Abstract
The CBF (C-repeat binding factor) pathway has a major role in plant cold acclimation, the process whereby certain plants increase in freezing tolerance in response to low nonfreezing temperatures. In Arabidopsis thaliana, the pathway is characterized by rapid cold induction of CBF1, CBF2, and CBF3, which encode transcriptional activators, followed by induction of CBF-targeted genes that impart freezing tolerance. At warm temperatures, CBF transcript levels are low, but oscillate due to circadian regulation with peak expression occurring at 8 h after dawn (zeitgeber time 8; ZT8). Here, we establish that the CBF pathway is also regulated by photoperiod at warm temperatures. At ZT8, CBF transcript levels in short-day (SD; 8-h photoperiod) plants were three- to fivefold higher than in long-day plants (LD; 16-h photoperiod). Moreover, the freezing tolerance of SD plants was greater than that of LD plants. Genetic analysis indicated that phytochrome B (PHYB) and two phytochrome-interacting factors, PIF4 and PIF7, act to down-regulate the CBF pathway and freezing tolerance under LD conditions. Down-regulation of the CBF pathway in LD plants correlated with higher PIF4 and PIF7 transcript levels and greater stability of the PIF4 and PIF7 proteins under LD conditions. Our results indicate that during the warm LD growing season, the CBF pathway is actively repressed by PHYB, PIF4, and PIF7, thus mitigating allocation of energy and nutrient resources toward unneeded frost protection. This repression is relieved by shortening day length resulting in up-regulation of the CBF pathway and increased freezing tolerance in preparation for coming cold temperatures.
Plants vary greatly in their ability to survive freezing temperatures. Whereas plants from tropical and subtropical regions are generally killed by the slightest freeze, plants from temperate regions exhibit varying degrees of freezing tolerance (1, 2). For instance, Arabidopsis thaliana (hereafter referred to as Arabidopsis) and wheat have a maximum freezing tolerance of about −10 °C and −20 °C, respectively, and hardy deciduous trees can survive freezing below −40 °C. However, the freezing tolerance of frost hardy plants is not a constant property; it changes over the course of the year in response to changing environmental conditions. The primary factor is low temperature (1, 2). When winter rye is grown at warm temperature, plants are killed upon freezing at about −5 °C, but upon exposure to low nonfreezing temperatures, they can survive freezing below −20 °C. The molecular basis for this phenomenon, known as cold acclimation, is not completely understood, but includes changes in membrane cryobehavior, the production of cryoprotective proteins, and the biosynthesis of low molecular weight cryoprotectants such as sucrose, raffinose, and proline (2, 3).
Many of the biochemical and metabolic changes that occur in response to low temperature and contribute to an increase in freezing tolerance involve changes in gene expression. The best understood cold regulatory pathway with a role in freezing tolerance is the CBF (C-repeat binding factor) pathway of Arabidopsis (4, 5). CBF1, CBF2, and CBF3 (also known as DREB1B, DREB1C, and DREB1A, respectively) encode closely related members of the AP2/ERF family of DNA-binding proteins that recognize the CRT/DRE DNA regulatory element, RCCGAC. Within minutes of transferring Arabidopsis plants to low temperature, CBF1, -2, and -3 are induced followed at about 3 h by induction of CBF-targeted cold-regulated (COR) genes, referred to as the CBF regulon. Constitutive overexpression of CBF1, -2, or -3 at warm temperature leads to constitutive expression of the CBF regulon and a marked increase in freezing tolerance (4, 5). Although the CBF pathway is not as well studied in other plant species, it has been established that cold-inducible CBF genes are highly conserved among higher plants and that CBF overexpression increases freezing tolerance in highly divergent plant species that are able to cold acclimate (6).
Photoperiod is another environmental factor that regulates freezing tolerance, a phenomenon that is well documented in woody deciduous trees (7, 8). As summer turns to fall, these plants sense the shortening day length and initiate developmental programs that result in the cessation of growth and an increase in freezing tolerance that can be more than 10 °C in some hardy species. As the season continues to progress and the temperatures become cold, the plants sense the low temperature and increase an additional 20 °C or more in freezing tolerance (7, 8). The molecular basis for photoperiodic regulation of freezing tolerance is not well understood, but appears to involve the action of phytochromes (9, 10). Phytochromes are light-absorbing photoreceptors that exist in two fundamental forms: the red (R)-light-absorbing form, designated Pr, and the far-red (FR)-light-adsorbing form, designated Pfr. The Pr form, which is inactive, is converted to the active Pfr form by exposure to R-light, and is converted back to the inactive Pr form by exposure to FR-light or through dark reversion (11). The increase in freezing tolerance that occurs in response to short-day in red-osier dogwood and other perennial woody tree species is prevented if the plants are exposed to R light during the nighttime, but not if the R-light exposure is followed by brief exposure to FR light (9, 10). These results are classic indicators of a phytochrome-mediated response (12) and suggest that an active Pfr phytochrome represses freezing tolerance.
Whereas photoperiodic regulation of freezing tolerance is recognized as a fundamental feature of cold acclimation in woody plants, there is little evidence for the phenomenon in herbaceous plants. Pietsch et al. (13) found that the freezing tolerance of Gaura coccinea, a perennial herbaceous species, increases about 3 °C when plants are exposed to short-day photoperiods, but beyond this, photoperiodic regulation of freezing tolerance at warm growth temperature is poorly documented in perennial and annual herbaceous species. However, similar to what has been reported in woody plants, Franklin and Whitelam (14) showed that phytochromes have a role in regulating freezing tolerance in Arabidopsis. When plants were grown at 16 °C under a 12-h photoperiod, the freezing tolerance and transcript levels for three CBF target genes—COR15a, COR15b, and KIN1—were greater in plants exposed to a low R/FR-light ratio than if they were exposed to a high R/FR-light ratio. Also, transferring plants to constant light and exposing them to a low R/FR-light ratio for 2 h in the morning resulted in increased transcript levels for CBF1, -2, and -3. These results suggested that a Pfr form of one or more phytochromes repressed the expression of the CBF pathway. Indeed, at 16 °C, under high R/FR light, both phyB and phyD mutations resulted in increased transcript levels for COR15a, a CBF target gene, and the phyD mutation resulted in greater freezing tolerance.
Arabidopsis has proven to be a powerful model plant to study the regulation of freezing tolerance by low temperature. Here we show that it is also a powerful model to study photoperiodic regulation of freezing tolerance. Our results indicate that Arabidopsis plants increase in freezing tolerance in response to a short-day photoperiod, that this regulation involves photoperiodic regulation of the CBF pathway, and that this regulation is mediated by the PHYB photoreceptor and two PIF transcription factors with which PHYB physically interacts, PIF4 and PIF7 (15).
Results
Freezing Tolerance Is Regulated by Photoperiod.
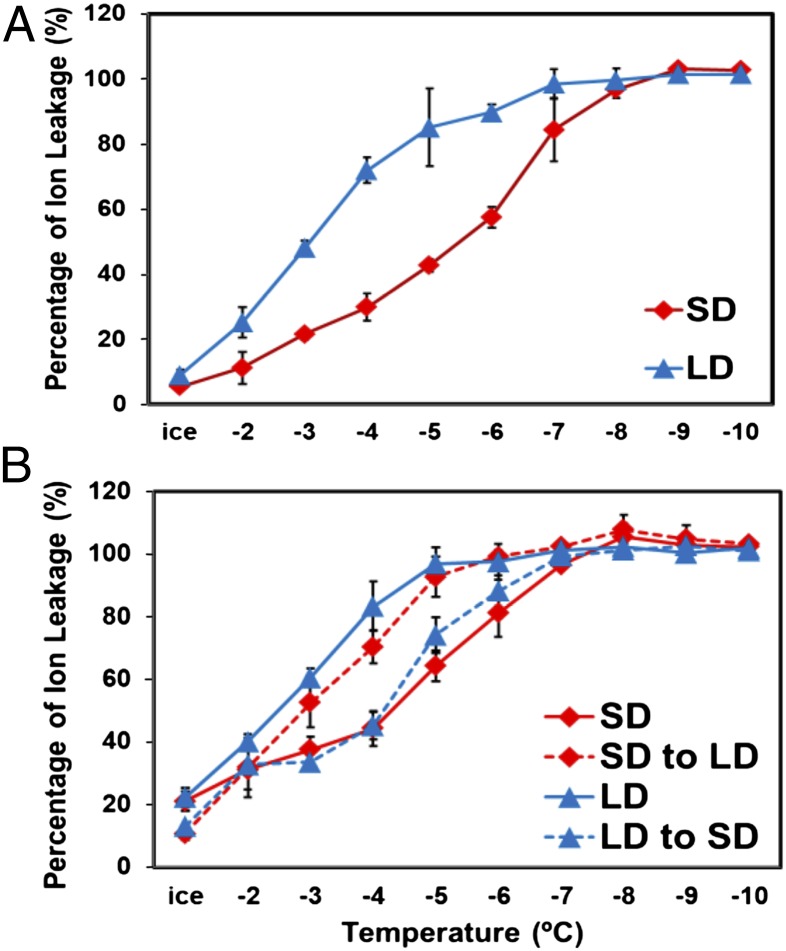
To determine whether the freezing tolerance of Arabidopsis (Col-0) is regulated by photoperiod, we grew plants under short days (SD; 8 h light, 16 h dark) and long days (LD; 16 h light, 8 h dark) and compared their freezing tolerance using the electrolyte leakage assay. The results indicated that the freezing tolerance of the SD plants was greater than that of the LD plants; whereas the EL50 (the temperature at which freezing damage results in leakage of 50% of the total cellular electrolytes) of the LD plants was about −3 °C, the SD plants had an EL50 of about −5.5 °C (Fig. 1A).
Fig. 1.
Arabidopsis freezing tolerance is regulated by photoperiod. (A) WT plants were grown under SD or LD conditions for 5 wk and 3 wk, respectively, and tested for freezing tolerance using the electrolyte leakage assay. (B) Plants grown under four conditions were tested for freezing tolerance: SD for 5 wk (SD); LD for 3 wk (LD); SD for 3 wk and transferred to LD for 2 wk (SD to LD); and LD for 2 wk and transferred to SD for 2 wk (LD to SD). The results are mean values from three independent experiments (error bars indicate SEM).
In these experiments, the SD and LD plants were tested at the point that they each had about eight true leaves. However, to produce this number of leaves, the SD and LD plants were grown for 5 wk and 3 wk, respectively. To address the possibility that differences in age were the cause of the observed differences in freezing tolerance, we grew plants under SD or LD conditions and then switched their photoperiod and tested their freezing tolerance (all plants again had about eight true leaves). When plants were grown under SD for 3 wk and transferred to LD for 2 wk, they had the same freezing tolerance as plants grown for 3 wk under LD (Fig. 1B). When plants were grown under LD for 2 wk and transferred to SD for 2 wk, they had the same freezing tolerance as plants grown under SD for 5 wk (Fig. 1B). Thus, regardless of the direction of the day-length shift or total age of the plants, the freezing tolerance of the plants was determined by the final 2-wk treatment; SD to LD produced the same freezing tolerance as constant LD treatment, and LD to SD produced the same freezing tolerance as constant SD treatment. From these results, we concluded that the freezing tolerance of Arabidopsis is regulated by photoperiod.
CBF Pathway Is Regulated by Photoperiod.
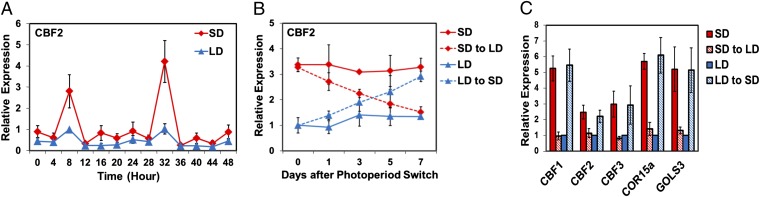
Given the prominent role of the CBF pathway in cold acclimation, we asked whether the CBF genes were expressed at different levels in SD and LD plants. Previous studies (16, 17) established that CBF1, -2, and -3 are regulated by the circadian clock and that the transcript levels for each gene peaks at about 8 h after dawn, a time referred to as zeitgeber time 8 (ZT8). Our results were consistent with these findings; the transcript levels for each CBF gene peaked at about ZT8 under both SD and LD conditions (results for CBF2 are shown in Fig. 2A, and those for CBF1 and CBF3 are shown in Fig. S1). However, the CBF transcript levels at ZT8 were about three- to fivefold higher in the SD plants compared with the LD plants. The transcript levels for two CBF regulon genes, COR15a and GOLS3, also oscillated, having peak expression between ZT8 and ZT12, and at their peak the transcript levels for these two genes were about fivefold higher in the SD plants (Fig. S1).
Fig. 2.
The CBF pathway is regulated by photoperiod. (A) WT plants were grown under SD or LD conditions and the transcript levels for CBF2 were determined at the indicated times. (B) Plants were grown under SD or LD conditions, further grown under the same photoperiod, or shifted from SD to LD or LD to SD for the indicated number of days, and the transcript levels for CBF2 were determined at ZT8. (C) Plants were grown as in B and the transcript levels for the indicated genes were determined at ZT8 (photoperiod shift was for 7 d). The results are mean values from three independent experiments (error bars indicate SEM).
To address the possibility that differences in plant age accounted for the differences in CBF and CBF regulon transcript levels in the SD and LD plants, we transferred LD plants to SD conditions and transferred SD plants to LD conditions, and determined their transcript levels. First, we determined the transcript levels of CBF2 at ZT8 at 0, 1, 3, 5, and 7 d after the switch in photoperiod. The results indicated that over the course of the week, the lower level of CBF2 transcripts initially observed in the LD plants rose to the level observed in the SD plants, and that the higher level of CBF2 transcripts initially observed in the SD plants decreased to the level observed in the LD plants (Fig. 2B). We then determined the transcript levels of CBF1, -2, and -3 and two CBF regulon genes, COR15a and GOLS3, at ZT8 in SD and LD plants, in SD plants transferred to LD for 7 d, and in LD plants transferred to SD for 7 d. The results indicated that with each gene, the lower transcript levels initially observed in the LD plants rose to those observed in the SD plants after transfer to SD, and that the higher transcript levels initially observed in the SD plants fell to those observed in the LD plants after transfer to LD (Fig. 2C). These results indicated that the CBF pathway is regulated by photoperiod and that the greater freezing tolerance of the SD plants was due, at least in part, to greater expression of the CBF pathway under SD conditions.
Photoperiodic Regulation of CBF2 Involves a G-box Motif Within the CBF2 Promoter.
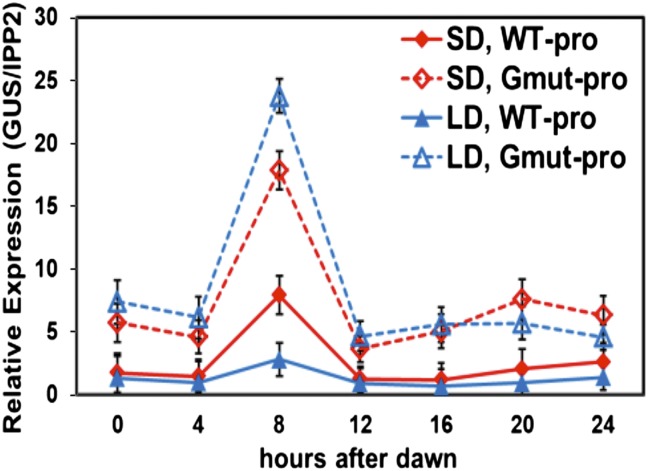
The photoperiodic control of CBF transcript levels could involve either transcriptional or posttranscriptional regulatory mechanisms or both. To determine whether transcriptional mechanisms were involved, we asked whether the CBF2 promoter included DNA regulatory elements that were responsive to photoperiod. Previous studies showed that the region of the CBF2 promoter from −189 to −35 relative to the transcription start site (this region is numbered −207 to −53 in the current TAIR10 database; hereafter, we use TAIR10 designations for sequence locations) included elements that could impart both cold (18) and circadian (19) regulation when fused the GUS reporter gene. We therefore tested this region for photoperiodic regulation. We fused the CBF2 promoter region from −207 to +134 to the GUS reporter gene (WT-pro) (Fig. S2A), transformed the construct into Arabidopsis, and determined the level of GUS transcripts in SD- and LD-grown plants (Fig. 3). The results indicated that the WT-pro construct produced peak levels of GUS transcripts at ZT8 in both SD and LD grown plants, but that the peak was about threefold greater in the SD plants. These results were consistent with the −207 to +134 CBF2 promoter fragment including a regulatory motif that was responsive to photoperiod.
Fig. 3.
A G-box motif within the CBF2 promoter confers photoperiod-regulated gene expression. Transgenic plants carrying WT (WT-pro) and mutant (Gmut-pro) versions of the CBF2 promoter fused to the GUS reporter gene were grown under SD or LD conditions and GUS transcript levels were determined at the indicated times. The WT-pro construct comprised CBF2 promoter sequences −207 to +134 and the Gmut-pro construct had CBF2 sequences −207 to +134 with the G-box motif at −112 to −107 mutated from CACGTG to GGTACC (Fig. S2A). The results are mean values from three independent experiments (error bars indicate SEM).
Previous studies showed that the CBF2 promoter region between −207 and −53 bp included a G-box motif, CACGTG (−112 to −107), that imparted negative regulation in plants that were grown at warm temperature and in plants that were exposed to low temperature (18, 20). Therefore, we asked whether this motif was also involved in photoperiodic regulation. We mutated the G-box sequence within the WT-pro construct (Gmut-pro) (Fig. S2A), transformed the construct into Arabidopsis, and determined the GUS transcript levels under SD and LD conditions. As with the WT-pro construct, the GUS levels for the Gmut-pro construct peaked at ZT8 in both SD- and LD-grown plants (Fig. 3). However, whereas the GUS transcript levels produced by the WT-pro construct were greater in SD plants, the levels produced by the Gmut-pro construct were approximately the same in the SD- and LD-grown plants (Fig. 3). These results were consistent with the G-box having a role in photoperiodic regulation of CBF2.
To confirm this result, we determined the GUS transcript levels at ZT8 for the WT-pro and Gmut-pro constructs in eight independent transgenic lines grown under SD and LD conditions (Fig. S2B). The results indicated that the GUS transcript levels obtained with the WT-pro construct were, on average, twofold higher in the SD plants compared with LD plants, and that this difference was eliminated when the G-box was mutated (Fig. S2C). In addition, the results indicated that mutation of the G-box resulted in higher-level expression of the reporter gene in both SD- and LD-grown plants (Fig. S2D); this finding was consistent with the element having a repressive effect.
PIF4 and PIF7 Repress Expression of the CBF Pathway Under LD Conditions.
Kidokoro et al. (20) found that the PIF7 transcription factor binds in vitro to the G-box within the CBF2 promoter (the G-box present in the WT-pro GUS fusion described above) and represses expression of the CBF genes during the subjective night phase in circadian regulation experiments. Thus, we considered PIF7 to be a candidate for mediating photoperiodic control of CBF2 expression. In addition, we considered PIF4 a candidate as it has been reported to physically interact with PIF7 (20) and, like other PIFs, it binds to G-box and related E-box (CANNTG) motifs (15).
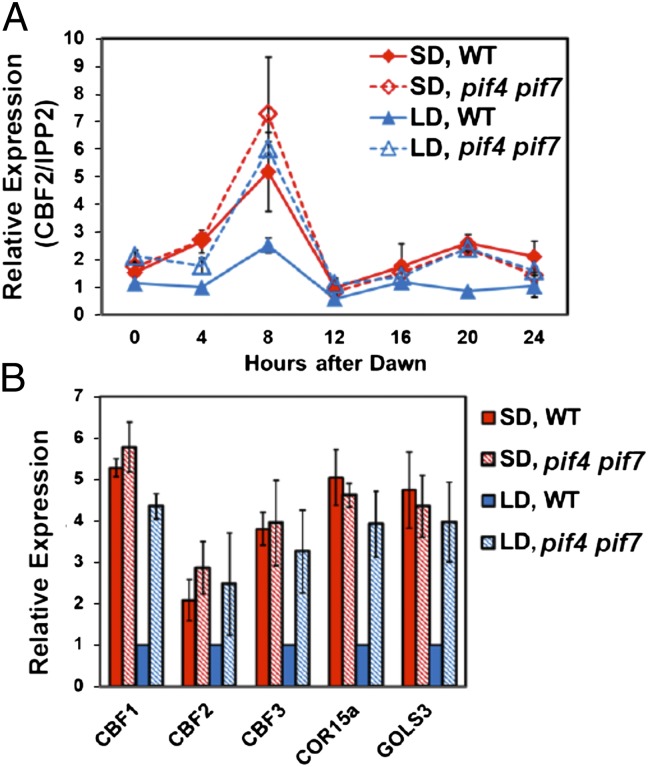
To test whether PIF4 or PIF7 were involved in photoperiodic regulation of CBF2, we asked whether pif4 or pif7 null mutations affected the patterns of CBF2 expression under LD or SD conditions. Our results indicated that neither of the single mutations had an effect (Fig. S3). However, the pif4 pif7 double mutation eliminated the photoperiodic regulation of CBF2; at ZT8, CBF2 transcript levels in pif4 pif7 double mutant plants grown under LD conditions were the same as in WT plants grown under SD conditions (Fig. 4A). The pif4 pif7 double mutation also eliminated the differences in transcript levels at ZT8 observed for CBF1, CBF3, COR15a, and GOLS3 in WT plants grown under SD and LD conditions (Fig. 4B). These results indicated that PIF4 and PIF7 function redundantly to repress expression of the CBF pathway under LD conditions.
Fig. 4.
Repression of the CBF pathway under LD conditions does not occur in pif4 pif7 double mutant plants. WT and pif4 pif7 double mutant plants were grown under SD or LD conditions. (A) The transcript levels for CBF2 were determined at the indicated times. (B) The transcript levels for the indicated genes were determined at ZT8. The results are mean values from three independent experiments (error bars indicate SEM).
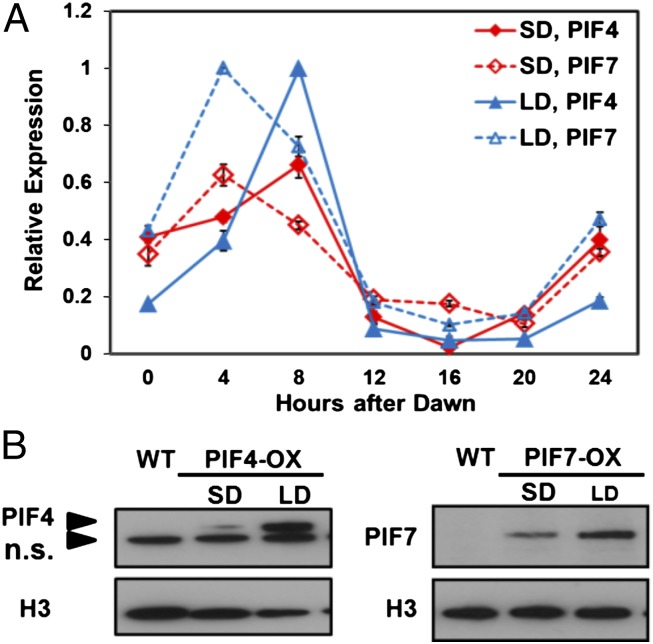
The lower level of CBF expression under LD conditions could have resulted from higher-level expression of PIF4 and PIF7 in plants grown under LD. Indeed, we found that the transcript levels for PIF4 and PIF7 oscillated over the course of the day under both LD and SD conditions, peaking at ZT8 and ZT4, respectively, and that the levels for both genes at these time points were nearly twofold higher under LD conditions (Fig. 5A). In addition, the stability of the PIF4 and PIF7 proteins was greater under LD conditions; this was determined by examining the levels of TAP-tagged PIF4 and CFP-tagged PIF7 in transgenic plants carrying these protein fusions placed under control of the constitutive CaMV 35S promoter. Whereas the transcript levels for the two transgenes were not affected by photoperiod (Fig. S4 B and D), the protein levels of both PIF4-TAP and PIF7-CFP at ZT8 were about twofold higher in LD plants compared with SD plants (Fig. 5B). Additional experiments indicated that PIF4-TAP and PIF7-CFP proteins were functional repressors; in the higher expressing lines, both PIF4-TAP and PIF7-CFP reduced CBF2 transcript levels by about 50% at ZT8 under SD conditions (Fig. S4 A and C); similar results were obtained testing CBF2 expression over a 24-h growth period (Fig. S5). Under LD conditions, PIF4-TAP also reduced CBF2 transcript levels by about 50%, whereas PIF7-CFP had little or no effect, suggesting that the endogenous PIF7 levels were saturating in regard to CBF2 repression under LD conditions (Fig. S5).
Fig. 5.
PIF4 and PIF7 are expressed at higher levels under LD conditions. (A) WT plants were grown under SD or LD conditions, and the transcript levels for PIF4 and PIF7 were determined at the indicated times. (B) Protein levels of PIF4-TAP (PIF4-OX) and PIF7-CFP (PIF7-OX) were determined in SD and LD transgenic plants at ZT8 using anti-myc or anti-GFP antibodies, respectively. Histone H3 protein was used as loading control and detected using rabbit anti-Histone H3 antibodies. n.s. indicates nonspecific signals. The results presented are representative of three experiments.
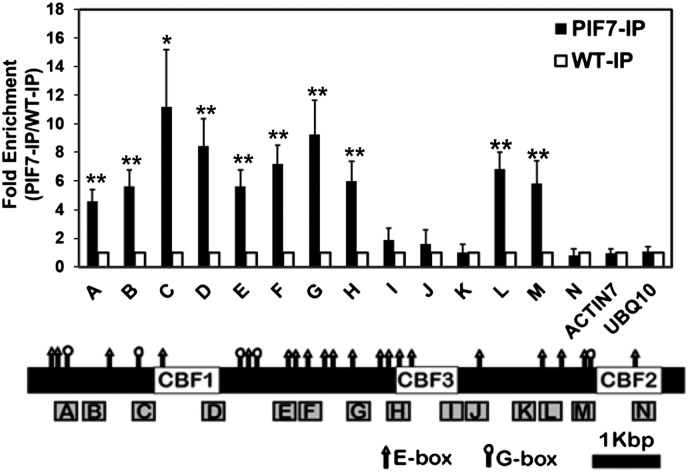
Using the electromobility shift assay we established that both PIF4 and PIF7 could bind to G-box motifs in the CBF1 and CBF2 promoters and the E-box motif in the CBF3 promoter (Fig. S6). Moreover, the results of chromatin immunoprecipitation (ChIP) experiments were consistent with PIF7 binding to the G-box within the CBF2 promoter that we determined to impart photoperiodic regulation; in test experiments using antibody that recognizes CFP (Fig. 6), but not in mock experiments using nonimmune serum (Fig. S7), significant enrichment of PIF7-CFP was detected at region M, which includes the functionally defined G-box (Fig. 3). In addition, in test (Fig. 6), but not mock (Fig. S7) experiments, PIF7-CFP was found to be enriched throughout the promoter regions of CBF1 and CBF3, which include a large number of G- and E-boxes, a result that is consistent with PIF7 also having a role in regulating expression of these genes.
Fig. 6.
PIF7 binds at the CBF locus. WT plants and transgenic plants overexpressing PIF7-CFP were grown under LD conditions, tissue was harvested at ZT8, and ChIP assays were performed using anti-GFP antibody (IP) (the results of mock experiments using rabbit IgG are presented in Fig. S7). Precipitated DNA sequences were quantified using primer sets across the CBF locus (boxes A through N). DNA sequences from ACTIN7 and UBQ10 were used as negative controls. The fold enrichment of precipitated DNA for each primer set in PIF7-OX samples (PIF7-IP, filled bars) are relative to the level in the WT samples (WT-IP, open bars). The locations and sequences of primer sets are listed in Table S1. Data are presented as mean ± SEM; n = 4 (*P < 0.05 and **P < 0.01, paired t test). The location of G-box (CACGTG, circle) and E-box (CANNTG, triangle) motifs are indicated.
PHYB Is Required for Photoperiodic Regulation of the CBF Pathway and Freezing Tolerance.
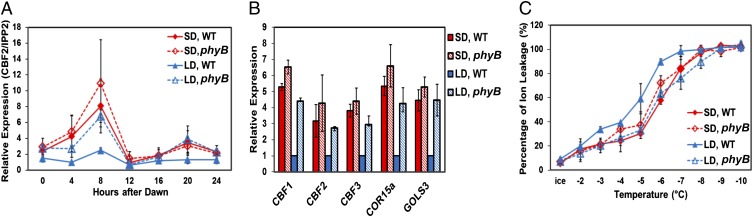
PHYB is known to physically interact with both PIF4 and PIF7 (15, 21). Thus, we asked whether PHYB was required for photoperiodic regulation of the CBF pathway. Our results indicated that it was. Under LD conditions, the transcript levels for CBF2 were about threefold higher at ZT8 in plants carrying a phyB null mutation than they were in WT plants, and matched the CBF2 transcript levels observed in WT plants grown under SD (Fig. 7A). The phyB mutation similarly affected CBF1, CBF3, and the CBF regulon genes COR15a and GOLS3 (Fig. 7B). Consistent with these results, the phyB mutation eliminated the photoperiodic regulation of freezing tolerance; the freezing tolerance of phyB plants grown under LD conditions was equal to that of WT plants grown under SD conditions (Fig. 7C).
Fig. 7.
PHYB is required for photoperiodic regulation of the CBF pathway and freezing tolerance. WT and phyB plants were grown under SD or LD conditions and tested for gene expression and freezing tolerance. (A) Relative transcript levels for CBF2 were determined at the indicated times. (B) Relative expression levels of the indicated genes were determined at ZT8. (C) Plant freezing tolerance was determined using the electrolyte leakage assay. The results in each test are mean values from three independent experiments (error bars indicate SEM).
Discussion
Here, we establish that Arabidopsis, like woody perennial trees, can sense shortening day length as a harbinger of coming cold temperatures and respond by increasing in freezing tolerance. Although the increase that we observed, about 2 °C, is modest in comparison with the increase that typically occurs in woody tree species, it is about the same as that reported for the perennial herbaceous species G. coccinea (13). Moreover, it is a considerable portion of the maximum increase in freezing tolerance that occurs in Arabidopsis in response to low temperature, which is about 6 °C (22, 23). Thus, it is likely that the SD-induced increase in freezing tolerance that occurs in Arabidopsis has adaptive value in nature protecting plants against sudden early autumn frosts. It will be of interest to determine whether there is significant natural variation in photoperiodic regulation of freezing tolerance among Arabidopsis ecotypes, and if there is, to understand the relationship of these differences to the environmental conditions that characterize the geographical locations from where the accessions originate. It will also be of interest to determine whether photoperiodic regulation of freezing tolerance has been overlooked as a common feature of cold acclimation in frost hardy herbaceous plants.
The results presented indicate that the difference in freezing tolerance that we observed between SD- and LD-grown Arabidopsis plants involves photoperiodic regulation of the CBF pathway. In particular, we show that the CBF pathway is repressed under LD conditions in warm-grown plants and that this repression is relieved under SD conditions resulting in an increase in freezing tolerance (Figs. 1 and 2). Down-regulation of the CBF pathway under LD conditions could have adaptive value, as it would diminish the allocation of energy and nutrient resources toward unneeded frost protection during the warm active growing season. In addition, it would mitigate CBF-induced retardation of growth. Achard et al. (24) have shown that activation of the CBF pathway results in up-regulation of gibberellin 2-oxidase genes causing a decrease in the levels of active gibberellins. This decrease, in turn, results in an increase in the levels of DELLA proteins, a small family of regulatory proteins that inhibit growth (25). Achard et al. (24) have presented evidence that the CBF-programmed repression of growth caused by the DELLA proteins contributes to the increase in freezing tolerance that occurs with cold acclimation.
Our genetic analysis demonstrates that repression of the CBF pathway under LD conditions involves action of PHYB and two PIFs with which PHYB interacts, PIF4 and PIF7. This conclusion is supported by multiple findings. First, whereas the peak transcript levels for CBF1, -2, and -3 and downstream CBF regulon genes were about three- to fivefold lower in LD WT plants than in SD WT plants, they were about the same in LD phyB plants, LD pif4 pif7 double mutant plants, and SD WT plants (Figs. 4 and 7); single pif4 and pif7 mutations did not affect expression of the CBF genes (Fig. S3), indicating that PIF4 and PIF7 act redundantly to repress expression of the CBF pathway. In addition, constitutive overexpression of PIF4-TAP and PIF7-CFP reduced the transcript levels of CBF2 at ZT8 in SD plants (Fig. S4 A and C); both PIF4 and PIF7 bound in vitro to G-box and E-box motifs that occur within the CBF locus (Fig. S6); the G-box at position −112 to −107 of the CBF promoter imparted photoperiod-regulated transcription of a reporter gene (Fig. 3 and Fig. S2); and ChIP experiments indicated that the PIF7-CFP protein bound at this G-box and elsewhere throughout the CBF locus in vivo (Fig. 6).
Our results also offer a partial explanation for why the CBF pathway is repressed to a greater extent under LD conditions than under SD conditions. Under LD conditions, the peaks in PIF4 and PIF7 transcript levels were nearly twofold greater, and the PIF4 and PIF7 proteins appeared to be about twofold more stable, than they were under SD conditions (Fig. 5). Combined, these differences in PIF4 and PIF7 transcript levels and protein stability could potentially account for three- to fourfold greater repression of the CBF genes under LD conditions, which is a considerable portion of repression that we observed in the LD plants. The mechanisms that underlie the differences in PIF4 and PIF7 expression and stability observed in response to photoperiod are unknown, but may involve action of PHYB as the phyb mutation eliminates the differences in CBF expression and freezing tolerance observed in WT plants grown under SD and LD conditions (Fig. 7).
The finding that PIF4 and PIF7 have roles in photoperiodic regulation of the CBF genes and freezing tolerance adds to the rapidly growing list of biological functions that these proteins have in Arabidopsis. For instance, PIF4 has recently been shown to promote hypocotyl elongation in response to high temperature by directly inducing genes involved in auxin synthesis (26, 27); to promote early flowering in response to high temperature by directly inducing expression of FLOWERING LOCUS T (28); to regulate stomatal development in response to light quantity (29); to control the rhythmic diurnal growth pattern of seedlings (30); and to have a role in the shade-avoidance response (31). Li et al. (32) have shown that PIF7 also has a role in regulating the shade-avoidance response and have presented a mechanism for this regulation. They showed that PIF7 is reversibly phosphorylated in response to light quality—the protein is phosphorylated in response to white-light and rapidly dephosphorylated in response to shade light—and that the dephosphorylated form of PIF7 binds to G-boxes in target promoters of auxin biosynthetic genes, induces their expression, and thus promotes rapid cell growth, a key feature of the shade-avoidance syndrome.
Our genetic and transcriptional analysis indicating that PIF4 has a role in repressing the transcription of CBF genes in the daytime is not what would be expected from what is generally known about PIF protein stability. It is well established that exposure of plants to white light converts the inactive Pr form of PHYB to the active Pfr form, which rapidly moves from the cytoplasm into the nucleus, where it interacts with PIF transcription factors to alter gene expression (15, 33). This interaction, however, generally results in degradation of the PIF proteins including PIF4. Indeed, Nozue et al. (30) showed that the level of PIF4 is much lower (although detectible) during the day than in the late evening, when it is involved in stimulating plant growth. Although it is true that the interaction of PHYB-Pfr with PIF7 does not lead to degradation of PIF7 (21), and thus, interaction of PHYB-Pfr with heterodimers of PIF4 and PIF7 might result in stable protein complexes, this would not explain our observed repression of the CBF genes in the pif7 mutant (Fig. S3). In our model, this repression would involve action of PIF4. A detailed analysis of the PIF protein complexes that are physically present at the CBF locus under SD and LD photoperiods will be required to resolve this issue.
A final point is that a key facet of PIF4 and PIF7 regulation of CBF expression is the concordant expression of these genes. The CBF genes are regulated by the circadian clock in plants exposed to normal warm growth temperature (17, 20, 34), and their peak expression is driven largely by the Myb transcription factors CCA1 and LHY (16), central components of the core circadian regulatory loop (35, 36). The CCA1 and LHY proteins, which have peak levels in the morning, bind to the Evening Element (EE) and related DNA regulatory motifs present within the CBF locus and induce high-level expression of the CBF genes at ZT8 (16). One simple model would be that PIF4 and PIF7 are also circadian-regulated and timed to peak in the morning hours. Indeed, the oscillation in PIF4 transcript levels is disrupted by constitutive overexpression of CCA1 (30), a classic indicator of circadian regulation. In addition, the promoters of PIF4 and PIF7 genes have EE motifs that could potentially drive their circadian regulation. Future experiments will be directed at testing this hypothesis and how output from the clock is integrated with photoperiodic regulation of the CBF pathway to condition freezing tolerance.
Materials and Methods
Plant Materials and Growth Conditions.
A. thaliana Columbia-0 WT and mutant derivatives were used in all experiments. Plants carrying the pif7-1, pif4-2, pif4-2 pif7-1, and phyb-9 null mutant alleles were kindly provided by Peter Quail (University of California, Berkeley) (21). Seeds were stratified for 3–5 d at 4 °C in the dark and then grown under either SD (8 h light, 16 h dark) or LD (16 h light, 8 h dark) conditions. For gene expression studies, plants were grown under SD (∼12 d) or LD (∼10 d) conditions on sterilized Gamborg’s B5 medium (Caisson Laboratories). For freezing-tolerance studies, plants were grown in soil as described (18) under SD (∼5 wk) or LD (∼3 wk) conditions unless indicated otherwise. All plants were grown at 22 °C under ∼100 μmol m−2 s−1 fluorescent white light.
Determination of Transcript Levels.
Transcript levels were determined using real-time quantitative RT-PCR (qRT-PCR) as described (18) with minor modifications in the amounts of total RNA and reaction volumes used. In the SD and LD experiments, the 8-h and 16-h time points, respectively, were taken during the light phase; in both experiments, the 24-h sample was taken during the light phase. IPP2 (ISOPENTENYL PYROPHOSPHATE-DIMETHYLALLYL PYROPHOSPHATE ISOMERASE 2) was used as the reference gene. Primers are listed in Table S1.
Gene Constructs.
The CBF2::GUS promoter (WT-pro and Gumt-pro), 35S::PIF7-CFP, and 35S::PIF4-TAP constructs were made as described in SI Materials and Methods and Table S2.
Protein Extraction and Immunoblots.
Protein was extracted from Arabidopsis seedlings and immunoblots prepared and processed as described in SI Materials and Methods.
ChIP.
The ChIP assays were performed as described (16) with minor modifications. The 35S::PIF7-CFP-HA line and wild-type (WT) 14-d-old seedlings were grown under LD and harvested at ZT8. For each biological replicate, IP and mock samples were normalized to the total input for each line, and the fold enrichment was relative to WT. The paired t test was applied to test the statistical significance of fold enrichment for each primer sets. The primers for qRT-PCR are listed in Table S1.
Supplementary Material
Acknowledgments
We thank Peter Quail for providing the pif7, pif4, pif4 pif7, and phyB mutants; Tiffany Liu for providing certain primers for ChIP; Beronda Montgomery for informative discussions about photoperiod and phytochrome-regulated gene expression; and Sarah Gilmour for helping prepare the manuscript. This research was funded primarily by National Science Foundation Plant Genome Project Grant DBI 0701709, but included infrastructure support provided by Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy and the Michigan Agricultural Experiment Station Grant DE-FG02-91ER20021.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211295109/-/DCSupplemental.
References
- 1.Sakai A, Larcher W. Frost Survival of Plants: Responses and Adaptation to Freezing Stress. Berlin: Springer; 1987. [Google Scholar]
- 2.Thomashow MF. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- 3.Guy C, Kaplan F, Kopka J, Selbig J, Hincha DK. Metabolomics of temperature stress. Physiol Plant. 2008;132:220–235. doi: 10.1111/j.1399-3054.2007.00999.x. [DOI] [PubMed] [Google Scholar]
- 4.Medina J, Catalá R, Salinas J. The CBFs: Three Arabidopsis transcription factors to cold acclimate. Plant Sci. 2011;180:3–11. doi: 10.1016/j.plantsci.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 5.Thomashow MF. Molecular basis of plant cold acclimation: Insights gained from studying the CBF cold response pathway. Plant Physiol. 2010;154:571–577. doi: 10.1104/pp.110.161794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chew YH, Halliday KJ. A stress-free walk from Arabidopsis to crops. Curr Opin Biotechnol. 2011;22:281–286. doi: 10.1016/j.copbio.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Harrison LC, Weiser CJ, Burke MJ. Environmental and seasonal factors affecting the frost-induced stage of cold acclimation in Cornus stolonifera Michx. Plant Physiol. 1978;62:894–898. doi: 10.1104/pp.62.6.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiser CJ. Cold resistance and injury in woody plants: Knowledge of hardy plant adaptations to freezing stress may help us to reduce winter damage. Science. 1970;169:1269–1278. doi: 10.1126/science.169.3952.1269. [DOI] [PubMed] [Google Scholar]
- 9.Williams BJ, Pellett NE, Klein RM. Phytochrome control of growth cessation and initiation of cold acclimation in selected woody plants. Plant Physiol. 1972;50:262–265. doi: 10.1104/pp.50.2.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKenzie JS, Weiser CJ, Burke MJ. Effects of red and far red-light on initiation of cold-acclimation in Cornus stolonifera Michx. Plant Physiol. 1974;53:783–789. doi: 10.1104/pp.53.6.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franklin KA, Quail PH. Phytochrome functions in Arabidopsis development. J Exp Bot. 2010;61:11–24. doi: 10.1093/jxb/erp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pietsch GM, Anderson NO, Li PH. Cold tolerance and short day acclimation in perennial Gaura coccinea and G. drummondii. Sci Hortic (Amsterdam) 2009;120:418–425. [Google Scholar]
- 14.Franklin KA, Whitelam GC. Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat Genet. 2007;39:1410–1413. doi: 10.1038/ng.2007.3. [DOI] [PubMed] [Google Scholar]
- 15.Leivar P, Quail PH. PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16:19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong MA, Farré EM, Thomashow MF. Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:7241–7246. doi: 10.1073/pnas.1103741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harmer SL, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 18.Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell. 2009;21:972–984. doi: 10.1105/tpc.108.063958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fowler SG, Cook D, Thomashow MF. Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 2005;137:961–968. doi: 10.1104/pp.104.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kidokoro S, et al. The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol. 2009;151:2046–2057. doi: 10.1104/pp.109.147033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leivar P, et al. The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell. 2008;20:337–352. doi: 10.1105/tpc.107.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilmour SJ, Hajela RK, Thomashow MF. Cold acclimation in Arabidopsis thaliana. Plant Physiol. 1988;87:745–750. doi: 10.1104/pp.87.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hannah MA, et al. Natural genetic variation of freezing tolerance in Arabidopsis. Plant Physiol. 2006;142:98–112. doi: 10.1104/pp.106.081141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Achard P, et al. The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell. 2008;20:2117–2129. doi: 10.1105/tpc.108.058941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun TP. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol. 2011;21:R338–R345. doi: 10.1016/j.cub.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 26.Franklin KA, et al. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA. 2011;108:20231–20235. doi: 10.1073/pnas.1110682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun J, Qi L, Li Y, Chu J, Li C. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet. 2012;8:e1002594. doi: 10.1371/journal.pgen.1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar SV, et al. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature. 2012;484:242–245. doi: 10.1038/nature10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casson SA, et al. Phytochrome B and PIF4 regulate stomatal development in response to light quantity. Curr Biol. 2009;19:229–234. doi: 10.1016/j.cub.2008.12.046. [DOI] [PubMed] [Google Scholar]
- 30.Nozue K, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 31.Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- 32.Li L, et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012;26:785–790. doi: 10.1101/gad.187849.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castillon A, Shen H, Huq E. Phytochrome Interacting Factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 2007;12:514–521. doi: 10.1016/j.tplants.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Bieniawska Z, et al. Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol. 2008;147:263–279. doi: 10.1104/pp.108.118059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 36.Pruneda-Paz JL, Kay SA. An expanding universe of circadian networks in higher plants. Trends Plant Sci. 2010;15:259–265. doi: 10.1016/j.tplants.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.