Abstract
Chromosomes must be accurately partitioned to daughter cells to prevent aneuploidy, a hallmark of many tumors and birth defects. Kinetochores are the macromolecular machines that segregate chromosomes by maintaining load-bearing attachments to the dynamic tips of microtubules. Here, we present the structure of isolated budding yeast kinetochore particles as visualized by electron microscopy (EM) and electron tomography of negatively stained preparations. The kinetochore appears as a ~126 nm particle containing a large central hub surrounded by multiple outer globular domains. In the presence of microtubules, some particles also have a ring that encircles the microtubule. Our data show that kinetochores bind to microtubules via multivalent attachments and lay the foundation to uncover the key mechanical and regulatory mechanisms by which kinetochores control chromosome segregation and cell division.
Introduction
The generation and survival of all organisms requires the precise partitioning of duplicated chromosomes to daughter cells. Defects in segregation lead to aneuploidy, the state where entire chromosomes are gained or lost. Aneuploidy is a hallmark of tumor cells and has been postulated to be a major factor in the evolution of cancer, and it is also the leading cause of spontaneous miscarriages and hereditary birth defects1,2. It is therefore critical to understand the mechanisms that ensure accurate chromosome segregation and thus maintain genomic stability.
Chromosome segregation requires forces generated by spindle microtubules that are translated into chromosome movement through interactions with kinetochores, highly conserved structures assembled from distinct subcomplexes3-6. The simplest kinetochore is in budding yeast where 38 core structural proteins assemble onto centromeric DNA to form a single microtubule-binding site (Figure 1a)7. Because most subcomplexes are present in multiple copies, the simplest kinetochore contains greater than 250 core proteins, as well as additional regulatory proteins. The majority of yeast kinetochore proteins are conserved, and it is thought that kinetochores in multicellular eukaryotes that bind to multiple microtubules may simply contain repeat units of the budding yeast kinetochore3,8,9. The inner kinetochore contains subcomplexes that directly bind to centromeric DNA, while the outer kinetochore is composed of subcomplexes that mediate microtubule attachment. The major microtubule binding activity of the kinetochore is mediated by KMN, an assembly of the KNL-1, Mis12 and Ndc80 subcomplexes that attaches to microtubules cooperatively10. The yeast-specific Dam1 complex also exhibits microtubule-binding activity, and it has been proposed that the vertebrate Ska1 complex may be an ortholog11-14.
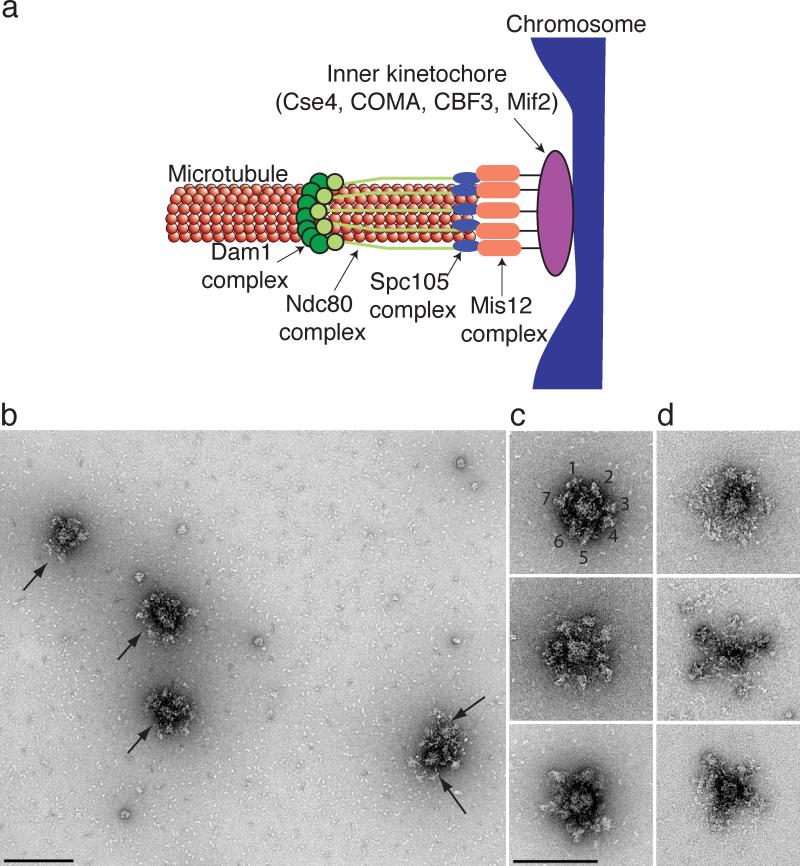
Figure 1. Kinetochore particles contain a central hub surrounded by a number of globular domains.
(a) A model for the budding yeast kinetochore shows that multiple copies of the Dam1, Ndc80, KNL-1 (Spc105) and Mis12 kinetochore subcomplexes mediate binding of the chromosome (blue) to the microtubule. The inner kinetochore contains one or more copies of the Cse4, Mif2, CBF3 and COMA subcomplexes. (b) A field of kinetochore particles in microtubule polymerization buffer was visualized by EM of negatively stained preparations. Five particles (arrows) and other small material are apparent. Note that two particles are touching. Scale bar = 200 nm. (c) Images of representative compact kinetochore particles in microtubule polymerization buffer with lower salt. The globular domains on a single particle in the top panel are numbered. (d). The particles are more extended in higher salt buffer used for purification. Scale bar = 150 nm. Additional particles are presented in Supplementary Figure 1.
Although a number of models have been proposed15-21, the structure of the kinetochore and the mechanism by which it attaches to microtubules is still not clear. Elegant fluorescence microscopy studies have shown that the overall positioning and stoichiometry of kinetochore components is highly conserved9,17,18,22, leading to a proposal for overall kinetochore architecture (Figure 1A). However, it has been difficult to obtain higher resolution information about complete kinetochores. The prevailing picture from electron microscopy studies on vertebrate cells revealed that the kinetochore is three-tiered structure23-26. More recent studies have visualized an outer kinetochore network connected to microtubules, supporting a model whereby multiple weak attachment sites mediate coupling activity21. In one study, peeling microtubule protofilaments can be seen attached to fibrils at the inner kinetochore, leading to the proposal that these fibrils could couple chromosome movement to microtubule depolymerization20. The Dam1 complex forms rings around microtubules in vitro at high concentrations, supporting proposals that envision rings as the major coupling mechanism27,28.
Visualizing the attachment state of kinetochores requires the isolation of large kinetochore assemblies that can be visualized at higher resolution. While progress has been made in elucidating the structure of recombinant kinetochore subcomplexes, they have not yet been reconstituted into larger assemblies suitable for structural work. We previously developed an assay to purify native budding yeast kinetochore particles that contain the majority of core structural components and can maintain attachments to microtubules under force29. Here, we set out to analyze these assemblies in both the presence and absence of microtubules by EM to obtain information about their structure.
Results
Kinetochores contain a hub surrounded by globular domains
Native kinetochore particles were isolated by affinity-capture of the kinetochore component Dsn1-Flag on beads and eluted with a Flag peptide29. To avoid potential cell cycle variability, kinetochores were purified from cells arrested in mitosis by the addition of the microtubule depolymerizing drug benomyl. Eluates were negatively stained and imaged on an electron microscope. The largest structures visible on the grids were approximately 126 +/- 13 nm in length end-to-end (n=88, Fig 1b, arrows). Smaller particles (that may or may not represent kinetochore subcomplexes) were also visible in the background due to the low stringency purification conditions needed to maintain functional and intact kinetochores29,30 (Fig 1b). As a negative control, we purified Dsn1-Flag from ndc80-1 mutant cells that abolish kinetochore function31. We previously found that these particles cannot bind to microtubules and lack most of the outer kinetochore when assayed by silver-stained SDS-PAGE29. Consistent with this, large particles were not detected in these eluates and we instead observed material of variable size and shape on the grids in the ndc80-1 samples (Supplementary Fig 1). Additional controls were prepared from cells lacking a Flag epitope-tagged protein or the Alk1-Flag protein that does not associate with kinetochores. The large particles were missing from these controls, consistent with the initial identification of the large structures as kinetochore particles (data not shown).
The kinetochores contain a 37 +/- 3 nm central hub (n=72) surrounded by a number of globular domains of variable shape with an average diameter of 21 +/- 2 nm (n=97, Figs 1b-d, Supplementary Fig 1). We analyzed the appearance of the kinetochores in two different buffer conditions and found that they appear more compact when incubated in a lower salt buffer compatible with microtubule polymerization (Fig 1c). The majority of kinetochores (n=54) contained five globular domains, although particles with as many as seven (n=7) were also seen (Figs 1c, d, Supplementary Table 1). Particles visualized in buffer containing a higher salt concentration appeared more extended and had a maximum of 5 globular domains radiating (Fig 1d). These data suggest that the particles either lose structural integrity at high salt concentrations, or that they are structurally flexible and can undergo large conformational rearrangements. All of the measurements reported in the manuscript were therefore performed on particles that had been incubated in the lower salt microtubule polymerization buffer.
Kinetochore Assemblies bound to Microtubules
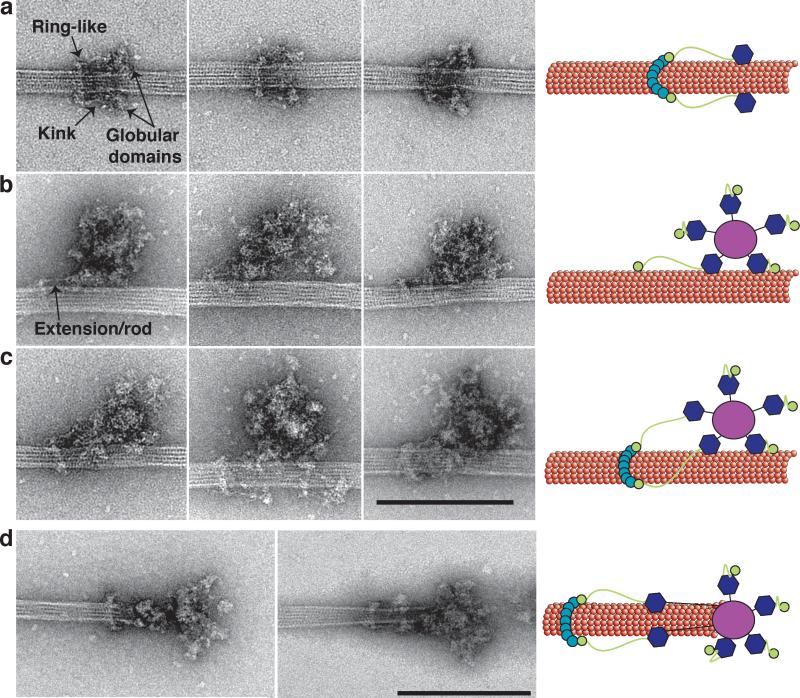
We next visualized the kinetochore particles bound to microtubules. When taxol-stabilized microtubules were incubated with Flag eluate, distinct assemblies became enriched on the microtubules (Fig 2 and Supplementary Fig 2). The first contained a rod-shaped structure oriented parallel to the microtubule with globular domains on one end and a ring-like structure oriented orthogonal to the microtubule at the opposite end (Fig 2a, Supplementary Fig 2 and Supplementary Table 2). The average length of the complex from the ring-like structure to each globular domain is 56 +/- 4 nm (n=128). The ring-like structure has an average outer diameter of 50 +/- 3 nm (n=99, Supplementary Table 2). There is a kink in the rod an average of 25 +/- 2 nm (n= 67) away from the ring (Fig 2a, arrow), and the globular domains often appear to contact the microtubule.
Figure 2. Kinetochore particles bound to taxol-stabilized microtubules.
(a) Representative images of fragments of kinetochore particles (56 nm long) bound to taxol-stabilized microtubules reveal a rod with a kink (arrow) connected to a ring on one end and a globular domain on the opposite end. (b) Large kinetochore particles (126 nm long) bind to microtubules through globular domains and an additional extension/rod (arrow) that emanates from one of the globular domains. (c) Large kinetochore particles bind to microtubules through multiple globular domains and contain an extension that connects to a ring. Scale bar is 200 nm. (d) Two selected images of kinetochores at the tip of taxol-stabilized microtubules. Globular domains extending 50 nm from the central hub bind to the microtubules and are connected to a distal ring 50 nm further. Scale bar is 200 nm. Cartoons on the left schematize the key features of the images.
The other assemblies detected on microtubules contained the 126 nm structures bound to microtubules in either the absence (Fig 2b) or presence (Fig 2c) of the ring-like structure shown in Fig 2a. In both cases, at least two of the globular domains that radiate from the central hub appear to contact the microtubule, suggesting that these domains contain microtubule-binding elements of the kinetochore. The contacts between the globular domains and the microtubules are more apparent in the 3D tomographic reconstructions (see below, Fig. 3). In sharp contrast, the central hub never appears to contact the microtubule directly. When the ring-like structure was missing, a rod-structure that appears similar to the rod in Fig 2a can be seen extending from the 126 nm particle (Fig 2b, arrow). When a ring-like structure is present, this rod touches it in a similar fashion as in the assemblies shown in Fig 2a. It is therefore most likely that the assemblies in Fig 2a result from a portion of the larger particle falling off the microtubule, or else represent a subset of smaller microtubule-binding kinetochore assemblies present in the eluate.
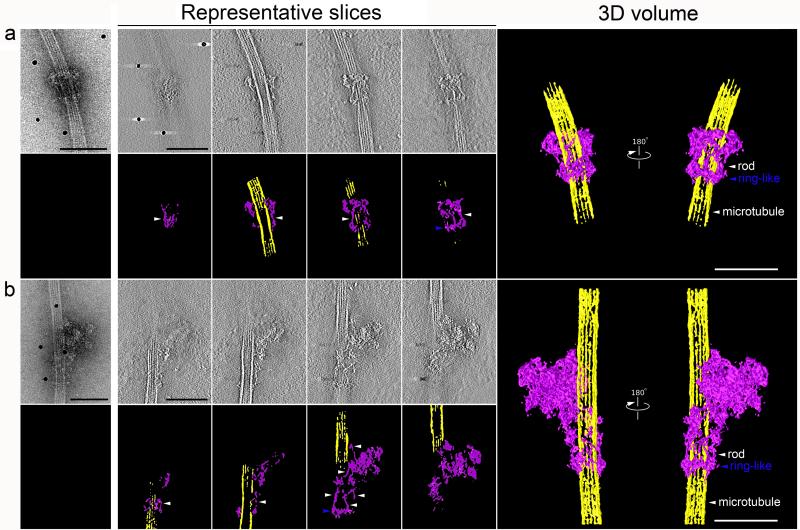
Figure 3. Three-dimensional structures of two types of kinetochore particles bound to a microtubule.
a and b, projection images of the two types of kinetochore assemblies on taxol-stabilized microtubules selected for tomographic reconstruction. Representative slices through the each particle are presented with the corresponding segmentation analysis in color underneath. Finally, 3D reconstruction of this complex by electron tomography is presented on the right. All slices are arranged from left to right traversing through the z-axis from bottom to top with the bottom defined at the grid surface (carbon layer). Both structures contain a ring-like structure (blue arrowhead) and multiple rods (white arrowhead). Scale bars for all panels represent 100 nm. Movies are presented in Supplementary Material.
We also observed a few kinetochores at microtubule tips (Fig 2d). In these cases, binding was only observed in the presence of a ring-like structure (n=8). The structure was similar to the lateral attachments shown in Fig 2c because they also contain a ring connected to a rod with a large globular domain. However, the particle appears to extend an additional ~50 nm from the globular domains to the tip of the microtubule. This suggests that the linkage between globular domains and the central hub is flexible and globular domains can extend outward from the central hub, consistent with our observation that the particles may possess flexible elements (Figs 1c and d).
Particles purified from mutants lack kinetochore structure
We next attempted to assign elements of the kinetochore to specific kinetochore subcomplexes. Repeated attempts at defining components by immuno-gold labeling and negative stain EM have so far not been successful (data not shown), so we used temperature sensitive mutants in kinetochore proteins to analyze the corresponding changes in appearance as an alternative approach. Although 13% of the large particles (n=410) bound to microtubules also contained a ring-like structure when purified from wild-type cells, particles purified from dad1-1 mutant cells that are defective in the Dam1 complex32 were indistinguishable from wild-type images but completely lacked ring-like structures when bound to microtubules (n= 175, Supplementary Table 3, Supplementary Fig 1). The lack of the ring-like structure in the dad1-1 mutant particles bound to microtubules is statistically significant (p=0.001) and strongly suggests that the rings correspond to the Dam1 complex. Consistent with this, previous studies have determined that the Dam1 complex is not required for lateral attachments to microtubules in vivo33 or for kinetochore particles to bind to microtubules in vitro29. Eluates from ndc80-1 mutant cells lacked all of the microtubule binding complexes that were visualized with WT particles in Figure 2 (Supplementary Fig 1). While WT eluates showed 0.68 particles per micrometer of microtubule (410 particles on a total of 493 micrometers observed), there were virtually none found for ndc80-1 (3 particles on a total of 585 micrometers observed), a reduction that is highly statistically significant (p< 0.0001) and consistent with the role of the outer kinetochore in making microtubule attachments.
Tomography reveals rings encircling the microtubule
We next used electron tomography and image processing to calculate the 3D reconstruction of two particles representative of those shown Figs 2a and c (Fig 3 and Supplementary Movies 1-4). Slices through these data identify at least two prominent features. First, a ring encircles the microtubule. Although it is not clear whether all the ring-like structures we observe by EM fully encircle the microtubule, this shows that at least these particles contain complete rings around the microtubule (Fig 3, blue arrowheads). Second, although only 2 rod-like structures were observed in projection (Fig 3a and b), multiple rod-like structures were connected to the ring-like structure in the tomogram. Although the stain flattens the assemblies so volumes for each component cannot be calculated accurately, the rods appear to be relatively evenly spaced around the ring-like structure.
Discussion
Here, we describe the structure of isolated kinetochore particles from budding yeast. The general architecture reveals a central hub surrounded by globular domains. When microtubules are present, at least one globular domain and an extension emanating from it make contact with the microtubule lattice. The extension is not seen in the absence of microtubules, consistent with a dynamic change in kinetochore structure in the presence of microtubules.
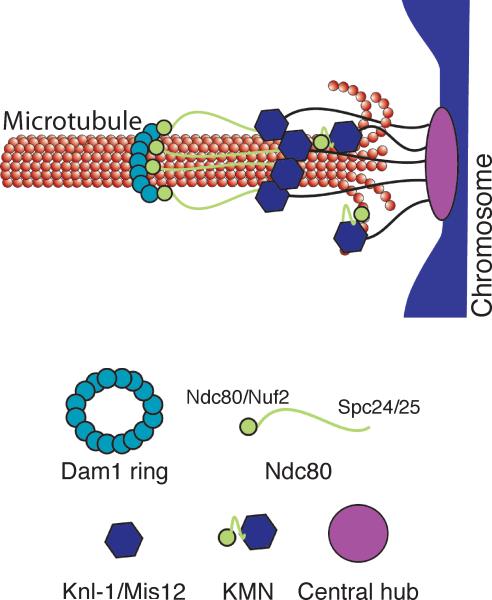
We propose the following model to describe the kinetochore particles we visualized by EM (Fig 4). Recombinant Dam1 complex forms rings with an outer diameter of 50 nm27,28 around microtubules, similar to the ring-like structures we observe in the presence of microtubules. These ring-like structures were never seen on the particles in the absence of microtubules, consistent with the substoichiometric amounts of Dam1 complex proteins in our kinetochore preparations and the requirement for microtubules in loading Dam1 on kinetochores12,29. It is likely that the eluates in our preparations contain some soluble Dam1 that can form ring-like structures around microtubules at very low concentrations in the presence of other kinetochore components.
Figure 4. Schematic of the proposed model of kinetochore architecture.
The central globular domain binds to the centromeric locus of the chromosome and globular domains containing the KMN complex extend to attach to the microtubule. The Ndc80 subcomplex makes an additional extension to contact a distal ring composed of the Dam1 subcomplex.
Given that Dam1 facilitates the function of the conserved Ndc80 complex on microtubules34,35, we propose that the extended rod-shaped structure connected to the ring is Ndc80. Consistent with this, the ends of the Ndc80 complex extend away from each other in the presence of microtubules in vivo18. The average length of the rod we observed is 56 nm and contains a kink, similar to what was observed for recombinant Ndc80 complex36,37. Although the kink we observe is in a different average position than recombinant Ndc80 complexes, our measurements are made in the presence of microtubules in the context of larger assemblies. When the orientation of the Ndc80 complex relative to the Dam1 ring in vivo is taken into account17, the Nuf2/Ndc80 head of the Ndc80 complex would be positioned at the ring. At this time, the resolution is not high enough for us to determine if the head is interior or exterior to the ring although our tomographic reconstructions suggest that the latter may be true. Our data are consistent with the possibility that the Ndc80 CH domain rather than the Ndc80 loop interacts with Dam138. This orientation means that the large globular entities at the opposite end of the rods would contain the Spc24/25 proteins; however, additional proteins must also be present to account for the size of the globular domain. We therefore propose that components of the Mis12 and/or KNL-1 subcomplexes that are known to bind to the Ndc80 complex to form KMN10, the core microtubule binding activity of the kinetochore, are located in these large globular domains. In this model, KMN would contain two microtubule binding sites, one composed of the Ndc80/Nuf2 head that can be seen extending from a subset of the globular domains to touch the microtubule, and the other containing Spc24/25 bound to Mis12/KNLl-1 complexes. This model is consistent with the cooperative microtubule binding behavior of KMN10. Given that the central hub never appears to associate with the microtubule, we propose that the chromosomal attachment site is mediated through this region. To date, we have not visualized nucleic acid in the particles by EM. At this time, it is not possible to estimate the stoichiometry of components within the particles, but we favor the possibility that each globular domain represents a single KMN complex. In the future, it will be important to identify the position of each kinetochore protein within the particles to precisely determine the precise architecture of the structures.
In summary, we have visualized isolated kinetochore particles and found that they appear as 126 nm particles containing a central hub with multiple outer globular domains. Our images exhibit some similarity to tomograms of laterally attached vertebrate kinetochores in vivo21. Because two or more of the globular domains bind to the microtubule, kinetochores appear to interact with microtubules via multivalent attachments that are flexible to move along microtubules, consistent with a biased diffusion mechanism39. We propose that when the particle is bound to a chromosome and the tip of a dynamic microtubule, a greater number of globular domains might bind to the microtubule to stabilize the interaction and maintain larger forces (Fig 4). In this case, the distance from the central hub to the outer region of the kinetochore would be consistent with measurements of tip-attached kinetochores in vivo17,18. The filaments observed at the ends of kinetochore microtubules in vivo20 could correspond to the rod-like structures we have observed along the microtubules, or connections between the globular domains and the central hub. Because yeast kinetochores bind to a single microtubule, a ring-based mechanism likely ensures processivity but is not required for direct attachment40. Together, these studies lay the foundation for future high-resolution structural and mechanistic studies aimed at understanding how the kinetochore ensures accurate chromosome segregation during cell division.
Online Methods
Yeast strains, plasmids and microbial techniques
Media and genetic and microbial techniques were essentially as described41. Mitotic cultures were prepared with benomyl as described30. For temperature sensitive mutants, cells were shifted to 37 °C for 3 hours. Yeast strains and plasmids used in this study are listed in Supplemental Table S4. The 3xFlag epitope tag strains were made using a PCR-based integration system and confirmed by PCR42-44. The 6xHis-3xFlag epitope tagging of the endogenous DSN1 gene was performed using a PCR-based integration system using primers SB2434 and SB2435 and plasmid pSB1590 as a template29. All tagged strains we constructed are functional in vivo and do not cause any detectable growth defects or temperature sensitivity. Specific primer sequences are available upon request.
Isolation of kinetochore particles
Native kinetochore particles were isolated from budding yeast as described29. Briefly, 2 L of yeast cells (SBY8253 or relevant strain) expressing Dsn1-Flag or Dsn1-His-Flag were arrested in mitosis with 60 μg/ml of microtubule depolymerizing drug benomyl for 3 hours and harvested. Cells were lysed in Buffer H (25 mM HEPES pH 8.0, 2 mM MgCl2, 0.1 mM EDTA, 0.5 mM EGTA, 150 mM KCl, 15% glycerol and 0.1% NP-40) supplemented with protease and phosphatase inhibitors, and kinetochore particles were captured with anti-Flag antibodies and eluted with 40 μl of Buffer H containing 0.5 mg/ml Flag peptide. The eluted material was used directly for negative stain EM studies as described below (note that this buffer resulted in less compact structures as shown in Figure 1d). Alternatively, the eluate was prepared for microtubule binding experiments as described below (note that these kinetochores appeared more compact as shown in Figure 1c). All measurements in the paper were done on particles that had been incubated for microtubule binding experiments.
Electron microscopy and image processing
All samples were prepared for negative stain EM as previously described45 with the following modifications. A 3 μl drop of Flag eluate was applied to a negatively charged carbon coated copper grid (Gilder 400 or 200 mesh) for 20 seconds and washed with a single drop of water, followed by 2 drops of freshly prepared 0.75% uranyl formate. Samples containing microtubules were treated similarly but were applied to positively charged copper grids. Specimens were screened on either a 100 kV transmission electron microscope (TEM) (Morgagni, FEI) or a 120 kV TEM (Spirit T12, FEI). Images were recorded using a Gatan slow scan bottom mount charge coupled device (CCD) camera at a nominal magnification of 18,000x at the specimen level. Measurements were taken either in the Digital Micrograph suite (Gatan, v 1.71.38) or ImageJ64. (v 1.43).
Electron tomography
Negatively stained samples prepared as above were coated with a second layer of carbon by evaporation and anti-mouse IgG-gold (5-10 nm) (Sigma-Aldrich) added as fiducial markers. Tilt series were collected using a Spirit T12 120 kV transmission electron microscope (FEI Company). Images were recorded using a Gatan slow scan 4K x 4K bottom mount CCD with a pixel size of 4.3Å at the sample level (52,000x). Tilt series were recorded from –70° to +70° with an increment of 2° at 2 μ m defocus. Three-dimensional reconstructions were calculated using Amira (v 5.3.1)46 and IMOD (v. 4.1.9)47 software.
Microtubule binding experiments
Taxol stabilized microtubules were prepared freshly as described48. A 200 μl aliquot was centrifuged at 58,000 r.p.m. (Beckman TLA 100.1 rotor), 37 °C for 10 minutes. The supernatant was decanted and the pellet was used for microtubule-kinetochore binding experiments as follows. 2 μl of Dsn1-Flag eluate was mixed with 7 μl of the microtubule pellet and incubated at RT for 10 minutes. The sample was then diluted with 200 μl warm BTAX (80 mM PIPES pH 6.9, 1 mM MgCl2, 1 mM EGTA and 10 μM Taxol, 37 °C). Grids for EM were prepared as described above. Note that excess tubulin dimers could be seen on the grids due to the microtubule polymerization buffer. Images were recorded on a CCD camera using either the 100 kV TEM or the 120 kV TEM at nominal magnification range of 14,000 – 36,000x at the specimen level. Measurements were taken either in the Digital Micrograph suite (Gatan, v 1.71.38) or ImageJ64. (v 1.43).
Quantification of MT binding
A total of 100 microtubules (for the dad1-1 mutant) or 200 microtubules (for WT and ndc80-1) ranging in size from 0.5 to 9 microns were assayed for the number of large particles bound in the presence or absence of a ring (Table 3). Eluates from SBY9047 or SBY7441 were used for all microtubule-binding experiments with WT kinetochore particles.
Supplementary Material
Acknowledgments
We are grateful to members of the Biggins and Gonen laboratories for valuable discussions and for comments on the manuscript. We are also grateful to Chip Asbury and Andy Powers (University of Washington), Barry Stoddard (Fred Hutchinson Cancer Research Center), and Jawdat Al-Bassam (University of California, Davis) for discussion and comments on the manuscript. The authors declare that none have a financial interest related to this work. This work was supported by NIH grants (GM078079 and GM064386 to SB), an NCI Cancer Center Support grant (CA015704 to SB), and the Howard Hughes Medical Institute (TG). Structure deposited to EMDB reference number XXXX.
Footnotes
Author Contributions
All authors contributed to designing the research. BA, ND and SB performed the kinetochore purifications. SG, MI and DS collected the EM data and did the EM analysis. SB and SG performed the microtubule binding experiments. TG and SB analyzed the data and wrote the manuscript.
References
- 1.Pfau SJ, Amon A. Chromosomal instability and aneuploidy in cancer: from yeast to man. EMBO Rep. 2012;13:515–27. doi: 10.1038/embor.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Compton DA. Mechanisms of aneuploidy. Curr Opin Cell Biol. 2011;23:109–13. doi: 10.1016/j.ceb.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–31. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 5.Takeuchi K, Fukagawa T. Molecular architecture of vertebrate kinetochores. Exp Cell Res. 2012;318:1367–74. doi: 10.1016/j.yexcr.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 6.DeLuca JG, Musacchio A. Structural organization of the kinetochore-microtubule interface. Curr Opin Cell Biol. 2012;24:48–56. doi: 10.1016/j.ceb.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu Rev Biochem. 2007;76:563–91. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- 8.Zinkowski RP, Meyne J, Brinkley BR. The centromere-kinetochore complex: a repeat subunit model. J Cell Biol. 1991;113:1091–110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joglekar AP, et al. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J Cell Biol. 2008;181:587–94. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–97. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann C, et al. Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J Cell Biol. 1998;143:1029–40. doi: 10.1083/jcb.143.4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, et al. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 2002;16:183–97. doi: 10.1101/gad.959402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Welburn JP, et al. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell. 2009;16:374–85. doi: 10.1016/j.devcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanisch A, Sillje HH, Nigg EA. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J. 2006;25:5504–15. doi: 10.1038/sj.emboj.7601426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asbury CL, Tien JF, Davis TN. Kinetochores’ gripping feat: conformational wave or biased diffusion? Trends Cell Biol. 2011;21:38–46. doi: 10.1016/j.tcb.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schittenhelm RB, et al. Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma. 2007;116:385–402. doi: 10.1007/s00412-007-0103-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joglekar AP, Bloom K, Salmon ED. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol. 2009;19:694–9. doi: 10.1016/j.cub.2009.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wan X, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–84. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welburn JP, Cheeseman IM. Toward a molecular structure of the eukaryotic kinetochore. Dev Cell. 2008;15:645–55. doi: 10.1016/j.devcel.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 20.McIntosh JR, et al. Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell. 2008;135:322–33. doi: 10.1016/j.cell.2008.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong Y, Vanden Beldt KJ, Meng X, Khodjakov A, McEwen BF. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat Cell Biol. 2007;9:516–22. doi: 10.1038/ncb1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston K, et al. Vertebrate kinetochore protein architecture: protein copy number. J Cell Biol. 2010;189:937–43. doi: 10.1083/jcb.200912022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brinkley BR, Stubblefield E. The fine structure of the kinetochore of a mammalian cell in vitro. Chromosoma. 1966;19:28–43. doi: 10.1007/BF00332792. [DOI] [PubMed] [Google Scholar]
- 24.Jokelainen PT. The ultrastructure and spatial organization of the metaphase kinetochore in mitotic rat cells. J Ultrastruct Res. 1967;19:19–44. doi: 10.1016/s0022-5320(67)80058-3. [DOI] [PubMed] [Google Scholar]
- 25.Roos UP. Light and electron microscopy of rat kangaroo cells in mitosis. II. Kinetochore structure and function. Chromosoma. 1973;41:195–220. doi: 10.1007/BF00319696. [DOI] [PubMed] [Google Scholar]
- 26.Rieder CL. The structure of cold stable kinetochore microtubules in metaphase PtK1 cells. Chromosoma. 1981;84:145–158. doi: 10.1007/BF00293368. [DOI] [PubMed] [Google Scholar]
- 27.Miranda JJ, De Wulf P, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol. 2005;12:138–43. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- 28.Westermann S, et al. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–90. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 29.Akiyoshi B, et al. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 2010;468:576–9. doi: 10.1038/nature09594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akiyoshi B, Nelson CR, Ranish JA, Biggins S. Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev. 2009;23:2887–99. doi: 10.1101/gad.1865909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wigge PA, et al. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol. 1998;141:967–77. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Enquist-Newman M, et al. Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity. Mol Biol Cell. 2001;12:2601–13. doi: 10.1091/mbc.12.9.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka K, et al. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–94. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- 34.Tien JF, et al. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J Cell Biol. 2010;189:713–23. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lampert F, Hornung P, Westermann S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J Cell Biol. 2010;189:641–9. doi: 10.1083/jcb.200912021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci U S A. 2005;102:5363–7. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang HW, et al. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J Mol Biol. 2008;383:894–903. doi: 10.1016/j.jmb.2008.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alushin GM, et al. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–10. doi: 10.1038/nature09423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hill TL. Theoretical problems related to the attachment of microtubules to kinetochores. Proc Natl Acad Sci U S A. 1985;82:4404–8. doi: 10.1073/pnas.82.13.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Westermann S, et al. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–9. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- 41.Rose MD, Winston F, Heiter P. Methods in yeast genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N. Y.: 1990. p. 198. [Google Scholar]
- 42.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–61. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 43.Gelbart ME, Rechsteiner T, Richmond TJ, Tsukiyama T. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol Cell Biol. 2001;21:2098–106. doi: 10.1128/MCB.21.6.2098-2106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stalling D, Westerhoff M, Hege H-C. The Visualization Handbook. Elsevier; 2005. [Google Scholar]
- 47.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–6. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 48.Franck AD, et al. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat Cell Biol. 2007;9:832–7. doi: 10.1038/ncb1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.