Abstract
Peroxisomes are ubiquitous organelles housing a variety of essential biochemical pathways. Peroxisome dysfunction causes a spectrum of human diseases known as peroxisome biogenesis disorders (PBD). While much is known regarding the mechanism of peroxisome biogenesis, it is still unclear how peroxisome dysfunction leads to the disease state. Several recent studies have shown that mutations in Drosophila peroxin genes cause phenotypes similar to those seen in humans with PBDs suggesting that Drosophila might be a useful system to model PBDs. We have analyzed the proteome of Drosophila to identify the proteins involved in peroxisomal biogenesis and homeostasis as well as metabolic enzymes that function within the organelle. The subcellular localization of five of these predicted peroxisomal proteins was confirmed. Similar to C. elegans, Drosophila appears to only utilize the peroxisome targeting signal (PTS) type 1 system for matrix protein import. This work will further our understanding of peroxisomes in Drosophila and add to the usefulness of this emerging model system.
Keywords: peroxisomes, drosophila, β-oxidation, α-oxidation, ether lipids
Peroxisomes are single membrane-bound organelles found in virtually all eukaryotic cells. Many essential reactions occur within the organelle including β-oxidation of fatty acids, α-oxidation of branched chain fatty acids, and ether lipid biosynthesis (1). Reactive oxygen species (ROS), such as H2O2, are generated within peroxisomes by acyl CoA-oxidases during β-oxidation and by other resident oxidases. Antioxidant enzymes, such as catalase, are present in the organelle to detoxify this stress.
Peroxisomes represent a semi-autonomous branch of the secretory pathway. Peroxisomes can form de novo by budding from the ER (2) and mature peroxisomes can grow and divide (3). Peroxisomal proteins are synthesized in the cytoplasm and trafficked to the organelle through three main routes. Most proteins are targeted to peroxisomes via a peroxisomal targeting signal type 1 (PTS1) at their carboxy-termini that matches SKL or a conserved variant (4). PTS1-containing proteins are bound by Pex5 in the cytoplasm, shuttled to the peroxisomal membrane, and translocated into the peroxisomal matrix by a poorly defined mechanism (5). Large proteins, even oligomers, can cross the peroxisomal membrane in a folded state via an unknown mechanism (6). Several models for import for matrix proteins have been proposed including modified endocytosis, static pore, and dynamic pore models (6). The Pex5 receptor itself has also been proposed to function as a dynamic pore and allow cargo to cross the membrane in addition to its role in targeting (7). Recent electrophysiological data suggests that a Pex5 complex that contains Pex14 can form membranous pores (8). Other peroxisomal proteins use a longer, less specific PTS type 2 signal at their amino-terminus that is recognized by the Pex7 receptor in the cytoplasm and shuttled to the peroxisome (9). Peroxisomal membrane proteins (PMPs) are targeted via a membrane protein targeting signal (mPTS) and interaction with the soluble Pex19 receptor (10). Most PMPs are thought to insert posttranslationally when Pex19 carries the PMP to Pex3 at the peroxisomal membrane (11). However, recent evidence suggests that all PMPs in yeast transit the endoplasmic reticulum en route to the peroxisome (12). Despite intense study, some basic features of peroxisome biology remain to be discovered.
A spectrum of human diseases, named peroxisome biogenesis disorders (PBDs), result from defects in organelle biogenesis or import of protein cargo (13). The most severe PBD, Zellweger syndrome, is characterized by neuronal dysfunction, craniofacial malformation, muscle weakness (hypotonia), renal cysts ultimately resulting in death in early childhood. There is currently no cure for PBDs and the treatments only reduce the severity of the symptoms. It is unclear how the loss of peroxisomes leads to the disease state, but low levels of ether lipids and high levels of very long chain fatty acids (VLCFAs) have been implicated as causative agents (13).
Mouse models for PBDs have been developed by targeted disruption of peroxin genes (14). These animals show many similarities to the clinical phenotypes including early death, VLCFA accumulation, neuronal migration defects, and locomotor problems. Despite these advances, effective therapies for PBD patients remain elusive.
Flies are emerging as valuable animal models for PBDs. Drosophila have been used to model many other human diseases including neurodegenerative diseases, mitochondrial diseases, and metabolic syndrome (15–17). Many pathways are conserved between humans and flies, with around 75% of human genes having fly homologs (18). Flies carrying mutations in peroxin genes have recently been shown to manifest some of the PBD clinical phenotypes including early death, VLCFA accumulation, neuronal defects, and locomotor problems (19–21). These studies have provided a platform for further investigations into the molecular consequences of peroxisome loss or dysfunction.
In this study, we have identified Drosophila proteins that are predicted to be involved in peroxisome formation, maintenance, and metabolism. The subcellular localization of some of these predicted peroxisomal proteins was determined by fluorescence confocal microscopy. The predicted peroxisomal pathways in Drosophila are compared to other organisms. This study provides foundational support for understanding the role peroxisomes play in the biology of Drosophila and may enhance their usefulness as animal models for PBDs.
Results and Discussion
We are developing tools to study peroxisome biogenesis and function in Drosophila melanogaster. Our first approach was to identify potential peroxisomal proteins in the Drosophila proteome by similarity with vertebrate homologs. To that end, we identified all human proteins in the Universal Protein Knowledgebase (UniprotKB, www.uniprot.com) that listed “peroxisome” in the “subcellular location” field. This search yielded 88 unique proteins (with 156 total isoforms). Additional candidates not found in the Uniprot database, but identified as peroxisomal in the literature (1, 22) were also added to the list, bringing the total to 112 sequences (Supplemental Table 1). Each human protein was used to query the Drosophila proteome at FlyBase (23) using the BLAST algorithm (24). These results were compared to a previous whole genome analysis of peroxisomal proteins, called PeroxIP that utilized the C-terminal PTS1 motif as a search criteria (25). Twenty proteins identified by BLAST searches were not found in the PeroxiP data set, and conversely, twenty six proteins in the PeroxiP dataset were not found by BLAST searching the Drosophila proteome (Supplemental Figure 3). The proteins unique to the PeroxiP set were further examined by the PProwler and PTS1 Prowler (26) algorithms to validate their PTS1 predictions (Supplemental Table 2). We chose to examine three major peroxisomal metabolic pathways in detail.
β-oxidation
The most ubiquitous function of all peroxisomes is the breakdown of fatty acids by β-oxidation. Drosophila possesses the enzymes required for peroxisomal and mitochondrial β-oxidation. It is likely that VLCFAs are preferentially broken down in fly peroxisomes. Flies with mutations in the peroxins pex10 or pex16 have elevated VLCFA levels, but normal levels of shorter chain fatty acids (19, 21).
Acyl-CoA synthetase
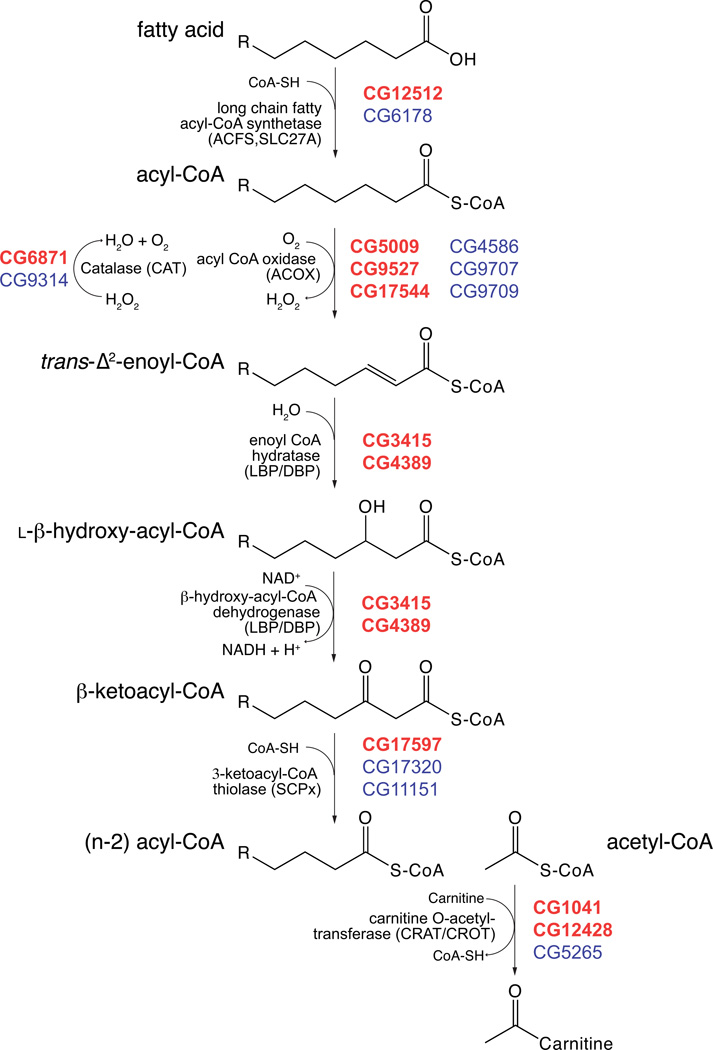
Figure 1 illustrates the pathway of peroxisomal β-oxidation and delineates the enzymatic activity and the corresponding fly CG numbers identified as potential homologs. Supplemental Table 1 lists each human protein involved in β-oxidation by Uniprot accession number, proposed function, and best BLAST hit in the fly proteome. Fatty acids are first activated by ligation to Co-enzyme A (CoA) by fatty acyl-Co synthetases. A large number of fatty acyl-CoA synthetases have been identified in all organisms with different subcellular locations as well as acyl-chain length specificities. Twenty six acyl-CoA synthetases (ACS)(27) have been identified in humans and have been subdivided into six families, largely based on chain length specificity. This report also identified 26 Drosophila genes (32 proteins including alternative splice variants) potentially encoding acyl-CoA synthetases (27). Chief among them in this context are two that terminate in PTS1 tripeptides, CG6178 and CG12512. Both of these candidates fall outside the conventional ACS classifications with CG6178 found in a fly/worm specific clade and CG12512 grouping with the ACSF2 clade (Acetyl-CoA Synthetase Family member 2). In most organisms, the location of CoA addition for peroxisomal β-oxidation is a matter of debate. Regardless, Drosophila peroxisomes likely contain at least two matrix localized ACSs.
Figure 1. Peroxisomal β-oxidation.
The steps of peroxisomal β-oxidation of fatty acids are illustrated with the CG numbers for each Drosophila protein listed at each step. The CG numbers are listed in descending order with the best hit(s) in bolded, red text. The percent identities of the Drosophila and human homologs are listed below. ACFS2/SLC27A2/SLC27A4: 22–36%, CAT: 57–65%, ACOX1/2/3: 43–22%, LBP/DBP: 27–53%, SCPx: 35–65%, CRAT/CROT: 25–34%.
One of the ACS subfamilies, specifically the very long chain ACS family, is composed of multispan membrane proteins of the solute carrier family 27 (SLC27) that are also thought to serve as lipid transporters (28). One of the few characterized Drosophila proteins with a predicted peroxisomal function is an ACS of this family called Bubblegum. Bubblegum (bgm) is a predicted very long chain fatty acid acyl CoA synthetase and mutations in the gene encoding this enzyme cause progressive neurodegeneration and VLCFA accumulation (29).
Acyl-CoA oxidase
Activated fatty acids are sequentially reduced by two carbon acetyl-CoA groups through the action of three enzymes. Acyl-CoA oxidase introduces a double bond between the alpha and beta carbon utilizing a bound flavin prosthetic group (FAD) and molecular oxygen. This reaction generates hydrogen peroxide, the hallmark metabolite of the organelle. Humans express three Acyl-CoA oxidases with differing chain-length specificity and preference for branched chain fatty acids. While BLAST searches with ACOX1 and ACOX2 both identified CG5009 as the best hit (43% and 34% identity respectively, Supplemental Table 1), five other proteins are highly similar (ranging from 22–38% identical). All six enzymes have a variant PTS1 tripeptide suggesting a peroxisomal localization. Two of the six putative acyl-CoA oxidases have been named in Flybase as Acox-57d (acyl-Coenzyme A oxidase at 57D distal, CG9709) and Acox-57p (acyl-Coenzyme A oxidase at 57D proximal, CG9707) but no additional characterization has been reported (30).
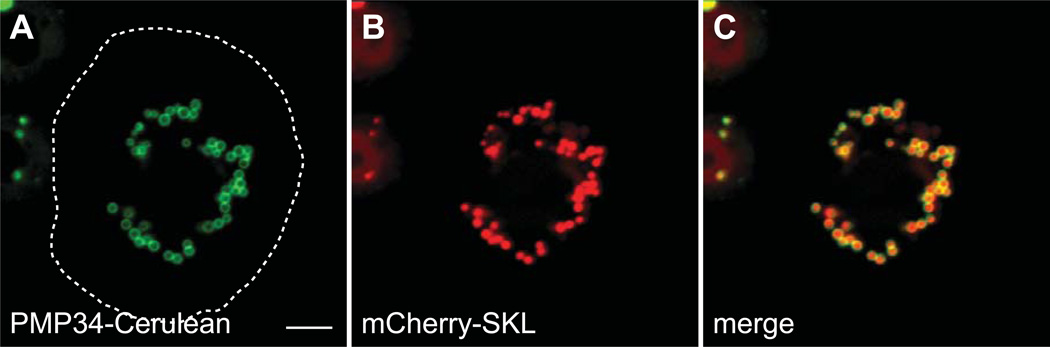
In addition to identifying potential peroxisomal pathways in Drosophila, we wanted to determine the subcellular localization of selected candidate proteins by transient expression of fluorescent fusion proteins in the Drosophila hemocyte cell line Schnieder-2 (S2) cells. Peroxisomes were visualized by expressing a matrix marker containing a PTS1 (mCherry-SKL) and a fluorescent protein fused to a peroxisomal membrane protein (PMP34-Cerulean) in Drosophila S2 cells. Figure 2 shows that both proteins colocalize in a punctate pattern, but PMP34-Cerulean is found at the edges of the puncta due its membrane localization.
Figure 2. mCherry-SKL and PMP34-Cerulean localize to peroxisomes.
Plasmids encoding mCherry-SKL and PMP34-Cerulean, under control of the actin 5c promoter, were cotransfected into S2 cells and imaged live by confocal microscopy. (A–C) The mCherry signal is present in a punctate pattern indicating peroxisomal matrix localization. The Cerulean signal is also punctate, but brighter at the edges of the puncta, indicating localization to the peroxisomal membrane. The approximate cell boundary is highlighted with a dashed white line. Scale bar equals 5 µM.
We first determined the localization of one of the putative peroxisomal acyl-CoA oxidases (CG17544). S2 cells were cotransfected with an mCherry-CG17544 plasmid and the peroxisomal membrane marker PMP34-Cerulean. Figure 3 illustrates that both PMP34-Cerulean and mCherry-CG17544 colocalize to peroxisomal puncta.
Figure 3. An mCherry-Acyl-CoA Oxidase (CG17544) Fusion Protein localizes to peroxisomes in S2 Cells.
Plasmids encoding mCherry-CG17544 and PMP34-Cerulean, under control of the actin 5c promoter, were cotransfected into S2 cells and imaged live by confocal microscopy. (A–C) Colocalizaion of mCherry and Cerulean indicates that mCherry-CG17544 localizes to peroxisomes. The approximate cell boundary is highlighted with a dashed white line. Scale bar equals 5 µM.
Bifunctional Protein
The next two reactions, enoyl-CoA hydration and β-hydroxy acyl-CoA dehydrogenation, are catalyzed by a single protein called peroxisomal Bifunctional Protein (also called peroxisomal Multifunction Enzyme (MFE)). Two distinct and significantly dissimilar proteins provide these activities in humans called LBP (l-Bifunctional Protein) and DBP (d-Bifunctional Protein). These enzymes prefer the l- or d-stereoisomer, respectively. LBP is primarily involved in straight-chain fatty acid metabolism while DBP prefers branched chain fatty acids such as pristanic acid and bile acid precursors (di- and trihydroxycholestanoic acid). Single homologs of LBP (CG4389) and DBP (CG3415) are found in the Drosophila proteome, each with a C-terminal PTS1. Interestingly, the LBP homolog encodes three splice variants with CG4389-PA containing a 39 amino acid N-terminal extension. This N-terminal sequence is predicted to encode a mitochondrial targeting peptide (mTP, 97% probability by MitoProtII v1.01 http://ihg2.helmholtz-muenchen.de/ihg/mitoprot.html, and 85% probability by TargetP v1.1, http://www.cbs.dtu.dk/services/TargetP-1.1/) (31, 32). While the expression of CG4389-PB and CG4389-PC likely result in an exclusively peroxisomal localization, the CG4389-PA isoform may be dually localized to both peroxisomes and mitochondria. When the coding region of the CG4389-PB gene is placed downstream of mCherry, this LBP homolog colocalizes with PMP34-Cerulean confirming its peroxisomal localization (Figure 4).
Figure 4. An mCherry-Bifunctional Protein (CG4389) Fusion Protein localizes to peroxisomes in S2 Cells.
Plasmids encoding mCherry-CG4389 and PMP34-Cerulean, under control of the actin 5c promoter, were cotransfected into S2 cells and imaged live by confocal microscopy. (A–C) Colocalization of mCherry and Cerulean indicates that mCherry-CG4389 localizes to peroxisomes. The approximate cell boundary is highlighted with a dashed white line. Scale bar equals 5 µM.
Thiolase/SCPx
The next step in β-oxidation is cleavage of the β-ketoacyl-CoA by the enzyme thiolase. This activity is provided by two classes of enzymes in humans, one a traditional β-ketoacyl-CoA thiolase encoded by the ACAA1 gene, and the other a multi-functional protein, Sterol Carrier Protein X (SCPx). SCPx has two domains, a C-terminal non-specific lipid transport protein as well as an N-terminal thiolase domain (33, 34). In mammals, the SCPx gene can produce three gene products. One is a full length ~58 kDa Thiolase-SCP2 fusion protein that can persist as a single protein or be proteolytically processed to an ~46 kDa thiolase domain and an ~13 kDa SCP2 domain. Additionally, the SCPx gene transcribes an ~15 kDa SCP2 protein that contains a 20 amino acid N-terminal presequence that is posttranslationally removed. All protein products of the SCPx gene are likely peroxisomal due to C-terminal PTS1 motifs in all sequences (33, 34).
Drosophila contains two homologs of SCPx (CG17320 (Scpx) and CG17597) that both contain PTS1 motifs. Scpx (CG17320) was originally identified as a transcript regionally expressed in the Drosophila embryonic midgut (35). While CG17320 is arranged much like human SCPx with an N-terminal Thiolase domain and a C-terminal SCP2 domain, CG17597 only contains the Thiolase domain without an attached SCP2, thus CG1730 is likely a peroxisomal thiolase. Two additional genes CG11151 and CG12269 appear to encode just an SCP2 domain. CG11151 terminates in AKL while CG12269 has no apparent PTS. Finally, an SCP2-like domain can be found at the C-terminus of CG5590, which is most similar (57% identical) to Hydroxysteroid dehydrogenase-like protein 2 (HSDL2, Supplemental Table 1). An SCP2 domain can also be found at the C-terminus of human DBP (34, 36), which is lacking in the fly homolog of DBP (CG3515).
Human peroxisomal thiolase, encoded by the ACAA1 gene, is localized to the peroxisome by virtue of an N-terminal PTS2 motif. The four closest BLAST hits with ACAA1 do not contain a canonical PTS1 or PTS2 motif. In fact, three of them (CG4600 (yip2), CG4581 (Thiolase), and CG10932) have readily identifiable mitochondrial targeting signals (mTP, >90%) and are predicted to be in the mitochondria, likely involved in mitochondrial β-oxidation. CG4581 is named Thiolase in Flybase in a systematic effort to identify mitochondrial proteins, but no additional characterization has been reported (37). Interestingly, CG10932 terminates in – EKL, perhaps a variant PTS1 sequence, that may yield a dual localization. This possibility remains to be experimentally validated.
Carnitine O-acetyl(octanoyl)transferase
While fatty acid uptake into the peroxisome does not utilize a carnitine shuttle like mitochondria (1), animal peroxisomes contain enzymes that produce acylcarnitine esters, likely for export to the mitochondria. The proteins CRAT and CROT are Carnitine O-acetyltransferase and Carnitine O-octanoyltransferases respectively. Three Drosophila proteins (CG5265, CG1041, and CG12428) show between 29–34% identity with either CRAT or CROT. All three proteins terminate in a PTS1 variant. In fact, we chose to use the final ten amino acids of CG1041 (-KNPPETKSKL) as our prototypical peroxisomal matrix marker (GFP-SKL in Figure 2).
Catalase
The peroxisome is defined by the presence of hydrogen peroxide, due to flavin-linked oxidases, and catalase, which decompose this toxic metabolite. There are two clear catalase homologs in Drosophila (CG6871 (Cat) and CG9314). Catalase (CG6871) is one of the few peroxisomal proteins to have been examined phenotypically in Drosophila. Drosophila cat null mutants are hypersensitive to H2O2 and display a decreased metabolic rate. (38, 39). Overexpression of cat and copper zinc superoxide dismutase (sod) extends lifespan in Drosophila (40). Catalase has also been localized to Drosophila peroxisomes by transmission electron microscopy and immunofluorescence (41, 42). CG9314 is predicted to be a testes specific catalase isoform (43, 44).
β-oxidation accessory enzymes
Different organisms utilize a variety of accessory enzymes to handle complex fatty acids including unsaturated and polyunsaturated fatty acids (Supplemental Figure 1). In humans, some of these genes include ECH1 (enoyl-CoA hydratase 1, also known as Δ3,5-Δ2,4-dienoyl-CoA isomerase), PECR (peroxisomal trans-Δ2-enoyl-CoA reductase), DECR2 (Δ2,4-dienoyl-CoA reductase), and PECI (trans-Δ3,2-enoyl-CoA isomerase). Homologs of PECI (CG13890, 34% identity) and ECH1 (CG9577, 45% identity) are readily apparent. However, the DECR2/PECR enzymes are more difficult to identify with confidence. There are several fly homologs similar to human DECR2/PECR but none stands out as a definitive homolog for each. These proteins (CG10672, CG5590, and CG3415) each contain a “short-chain dehydrogenase/reductase SDR” motif (IPR002198) of which there are 45 members in Drosophila melanogaster.
α-oxidation
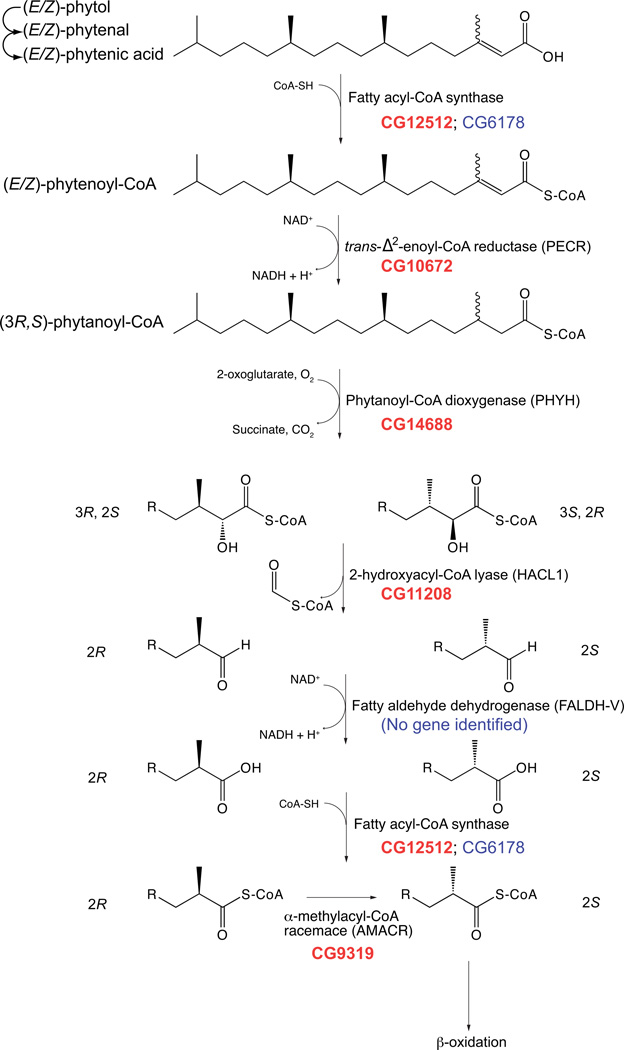
Another important peroxisomal pathway of lipid metabolism is α-oxidation. In humans, this pathway is primarily responsible for the catabolism of 3-methyl branched-chain fatty acids such as phytol (1, 45–47). Figure 5 illustrates the pathway of peroxisomal α-oxidation of phytol and delineates the enzymatic activity and the corresponding fly CG number identified as potential homologs. Supplemental Table 1 lists each human protein involved in α-oxidation by Uniprot accession number, proposed function, and best BLAST hit in the fly proteome. Phytol is reduced to phytenic acid by extraperoxisomal enzymes then thioesterified to its CoA derivative phytenoyl-CoA, perhaps by peroxisomal acyl-CoA synthetases. The double bond between the α β by PECR, and the α-carbon is hydroxylated by phytanoyl-CoA dioxygenase (PHYH). The best Drosophila homolog of PHYH, CG14688, is only 13% identical, but BLAST searches against the human genome with CG14688 identified only one protein, human PHYH. PHYH is targeted to the peroxisome in humans by an N-terminal PTS2 motif, however, the fly homolog does not possess a PTS2 or PTS1 sorting signal. Comparative analysis of PHYH proteins from 18 organisms shows that a PTS2 signal is present in vertebrates, but no PTS is readily identifiable from insects, worms, plants and slime molds. The mechanism of PHYH sorting in Drosophila is currently unknown.
Figure 5. Peroxisomal α-oxidation Pathway.
The steps of peroxisomal α-oxidation are illustrated with the CG numbers for each Drosophila protein listed at each step. The CG numbers are listed in descending order with the best hit(s) in bolded, red text. The percent identities of the Drosophila and human homologs are listed below. ACFS2/SLC27A2/4: 22–36%, PECR: 27%, PHYH: 13%, HACL1: 53%, AMACR: 49%.
The racemically-mixed 2-hydroxyacyl-CoA is cleaved by 2-hydroxyacyl-CoA lyase 1 (HACL1) yielding formyl-CoA and a fatty aldehyde chain-shortened by one carbon. This fatty aldehyde is oxidized to a carboxylic acid by a fatty aldehyde dehydrogenase. In humans, this reaction is catalyzed by a specific splice variant of the aldehyde dehydrogenase 3 family, member A2 gene (ALDH3A2), a fatty aldehyde dehydrogenase isozyme (FALDH-V) (46). There is no clear Drosophila homolog of FALDH-V. The (2S) stereoisomer is then further metabolized by β-oxidation, following activation by acyl-CoA synthetase. However, the (2R) stereoisomer in not recognized by acyl-CoA oxidase and must be converted to (2S) by α-methyl acyl-CoA racemase (AMACR)(46). AMACR is encoded by the CG9319 gene in Drosophila. AMACR is dually-localized to mitochondria and peroxisomes in humans. A single transcript produces a protein with an N-terminal mitochondrial targeting signal (mTP) and a C-terminal PTS1 (46, 48, 49). CG9319 terminates in AKL, a prototypical PTS1, but does not include an N-terminal mTP suggesting that AMACR is only peroxisomal in flies. Interestingly, the AMACR gene is overexpressed in many cancers (46, 50).
Ether lipid synthesis
A third key peroxisomal metabolic pathway is the production of ether-linked lipids (Figure 6). This class of compounds is used to produce plasmalogens, bioactive lipids such as platelet activating factor (PAF), as well as lipid attachments for some Glycosylphosphatidylinositide (GPI) anchors (1, 51). The enzymology of ether lipid synthesis is well established for animals. The glycolic intermediate dihydroxyacetone phosphate is fatty esterified at the sn1 position by the addition of a fatty acid by the peroxisomal enzyme glyceronephosphate O-acyltransferase (GNPAT). GNPAT (also known as dihydroxyacetone phosphate acyltransferase, DHAP-AT) is encoded by CG4625 in Drosophila and has been named Dhap-at in Flybase, but no additional characterization has been reported (52). Next, the characteristic ether linkage is introduced by the enzyme alkylglycerone phosphate synthase (AGPS), which exchanges the fatty acid added by GNPAT with a long-chain fatty alcohol produced by a fatty acyl-CoA reductase. The Drosophila AGPS homolog CG10253 is 50% identical to human AGPS (Supplemental Figure 2). AGPS is localized to the peroxisome in humans by virtue of its N-terminal PTS2 sequence. Drosophila AGPS does not contain a PTS2 motif, but ends with the C-terminal PTS1 sequence AKL specifying a peroxisomal localization. The long chain fatty alcohol needed for ether lipid synthesis is produced by fatty acyl-CoA reductase, the product of the FAR1 and FAR2 genes in human. These membrane proteins are localized to the peroxisome in animals (53). The best Drosophila homolog for both FAR1 and FAR2 is CG5065 (Supplemental Table 1); however, closer inspection of the fly proteome reveals an additional 16 genes that share significant similarity to the human FAR genes. In total, these 17 genes are predicted to code for 29 proteins accounting for all splice variants. All of these homologs are likely to encode FAR activity since the most dissimilar member, CG34342-PC with 25% identity, only identifies FAR1 and FAR2 when BLASTed against the translated human genome. Since the membrane peroxisomal targeting signal (mPTS) for FAR1 or FAR2 has not been identified, it is difficult to predict the localization of the fly proteins. Given that fatty alcohol products of the FAR proteins are used for other biosynthetic processes including wax production (53), it is likely that the Drosophila isoforms may localize to many organelles.
Figure 6. Peroxisomal ether lipid synthesis Pathway.
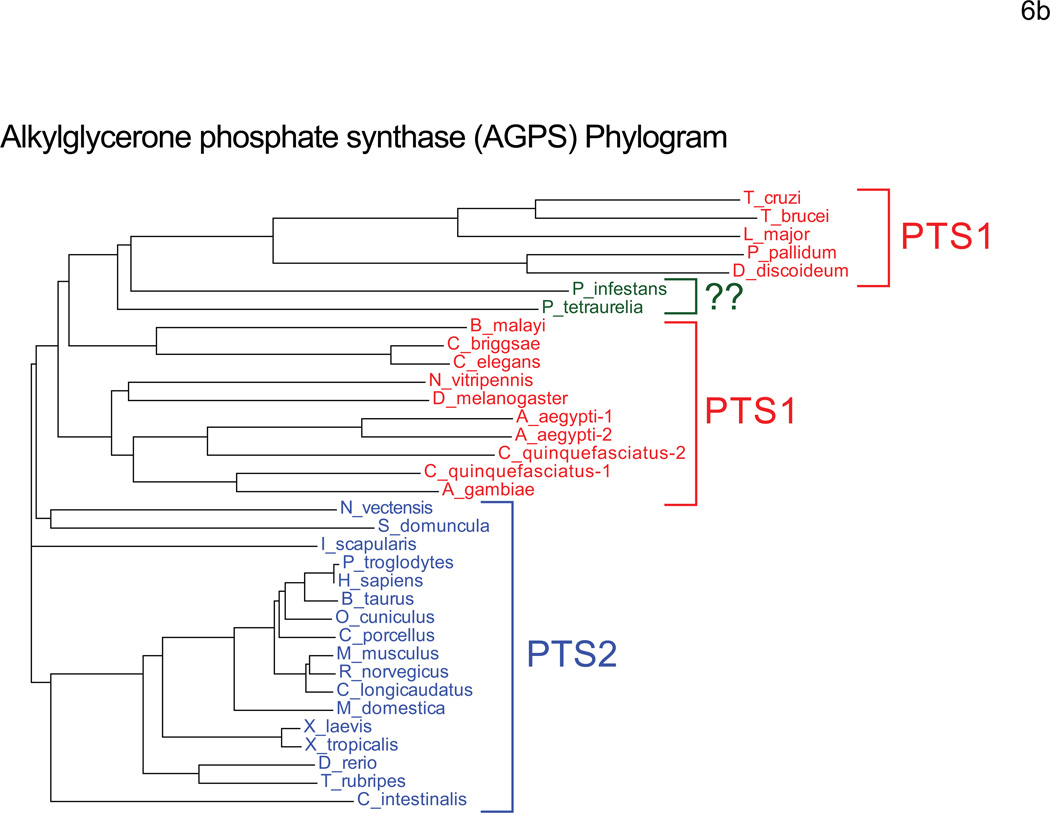
A. The steps of peroxisomal ether lipid synthesis are illustrated with the CG numbers for each Drosophila protein listed at the right of each step. The CG numbers are listed in descending order with the best hit(s) in bolded, red text. No homolog of 1-alkyldihydroxyacetone 3-phosphate reductase could be identified in the Drosophila genome. B. A Phylogram of alkylglycerone phosphate synthase (AGPS) from multiple species was generated with ClustalW2. All vertebrate AGPS sequences analyzed contain a PTS2, while most non-vertebrate AGPS sequences, including Drosophila, have a PTS1. P. tetraurelia and P. infestans do not have identifiable PTS motifs. The percent identities of the Drosophila and human homologs are listed below. GNPAT: 25%, FAR1/2: 25–38%, AGPS: 50%.
Peroxisome Biogenesis
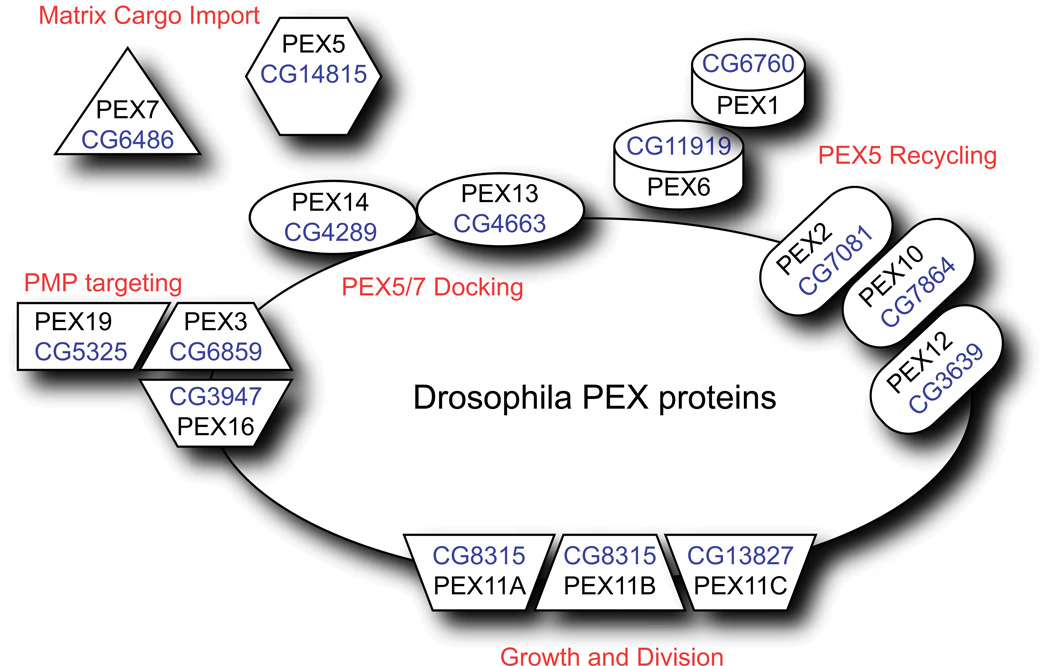
We also searched the Drosophila proteome for proteins known to be involved in peroxisome biogenesis or peroxin (PEX) proteins (54). We identified 15 PEX genes including three isoforms of PEX11 (Figure 7, Supplemental Table 1). The identity of PEX homologs in Drosophila was also recently reported in an analysis of ring finger peroxins PEX2 and PEX10 (19). The Drosophila PEX genes range from 22–43% identical to their human counterparts. A recent study (20) demonstrated that knockdown of pex genes in Drosophila S2 cells inhibits PTS1 import.
Figure 7. Drosophila Peroxins.
Proteins involved in peroxisome biogenesis, protein import, and growth and division are illustrated. CG numbers for the Drosophila homologs are shown.
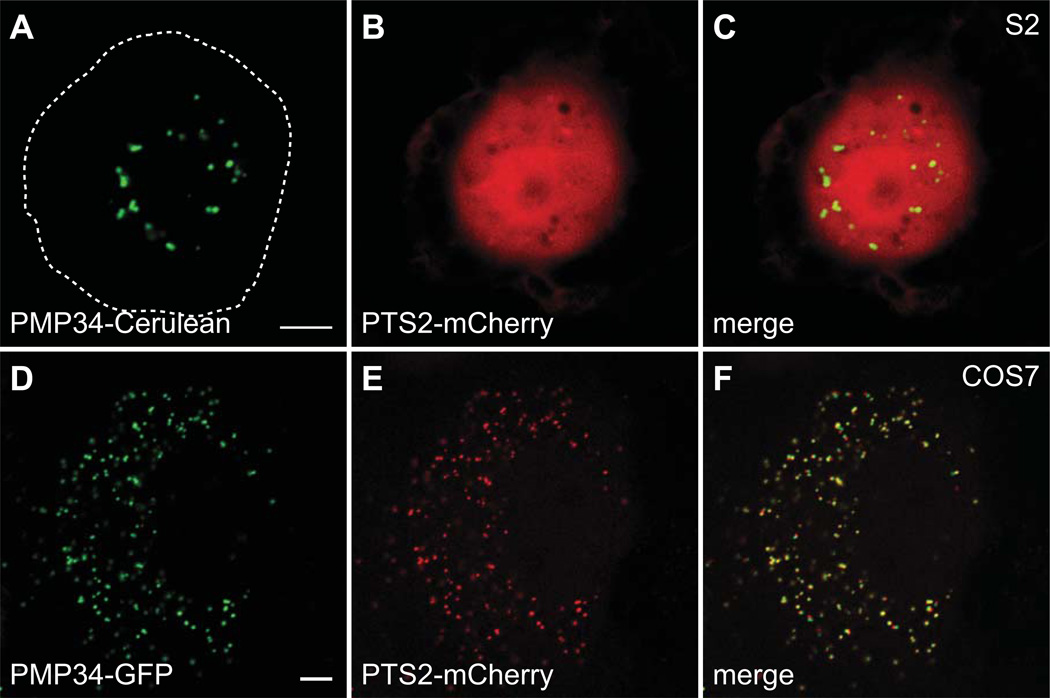
It is currently unclear if Drosophila possesses a functional PTS2 import system. A PEX7 homolog is observed (CG6486, 42% identical), but its function has not been confirmed. Knockdown of pex7 in S2 cells does not affect the import of PTS1-containing cargo, but its effect on PTS2 import in unknown (20). In humans and plants, Pex7 interacts with a long splice form of Pex5 during PTS2 import (55, 56). The Drosophila Pex5 homolog is unlikely to interact with Pex7 since it lacks the region required for this interaction. Additionally, no clear homologs of the fungal-specific peroxins involved in Pex7-mediated import (Pex18, Pex20, and Pex21)(57–59) are revealed by BLAST searches. No PTS2-containing cargo proteins can be identified in the Drosophila proteome and proteins that contain PTS2 signals in other organisms, such as AGPS, have a PTS1 in Drosophila (Supplemental Figure 2). Interestingly, there is a Drosophila homolog of the human peroxisome leader peptide-processing protease (Tysnd1), which cleaves the PTS2 sequence from cargo after import (60). However, this enzyme could have other substrates such as β-oxidation enzymes, as has been demonstrated in mammals (61). The canonical PTS2 sequence from rat thiolase is not sufficient to localize a fluorescent protein to peroxisomes in S2 cells (Figure 8).
Figure 8. PTS2-mCherry localizes to the cytoplasm in S2 cells and peroxisomes in COS7 cells.
Plasmids encoding PTS2-mCherry and PMP34-Cerulean, under control of the actin 5c promoter, were cotransfected into S2 cells and imaged live by confocal microscopy. (A–C) The mCherry signal does not colocalize with Cerulean and is diffuse throughout the cell indicating that PTS2-mCherry localizes to the cytoplasm. Plasmids encoding PTS2-mCherry and PMP34-GFP, under control of the CMV promoter, were cotransfected into COS7 cells and imaged live by confocal microscopy. (D–F) The PTS2-mCherry signal colocalizes with PMP34-GFP, indicating peroxisomal localization. The approximate S2 cell boundary is highlighted with a dashed white line. Scale bar equals 5 µM.
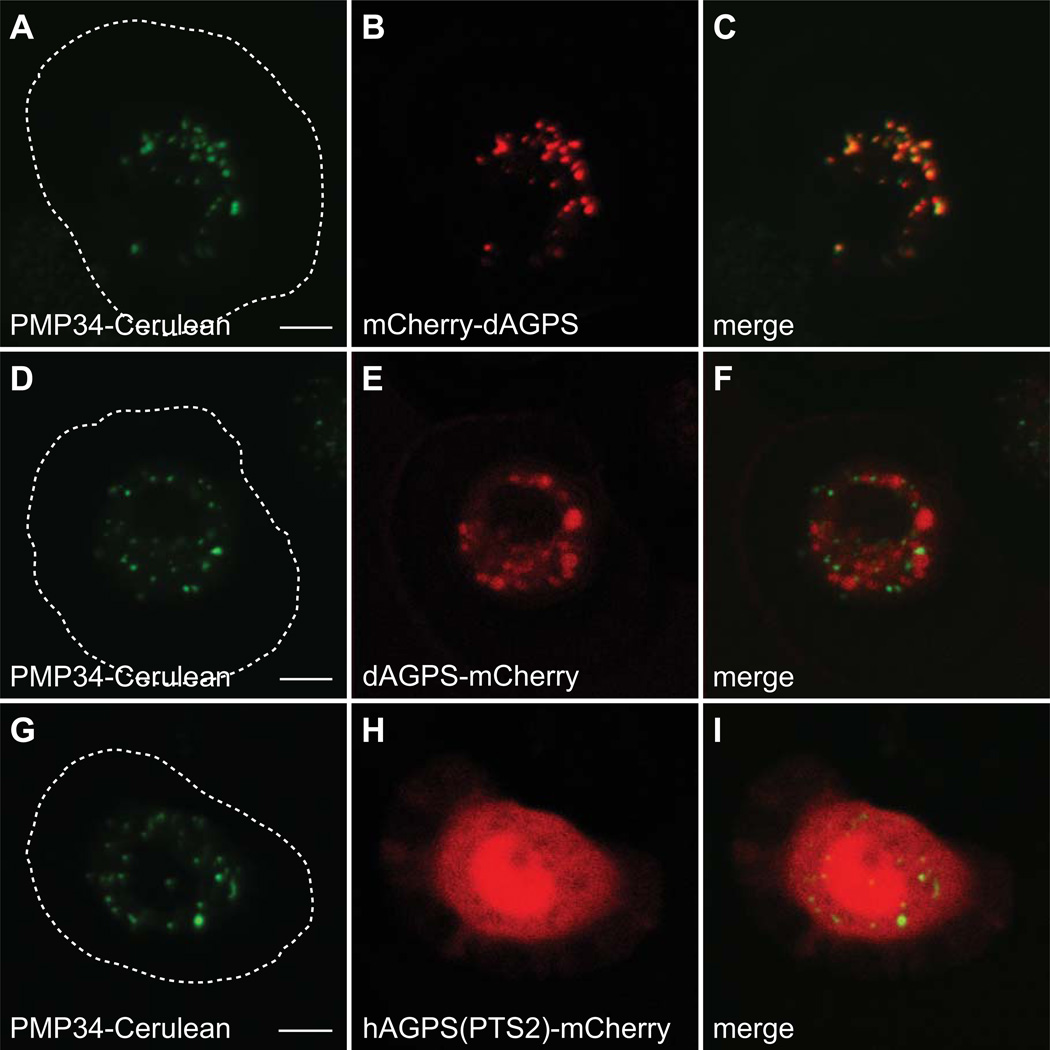
We further examined PTS2 function is S2 cells by confirming the peroxisomal localization signal of alkylglycerone phosphate synthase (AGPS). Human AGPS utilizes an N-terminal PTS2, while Drosophila AGPS appears to target with a PTS1 C-terminal tripeptide. We analyzed the localization of both Drosophila and Human AGPS in S2 as well as COS7 cells by fluorescence confocal microscopy (Figures 9 and 10). Full length Drosophila AGPS (dAGPS) expressed at the C-terminus of mCherry to expose a C-terminal PTS1 was peroxisomal in both S2 cells (Figure 9 A–C) and COS7 cells (Figure 10 A–C). When dAGPS was fused to the N-terminus of mCherry to reveal a potential PTS2 motif, the chimera remained cytoplasmic in both S2 cells (Figure 9 D–F) and COS7 cells (Figure 10 D–F), indicating that no PTS2 motif is present in this protein. We also targeted mCherry to the peroxisome in COS7 cells with the PTS2 motif of humans AGPS (hAGPS). This chimera, containing the N-terminal 72 amino acids of hAGPS (Supplemental Figure 2), fused to the N-terminus of mCherry, was sufficient to localize the fusion protein to peroxisomes in COS7 cells (Figure 10, G–I), but not in S2 cells (Figure 9 G–I). These data confirm that dAGPS sorts to peroxisomes via a PTS1 motif and suggest that S2 cells do not possess a functional PTS2 import system. Like C. elegans, Drosophila may have lost its PTS2 system entirely (62). It is also possible that Drosophila uses a unique PTS2 motif that is not identifiable by sequence comparison.
Figure 9. Alkylglycerone phosphate synthase (AGPS) localization in S2 cells.
Plasmids encoding mCherry-dAGPS, dAGPS-mCherry, or hAGPS(PTS2)-mCherry, under control of the actin 5c promoter, were transfected into S2 cells stably expressing PMP34-Cerulean and imaged live by confocal microscopy. (A–C) The mCherry-dAGPS signal colocalizes with Cerulean, indicating peroxisomal localization due to the PTS1 (AKL) of dAGPS. (D–F) The dAGPS-mCherry signal does not colocalize with Cerulean, indicating that dAGPS does not sort to peroxisomes via an N-terminal PTS2 motif. (G–I) The hAGPS(PTS2)-mCherry signal does not colocalize with Cerulean, indicating that the PTS2 of hAGPS is not sufficient to target mCherry to peroxisomes in S2 cells. The approximate cell boundary is highlighted with a dashed white line. Scale bar equals 5 µM.
Figure 10. Alkylglycerone phosphate synthase (AGPS) localization in COS7 cells.
Plasmids encoding PMP34-GFP and either mCherry-dAGPS, dAGPS-mCherry, or hAGPS(PTS2)-mCherry, under control of the CMV promoter, were cotransfected into COS7 cells and imaged live by confocal microscopy. (A–C) The mCherry-dAGPS signal colocalizes with GFP, indicating peroxisomal localization due to the PTS1 (AKL) of dAGPS. (D–F) The dAGPS-mCherry signal does not colocalize with GFP, indicating that dAGPS does not sort to peroxisomes via an N-terminal PTS2 motif. (G–I) The hAGPS(PTS2)-mCherry signal colocalizes with Cerulean, indicating that the PTS2 of hAGPS is sufficient to target mCherry to peroxisomes in COS7 cells. Scale bar equals 5 µM.
Reactive oxygen metabolism
Peroxisomes represent a source of ROS and a source of antioxidant enzymes (63). Many oxidases, including those involved in β-oxidation, generate hydrogen peroxide (H2O2) and multiple peroxidases, such as catalase are present in the organelle to metabolize H2O2. Other sources of ROS include superoxide (O2•−) generated by Xanthine oxidase. O2•− can be metabolized by Superoxide dismutase. Manganese Sod (MnSod) localizes to the cytoplasm and mitochondria, while Copper zinc Sod (Cu/ZnSod) has been detected in the cytoplasm and peroxisomes (64, 65). Mammalian Cu/ZnSod does not contain a canonical PTS, but “piggybacks” into the organelle by interacting with a PTS1-containing protein, Copper chaperone for Sod (CCS, (66)). Drosophila Cu/ZnSod (CG11793, Sod) terminates in the C-terminal tripeptide -AKV suggesting that it contains its own PTS1. In fact, the 10 C-terminal amino acids of Sod are sufficient to dually localize a fluorescent protein to the cytoplasm and peroxisomes in S2 cells (Figure 11 A–C). If the PTS1 is altered from –AKV to –AKL the localization of the fusion protein shifts to almost completely peroxisomal (Figure 11 D–F). If the last three amino acids are removed (ΔAKV), the fusion protein stays in the cytoplasm (Figure 11 G–I). This motif is non-canonical and may interact weakly with the Pex5 receptor. If so, this weak interaction could be responsible for its dual localization. The Drosophila CCS homolog (CG17753) has a C-terminal –QKL sequence, which may also function as a PTS1. When fused to the C-terminus of mCherry, the 10 C-terminal amino acids of dCCS are sufficient to dually localize the fusion protein to the cytoplasm and peroxisomes (Figure 11 J–L). Peroxisomal localization with this sequence (QKL) is somewhat surprising and suggests that the PTS1 motif in Drosophila may display some organism-specific plasticity. Furthermore, these data indicate that Sod localizes to peroxisomes via a PTS1 motif. It is unclear if Sod can “piggyback” into peroxisome by interacting with dCCS, but this combination of “weak” PTS1 sequences may be enhanced by dimerization prior to translocation.
Figure 11. Cu/Zn superoxide dismutase carries a PTS1 motif that partially localizes mCherry to peroxisomes.
Plasmids encoding PMP34-Cerulean and either mCherry-Sod-PTS1(wild type), mCherry-Sod-PTS1(AKL), or mCherry-Sod-PTS1(ΔAKV) under the control of the actin 5c promoter were cotransfected into S2 cells and imaged by live confocal microscopy. (A–C) The wild-type PTS1 of Cu/Zn Sod localizes mCherry to both the cytoplasm and peroxisomes. Changing the PTS1 to AKL (D–F), shifts mCherry localization to the peroxisome exclusively. Removing the PTS1 (G–I) results in a cytoplasmic localization of mCherry. The PTS1 of Cu/Zn Sod may be a non-optimal sequence and lead to dual localization of this protein. (J–L) Plasmids encoding mCherry-SKL and GFP-CCS-PTS1 under the control of the actin 5c promoter were cotransfected into S2 cells and imaged by live confocal microscopy. The 10 C-terminal amino acids of Copper chaperone for Sod (CCS, CG17753) fused to mCherry is also dually localized to the cytoplasm and peroxisomes. The approximate cell boundary is highlighted with a dashed white line. Scale bar equals 5 µM.
Other peroxisomal functions
Peroxisomes also conduct other metabolic business in addition to the selected pathways highlighted above. This includes metabolism of amino acid, nitrogen, polyamines, glyoxylate and perhaps isoprenes (1, 67, 68). Supplemental Table 1 lists 57 other proteins implicated in peroxisomal functions. While many have canonical PTS1 sorting signals, others do not (63, 69). This observation suggests that some peroxisomal pathways may not be conserved between flies and humans
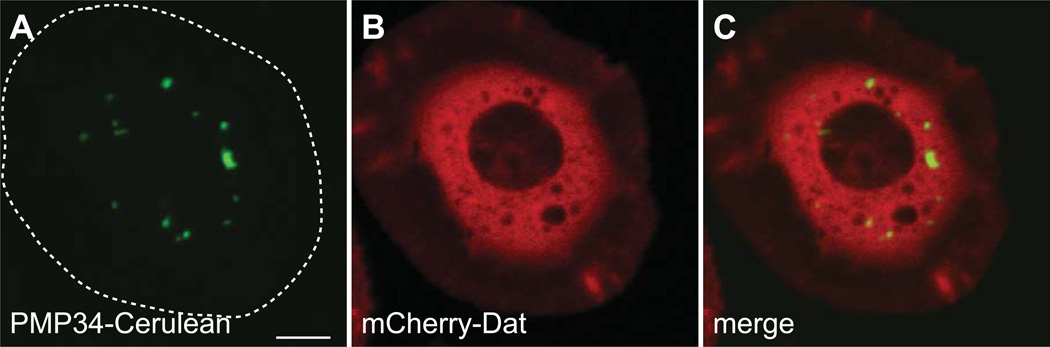
The variability of the canonical PTS1 targeting motif has not been examined in Drosophila making precise identification based on sequence comparison more difficult. The relatively loose motif ([A/C/H/K/N/P/S/T]–[H/K/N/Q/R/S]–[A/F/I/L/M/V]) used in previous PTS1 predictions identified some protein that may or may not be peroxisomal. This possibility is further exemplified by the partial peroxisomal localization of dCCS (Figure 11 J–L) by the PTS1 variant QKL. To better contextualize the PTS1 signal in flies, we determined the subcellular localization of another predicted weak PTS1, TKL, in the protein Dopamine N-acetyltransferase, or Dat (CG3318). An mCherry fusion to Dat localized primarily to the cytoplasm with little or no colocalization with GFP-SKL (Figure 12).
Figure 12. A mCherry-Dopamine N-acetyltransferase (CG3318) Fusion Protein localizes to the cytoplasm in S2 Cells.
Plasmids encoding mCherry-CG3318 and PMP34-Cerulean, under control of the actin 5c promoter, were cotransfected into S2 cells and imaged live by confocal microscopy. (A–C) The mCherry signal does not colocalize with Cerulean and is diffuse throughout the cell indicating that mCherry-CG3318 localizes to the cytoplasm. The approximate cell boundary is highlighted with a dashed white line. Scale bar equals 5 µM.
We have identified all of the major vertebrate peroxisomal pathways in Drosophila and confirmed peroxisomal localization of five candidate enzymes. Analysis of PTS1 sorting suggests that the prototypical PTS1 signals –SKL, -AKL and -AKV direct proteins to the peroxisome. Additionally, and unpredicted variant (–QKL) sorts to the peroxisome, but a loose variant in other systems (-TKL) does not. Finally, we conclude that Drosophila does not utilize a PTS2 motif given the absence of PTS2-containing proteins and the cytoplasmic localization of two independent PTS2-mCherry chimeras.
Materials and Methods
Bioinformatics
Sequences were aligned and phylograms were generated by ClustalW2 hosted by the European bioinformatics Institute (EBI http://www.ebi.ac.uk/Tools/msa/clustalw2/).
Cloning
To generate pJM664 (mCherry-SKL in pAc5.1/V5-His), mCherry was amplified by PCR using the primers KpnI-Kozak-GFP (GAGGTACCAACATGGTGAGCAAGGGCGAG) and eYFP-PTS1-XbaI (GCTCTAGATTACAACTTCGAC TTAGTCTCAGGCGGGTTCTTCTTGTACAGCTCGTCCATG). The mCherry PCR product was digested with KpnI and XbaI and ligated into pAc5.1/V5-His cut with the same enzymes. pJM664 expresses mCherry with the C-terminal 10 amino acids of CG1041 (KNPPETKSKL)
To generate pJM621 (PMP34-cerulean in pAc5.1/V5-His), cerulean was amplified by PCR using the primers NotI-GFP (GCGGCCGCAACCATGGTGAGCAAGG) and GFP-XhoI (CTCGAGTTACTTGTACAGCTCGTCC) and cloned into pCR-Blunt II-TOPO (Invitrogen). Drosophila PMP34 (CG32250) was amplified by PCR from the Drosophila Genomic Resource Center (DGRC) clone RE36975 using the primers KpnI-EcoRI-Dm_PMP34 (GGTACCGAATTCACAAAATGGTGGCCCCCTCG) and Dm_PMP34-NotI (GCGGCCGCGTTGCGCTTAA GCAGC) and cloned into pCR-Blunt II-TOPO (Invitrogen). Cerulean was cut from pCR-Blunt II-TOPO with NotI and XhoI and ligated into pAc5.1/V5-His cut with the same enzymes. PMP34 was cut from pCR-Blunt II-TOPO with KpnI and NotI and ligated into Cerulean-pAc5.1/V5-His cut with the same enzymes.
To generate pJM659 (PTS2-mCherry in pAc5.1/V5-His), 5’ phosphorylated oligos EcoRI-PTS2-NotI_Top (AATTC ACAAAATGCACCGCCTGCAGGTGGTGCTGGGCCACCTGGC) and EcoRI-PTS2-NotI_Bottom (GGCCGCC AGGTGGCCCAGCACCACCTGCAGGCGGTGCATTTTGTG) were annealed and ligated into pAc5.1/V5-His cut with EcoRI and NotI to generate pJM648. mCherry was amplified by PCR using the primers NotI-GFP and GFP-XhoI, cut with NotI and XhoI, and ligated into pJM648 cut with the same enzymes.
To generate the mCherry fusions with predicted peroxisomal genes, mCherry was amplified by PCR using the primers GFP-No Stop-KpnI (CGGGTACCCTTGTACAGCTCGTCC) and HindIII-Kozak-GFP (GCAAGCTTCAAAATGGTGAGCAAGG GCGAG), cut with HindIII and KpnI, and ligated into pAc5.1/V5-His cut with the same enzymes. The acyl-CoA oxidase (CG17544) ORF was amplified by PCR from the DGRC clone SD05719 using the primers KpnI-CG17544 (GAGGTACCATGGGCAGCGAGGACAC AAG) and CG17544-NotI (GTGCGGCCGCTCAA AGCTTGGACTGGG). The dopamine N-acetyltransferase (CG3318) ORF was amplified by PCR from the DGRC clone GH12636 using the primers KpnI-CG3318 (GAGGTACCATGGAAGTGCAGAAGC TGCCG) and CG3318-XhoI (CGCTCGAGCTA CAGCTTGGTCTGCGC). The L-bifunctional protein (CG4389) ORF was amplified by PCR from the DGRC clone GH12558 using the primers KpnI-CG4389-PB (GAGGTACCATGTCCACGAATCCCGCACCGG) and CG4389-XhoI (CGCTCGAGCTACAACTTCGAGGA GCCAG). PCR products were digested with KpnI and either XhoI or NotI and ligated into mCherry-pAc5.1/V5-His cut with the same enzymes.
To generate the mCherry-Sod(PTS1) and mCherry-CCS(PTS1) clones, mCherry was amplified by PCR using and the forward primer KpnI-Kozak-GFP (GAGGTACCAACATGGTGAGCAAGGGCGAG) and each of the reverse primers GFP_SOD-AKV_XbaI (CGTCTAGACTAGACCTTGGCAATGCCAATAACGCCGCACCCCTTGTACAGCTCGTCC), GFP_SOD-AKL_XbaI (CGTCTAGACTACAGCTTGGCAATGCCAATAACGCCGCACCCCTTGTACAGCTCGTCC), and GFP_SOD∆AKV_XbaI (CGTCTAGACTAAATGCCAATAACGCCGCACCCCTTGTACAGCTCGTCC). GFP was amplified by PCR using the primers KpnI-Kozak-GFP and GFP_CCS-QKL_XbaI (CGTCTAGACTACAGCTTTTGTGAGCGCTCCTTGCCAGCCAGCTTGTACAGCTCGTCC). PCR products were digested with KpnI and XbaI and ligated into pAc5.1/V5-His cut with the same enzymes.
To generate pJM896 (mCherry-dAGPS in pAC5.1/V5-His), mCherry was amplified by PCR using the primers EagI-mCherry (TAcggccgATGGTGAGCAAGGGC) and mCherry-NS-XhoI (TACTCGAGCTTGTACAGCTCGTCCATGC) and ligated into pGEM-T Easy (Promega) using manufacturer’s protocol. mCherry was digested from pGEM-T Easy using EagI and XhoI and ligated into pAC5.1/V5-His digested with NotI and XhoI to generate pJM894. dAGPS was amplified by PCR using the primers XhoI-dAGPS (TActcgagATGGCAGCCAAGCGG) and dAGPS-S-XbaI (GCTCTAGACTACAATTTGGCCTTTGGTGGTG) and ligated into pGEM-T Easy. dAGPS was digested from pGEM-T Easy with XhoI and XbaI and ligated into pJM894.
To generate pJM901 (dAGPS-mCherry in pAC5.1/V5-His), mCherry was amplified by PCR using the primers XhoI-mCherry (TActcgagATGGTGAGCAAGGGC) and mCherry-S-XbaI (TTTCTAGATTACTTGTACAGCTCGTCCATGC), digested with XhoI and XbaI and ligated into pAC5.1/V5-His cut with the same enzymes to generate pJM899. dAGPS was amplified by PCR using the primers EagI-dAGPS (TAcggccgATGGCAGCCAAGCGG) and dAGPS-NS-XhoI (GCCTCGAGCAATTTGGCCTTTGGTGGTG) and ligated into pGEM-T Easy. dAGPS was digested from pGEM-T Easy with EagI and XhoI and ligated into pJM899 digested with NotI and XhoI.
To generate pJM961 (hAGPS(PTS2) in pAC5.1/V5-His), the PTS2 sequence of hAGPS (cggccgATGGCGGAGGCGGCGGCTGCAGCGGGTGGGACTGGCTTGGGCGCGGGCGCGAGCTACGGGTCTGCAGCGGACCGGGACCGGGACCCGGACCCGGACCGCGCCGGGCGGAGGCTGCGGGTTCTCTCTGGCCATCTGCTGGGCCGGCCCCGGGAGGCTCTGAGTACCAATGAGTGCAAAGCGCGGAGAGCCGCGTCGGCGGCCACGGCAGCGCCCACGctcgag) was synthesized by IDT and supplied in pIDTSMART-AMP (pJM959). The hAGPS(PTS2) was digested from pJM959 with EagI and XhoI and ligated into pJM899 cut with NotI and XhoI.
To generate pJM933 (mCherry-dAGPS in pcDNA3), pJM896 was digested with EagI and XbaI and ligated into pcDNA3 cut with NotI and XbaI. To generate pJM926 (dAGPS-mCherry in pcDNA3), pJM901 was digested with EagI and XbaI and ligated into pcDNA3 cut with NotI and XbaI. To generate pJM940 (PTS2-mCherry in pcDNA3.1), pJM659 was digested with KpnI and XhoI and ligated into pJM850 (Venus-Atlastin in pCDNA 3.1) digested with the same enzymes. mCherry was digested from pJM899 with XhoI and XbaI and ligated pcDNA3 cut with the same enzymes to generate pJM944. To generate pJM962 (hAGPS(PTS2)-mCherry in pcDNA3) hAGPS(PTS2) was cut from pJM959 with EagI and XhoI and ligated into pJM944 cut with NotI and XhoI.
The Human PMP34 in pEGFP-N1 plasmid was a gift from Dr. Peter Kim.
In vivo imaging of predicted peroxisomal proteins
Plasmids were transfected into S2 cells (DGRC) using Fectofly II (Polyplus) or TransIT-2020 (Mirus) as per manufacturer’s protocols. Stable cell lines were cotransfected with pCoBlast (Invitrogen) and grown in Blasticidin-containing media. Cells were adhered to a Concanavalin A-coated coverslip and imaged live on a Zeiss LSM 510 confocal microscope using a 63× oil immersion objective. mCherry was excited with a 543 nm helium-neon laser and a HFT UV/488/543/633 primary dichroic. mCherry emission was filtered with a LP560 filter before collection. Cerulean was excited with a 458 nm argon laser and a HFT 458/514 primary dichroic. Cerulean emission was filtered with NFT 545 and BP 480–520 filters before collection. GFP was excited with a 488 nm argon laser and a HFT 488 primary dichroic. GFP emission was filtered with NFT 545 and BP 500–530 IR filters before collection.
Plasmids were transfected into COS7 cells (ATCC) using TransIT-COS (Mirus) transfection reagent as per manufacturer’s protocol. The cells were plated at 60,000 cells per mL the day before transfection on a coverslip in a 24-well plate and grown in DMEM with 10% FBS and penicillin 100IU/streptromycin 100 µg/mL. Cells were imaged live on a Zeiss LSM 710 confocal microcope using a 100X oil immersion objective. mCherry was excited with a 543 helium-neon laser. The emission was filtered with a main beam splitter 458/543 and 578–696nm wavelengths were collected. GFP was excited at 488nm. The emission was filtered with a main bean splitter 488 and 493–598nm wavelengths were collected. Cerulean was excited with 405-30 diode laser. The emission was filtered with a main beam splitter 405nm and 454–581 wavelengths were collected.
Images were processed in Fiji (http://fiji.sc) and Illustrator (Adobe).
Supplementary Material
Acknowledgments
We would like to thank Dr. Peter Kim for the PMP34-GFP plasmid and Dr. Michael Stern, James Summerville, Tyler Moss, and Tanvi Desai for helpful comments on the manuscript. Work in the McNew lab is supported by funds from the National Institutes of Health (Grant GM071832), the G. Harold and Leila Y. Mathers Foundation, and the National Science Foundation, Center for Biological and Environmental Nanotechnology (EEC-0647452). Confocal microscopy of COS7 cells was performed on equipment obtained through a Shared Instrumentation Grant from the National Institute of Health (S10RR026399-01).
Footnotes
Supporting Information
Figure S1. Peroxisomal β-oxidation accessory pathways. peroxisomal β-oxidation accessory pathways are illustrated with the CG numbers for each Drosophila protein listed at each step. The CG numbers are listed in descending order with the best hit(s) in bolded, red text. The percent identities of the Drosophila and human homologs are listed below. PECI: 34%, ECH1: 45%, LBP/DBP: 27–53%, DECR2/PECR: 13–27%.
Figure S2. Drosophila and many invertebrates use a PTS1 motif for targeting AGPS to peroxisomes. ClustalW2 was used to align AGPS protein sequences from many species. All vertebrates and a few invertebrates use a PTS2 motif for peroxisomal targeting of AGPS. Drosophila and most invertebrates use a PTS1 motif instead. Accession numbers used in the alignment are available upon request.
Figure S3. Comparision of PeroxiP with the results from BLAST searches. Many predicted peroxisomal proteins identified by PeroxiP were also found using BLAST. However, each technique produced unique results.
References
- 1.Wanders RJ, Waterham HR. Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem. 2006;75:295–332. doi: 10.1146/annurev.biochem.74.082803.133329. [DOI] [PubMed] [Google Scholar]
- 2.Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 2005;122(1):85–95. doi: 10.1016/j.cell.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 3.Platta HW, Erdmann R. Peroxisomal dynamics. Trends Cell Biol. 2007;17(10):474–484. doi: 10.1016/j.tcb.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Gould SG, Keller GA, Subramani S. Identification of a peroxisomal targeting signal at the carboxy terminus of firefly luciferase. J Cell Biol. 1987;105(6 Pt 2):2923–2931. doi: 10.1083/jcb.105.6.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dammai V, Subramani S. The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell. 2001;105(2):187–196. doi: 10.1016/s0092-8674(01)00310-5. [DOI] [PubMed] [Google Scholar]
- 6.McNew JA, Goodman JM. An oligomeric protein is imported into peroxisomes in vivo. J Cell Biol. 1994;127(5):1245–1257. doi: 10.1083/jcb.127.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erdmann R, Schliebs W. Peroxisomal matrix protein import: the transient pore model. Nat Rev Mol Cell Biol. 2005;6(9):738–742. doi: 10.1038/nrm1710. [DOI] [PubMed] [Google Scholar]
- 8.Meinecke M, Cizmowski C, Schliebs W, Kruger V, Beck S, Wagner R, Erdmann R. The peroxisomal importomer constitutes a large and highly dynamic pore. Nat Cell Biol. 2010 doi: 10.1038/ncb2027. [DOI] [PubMed] [Google Scholar]
- 9.Swinkels BW, Gould SJ, Bodnar AG, Rachubinski RA, Subramani S. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 1991;10(11):3255–3262. doi: 10.1002/j.1460-2075.1991.tb04889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones JM, Morrell JC, Gould SJ. PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. The Journal of Cell Biology. 2004;164(1):57–67. doi: 10.1083/jcb.200304111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang Y, Morrell JC, Jones JM, Gould SJ. PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins. J Cell Biol. 2004;164(6):863–875. doi: 10.1083/jcb.200311131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Zand A, Braakman I, Tabak HF. Peroxisomal Membrane Proteins Insert into the Endoplasmic Reticulum. Mol Biol Cell. 2010 doi: 10.1091/mbc.E10-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinberg SJ, Dodt G, Raymond GV, Braverman NE, Moser AB, Moser HW. Peroxisome biogenesis disorders. Biochimica Et Biophysica Acta. 2006;1763(12):1733–1748. doi: 10.1016/j.bbamcr.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Baes M, Van Veldhoven PP. Generalised and conditional inactivation of Pex genes in mice. Biochim Biophys Acta. 2006;1763(12):1785–1793. doi: 10.1016/j.bbamcr.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 15.Bilen J, Bonini NM. Drosophila as a model for human neurodegenerative disease. Annu Rev Genet. 2005;39:153–171. doi: 10.1146/annurev.genet.39.110304.095804. [DOI] [PubMed] [Google Scholar]
- 16.Sánchez-Martínez A, Luo N, Clemente P, Adán C, Hernández-Sierra R, Ochoa P, Fernández-Moreno MA, Kaguni LS, Garesse R. Modeling human mitochondrial diseases in flies. Biochimica Et Biophysica Acta. 2006;1757(9–10):1190–1198. doi: 10.1016/j.bbabio.2006.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arquier N, Leopold P. Fly foie gras: modeling fatty liver in Drosophila. Cell Metab. 2007;5(2):83–85. doi: 10.1016/j.cmet.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Reiter LT, Potocki L, Chien S, Gribskov M, Bier E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Research. 2001;11(6):1114–1125. doi: 10.1101/gr.169101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, Liu Z, Huang X. Drosophila models of peroxisomal biogenesis disorder: peroxins are required for spermatogenesis and very-long-chain fatty acid metabolism. Hum Mol Genet. 2010;19(3):494–505. doi: 10.1093/hmg/ddp518. [DOI] [PubMed] [Google Scholar]
- 20.Mast FD, Li J, Virk MK, Hughes SC, Simmonds AJ, Rachubinski RA. A Drosophila model for the Zellweger spectrum of peroxisome biogenesis disorders. Disease Models & Mechanisms. 2011 doi: 10.1242/dmm.007419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakayama M, Sato H, Okuda T, Fujisawa N, Kono N, Arai H, Suzuki E, Umeda M, Ishikawa HO, Matsuno K. Drosophila carrying pex3 or pex16 mutations are models of zellweger syndrome that reflect its symptoms associated with the absence of peroxisomes. PLoS One. 2011;6(8):e22984. doi: 10.1371/journal.pone.0022984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schluter A, Real-Chicharro A, Gabaldon T, Sanchez-Jimenez F, Pujol A. PeroxisomeDB 2.0: an integrative view of the global peroxisomal metabolome. Nucleic Acids Res. 2010;38(Database issue):D800–D805. doi: 10.1093/nar/gkp935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, Zhang H. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37(Database issue):D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Emanuelsson O, Elofsson A, von Heijne G, Cristobal S. In silico prediction of the peroxisomal proteome in fungi, plants and animals. J Mol Biol. 2003;330(2):443–456. doi: 10.1016/s0022-2836(03)00553-9. [DOI] [PubMed] [Google Scholar]
- 26.Hawkins J, Mahony D, Maetschke S, Wakabayashi M, Teasdale RD, Boden M. Identifying novel peroxisomal proteins. Proteins. 2007;69(3):606–616. doi: 10.1002/prot.21420. [DOI] [PubMed] [Google Scholar]
- 27.Watkins PA, Maiguel D, Jia Z, Pevsner J. Evidence for 26 distinct acyl-coenzyme A synthetase genes in the human genome. J Lipid Res. 2007;48(12):2736–2750. doi: 10.1194/jlr.M700378-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Jia Z, Pei Z, Maiguel D, Toomer CJ, Watkins PA. The fatty acid transport protein (FATP) family: very long chain acyl-CoA synthetases or solute carriers? J Mol Neurosci. 2007;33(1):25–31. doi: 10.1007/s12031-007-0038-z. [DOI] [PubMed] [Google Scholar]
- 29.Min KT, Benzer S. Preventing neurodegeneration in the Drosophila mutant bubblegum. Science. 1999;284(5422):1985–1988. doi: 10.1126/science.284.5422.1985. [DOI] [PubMed] [Google Scholar]
- 30.Conley C, Rohwer-Nutter D, Blair S. Program and Abstracts 40th Annual Drosophila Research Conference. Vol. 1999. Bellevue: 1999. Identification of the Drosophila acyl-CoenzymeA oxidase gene; p. 403C. [Google Scholar]
- 31.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2(4):953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 32.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241(3):779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 33.Seedorf U, Ellinghaus P, Roch Nofer J. Sterol carrier protein-2. Biochim Biophys Acta. 2000;1486(1):45–54. doi: 10.1016/s1388-1981(00)00047-0. [DOI] [PubMed] [Google Scholar]
- 34.Stolowich NJ, Petrescu AD, Huang H, Martin GG, Scott AI, Schroeder F. Sterol carrier protein-2: structure reveals function. Cell Mol Life Sci. 2002;59(2):193–212. doi: 10.1007/s00018-002-8416-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomiichiro K, Satoru K, Masukichi O. Regional expression of the transcript encoding sterol carrier protein x-related thiolase and its regulation by homeotic genes in the midgut of<i>Drosophila</i>embryos. Development Growth & Differentiation. 1996;38(4):373–381. doi: 10.1046/j.1440-169X.1996.t01-3-00005.x. [DOI] [PubMed] [Google Scholar]
- 36.Leenders F, Tesdorpf JG, Markus M, Engel T, Seedorf U, Adamski J. Porcine 80-kDa protein reveals intrinsic 17 beta-hydroxysteroid dehydrogenase, fatty acyl-CoA-hydratase/dehydrogenase, and sterol transfer activities. J Biol Chem. 1996;271(10):5438–5442. doi: 10.1074/jbc.271.10.5438. [DOI] [PubMed] [Google Scholar]
- 37.Sardiello M, Licciulli F, Catalano D, Attimonelli M, Caggese C. MitoDrome: a database of Drosophila melanogaster nuclear genes encoding proteins targeted to the mitochondrion. Nucleic Acids Res. 2003;31(1):322–324. doi: 10.1093/nar/gkg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mackay WJ, Bewley GC. The genetics of catalase in Drosophila melanogaster: isolation and characterization of acatalasemic mutants. Genetics. 1989;122(3):643–652. doi: 10.1093/genetics/122.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orr WC, Arnold LA, Sohal RS. Relationship between catalase activity, life span and some parameters associated with antioxidant defenses in Drosophila melanogaster. Mech Ageing Dev. 1992;63(3):287–296. doi: 10.1016/0047-6374(92)90006-y. [DOI] [PubMed] [Google Scholar]
- 40.Orr WC, Sohal RS. Extension of life-span by overexpression of superoxide dismutase and catalase in Drosophila melanogaster. Science. 1994;263(5150):1128–1130. doi: 10.1126/science.8108730. [DOI] [PubMed] [Google Scholar]
- 41.Beard ME, Holtzman E. Peroxisomes in wild-type and rosy mutant Drosophila melanogaster. Proc Natl Acad Sci U S A. 1987;84(21):7433–7437. doi: 10.1073/pnas.84.21.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Southall TD, Terhzaz S, Cabrero P, Chintapalli VR, Evans JM, Dow JA, Davies SA. Novel subcellular locations and functions for secretory pathway Ca2+/Mn2+-ATPases. Physiol Genomics. 2006;26(1):35–45. doi: 10.1152/physiolgenomics.00038.2006. [DOI] [PubMed] [Google Scholar]
- 43.Gelbart WM, Emmert DB. FlyBase High Throughput Expression Pattern Data Beta Version. 2010 [Google Scholar]
- 44.Wasbrough ER, Dorus S, Hester S, Howard-Murkin J, Lilley K, Wilkin E, Polpitiya A, Petritis K, Karr TL. The Drosophila melanogaster sperm proteome-II (DmSP-II) J Proteomics. 2010;73(11):2171–2185. doi: 10.1016/j.jprot.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 45.Jansen GA, Wanders RJ. Alpha-oxidation. Biochim Biophys Acta. 2006;1763(12):1403–1412. doi: 10.1016/j.bbamcr.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 46.Lloyd MD, Darley DJ, Wierzbicki AS, Threadgill MD. alpha-Methylacyl-CoA racemase - an 'obscure' metabolic enzyme takes centre stage. FEBS J. 2008 doi: 10.1111/j.1742-4658.2008.06290.x. [DOI] [PubMed] [Google Scholar]
- 47.Mukherji M, Schofield CJ, Wierzbicki AS, Jansen GA, Wanders RJ, Lloyd MD. The chemical biology of branched-chain lipid metabolism. Prog Lipid Res. 2003;42(5):359–376. doi: 10.1016/s0163-7827(03)00016-x. [DOI] [PubMed] [Google Scholar]
- 48.Amery L, Fransen M, De Nys K, Mannaerts GP, Van Veldhoven PP. Mitochondrial and peroxisomal targeting of 2-methylacyl-CoA racemase in humans. J Lipid Res. 2000;41(11):1752–1759. [PubMed] [Google Scholar]
- 49.Ferdinandusse S, Denis S, L IJ, Dacremont G, Waterham HR, Wanders RJ. Subcellular localization and physiological role of alpha-methylacyl-CoA racemase. J Lipid Res. 2000;41(11):1890–1896. [PubMed] [Google Scholar]
- 50.Jiang Z, Fanger GR, Woda BA, Banner BF, Algate P, Dresser K, Xu J, Chu PG. Expression of alpha-methylacyl-CoA racemase (P504s) in various malignant neoplasms and normal tissues: astudy of 761 cases. Hum Pathol. 2003;34(8):792–796. doi: 10.1016/s0046-8177(03)00268-5. [DOI] [PubMed] [Google Scholar]
- 51.Brites P, Waterham HR, Wanders RJ. Functions and biosynthesis of plasmalogens in health and disease. Biochim Biophys Acta. 2004;1636(2–3):219–231. doi: 10.1016/j.bbalip.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 52.Chase BA, Kunwar PS, Markopoulou K. Drosophila as a model system to study disease associated with defects in peroxisomal assembly and function; Program and Abstracts 42nd Annual Drosophila Research Conference; Washington DC. 2001. p. 763A. [Google Scholar]
- 53.Cheng JB, Russell DW. Mammalian wax biosynthesis. I. Identification of two fatty acyl-Coenzyme A reductases with different substrate specificities and tissue distributions. J Biol Chem. 2004;279(36):37789–37797. doi: 10.1074/jbc.M406225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Distel B, Erdmann R, Gould SJ, Blobel G, Crane DI, Cregg JM, Dodt G, Fujiki Y, Goodman JM, Just WW, Kiel JA, Kunau WH, Lazarow PB, Mannaerts GP, Moser HW, et al. A unified nomenclature for peroxisome biogenesis factors. J Cell Biol. 1996;135(1):1–3. doi: 10.1083/jcb.135.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woodward AW, Bartel B. The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol Biol Cell. 2005;16(2):573–583. doi: 10.1091/mbc.E04-05-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Otera H, Harano T, Honsho M, Ghaedi K, Mukai S, Tanaka A, Kawai A, Shimizu N, Fujiki Y. The mammalian peroxin Pex5pL, the longer isoform of the mobile peroxisome targeting signal (PTS) type 1 transporter, translocates the Pex7p.PTS2 protein complex into peroxisomes via its initial docking site, Pex14p. The Journal of Biological Chemistry. 2000;275(28):21703–21714. doi: 10.1074/jbc.M000720200. [DOI] [PubMed] [Google Scholar]
- 57.Stein K, Schell-Steven A, Erdmann R, Rottensteiner H. Interactions of Pex7p and Pex18p/Pex21p with the peroxisomal docking machinery: implications for the first steps in PTS2 protein import. Mol Cell Biol. 2002;22(17):6056–6069. doi: 10.1128/MCB.22.17.6056-6069.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sichting M, Schell-Steven A, Prokisch H, Erdmann R, Rottensteiner H. Pex7p and Pex20p of Neurospora crassa function together in PTS2-dependent protein import into peroxisomes. Mol Biol Cell. 2003;14(2):810–821. doi: 10.1091/mbc.E02-08-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Einwachter H, Sowinski S, Kunau WH, Schliebs W. Yarrowia lipolytica Pex20p, Saccharomyces cerevisiae Pex18p/Pex21p and mammalian Pex5pL fulfil a common function in the early steps of the peroxisomal PTS2 import pathway. EMBO Rep. 2001;2(11):1035–1039. doi: 10.1093/embo-reports/kve228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kurochkin IV, Mizuno Y, Konagaya A, Sakaki Y, Schonbach C, Okazaki Y. Novel peroxisomal protease Tysnd1 processes PTS1- and PTS2-containing enzymes involved in beta-oxidation of fatty acids. EMBO J. 2007;26(3):835–845. doi: 10.1038/sj.emboj.7601525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okumoto K, Kametani Y, Fujiki Y. Two proteases, trypsin domain-containing 1 (Tysnd1) and peroxisomal lon protease (PsLon), cooperatively regulate fatty acid beta-oxidation in peroxisomal matrix. J Biol Chem. 2011;286(52):44367–44379. doi: 10.1074/jbc.M111.285197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Motley AM, Hettema EH, Ketting R, Plasterk R, Tabak HF. Caenorhabditis elegans has a single pathway to target matrix proteins to peroxisomes. EMBO Rep. 2000;1(1):40–46. doi: 10.1093/embo-reports/kvd010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schrader M, Fahimi HD. Peroxisomes and oxidative stress. Biochim Biophys Acta. 2006;1763(12):1755–1766. doi: 10.1016/j.bbamcr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 64.Keller GA, Warner TG, Steimer KS, Hallewell RA. Cu,Zn superoxide dismutase is a peroxisomal enzyme in human fibroblasts and hepatoma cells. Proc Natl Acad Sci U S A. 1991;88(16):7381–7385. doi: 10.1073/pnas.88.16.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dhaunsi GS, Gulati S, Singh AK, Orak JK, Asayama K, Singh I. Demonstration of Cu-Zn superoxide dismutase in rat liver peroxisomes. Biochemical and immunochemical evidence. J Biol Chem. 1992;267(10):6870–6873. [PubMed] [Google Scholar]
- 66.Islinger M, Li KW, Seitz J, Volkl A, Luers GH. Hitchhiking of Cu/Zn Superoxide Dismutase to Peroxisomes-Evidence for a Natural Piggyback Import Mechanism in Mammals. Traffic. 2009 doi: 10.1111/j.1600-0854.2009.00966.x. [DOI] [PubMed] [Google Scholar]
- 67.Angermuller S, Islinger M, Volkl A. Peroxisomes and reactive oxygen species, a lasting challenge. Histochem Cell Biol. 2009;131(4):459–463. doi: 10.1007/s00418-009-0563-7. [DOI] [PubMed] [Google Scholar]
- 68.Wanders RJ, Ferdinandusse S, Brites P, Kemp S. Peroxisomes, lipid metabolism and lipotoxicity. Biochim Biophys Acta. 2010;1801(3):272–280. doi: 10.1016/j.bbalip.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 69.Bonekamp NA, Volkl A, Fahimi HD, Schrader M. Reactive oxygen species and peroxisomes: struggling for balance. Biofactors. 2009;35(4):346–355. doi: 10.1002/biof.48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.