Abstract
The conserved diagonal docking mode observed in structures of T-cell receptors (TCRs) bound to peptide–MHC ligands is believed to reflect coevolution of TCR and MHC genes. This coevolution is supported by the conservation of certain interactions between the germ-line–encoded complementarity-determining region (CDR)1 and CDR2 loops of TCR and MHC. However, the rules governing these interactions are not straightforward, even when the same variable (V) region recognizes the same MHC molecule. Here, we demonstrate that the somatically generated CDR3 loops can markedly alter evolutionarily selected contacts between TCR and MHC (“CDR3 editing”). To understand CDR3 editing at the atomic level, we determined the structure of a human melanoma-specific TCR (G4) bound to the MHC class II molecule HLA-DR1 and an epitope from mutant triose phosphate isomerase (mutTPI). A comparison of the G4–mutTPI–DR1 complex with a complex involving a TCR (E8) that uses the same Vα region to recognize the same mutTPI–DR1 ligand as G4 revealed that CDR1α adopts markedly different conformations in the two TCRs, resulting in an almost entirely different set of contacts with MHC. Based on the structures of unbound G4 and E8, the distinct conformations of CDR1α in these TCRs are not induced by binding to mutTPI–DR1 but result from differences in the length and sequence of CDR3α that are transmitted to CDR1α. The editing of germ-line–encoded TCR–MHC interactions by CDR3 demonstrates that these interactions possess sufficient intrinsic flexibility to accommodate large structural variations in CDR3 and, consequently, in the TCR-binding site.
T-cell receptors (TCRs) bind peptide–MHC (pMHC) via their six complementarity-determining region (CDR) loops, three from the variable alpha (Vα) domain and three from Vβ. The first and second CDRs (CDR1 and CDR2) are encoded within the TCR Vα and Vβ gene segments; the third CDR (CDR3) is formed by DNA recombination involving juxtaposition of Vα and Jα segments for the α chain genes, and of Vβ, D, and Jβ segments for the β chain genes. As a result, the somatically generated Vα and Vβ CDR3 loops account for most of the variability of TCR-binding sites, whereas variability contributed by CDR1 and CDR2 is restricted to that already existing in the germ line.
Structural studies of numerous (>25) TCR–pMHC complexes have demonstrated remarkable similarities in the overall topology of TCR binding, irrespective of MHC class I or class II restriction (1–3). Typically, the TCR is positioned diagonally over the center of the composite surface created by the peptide and the MHC α-helices that flank the peptide-binding groove, with the Vα domain situated over the N-terminal half of the peptide and the Vβ domain over the C-terminal half, although the exact angle and pitch of TCR engagement vary (1). In addition, these studies revealed the conservation of specific TCR–MHC interactions in complexes involving common V segments and MHC alleles (2, 4–6). For example, crystal structures of six mouse TCRs expressing Vβ8.2 bound to I-Ab, I-Ak, or I-Au MHC class II molecules showed a close convergence of CDR1β and CDR2β contacts with the α1-helix of these I-A alleles (4, 5). This pairwise interaction motif comprises TCR residues CDR1β Asn31, CDR2β Tyr48, and CDR2β Glu54 contacting I-A residues Gln61α, Gln57α, and Lys39α, respectively. These and related findings demonstrating conserved interactions between germ-line–encoded CDR1 and CDR2 loops and particular MHC alleles strongly support the hypothesis that the canonical diagonal docking orientation of TCR on MHC observed in TCR–pMHC complexes is the result, at least in part, of coevolution of TCR and MHC molecules (2, 6–11). This conserved orientation probably also reflects the requirement to form a TCR–pMHC–CD4 or TCR–pMHC–CD8 ternary complex with the CD4 or CD8 coreceptor that is geometrically competent to deliver a maturation signal to double-positive thymocytes during T-cell selection (8, 11–17).
At the same time, it is evident from the structural database that the rules governing evolutionarily selected interactions are not straightforward, even when a particular V region recognizes the same MHC allele (2). For example, structures of a single TCR in complex with different peptides bound to H-2Kb showed that the detailed interactions made by CDR1 and CDR2 with the MHC helices differed considerably, because of repositioning of the TCR (18). The alternative TCR–MHC contacts were attributed to selection by the peptide of the germ-line–encoded interactions that were optimal for that particular peptide (“peptide editing”) (6). Similarly, different peptides were found to substantially alter the docking mode of another TCR on H-2Ld (8).
In addition to peptide editing, Garcia and coworkers have proposed that CDR3 can modulate germ-line–encoded interactions between CDR1 and CDR2 and MHC (“CDR3 editing”) (4, 6, 19). In support of this idea, a mutational analysis of CDR3β of a TCR cross-reactive with multiple I-A alleles revealed that many CDR3β sequences, even some with no similarity to wild type, were compatible with MHC reactivity, although different CDR3β sequences conferred varying degrees of MHC promiscuity (20). However, in all cases, MHC recognition was dependent on CDR1β and CDR2β. These results suggested that germ-line TCR V regions are inherently reactive with MHC but that this reactivity is fine-tuned, or edited, by the somatically generated CDR3 loops. However, the structural basis for CDR3 editing remains to be elucidated.
With the goal of understanding CDR3 editing at the atomic level, and to address whether CDR3 can alter germ-line–encoded interactions between TCR and MHC, we determined the crystal structure of a human melanoma-specific TCR (G4) bound to the MHC class II molecule HLA-DR1 and an altered self-peptide from triose phosphate isomerase (TPI). A naturally occurring point mutation in TPI found in a patient with melanoma replaced an isoleucine residue for threonine in the recognized epitope (TPI 23–37; Thr28Ile) (21). This substitution resulted in greatly enhanced recognition of mutant TPI (mutTPI) relative to wild-type TPI, effectively disclosing this altered self-peptide to T cells. We previously reported the structure of another melanoma-specific TCR (E8) that recognizes the same mutTPI–DR1 ligand as G4 (22). TCRs G4 and E8 use the same Vα region (AV13.1) but have different CDR3α sequences (Fig. S1). A comparison of the G4–mutTPI–DR1 and E8–mutTPI–DR1 complexes, in conjunction with the structures of the unbound TCRs, revealed that CDR1α adopts different conformations in the two TCRs, resulting in a different set of contacts with MHC. The distinct conformations of CDR1α in G4 and E8 are not attributable to pMHC binding but, instead, result from differences in CDR3α. The editing of evolutionarily selected interactions between TCR and MHC by CDR3 reported here demonstrates that these interactions possess the capacity to accommodate structural variability in the TCR-binding site. In addition, the G4–mutTPI–DR1 structure explains how a natural mutation in TPI rendered a normally cryptic self-protein visible to the immune system through creation of a tumor-specific neoepitope.
Results
Overview of the G4–mutTPI–DR1 Complex.
The mutTPI-specific T-cell clone G4 was derived from tumor-infiltrating lymphocytes from a metastatic melanoma lesion (SI Materials and Methods). The very low affinity of TCR G4 for mutTPI–DR1 (KD > 300 μM) (22) precluded crystallizing the G4–mutTPI–DR1 complex using the individual proteins. To stabilize the complex for crystallization, we covalently attached the mutTPI peptide to the TCR β-chain via an octapeptide linker (23). We loaded the fused the mutTPI peptide into the binding groove of invariant chain peptide-bound (CLIP-bound) HLA-DR1 molecules using the peptide exchange catalyst HLA-DM to generate a stable G4–mutTPI–DR1 complex. We determined the structure of the complex to 2.6-Å resolution (Table 1 and Fig. 1A). The interface between TCR and pMHC was in unambiguous electron density for each of the two molecules in the asymmetric unit of the crystal. A 2Fo – Fc electron-density map of the isoleucine residue at position P3 of mutTPI (P3 Ile) and its vicinity is shown in Fig. S2. The average B factors of the mutTPI peptide (42.2 Å2); Ile P3 of mutTPI (34.2 Å2); CDR1, -2, and -3 of Vα (29.7 Å2, 31.4 Å2, and 38.6 Å2, respectively); and CDR1, -2, and -3 of Vβ (43.1 Å2, 47.0 Å2, and 36.0 Å2, respectively) are all lower than the overall average B factor (49.0 Å2) of the G4–mutTPI–DR1 complex, indicating that the mutTPI peptide and the G4-binding site are well ordered in the crystal.
Table 1.
Data collection and refinement statistics
| G4–mutTPI–DR1 | G4 | |
| Data processing statistics | ||
| Resolution limit, Å* | 2.60 (2.70–2.60) | 2.70 (2.80–2.70) |
| Space group | C2 | C2 |
| Cell dimensions, Å, ° | a = 126.1, b = 175.6, c = 88.6 β = 110.8 | a = 168.1, b = 75.4, c = 130.8 β = 123.5 |
| Unique reflections* | 53,716 (5,739) | 36,498 (6,064) |
| Completeness, %* | 97.2 (83.8) | 99.9 (99.9) |
| Rmerge, %* | 7.6 (26.9) | 6.1 (23.6) |
| I/σI* | 19.3 (2.6) | 26.4 (16.3) |
| Refinement statistics | ||
| Rwork, % | 19.7 | 20.0 |
| Rfree, %† | 26.4 | 26.2 |
| rmsd from ideality | ||
| Bond lengths, Å | 0.011 | 0.011 |
| Bond angles, ° | 1.687 | 1.440 |
| Ramachandran plot statistics | ||
| Most favored, % | 88.0 | 89.5 |
| Additionally allowed, % | 11.3 | 10.1 |
| Generously allowed, % | 0.7 | 0.4 |
| Disallowed, % | 0 | 0 |
*Values in the parentheses are statistics of the highest resolution shell.
†Rfree is calculated for a randomly selected 5.0% of reflections not included in the refinement.
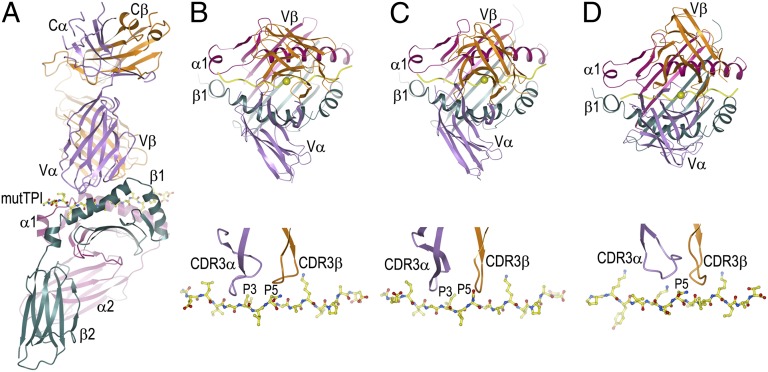
Fig. 1.
Structure of the G4–mutTPI–DR1 complex and comparison with other TCR–peptide–DR1 complexes. (A) Side view of the G4–mutTPI–DR1 complex (ribbon diagram). TCR α-chain, violet; TCR β-chain, orange; MHC α-chain, magenta; MHC β-chain, dark green; mutTPI peptide, yellow. (B, Upper) Top view of the G4–mutTPI–DR1 complex. The central P5 residue of the peptide is shown as a yellow sphere. (Lower) Position of the TCR CDR3 loops over the mutTPI peptide. The peptide is drawn in ball-and-stick representation, with carbon atoms in yellow, nitrogen atoms in blue, and oxygen atoms in red. The CDR3 loops are positioned above the P3 residue of the peptide. (C) Antitumor E8–mutTPI–DR1 complex (PDB ID code 2IAM) (22). (D) Antimicrobial HA1.7–HA–DR1 complex (PDB ID code 1FYT) (23). The CDR3 loops of G4 and E8 are centered over peptide residue P3, whereas those of HA1.7 are positioned over P5.
The root mean squared deviation (rmsd) in α-carbon positions for the TCR VαVβ and MHC α1β1 modules, including the mutTPI peptide, is 0.3 Å for the two complex molecules in the asymmetric unit. Based on this close similarity, the following description of TCR–pMHC interactions applies to both complexes.
The overall binding topology of the G4–mutTPI–DR1 complex is similar to that of the E8–mutTPI–DR1 complex (Fig. 1 B and C) (22). In both cases, the TCR docks on pMHC in the canonical diagonal orientation (1–3), with the Vα domain over the β1-helix of HLA-DR1 and the Vβ domain over the α1-helix. The crossing angle of G4 to mutTPI–DR1 is 67°, which differs from that of E8 (71°) by a rotation of 4°. In common with E8, the position of G4 along the MHC peptide-binding groove is shifted toward the N terminus of the bound peptide, compared with antimicrobial TCRs such HA1.7, which recognizes an influenza hemagglutinin (HA) peptide bound to HLA-DR1 (Fig. 1D) (23). In addition, G4, like E8, is tilted toward the HLA-DR1 β-chain relative to HA1.7.
The much lower affinity of G4 than E8 for mutTPI–DR1 (22) may be understood in terms of the complex structures. Although G4–mutTPI–DR1 and E8–mutTPI–DR1 bury very similar solvent-accessible surfaces (2,122 Å2 and 2,244 Å2, respectively), they differ in other structural parameters that are known to affect affinity. Thus, G4 makes 107 total contacts with mutTPI–DR1, of which 33 are with the peptide and 74 with MHC. By contrast, E8 makes substantially more contacts, 166, of which 62 are with the peptide and 104 with MHC. The relative paucity of contacts in the G4–mutTPI–DR1 interface contributes to its poor shape complementarity, based on a calculated shape correlation statistic (Sc) (24) of 0.47 (Sc = 1.0 for perfect fits), compared with 0.69 for the interface between E8 and mutTPI–DR1. Although G4 makes 1 more salt bridge with mutTPI than E8, G4 makes only 7 hydrogen bonds with mutTPI–DR1, compared with 14 for E8. These differences (fewer van der Waals contacts, poorer shape complementarity, and fewer hydrogen bonds) probably explain the weaker binding of G4 than E8 to mutTPI–DR1, which correlated with reduced levels of cytokine secretion at a given peptide concentration in T-cell stimulation assays (22).
Peptide Recognition by TCRs G4 and E8.
To understand the basis for TCR recognition of the tumor-specific mutTPI peptide, we examined the interaction of G4 with mutTPI and compared this interaction with recognition by E8. As in the case of E8, the CDR3 loops of G4 are positioned directly over P3 Ile of mutTPI (Fig. 1 B and C), the site of the Thr28Ile mutation in TPI that resulted in much higher TCR affinity for mutTPI–DR1 than for TPI–DR1 (22). Nevertheless, G4 and E8 recognize mutTPI in markedly different ways. In the G4–mutTPI–DR1 complex, peptide recognition is mediated almost entirely by CDR3α and CDR3β (Fig. 2 A and C), which together account for 85% (28 of 33) of the total contacts to mutTPI. The remaining contacts are contributed by CDR1β and CDR2β (3% and 12%, respectively); CDR1α and CDR2α do not participate in peptide recognition. In the E8–mutTPI–DR1 complex, by contrast, the CDR1 and CDR3 loops of both Vα and Vβ make approximately equal contributions to contacts with mutTPI (Fig. 2 B and D).
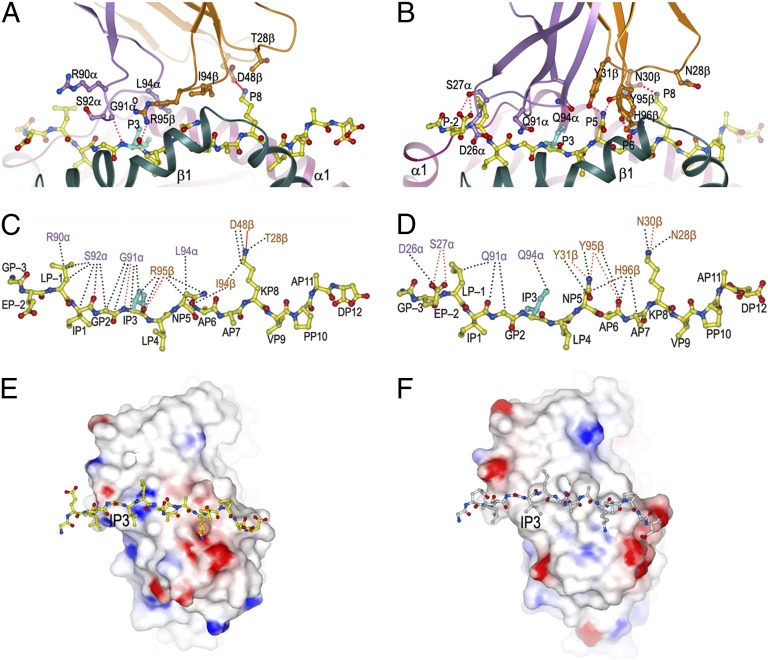
Fig. 2.
Interactions of TCRs G4 and E8 with mutTPI peptide. (A) G4 residues in contact with mutTPI. The colors are as in Fig. 1, except that the substituted TPI isoleucine residue at P3 is cyan. Hydrogen bonds are red dotted lines; salt bridge is a red solid line. TCR residues are identified by a one-letter amino acid designation, followed by position number and chain designation. (B) E8 residues in contact with mutTPI. (C) Interactions between TCR G4 and mutTPI. Peptide residues are identified by a one-letter amino acid designation, followed by position (P) number. van der Waals contacts are black dotted lines. (D) Interactions between TCR E8 and mutTPI. (E) Electrostatic potential surface of TCR G4 opposite the mutTPI peptide. Solvent-accessible surface is colored according to electrostatic potential, with positively charged regions in blue and negatively charged regions in red. Electrostatic surface potentials were calculated with the program GRASP (36). (F) Electrostatic potential surface of TCR E8 opposite the mutTPI peptide.
Superposition of the HLA-DR1 α1β1 domains of mutTPI–DR1 in the G4–mutTPI–DR1 and E8–mutTPI–DR1 complexes (Fig. S3) revealed that two mutTPI residues, P3 Ile and P12 Asp, adopt different conformations in the two structures (Fig. 2 C and D). In the G4–mutTPI–DR1 complex, the δ1 methyl group of P3 Ile does not contact TCR but is, instead, buried in the peptide-binding groove of HLA-DR1 (Fig. 2 A and C), where it interacts extensively with Phe22α, Phe54α, Gly58α, Ala59α, and Asn62α, as in the structure of unbound mutTPI–DR1 (25). The position of the δ1 methyl group of P3 Ile is clearly defined in the electron density (Fig. S2). In the E8–mutTPI–DR1 complex, by contrast, the side chain of P3 Ile has undergone a rotation of ∼80° about the Cα–Cβ axis, bringing the δ1 methyl group into contact with CDR3α Gln94 (Fig. 2 B and D). Regarding P12 Asp, its side chain orientation differs by a rotation of ∼180° in the G4–mutTPI–DR1 compared with the E8–mutTPI–DR1 complex (Fig. S3). However, in neither complex does P12 Asp contact TCR.
The electrostatic surface potentials of G4 and E8 illustrate how these TCRs use different chemistries in their interactions with mutTPI. As depicted in Fig. 2E, the binding surface of G4 presents an electropositive patch opposite P3 Ile in the complex interface, which would make rotating the hydrophobic side chain of P3 Ile toward G4 energetically unfavorable. Instead, P3 Ile forms three hydrogen bonds, via its main-chain atoms, with the main-chain oxygen of CDR3α Gly91 and side chain of CDR3β Arg95 (Fig. 2A). In contrast, the CDR3 loops of E8 form a hydrophobic pocket that accommodates the δ1 methyl group of P3 Ile (Fig. 2 B and F). As shown in Fig. 2F, the portion of the E8-binding surface that juxtaposes P3 Ile lacks the electropositive patch found in G4 and is, instead, hydrophobic (Fig. 2E).
We showed previously that replacement of threonine with isoleucine at position P3 in wild-type TPI resulted in much higher affinity of both G4 and E8 for mutTPI–DR1 than for wild-type TPI–DR1 (22). A combination of additional van der Waals contacts with TCR and improved shape complementarity at the mutation site accounted for increased binding of E8 to mutTPI–DR1. Although the structure of G4 bound to wild-type TPI–DR1 is unknown, the effect of the Thr28Ile mutation in TPI on the binding interface with G4 may be addressed by superposing wild-type TPI–DR1 (25) onto mutTPI–DR1 in the G4–mutTPI–DR1 structure to construct a hypothetical wild-type complex. In the modeled G4–TPI–DR1 complex, the side chain of P3 Thr is completely buried against the HLA-DR1 α-chain, rendering it inaccessible to TCR (Fig. S4). In the G4–mutTPI–DR1 structure, by contrast, the Cγ2 atom of P3 Ile points toward G4 and makes two van der Waals contacts with CDR3α Gly91 and CDR3β Arg95 (Fig. 2C). This difference is attributable to a rotation of the mutant P3 Ile side chain about the Cα–Cβ axis relative to that of wild-type P3 Thr. An additional effect of the Thr28Ile substitution in TPI is to increase the amount of apolar buried surface at the mutation site. After complex formation, P3 Ile contributes 62 Å2 of apolar buried surface area to the interface with G4, whereas P3 Thr contributes only 38 Å2 in the modeled G4–TPI–DR1 complex (Fig. S4). Together, these structural differences, albeit subtle, probably account for the tighter binding of G4 to mutTPI–DR1 than to wild-type TPI–DR1 (22). Moreover, they explain how a natural mutation in TPI disclosed a normally invisible self-protein to T cells through formation of an altered self-epitope.
Interaction of TCRs G4 and E8 with HLA-DR1.
The overall footprint of G4 on mutTPI-DR1 is similar to that of E8 (Fig. 1 B and C). However, G4 employs the germ-line–encoded CDR1 and CDR2 loops for most van der Waals contacts with the HLA-DR1 α-helices (48 of 74; 65%), whereas E8 relies more on the somatically generated CDR3 loops for MHC recognition (56% of contacts). Because G4 is tilted toward the HLA-DR1 β-chain (Fig. 1B), CDR1α and CDR2α, rather than CDR1β and CDR2β, dominate the interactions with MHC (48 versus 26 contacts). Overall, 12 Vα and 6 Vβ residues interact with 13 MHC residues of which 11 are contacted by E8 and 10 by HA1.7 (Table S1).
Because G4 and E8 use the same Vα region (AV13.1) to bind identical pMHC ligands (Fig. S1), one might have expected the CDR1α and CDR2α loops to mediate similar interactions with HLA-DR1. Indeed, CDR2α displays nearly identical conformations in the G4–mutTPI–DR1 and E8–mutTPI–DR1 complexes (rmsd in α-carbon positions of 0.3 Å for residues 47–53) and makes similar interactions with MHC (Fig. 3 A and B and Table 2). Most notable is a salt bridge between CDR2α Lys53 and HLA-DR1 Asp69β that is conserved in the E8–mutTPI–DR1 complex and that may be critical for anchoring G4 and E8 to the MHC β1 α-helix.
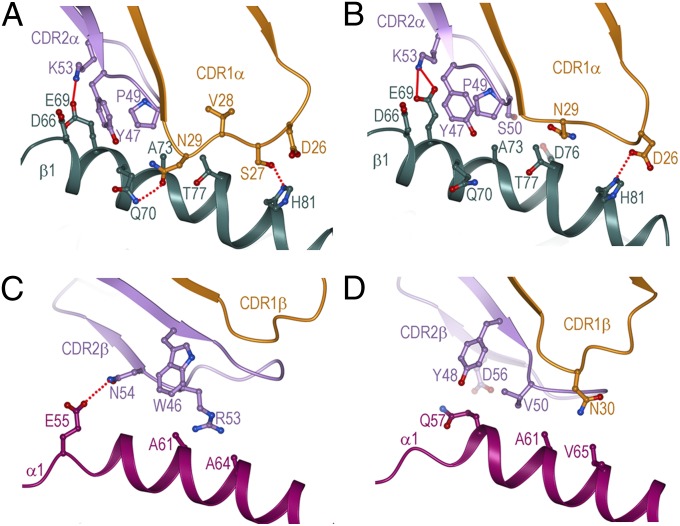
Fig. 3.
Comparisons of interactions of germ-line–encoded TCR CDR1 and CDR2 loops with MHC class II. (A) Interactions between CDR1α (orange) and CDR2α (violet) of G4 and the HLA-DR1 β1 α-helix (dark green) in the G4–mutTPI–DR1 complex. The side chains of contacting residues are drawn in ball-and-stick representation, with carbon atoms in orange (CDR1α) or violet (CDR2α), nitrogen atoms in blue, and oxygen atoms in red. Hydrogen bonds are indicated by red dotted lines and salt bridges by red solid lines. (B) Interactions between CDR1α and CDR2α of E8 and the HLA-DR1 β1 α-helix in the E8–mutTPI–DR1 complex (22). (C) Interactions between CDR1β (orange) and CDR2β (violet) of G4 and the HLA-DR1 α1 α-helix (magenta). (D) Interactions between CDR1β and CDR2β of E8 and the HLA-DR1 α1 α-helix.
Table 2.
Interactions between CDR1α and CDR2α and MHC in the G4–mutTPI–DR1 and E8–mutTPI–DR1 complexes
| G4–mutTPI–DR1 |
E8–mutTPI–DR1 |
|||
| Hydrogen bonds and salt bridges (bold) | van der Waals contacts | Hydrogen bonds and salt bridges (bold) | van der Waals contacts | |
| CDR1α | ||||
| D26α | H81β | D26α(Oδ2) H81β(Nδ1) | H81β | |
| S27α | S27α(Oγ) H81β(Nδ1) | H81β T77β | ||
| V28α | T77β | |||
| N29α | N29α(Nδ2) Q70β(Oε2) | Q70β | T77β | |
| CDR2α | ||||
| Y47α | D66β Q70β | D66β E69β Q70β | ||
| P49α | A73β | E69β A73β | ||
| S50α | A73β | A73β D76β | ||
| K53α | K53α E69β | E69β | K53α E69β | E69β |
In sharp contrast to CDR2α, CDR1α adopts different conformations in the two TCRs (Fig. 3 A and B), with an rmsd in α-carbon positions of 2.3 Å for residues 26–30. As a result, CDR1α engages MHC through an almost entirely different set of contacts in the G4–mutTPI–DR1 and E8–mutTPI–DR1 complexes (Table 2). Indeed, the only conserved interaction is between CDR1α Asp26 and HLA-DR1 His81β (Fig. 3 A and B). By contrast, CDR1α Asn29 contacts HLA-DR1 Gln70β in G4–mutTPI–DR1 but Thr77β in E8–mutTPI–DR1. Additionally, CDR1α Ser27 and Val28 of G4 engage HLA-DR1 His81β and Gln70β via a side-chain–side-chain hydrogen bond and multiple van der Waals contacts; these interactions are totally absent in the E8–mutTPI–DR1 complex (Table 2). These differences in CDR1α structure and contacts are attributable to the CDR3α loops of G4 and E8, as described below.
The CDR2β loops of G4 and E8, which use different Vβ regions (BV5S8 and BV13S6, respectively) (Fig. S1), recognize different sets of residues on the MHC α1 α-helix, with the sole exception of HLA-DR1 Ala61α (Fig. 3 C and D and Table S1). This residue contacts CDR2β Arg53 of G4 and CDR2β Val50 of E8. No interactions are observed between CDR1β of G4 and HLA-DR1, whereas CDR1β Asn30 of E8 contacts HLA-DR1 Val65α.
Influence of CDR3α on CDR1α Loop Conformation.
The different conformations of CDR1α seen in the G4–mutTPI–DR1 and E8–mutTPI–DR1 complexes could be attributable to structural rearrangements in the TCRs upon binding pMHC. Alternatively, the conformational differences in CDR1α could preexist in the unbound TCRs. To distinguish between these possibilities, we determined the structure of G4 in free form to a resolution of 2.7 Å (Table 1). Superposition of the VαVβ domains of unbound G4 onto those of G4 in complex with mutTPI–DR1 gave an rmsd in α-carbon positions of 0.8 Å, indicating close overall similarity. In particular, CDR1α of G4 displays nearly identical conformations before and after binding mutTPI–DR1 (rmsd in α-carbon positions of 0.3 Å for residues 26–30), except for several adjustments in side-chain orientation that allow formation of two side-chain–side-chain hydrogen bonds with the MHC β1 α-helix: CDR1α Ser27 Oγ–Nδ DR1 His81β and CDR1α Asn29 Nδ2–Oε2 DR1 Gln70β (Fig. 4A and Table 2). The near identity of CDR1α loop conformation occurs despite very different crystallization conditions for free versus bound G4 (Materials and Methods). Likewise, a comparison of E8 in free and bound forms showed that the overall conformation of CDR1α is maintained following ligation (rmsd in α-carbon positions of 0.4 Å for residues 26–30), although the loop does undergo a slight movement to accommodate the DR1 β1 α-helix (Fig. 4B). Therefore, the distinct conformations of CDR1α in G4 and E8 are intrinsic to these TCRs and are not induced by pMHC binding.
Fig. 4.
Conformational changes upon complex formation. (A) Structural rearrangements in CDR1α and CDR2α of G4 induced by binding to mutTPI–DR1. Bound CDR1α, orange; bound CDR2α, violet; unbound CDR1α and CDR2α, cyan; HLA-DR1 β1 helix, dark green. (B) Structural rearrangements in CDR1α and CDR2α of E8 induced by binding to mutTPI–DR1.
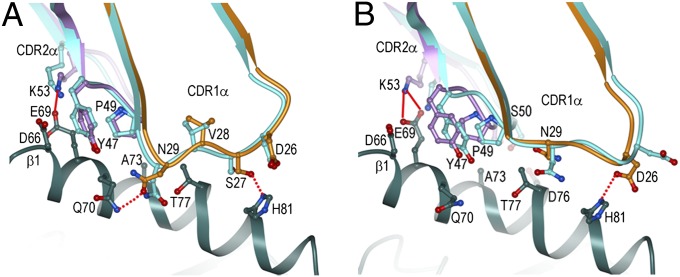
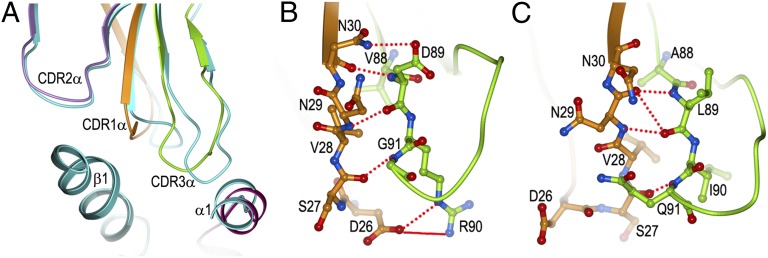
The different conformations adopted by CDR1α in G4 and E8 are best explained by the CDR3α loops of these TCRs, which differ in both sequence and length (VDRGSTLGR for G4; ALIQGAQK for E8) (Fig. S1). As a result, CDR3α displays markedly different conformations in the two TCRs (Fig. 5A). These structural differences are transmitted to CDR1α via its extensive interactions with CDR3α, which include five hydrogen bonds and one salt bridge in G4 (Fig. 5B) and four main-chain–main-chain hydrogen bonds in E8 (Fig. 5C). In G4, the tip of CDR3α points toward CDR1α, displacing CDR1α toward CDR2α relative to its position in E8, the CDR3α of which points away from CDR1α (Fig. 5A). Consequently, major steric clashes with G4 CDR3α would prevent CDR1α from assuming the conformation found in E8, thereby necessitating substantial adjustments in CDR1α that alter its interactions with HLA-DR1.
Fig. 5.
Influence of CDR3α on the conformation of CDR1α. (A) Superposition of the CDRα loops of G4 (CDR1α, orange; CDR2α, violet; CDR3α, green; HLA-DR1 α1 helix, magenta; HLA-DR1 β1 helix, dark green) onto those of E8 (cyan). (B) Interactions between CDR1α (orange) and CDR3α (green) of G4. (C) Interactions between CDR1α (orange) and CDR3α (green) of E8.
Discussion
Previous studies have shown that different peptides can modify germ-line–encoded TCR–MHC interactions by repositioning the TCR on MHC (8, 18). Here, we have shown that CDR3α, which abuts CDR1α in the TCR-binding site, can alter TCR–MHC contacts through its effects on CDR1α. The distinct conformations of CDR1α found in TCRs G4 and E8 are not attributable to pMHC engagement but are, instead, required to accommodate large structural differences in CDR3α. In other cases, CDR3α might exert more subtle, albeit still functionally important, effects on CDR1α that may not involve conformational rearrangements but changes in CDR1α dynamics or stability. Similar considerations probably apply to CDR3β and CDR1β. In contrast to peptide editing, whereby the peptide selects germ-line–encoded TCR–MHC interactions that are energetically optimal for that particular peptide (6, 8), CDR3 editing involves the selection of TCR–MHC interactions by differences in CDR3 sequence or length. The flexibility of germ-line–encoded contacts between TCR and MHC implied by these two mechanisms allows TCRs to maintain their roughly conserved diagonal docking mode, despite variations in the antigenic peptide and the topology of the TCR-binding site. More generally, our study illustrates how the malleability of protein–protein interfaces permits considerable wobble in evolutionarily selected interactions that is essential for maintaining function (in this case, MHC specificity) through the accommodation of large structural changes in one or both of the binding partners.
Although we have focused here on the flexibility of some germ-line–encoded interactions between TCR and MHC, it is important to emphasize that other germ-line–encoded interactions are maintained in the G4–mutTPI–DR1 and E8–mutTPI–DR1 complexes, in agreement with the TCR–MHC coevolution hypothesis (2, 6–11). In particular, CDR2α makes nearly identical contacts with MHC in the two complexes (Table 2). These conserved interactions serve as anchor points to enable G4 and E8 to dock onto mutTPI–DR1 in similar overall orientations, even though these TCRs use different Vβ regions. At the same time, the ability of CDRs to bind MHC in flexible ways, as exemplified by CDR1α of G4 and E8, permits TCRs to maintain specificity for MHC while accommodating differences in CDR3 loops and MHC-bound peptides. Regarding the TCR β chain, G4 uses a Vβ region (BV5S8) belonging to the same family as the Vβ used by TCR 3A6 (BV5S1), which recognizes a myelin-derived peptide (MBP) bound to HLA-DR2a (26). A comparison of the G4–mutTPI–DR1 and 3A6–MBP–DR2a revealed several conserved interactions between CDR2β and these MHC alleles (CDR1β of G4 does not contact MHC). These germ-line–encoded interaction pairs include CDR2β residues Arg53 and Asn54 contacting HLA-DR residues Ala61α and Glu55α, respectively. Such interactions support the idea that coevolution of TCR and MHC molecules is a major contributor to the canonical diagonal orientation observed in TCR–pMHC complexes (2, 6–11). However, this conserved orientation probably also reflects a role for the CD4 and CD8 coreceptors in limiting TCR-docking options on pMHC during T-cell selection (8, 11–17). In this view, TCR-docking topology is restricted by the requirement to form a TCR–pMHC–CD4 or TCR–pMHC–CD8 ternary complex that is geometrically competent to deliver a maturation signal to CD4+CD8+ thymocytes by proper intracellular juxtaposition of coreceptor-bound Lck with CD3 immunoreceptor tyrosine activation motifs (ITAMs).
Unexpectedly, we observed that two unique TCRs specific for the same tumor neoantigen had very weak affinities for pMHC (KD > 300 μM) (22). This illustrates an important mechanism of immune tolerance underpinning tumor–host interactions. Treatment strategies designed to activate T cells bearing suboptimal TCRs such as G4 and E8 to achieve an in vivo activation threshold include agents blocking T-cell coinhibitory receptors such as anti–CTLA-4 and anti–PD-1, which have already shown clinical promise (27). It is the complex integration of TCR signals, which are endogenous and not easily manipulated, and coreceptor signals, which can be pharmacologically manipulated, that determines T-cell functional outcomes and ultimately the fate of the tumor–host equilibrium.
Materials and Methods
Protein Preparation.
Soluble HLA-DR1 loaded with CLIP 87–101 peptide (PVSKMRMATPLIMQA) was prepared by in vitro folding from inclusion bodies produced in Escherichia coli as described (25). The gene encoding residues Ile1–Ser203 of the G4 α-chain was inserted into the pET-28a expression vector (Novagen). The β-chain construct, which encoded a fusion protein comprising mutTPI (GELIGILNAAKVPAD) connected by an octapeptide linker (GGSGGGGG) to residues Gly1–Asp239 of the G4 β chain, was cloned into the same vector. To increase yields of paired αβ heterodimers, we engineered a CαCys157–CβCys166 interchain disulfide (28). The mutated TCR α- and β-chains were expressed separately as inclusion bodies in BL21(DE3) E. coli cells (Novagen). The inclusion bodies were dissolved in 6 M guanidine, 10 mM DTT, and 10 mM EDTA. For in vitro folding, the G4 α- and β-chains were mixed in a 1.2:1 molar ratio and diluted into folding buffer containing 5 M urea, 0.4 M l-arginine-HCl, 100 mM Tris-HCl (pH 8.2), 3.7 mM cystamine, and 6.6 mM cysteamine to a final protein concentration of 70 mg/L. After 72 h at 4 °C, the folding mixture was concentrated 50-fold and dialyzed against 20 mM Tris-HCl (pH 8.0). Disulfide-linked G4 heterodimer was purified using a MonoQ FPLC column (GE Healthcare). TCR G4 without an attached mutTPI peptide was prepared similarly.
The linked G4–mutTPI–DR1 complex was assembled by loading the mutTPI peptide, fused to TCR G4, into CLIP–DR1 using soluble HLA-DM. TCR, CLIP–DR1, and HLA-DM were incubated at a molar ratio of 6:4:1 for 15 h at 37 °C in 50 mM sodium citrate (pH 5.9), and 0.1 M NaCl. Linked complexes were separated from aggregates, HLA-DM, and MHC, and TCR not in complex with a Superdex 200 column (GE Healthcare). Final purification was carried out using a MonoQ column.
Crystallization and Data Collection.
Crystals of the G4–mutTPI–DR1 complex were grown in 20% (wt/vol) polyethylene glycol 1000, 0.2 M calcium acetate, and 0.1 M imidazole (pH 8.0) at a protein concentration of 10 mg/mL. Ethylene glycol [20% (vol/vol)] was used as a cryoprotectant. Unbound TCR G4 crystallized in 1.5 M sodium nitrate and 0.1 M sodium citrate (pH 5.0); 6 M sodium nitrate was used as a cryoprotectant. X-ray diffraction data were collected at beamline ×29 of the Brookhaven National Synchrotron Light Source with an ADSC Quantum-315 CCD detector. All data were indexed, integrated, and scaled with the program HKL2000 (29). Data collection statistics are presented in Table 1.
Structure Determination and Refinement.
The structure of the G4–mutTPI–DR1 complex was determined by molecular replacement with the program Phaser (30) using the E8–mutTPI–DR1 complex (PDB ID code 2IAM) (22) as the search model. Unambiguous solutions were found in the cross rotation and translation functions for the two complex molecules in the asymmetric unit. The mutant TPI peptide was omitted in the initial refinement. After refinement with RefMac 5.0 (31), the resulting electron-density maps clearly showed the mutant TPI peptide in the DR1-binding groove. The peptide, including the Thr28Ile mutation, was unambiguously built into electron density using XtalView (32). The structure of unbound TCR G4 was solved by molecular replacement with Phaser (30) using the refined structure of bound G4 as the search model. The CDR loops were deleted in the initial refinement and built into where unambiguous electron density was shown with the program COOT (33). Refinement statistics are given in Table 1.
Contact residues in the G4–mutTPI–DR1 complex were identified using the program CONTACT (34) and were defined as residues containing an atom ≤4.0 Å from a residue of the binding partner. Buried surface areas were calculated using SURFACE (34) with a 1.4-Å probe radius. The programs BobScript (35), POV-Ray (http://www.povray.org), and GLR (http://convent.nci.nih.gov/glr/glrhome.html) were used to prepare figures.
Supplementary Material
Acknowledgments
We thank H. Robinson (Brookhaven National Synchrotron Light Source) for X-ray data collection. This work was supported by National Institutes of Health Grants AI073654 (to R.A.M. and S.L.T.) and AI036900 (to R.A.M.). Beamline X29 is supported by the Department of Energy Offices of Biological and Environmental Research and Basic Energy Sciences and by the National Center for Research Resources of the National Institutes of Health. L.D. was supported by the Irvington Institute Fellowship Program of the Cancer Research Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4E41 and 4E42).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207186109/-/DCSupplemental.
References
- 1.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 2.Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wucherpfennig KW, Call MJ, Deng L, Mariuzza RA. Structural alterations in peptide-MHC recognition by self-reactive T cell receptors. Curr Opin Immunol. 2009;21:590–595. doi: 10.1016/j.coi.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 5.Dai S, et al. Crossreactive T Cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules. Immunity. 2008;28:324–334. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia KC, Adams JJ, Feng D, Ely LK. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol. 2009;10:143–147. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott-Browne JP, White J, Kappler JW, Gapin L, Marrack P. Germline-encoded amino acids in the alphabeta T-cell receptor control thymic selection. Nature. 2009;458:1043–1046. doi: 10.1038/nature07812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams JJ, et al. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity. 2011;35:681–693. doi: 10.1016/j.immuni.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott-Browne JP, et al. Evolutionarily conserved features contribute to αβ T cell receptor specificity. Immunity. 2011;35:526–535. doi: 10.1016/j.immuni.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia KC, et al. A closer look at TCR germline recognition. Immunity. 2012;36:887–888. doi: 10.1016/j.immuni.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia KC. Reconciling views on T cell receptor germline bias for MHC. Trends Immunol. 2012 doi: 10.1016/j.it.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazza C, Malissen B. What guides MHC-restricted TCR recognition? Semin Immunol. 2007;19:225–235. doi: 10.1016/j.smim.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Buslepp J, Wang H, Biddison WE, Appella E, Collins EJ. A correlation between TCR Valpha docking on MHC and CD8 dependence: Implications for T cell selection. Immunity. 2003;19:595–606. doi: 10.1016/s1074-7613(03)00269-3. [DOI] [PubMed] [Google Scholar]
- 14.Van Laethem F, et al. Deletion of CD4 and CD8 coreceptors permits generation of alphabetaT cells that recognize antigens independently of the MHC. Immunity. 2007;27:735–750. doi: 10.1016/j.immuni.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Tikhonova AN, et al. αβ T cell receptors that do not undergo major histocompatibility complex-specific thymic selection possess antibody-like recognition specificities. Immunity. 2012;36:79–91. doi: 10.1016/j.immuni.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Laethem F, Tikhonova AN, Singer A. MHC restriction is imposed on a diverse T cell receptor repertoire by CD4 and CD8 co-receptors during thymic selection. Trends Immunol. 2012 doi: 10.1016/j.it.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin Y, Wang XX, Mariuzza RA. Crystal structure of a complete ternary complex of T-cell receptor, peptide-MHC, and CD4. Proc Natl Acad Sci USA. 2012;109:5405–5410. doi: 10.1073/pnas.1118801109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazza C, et al. How much can a T-cell antigen receptor adapt to structurally distinct antigenic peptides? EMBO J. 2007;26:1972–1983. doi: 10.1038/sj.emboj.7601605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colf LA, et al. How a single T cell receptor recognizes both self and foreign MHC. Cell. 2007;129:135–146. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 20.Rubtsova K, et al. Many different Vbeta CDR3s can reveal the inherent MHC reactivity of germline-encoded TCR V regions. Proc Natl Acad Sci USA. 2009;106:7951–7956. doi: 10.1073/pnas.0902728106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pieper R, et al. Biochemical identification of a mutated human melanoma antigen recognized by CD4 T cells. J Exp Med. 1999;189:757–766. doi: 10.1084/jem.189.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng L, et al. Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor. Nat Immunol. 2007;8:398–408. doi: 10.1038/ni1447. [DOI] [PubMed] [Google Scholar]
- 23.Hennecke J, Carfi A, Wiley DC. Structure of a covalently stabilized complex of a human alphabeta T-cell receptor, influenza HA peptide and MHC class II molecule, HLA-DR1. EMBO J. 2000;19:5611–5624. doi: 10.1093/emboj/19.21.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 25.Sundberg EJ, et al. Minor structural changes in a mutated human melanoma antigen correspond to dramatically enhanced stimulation of a CD4+ tumor-infiltrating lymphocyte line. J Mol Biol. 2002;319:449–461. doi: 10.1016/S0022-2836(02)00370-4. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, et al. Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule. EMBO J. 2005;24:2968–2979. doi: 10.1038/sj.emboj.7600771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Topalian SL, Weiner GJ, Pardoll DM. Cancer immunotherapy comes of age. J Clin Oncol. 2011;29:4828–4836. doi: 10.1200/JCO.2011.38.0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boulter JM, et al. Stable, soluble T-cell receptor molecules for crystallization and therapeutics. Protein Eng. 2003;16:707–711. doi: 10.1093/protein/gzg087. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 30.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 31.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 32.McRee DE. XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collaborative Computational Project No. 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:240–255. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 35.Esnouf RM. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J Mol Graph Model. 1997;15:132–134, 112–113. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 36.Nicholls A, Sharp KA, Honig B. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.