] are shown. Some metabolites in this heat map were identified and shown in Fig. 4. On the heat maps, magenta tiles indicate a high quantity of substances and green tiles indicate a low quantity in plasma. Metabolites are sorted according to their molecular peak time.
Body-Time Estimation with Metabolic Molecular Timetable.
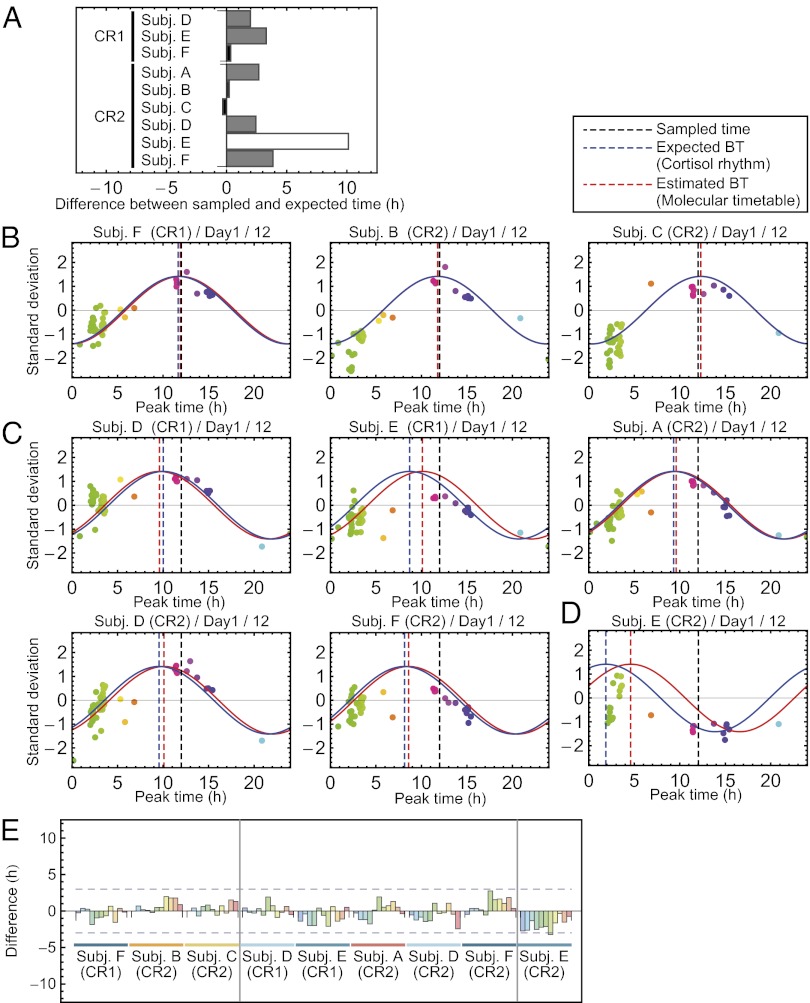
We analyzed the remaining nine blood sample sets (three sample sets from subjects D–F in CR1 and all six sample sets from subjects A–F in CR2) to test the accuracy of our molecular timetable method. To evaluate its accuracy, we determined the “expected body time” for each sample set by a conventional method (Fig. 1A) using cosine fitting to time-course data of cortisol abundance over 1.5 d (Fig. 1E, Lower). Although we used cortisol peak time for this purpose, we did not detect a significant difference when we determined the expected body time based on dim-light melatonin onset (DLMO) (SI Results and Discussion). Each sample showed a wide range of expected body time. For example, some samples showed expected body time that is almost identical to the sampled time (Fig. 3A, black bars), whereas one showed expected body time that was almost antiphasic to the sampled time (Fig. 3A, white bar). Remaining samples showed expected body time with an intermediate difference from the sampled time (Fig. 3A, gray bars). The observed variability did not result from errors of the conventional method, but reflect individual differences of internal body time because each sample showed similar expected body time when calculated from the 1.5-d time-course abundance data of melatonin (Fig. S2A). Accordingly, we categorized nine sample sets into three groups on the basis of the magnitude of the differences between expected body time based on a cortisol rhythm and sampled times: small shift group (CR1 of subject F; CR2 of subjects B and C), moderate shift group (CR1 of subjects D and E; CR2 of subjects A, D, and F), and large shift group (CR2 of subject E).
Fig. 3.
Estimation of human body time. (A) Difference between expected body time and sampled time. Nine sample sets for body-time estimation were classified into three groups by the magnitude of the difference: small (black), moderate (gray), and large (white). (B–D) Body-time estimation at 12 h on the first day for nine sample sets in small (B), moderate (C), and large (D) body-time difference groups. Colors of the dots indicate the molecular peak times of each metabolite. Peak time of the red cosine curves indicates estimated body time and peak time of the blue indicates the time of the expected body time. The smaller the distance between red and blue curves, the greater the accuracy of the measurement. Dashed vertical lines show the estimated body time (red), expected body time (blue), or the time the sample was taken (black). In all subjects, expected body time (based on cortisol rhythm) and estimated body time (based on molecular timetable) were similar, indicated by the close proximity of the blue and red dashed lines. (E) Summary of body-time estimation. Difference between expected and estimated body time for all estimated samples is shown. The leftmost three samples were ones in the small difference group, the middle five samples were in the moderate difference group, and the rightmost sample was in the large difference group. All samples estimated internal body time within or around 3-h differences between expected and estimated body time. BT, body time.
Next, we calculated the “estimated body time” for each sample set (Fig. 3 B–E and Table S1 and Figs. S2 B and C) by our molecular timetable method using the constructed reference metabolite timetable of human blood (Fig. 2). For body-time estimation, we used only two samples of blood drawn 12 h apart. We derived 11 estimated body-time values for each sample set consisting of 17 samples (Fig. 3E) and then compared the estimated body time with the predetermined expected body time (via conventional method) to evaluate the accuracy of our molecular timetable method. In the small shift group, the differences between estimated and expected body time were within 2 h (Fig. 3 B and E). The result was comparable to the previous study using mouse blood samples (12). In the moderate shift group, the differences were within 3 h (Fig. 3 C and E) and in the large shift group, the differences were about 3 h (Fig. 3 D and E). In these two groups, the differences were larger than the small shift group. However, in the moderate shift groups, only 3 time points (out of 55) differed by more than 2 h. The time difference of this method (<3 h) is less than the range of the internal body time of shift workers (27, 28). These results suggest that our molecular timetable method can correctly detect shifts of internal body time.
Identification of Metabolites with Circadian Oscillation.
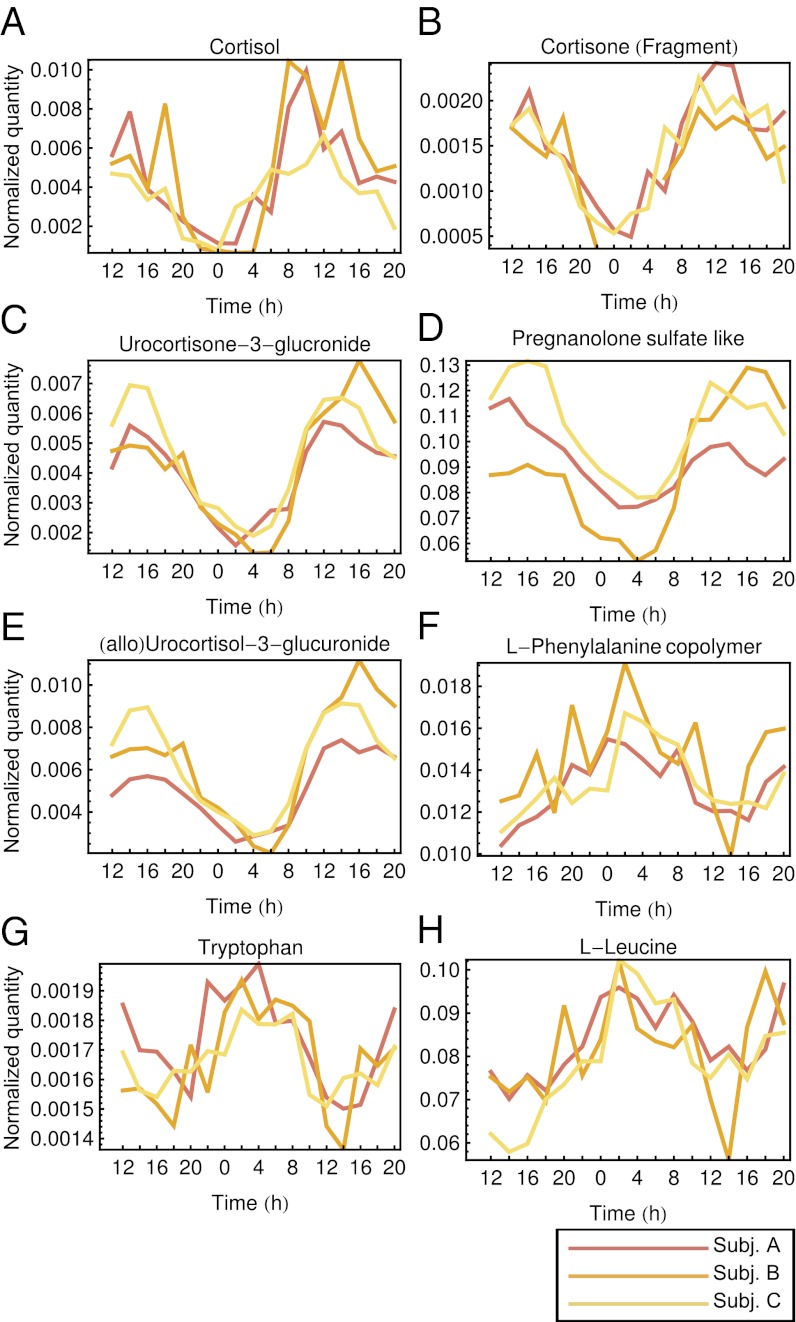
Identification of the circadian-oscillating metabolites is important for connecting our current metabolite timetable of human blood to previously reported circadian biology in humans. Identification is also important for further development of our method because once a metabolite is identified, we can update the circadian-metabolite reference list in our human blood timetable, which will improve the sensitivity and specificity of the molecular timetable (12, 42). Therefore, we attempted to identify several circadian-oscillating metabolites in constructing our metabolite timetable and found that a large fraction belong to the steroid hormone metabolism pathway (Fig. 4 and Fig. S3 A and B). These include cortisol (Fig. 4A), cortisone (Fig. 4B), urocortisone-3-glucronide (Fig. 4C), pregnanolone sulfate-like metabolite (Fig. 4D), and urocoritsol-3-glucronide (Fig. 4E) as well as the amino acids, phenylalanine (Fig. 4F), tryptophan (Fig. 4G), and leucine (Fig. 4H). Cortisol is a well-known circadian-oscillating metabolite in the blood (11), and cortisone is a direct metabolite of the cortisol also found in the blood (49). Urocortisone-3-glucuronide and urocortisol-3-glucronide are glucuronide forms of urocortisone and urocortisol, respectively, which are downstream metabolites of cortisol and cortisone. A pregnanolone isomer and its precursor (progesterone) were also found as circadian-oscillating metabolites in the brain and blood (50). The circadian oscillations in the human blood of cortisol and cortisone were also detected in the recent report about the human circadian metabolome (13). The peak times (PT) of the cortisol, cortisone, and urocortisone-3-glucronide were around the afternoon (PT12–PT15; we defined PT12 as noon), and peak times were aligned catabolically according to the steroid metabolism pathway from upstream to downstream (Fig. S3E). The peak time of the corticosterone in the mouse plasma samples was CT11.7 (corresponding to PT17.7 in this human study, because both CT6 and PT12 indicate noon) in our previous study (12), and is ∼3–6 h later than human peak time. The amino acids, phenylalanine, tryptophan, and leucine were also identified as oscillating metabolites in our mouse plasma study (12), and peak times in mouse and human plasma were ZT18.7–18.9 (corresponding to PT0.7–0.9 in this human study because both ZT6 and PT12 indicate noon), and PT2.2–3.1, respectively, which might reflect the similarity in the phase of the central clock between diurnal and nocturnal animals (51–56).
Fig. 4.
Identified circadian-oscillating metabolites. Abundance is shown of cortisol (A), cortisone (B), urocortisone-3-glucronide (C), pregnanolone sulfate-like metabolite (D), (allo)urocortisol-3-glucronide (E), l-phenylalanine (F), tryptophan (G), and l-leucine (H) in the blood sampled from three subjects (subject A, subject B, and subject C) whose CR1 samples were used in the metabolite timetable construction. X axes show the sampled time from 12 h on day 2 to 20 h on day 3 in CR1, and y axes show area of the corresponding peak in the LC–MS analysis.
We also found several lipids among oscillating metabolites, although these metabolites had weak amplitude and/or high variability among individuals (Fig. S3 A–D). These lipids include PG(18:1(9Z)/0:0), a glycerophospholipid (Fig. S3C), and LysoPC(16:0), a lysophospholipid (Fig. S3D). Peak times of these metabolites differed: PT12.0 [PG(18:1(9Z)/0:0)], and PT3.2 [LysoPC(16:0)]. Lysophospholipids also showed circadian oscillation in mouse plasma (12), but their peak time was almost antiphasic (ZT8 in mice corresponding to PT14 in this study), which might reflect the differences between human diurnality and murine nocturnality.
The comparison of our identified circadian-oscillating metabolites to recent studies (13, 14) is explained in SI Results and Discussion.
Discussion
In this study, we constructed a human blood metabolite timetable from three individuals, and applied it to these individuals and three other individuals after a forced desynchronization protocol. We successfully detected internal body time in all individuals, even in the different experimental conditions (Fig. S4), which suggests the robustness of our metabolite-timetable method against individual genetic differences. Interestingly, some individuals such as subjects E and F showed 6.8-h and 3.5-h delayed internal body time in constant routine 2 after they experienced a semiconstant routine of enforced 28-h sleep–wake cycles, in comparison with those in constant routine 1 (Fig. 3A). Importantly, these delays after enforced sleep–wake cycles were successfully detected by using our metabolite timetable method (Fig. 3 C and D). These results suggest that the molecular timetable method has potential to be a body-time estimation tool for humans in both normal and abnormal environments like enforced sleep–wake cycles.
Abnormal environments such as shift working, jet lag, and other irregular lifestyles, cause changes in the internal body time of individuals (57). In addition to abnormal environments, genetic differences such as familial advanced sleep phase syndrome (FASPS) also cause changes in the internal body time of individuals (58–60). Brown et al. recently developed a method to potentially detect the circadian rhythm disorders caused by genetic differences (61, 62). They collected skin samples from human subjects, cultured these cells, and transfected them with a clock-controlled reporter to characterize the features of the molecular clock in these tissues. They found that the circadian rhythmicity in isolated cells is correlated with the chronotypes of the subjects, implying that the method could detect circadian rhythm disorders caused by genetic differences. Our molecular timetable method can detect both genetic and environmental circadian rhythm disorders, but cannot distinguish between them, whereas Brown et al.’s method can detect genetic but not acquired disorders. These two methods are therefore complementary for detecting circadian rhythm disorders. Our metabolite timetable method may provide a good platform for the initial screening of body-time abnormality caused by either abnormal environments or genetic differences, because our method is minimally invasive and less labor intensive compared with the conventional method or biopsy adopted in Brown et al. (61, 62). Moreover, the metabolite timetable method can track the transient dynamics of internal body time, which cannot be detected by conventional time-course sampling method. It will be interesting in the future to determine whether our timetable method can detect internal body time differences caused by such abnormal environments or genetic differences.
In this study, we used metabolites in blood to detect the internal body time. We note that a molecular timetable concept can be applied not only for circadian-oscillating metabolites, but also for other circadian-oscillating substances (12, 42) and in other tissues like human hair follicle cells from the head or chin (63). Comparing molecular timetable methods with different reference molecules such as clock-controlled RNAs in humans will be of interest to reveal internal synchronization (or desynchronization) between different peripheral clocks such as blood clocks and skin clocks.
In summary, our study shows the possibility that a metabolic molecular timetable can be a convenient diagnostic tool. A minimally invasive internal body-time detection method is essential for facilitating the development of chronotherapy to realize tailored medication regimens as well as for efficient time-restricted feeding to avoid or cure obesity. To measure the abundance of metabolites for body-time estimation, we used LC–MS analysis, which is rarely implemented in current clinical hospitals. However, bench-top MS machines are beginning to appear, and they might be available at hospitals in the future as a routine diagnostic tool; additionally, it may be possible to develop a simpler diagnosis kit using antibodies against a subset of selected circadian metabolites. In any case, our method is more pragmatic than serial time-course sampling because only a few samples are required for body-time estimation and because blood is an easy tissue to sample and analyze. Moreover, our timetable is robust against personal differences because we selected metabolites that oscillated independently of the individual, food intake, sleep, light, and temperature. Thus, the metabolite timetable proposed in this study will provide a convenient way to estimate human body time and may lead to highly optimized and personalized medicine in the future.
Materials and Methods
Subjects.
Six healthy, male volunteers, aged 20–23 y (mean age 21.5 y) participated in this study. Subjects had regular sleep–wake patterns, normal hematology, and urinalysis data, and no history of psychiatric disease (assessed by the Mini-International Neuropsychiatric Interview) or severe physical disease. Written informed consent was obtained from each subject. See SI Materials and Methods for details.
Measurement of Metabolites.
The LC system used was an Agilent 1290 infinity HPLC (Agilent Technologies) with the KEIO MasterHands software (64). See SI Materials and Methods for details about the sample preparation, measurements, and data preprocessing.
Construction of Molecular Timetable.
We chose peaks that were detected and associated at nine or more time points in all three subjects (subjects A–C) in CR1. For each chosen peak, we performed two statistical tests for selecting candidates for the timetable. See SI Materials and Methods for details.
Body-Time Estimation.
Body-time estimation was performed as described for the previous mouse blood study (12). Expected body time of nine sample sets for body-time estimation (subjects D–F during CR1, and all six subjects during CR2) were determined on the basis of peak times of the cortisol rhythms (Fig. 1D, Lower). See SI Materials and Methods for details.
MS Identification of Oscillating Metabolites.
LC MS/MS analysis was performed for unknown peak identification based on the method described previously (12) with slight modification. See SI Materials and Methods for details.
Ethics.
The study was approved by RIKEN Kobe Institute Research Ethics Committee (KOBE-IRB-09-07) and the ethics committee of the National Center of Neurology and Psychiatry (22–1-4). All participants have given written consent for the use of material for this research purpose.
Supplementary Material
Acknowledgments
We thank Shinobu Abe and Yasuko Sakaguchi at the Institute for Advanced Biosciences (IAB) for metabolomic analysis; Masahiro Hayashi, Hiroshi Fujishima, and Tomoyo Hashiguchi at RIKEN Center for Developmental Biology (CDB) for handling samples; Hiroyuki Tamiya and Ryohei Narumi at RIKEN CDB for helpful discussion; Arthur Millius at RIKEN CDB for helpful discussion and for proofreading of the manuscript; and Takuro Ito at IAB, Keio University for providing materials used in the identification of oscillating metabolites. This research was supported by an intramural grant-in-aid from the RIKEN CDB (to H.R.U.), research funds from New Energy Developing Organization (to H.R.U.), research funds from the Yamagata Prefectural Government and the city of Tsuruoka (to M.S., M.M., and T.S.), a grant-in-aid for the Strategic Research Program for Brain Sciences from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to K.M.), an Intramural Research Grant (No. 23-3) for Neurological and Psychiatric Disorders of National Center of Neurology and Psychiatry, and a Grant-in Aid for Scientific Research (No. 21390335) from Japan Society for the Promotion of Science (to K.M. and A.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207768109/-/DCSupplemental.
References
- 1.Dunlap JC, Loros JJ, DeCoursey PJ, editors. Chronobiology: Biological Timekeeping. Sunderland, MA: Sinauer; 2004. [Google Scholar]
- 2.Hastings MH, Maywood ES, O’Neill JS. Cellular circadian pacemaking and the role of cytosolic rhythms. Curr Biol. 2008;18:R805–R815. doi: 10.1016/j.cub.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 3.Akhtar RA, et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 4.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 5.Storch KF, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 6.Ueda HR, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 7.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 8.Ueda HR. Systems biology of mammalian circadian clocks. Cold Spring Harb Symp Quant Biol. 2007;72:365–380. doi: 10.1101/sqb.2007.72.047. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kennaway DJ, Voultsios A, Varcoe TJ, Moyer RW. Melatonin in mice: Rhythms, response to light, adrenergic stimulation, and metabolism. Am J Physiol Regul Integr Comp Physiol. 2002;282:R358–R365. doi: 10.1152/ajpregu.00360.2001. [DOI] [PubMed] [Google Scholar]
- 11.Weitzman ED, et al. Twenty-four hour pattern of the episodic secretion of cortisol in normal subjects. J Clin Endocrinol Metab. 1971;33:14–22. doi: 10.1210/jcem-33-1-14. [DOI] [PubMed] [Google Scholar]
- 12.Minami Y, et al. Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci USA. 2009;106:9890–9895. doi: 10.1073/pnas.0900617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci USA. 2012;109:2625–2629. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckel-Mahan KL, et al. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci USA. 2012;109:5541–5546. doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fustin JM, et al. Rhythmic nucleotide synthesis in the liver: temporal segregation of metabolites. Cell Rep. 2012;1:341–349. doi: 10.1016/j.celrep.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Halberg F. Chronobiology. Annu Rev Physiol. 1969;31:675–725. doi: 10.1146/annurev.ph.31.030169.003331. [DOI] [PubMed] [Google Scholar]
- 17.Labrecque G, Bélanger PM. Biological rhythms in the absorption, distribution, metabolism and excretion of drugs. Pharmacol Ther. 1991;52:95–107. doi: 10.1016/0163-7258(91)90088-4. [DOI] [PubMed] [Google Scholar]
- 18.Lemmer B, Scheidel B, Behne S. Chronopharmacokinetics and chronopharmacodynamics of cardiovascular active drugs. Propranolol, organic nitrates, nifedipine. Ann N Y Acad Sci. 1991;618:166–181. doi: 10.1111/j.1749-6632.1991.tb27245.x. [DOI] [PubMed] [Google Scholar]
- 19.Reinberg A, Halberg F. Circadian chronopharmacology. Annu Rev Pharmacol. 1971;11:455–492. doi: 10.1146/annurev.pa.11.040171.002323. [DOI] [PubMed] [Google Scholar]
- 20.Reinberg A, Smolensky M, Levi F. Aspects of clinical chronopharmacology. Cephalalgia. 1983;3(Suppl 1):69–78. doi: 10.1177/03331024830030S108. [DOI] [PubMed] [Google Scholar]
- 21.Bocci V. Administration of interferon at night may increase its therapeutic index. Cancer Drug Deliv. 1985;2:313–318. doi: 10.1089/cdd.1985.2.313. [DOI] [PubMed] [Google Scholar]
- 22.Ohdo S, Koyanagi S, Suyama H, Higuchi S, Aramaki H. Changing the dosing schedule minimizes the disruptive effects of interferon on clock function. Nat Med. 2001;7:356–360. doi: 10.1038/85507. [DOI] [PubMed] [Google Scholar]
- 23.Lévi F, Zidani R, Misset JL. International Organization for Cancer Chronotherapy Randomised multicentre trial of chronotherapy with oxaliplatin, fluorouracil, and folinic acid in metastatic colorectal cancer. Lancet. 1997;350:681–686. doi: 10.1016/s0140-6736(97)03358-8. [DOI] [PubMed] [Google Scholar]
- 24.Hrushesky WJ. Circadian timing of cancer chemotherapy. Science. 1985;228:73–75. doi: 10.1126/science.3883493. [DOI] [PubMed] [Google Scholar]
- 25.Wright KP, Jr, Gronfier C, Duffy JF, Czeisler CA. Intrinsic period and light intensity determine the phase relationship between melatonin and sleep in humans. J Biol Rhythms. 2005;20:168–177. doi: 10.1177/0748730404274265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hasan S, et al. Assessment of circadian rhythms in humans: Comparison of real-time fibroblast reporter imaging with plasma melatonin. FASEB J. 2012;26:2414–2423. doi: 10.1096/fj.11-201699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horowitz TS, Cade BE, Wolfe JM, Czeisler CA. Efficacy of bright light and sleep/darkness scheduling in alleviating circadian maladaptation to night work. Am J Physiol Endocrinol Metab. 2001;281:E384–E391. doi: 10.1152/ajpendo.2001.281.2.E384. [DOI] [PubMed] [Google Scholar]
- 28.Smith MR, Eastman CI. Night shift performance is improved by a compromise circadian phase position: Study 3. Circadian phase after 7 night shifts with an intervening weekend off. Sleep. 2008;31:1639–1645. doi: 10.1093/sleep/31.12.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW. Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring) 2009;17:2100–2102. doi: 10.1038/oby.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatori M, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15:848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rutter J, Reick M, McKnight SL. Metabolism and the control of circadian rhythms. Annu Rev Biochem. 2002;71:307–331. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- 32.Yang X, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 33.Froy O. The relationship between nutrition and circadian rhythms in mammals. Front Neuroendocrinol. 2007;28:61–71. doi: 10.1016/j.yfrne.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 34.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alenghat T, et al. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le Martelot G, et al. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vollmers C, et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang EE, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Froy O. Metabolism and circadian rhythms—implications for obesity. Endocr Rev. 2010;31:1–24. doi: 10.1210/er.2009-0014. [DOI] [PubMed] [Google Scholar]
- 41.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ueda HR, et al. Molecular-timetable methods for detection of body time and rhythm disorders from single-time-point genome-wide expression profiles. Proc Natl Acad Sci USA. 2004;101:11227–11232. doi: 10.1073/pnas.0401882101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masumoto KH, et al. Acute induction of Eya3 by late-night light stimulation triggers TSHβ expression in photoperiodism. Curr Biol. 2010;20:2199–2206. doi: 10.1016/j.cub.2010.11.038. [DOI] [PubMed] [Google Scholar]
- 44.Plumb R, et al. Metabonomic analysis of mouse urine by liquid-chromatography-time of flight mass spectrometry (LC-TOFMS): Detection of strain, diurnal and gender differences. Analyst (Lond) 2003;128:819–823. doi: 10.1039/b304296k. [DOI] [PubMed] [Google Scholar]
- 45.Wilson ID, et al. High resolution “ultra performance” liquid chromatography coupled to oa-TOF mass spectrometry as a tool for differential metabolic pathway profiling in functional genomic studies. J Proteome Res. 2005;4:591–598. doi: 10.1021/pr049769r. [DOI] [PubMed] [Google Scholar]
- 46.Tolstikov VV, Lommen A, Nakanishi K, Tanaka N, Fiehn O. Monolithic silica-based capillary reversed-phase liquid chromatography/electrospray mass spectrometry for plant metabolomics. Anal Chem. 2003;75:6737–6740. doi: 10.1021/ac034716z. [DOI] [PubMed] [Google Scholar]
- 47.Mills JN, Minors DS, Waterhouse JM. Adaptation to abrupt time shifts of the oscillator(s) controlling human circadian rhythms. J Physiol. 1978;285:455–470. doi: 10.1113/jphysiol.1978.sp012582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Czeisler CA, et al. Stability, precision, and near-24-hour period of the human circadian pacemaker. Science. 1999;284:2177–2181. doi: 10.1126/science.284.5423.2177. [DOI] [PubMed] [Google Scholar]
- 49.Nuller YL, Morozova MG, Kushnir ON, Hamper N. Effect of naloxone therapy on depersonalization: A pilot study. J Psychopharmacol. 2001;15:93–95. doi: 10.1177/026988110101500205. [DOI] [PubMed] [Google Scholar]
- 50.Corpéchot C, et al. Brain neurosteroids during the mouse oestrous cycle. Brain Res. 1997;766:276–280. doi: 10.1016/s0006-8993(97)00749-x. [DOI] [PubMed] [Google Scholar]
- 51.Inouye ST, Kawamura H. Persistence of circadian rhythmicity in a mammalian hypothalamic “island” containing the suprachiasmatic nucleus. Proc Natl Acad Sci USA. 1979;76:5962–5966. doi: 10.1073/pnas.76.11.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kubota A, Inouye ST, Kawamura H. Reversal of multiunit activity within and outside the suprachiasmatic nucleus in the rat. Neurosci Lett. 1981;27:303–308. doi: 10.1016/0304-3940(81)90447-x. [DOI] [PubMed] [Google Scholar]
- 53.Nunez AA, Bult A, McElhinny TL, Smale L. Daily rhythms of Fos expression in hypothalamic targets of the suprachiasmatic nucleus in diurnal and nocturnal rodents. J Biol Rhythms. 1999;14:300–306. doi: 10.1177/074873099129000713. [DOI] [PubMed] [Google Scholar]
- 54.Schwartz MD, Nunez AA, Smale L. Differences in the suprachiasmatic nucleus and lower subparaventricular zone of diurnal and nocturnal rodents. Neuroscience. 2004;127:13–23. doi: 10.1016/j.neuroscience.2004.04.049. [DOI] [PubMed] [Google Scholar]
- 55.Ramanathan C, Nunez AA, Martinez GS, Schwartz MD, Smale L. Temporal and spatial distribution of immunoreactive PER1 and PER2 proteins in the suprachiasmatic nucleus and peri-suprachiasmatic region of the diurnal grass rat (Arvicanthis niloticus) Brain Res. 2006;1073–1074:348–358. doi: 10.1016/j.brainres.2005.11.082. [DOI] [PubMed] [Google Scholar]
- 56.Ramanathan C, Stowie A, Smale L, Nunez AA. Phase preference for the display of activity is associated with the phase of extra-suprachiasmatic nucleus oscillators within and between species. Neuroscience. 2010;170:758–772. doi: 10.1016/j.neuroscience.2010.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duffy JF, Czeisler CA. Effect of light on human circadian physiology. Sleep Med Clin. 2009;4:165–177. doi: 10.1016/j.jsmc.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jones CR, et al. Familial advanced sleep-phase syndrome: A short-period circadian rhythm variant in humans. Nat Med. 1999;5:1062–1065. doi: 10.1038/12502. [DOI] [PubMed] [Google Scholar]
- 59.Toh KL, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 60.Xu Y, et al. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 61.Brown SA, et al. The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS Biol. 2005;3:e338. doi: 10.1371/journal.pbio.0030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown SA, et al. Molecular insights into human daily behavior. Proc Natl Acad Sci USA. 2008;105:1602–1607. doi: 10.1073/pnas.0707772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Akashi M, et al. Noninvasive method for assessing the human circadian clock using hair follicle cells. Proc Natl Acad Sci USA. 2010;107:15643–15648. doi: 10.1073/pnas.1003878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010;6:78–95. doi: 10.1007/s11306-009-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.