Summary
Originally described as a repressor of gene expression in the stationary phase of growth, CsrA (RsmA) regulates primary and secondary metabolic pathways, biofilm formation, motility, virulence circuitry of pathogens, quorum sensing, and stress response systems by binding to conserved sequences in its target mRNAs and altering their translation and/or turnover. While the binding of CsrA to RNA is understood at an atomic level, new mechanisms of gene activation and repression by this protein are still emerging. In the γ-proteobacteria, small noncoding RNAs (sRNAs) use molecular mimicry to sequester multiple CsrA dimers away from mRNA. In contrast, the FliW protein of Bacillus subtilis inhibits CsrA activity by binding to this protein, thereby establishing a checkpoint in flagellum morphogenesis. Turnover of CsrB and CsrC sRNAs in Escherichia coli requires a specificity protein of the GGDEF-EAL domain superfamily, CsrD, in addition to the housekeeping nucleases RNase E and PNPase. The Csr system of E. coli contains extensive autoregulatory circuitry, which governs the expression and activity of CsrA. Interaction of the Csr system with transcriptional regulatory networks results in a variety of complex response patterns. This minireview will highlight basic principles and new insights into the workings of these complex eubacterial regulatory systems.
Keywords: global regulation, CsrA, stringent response, ppGpp, posttranscriptional regulation, regulatory RNAs
Introduction
A plethora of posttranscriptional regulators
DNA binding proteins and other transcription factors play important roles in the global regulatory networks that coordinate bacterial metabolism and physiology in response to the environment (reviewed in Ishihama, 2010). In addition, posttranscriptional regulation plays a major part in coordinating gene expression networks. Two kinds of posttranscriptional systems predominate in the γ-proteobacteria: trans-acting RNAs that use an RNA chaperone protein, Hfq, to pair with mRNA targets (Gottesman and Storz, 2011), and the Csr (carbon storage regulator) or Rsm (repressor of stationary phase metabolites) systems, which exploit a sequence-specific RNA binding protein, referred to as CsrA, RsmA, or RsmE, to alter the translation and/or stability of mRNA targets (Babitzke and Romeo, 2007; Romeo, 1998). Noncoding sRNAs (CsrB and CsrC in E. coli) containing multiple CsrA binding sites, which mimic the binding sites of mRNAs, sequester and antagonize CsrA (Liu et al., 1997; Weilbacher et al., 2003). The mRNA binding protein Crc (catabolite repression control) of pseudomonads is also a global regulator of carbon metabolism and other processes (Moreno et al., 2007) whose activity is governed by binding to antagonistic sRNAs (Sonnleitner et al., 2009; Moreno et al., 2012). Because CsrA and Crc share no amino acid sequence similarity and their binding specificities are unrelated, this efficient strategy for global regulation has apparently evolved more than once. RNA-based regulatory mechanisms such as ligand-binding riboswitches, attenuators, and cis-acting sRNAs mediate localized effects on gene expression (Waters and Storz, 2009; Breaker, 2011; Gollnick and Babitzke, 2002). Recent studies have identified the CRISPR-Cas systems (clustered regularly interspaced short palindromic repeats-CRISPR associated genes) as adaptive immune systems of bacteria and archea (Bhaya et al., 2011). These systems incorporate short DNA fragments from invading bacteriophage or plasmids into the bacterial CRISPR locus. Transcripts derived from the CRISPR locus are processed into small noncoding RNAs that can direct an attack on invading elements. Host genetic information is occasionally incorporated into CRISPR loci, leading to speculation that CRISPR-Cas systems may also contribute to posttranscriptional regulation (Bhaya et al., 2011).
γ-proteobacterial Csr/Rsm systems: Once novel, still unfolding regulatory paradigm
In the natural environment, bacteria must cope with wide swings in nutrient availability (Matin et al, 1989; Kolter et al., 1993). This feast or famine existence requires readjustments in global gene expression patterns that favor rapid growth or stress resistance, respectively. Early attempts to understand the underlying mechanisms for the response of E. coli to stationary phase growth conditions led to the recognition of the sigma factor RpoS or σs, which serves as a central activator of general stress responses and stationary phase gene expression (Battesti et al., 2011; Klauk et al., 2007), and its apparent doppelgänger or “evil twin” CsrA, which represses stationary phase gene expression and activates genes needed for growth (Romeo et al., 1993; Sabnis et al., 1995; Yang et al., 1996; Wei et al., 2001). Shortly after CsrA was discovered, its orthologue, RsmA of Erwinia carotovora, was found to be involved in host interactions of this plant pathogen, quorum sensing pathways, and motility (Chatterjee et al., 1995; Mukherjee et al., 1996). These findings opened the way for studies of diverse regulatory roles of CsrA in many species of bacteria.
A few seminal observations provided the basic understanding of how CsrA works as a posttranscriptional regulator: It greatly accelerates the turnover of mRNA for glycogen biosynthesis, glgC, utilizing a cis-acting region that overlaps the ribosome binding site of this transcript (Liu et al., 1995). Purified CsrA binds specifically to glgC mRNA and inhibits its translation in vitro (Liu and Romeo, 1997; Baker et al., 2002). In addition, the purified CsrA protein was found to reside in large ribonucleoprotein complexes with a noncoding RNA, CsrB, which sequesters and antagonizes CsrA (Liu et al., 1997). Thus, the basic model for the γ-proteobacterial Csr system was established (Romeo, 1998; Fig. 1).
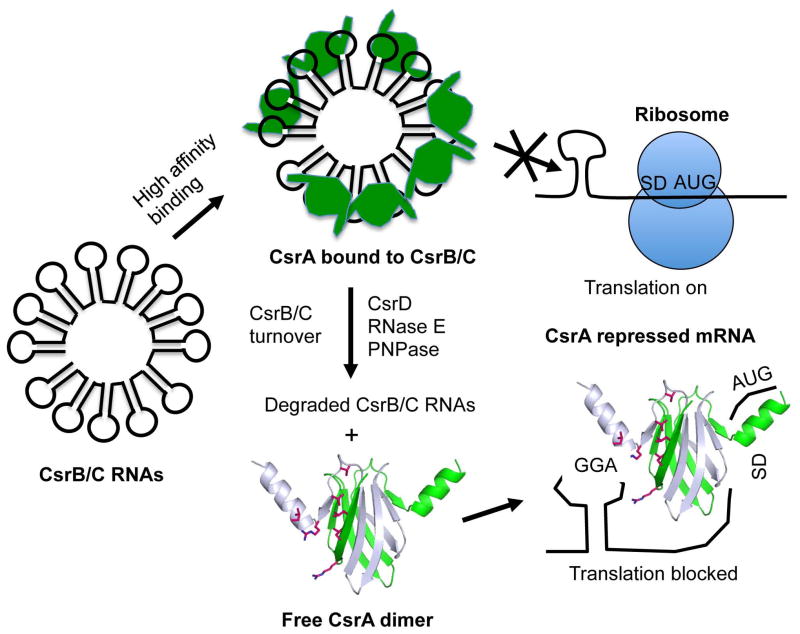
Fig. 1.
Outline of the E. coli Csr system. Binding of the CsrA dimer to an mRNA can block translation (shown), destabilize, or stabilize the mRNA. Noncoding RNAs (CsrB and CsrC) containing multiple CsrA binding sites within the loops of predicted stem-loops compete with mRNAs for CsrA binding, thus antagonizing CsrA activity. Free CsrA is regenerated by the turnover of CsrB/C RNAs, which requires RNase E, polynucleotide phosphorylase (PNPase) and CsrD, an atypical GGDEF-EAL domain protein.
CsrA-RNA interactions
Soon after the identification of CsrA as a pleiotropic regulator of carbon metabolism and cell surface properties (Romeo et al., 1993; Sabnis et al., 1995), it was found to be an mRNA binding protein (Liu and Romeo, 1997). CsrA binds to several sites in the CsrB and CsrC sRNAs (Liu et al., 1997; Weilbacher et al., 2003) and multiple sites in most of its target mRNAs (e.g. Baker et al., 2002; Dubey et al., 2003; reviewed in Babitzke and Romeo, 2007). Although considerable sequence variation exists among the known CsrA binding sites, a GGA motif was identified as a highly conserved element. The critical nature of the GGA motif was confirmed by studies using SELEX (systematic evolution of ligands by exponential enrichment). The SELEX-derived consensus was determined to be RUACARGGAUGU, with the GGA motif being 100% conserved (Dubey et al., 2005). Each of the SELEX-derived RNAs contained the GGA motif in the loop of a predicted RNA hairpin (Dubey et al., 2005), an arrangement that has been observed for several naturally occurring CsrA binding sites (Liu et al., 1997; Babitzke and Romeo, 2007).
CsrA functions as a homodimer with an identical RNA binding surface located on each side of the dimer (Dubey et al., 2003; Mercante et al., 2006; Schubert et al., 2007). CsrA comprises a novel RNA-binding fold, formed by the interdigitation of the five β-strands of each of the subunits, and stabilized by a hydrophobic core (Gutierrez et al., 2005; Rife et al., 2005). Two identical RNA binding surfaces of CsrA are formed primarily by the first and last β-strands of opposite polypeptides, which lie parallel to each other on opposite sides of the dimer. Several amino acid residues of the β-strands are involved in RNA binding, with R44 being the most important (Mercante et al., 2006). An NMR structure of a CsrA orthologue (RsmE) complexed with RNA confirmed and extended the findings from several previous biochemical studies, including the identification of amino acid contacts with the conserved GGA motif. The RsmE-RNA structure revealed a protein dimer bound to two separate RNA molecules (Schubert et al., 2007). It was recently shown that CsrA is capable of binding simultaneously to two sites within a single transcript if the two sites were separated by 10 to >63 nt, although binding was weak with <18 nt spacing. The ability of a CsrA dimer to bridge two sites within a single transcript was shown to be important for controlling gene expression (Mercante et al., 2009). This property of CsrA is relevant for virtually all CsrA-dependent regulatory mechanisms and is crucial for interactions with complex RNA structures.
CsrA-mediated Repression and Activation Mechanisms
CsrA-mediated repression typically involves CsrA binding to multiple sites in the untranslated leader and/or initially translated region of target transcripts, one of which overlaps the cognate Shine-Dalgarno (SD) sequence (Babitzke and Romeo, 2007). Thus, bound CsrA represses translation by competing with ribosome binding (Baker et al., 2002; Dubey et al., 2003). Translational repression by CsrA may lead to destabilization of the mRNA, although this is not always the case (Baker et al., 2007; Pannuri et al. 2012). CsrA-mediated activation has not been extensively studied, but also involves binding to the untranslated mRNA leader, with consequent effects on RNA turnover and/or translation (Wei et al., 2001; unpublished studies).
General mechanism of CsrA-mediated translational repression
The general mechanism of CsrA-mediated repression was first identified for the glycogen biosynthetic gene, glgC (Baker et al., 2002; Mercante et al., 2009). CsrA binds to four sites in the untranslated leader of the glgCAP operon transcript, with the last (promoter distal) site overlapping the glgC SD sequence. Bound CsrA represses translation by competing with ribosome binding, leading to rapid degradation of the polycistronic transcript (Romeo, 1998; Baker et al., 2002; Mercante et al., 2009). Of particular interest, CsrA inhibits its own expression by essentially the same translation repression mechanism (Yakhnin et al., 2011b).
While mechanistic information on CsrA-mediated translational repression has largely been derived from E. coli studies, it appears that this general repression mechanism is conserved in several other Gram-negative organisms. For example, CsrA of Salmonella enterica serotype Typhimurium (S. Typhimurium) represses translation of hilD by binding to this gene’s SD sequence and translation initiation codon, leading to accelerated mRNA turnover (Martínez et al., 2011). As HilD activates expression of hilA and ssrAB, the genes encoding critical regulators of Salmonells pathogenicity islands 1 (SPI-1) and 2 (SPI-2), CsrA plays an important role in controlling S. Typhimurium pathogenesis (Martínez et al., 2011). In another example, the CsrA homologs of Pseudomonas fluorescens, RsmA and RsmE, repress translation of hcnA, a gene encoding hydrogen cyanide synthase subunit A (Lapouge et al., 2007). Although not studied in mechanistic detail, it appears that CsrA-mediated translational repression is a common regulatory mechanism in a variety of organisms.
Several deviations from the general translational repression mechanism have been identified. For example, CsrA inhibits hfq translation by binding to a single site overlapping its SD sequence (Baker et al., 2007). Interestingly, Hfq is another global posttranscriptional regulatory factor. This protein acts as an RNA chaperone to promote sRNA-mRNA base pairing, thereby affecting the stability and/or translation of the target mRNAs (reviewed in Gottesman, 2004; Vogel and Luisi, 2011). cstA, a gene encoding a peptide transporter that is induced upon carbon starvation, is regulated by a slightly different mechanism. In this case, two of the four CsrA binding sites in cstA mRNA lie downstream from the translation initiation codon (Dubey et al., 2003). As described below, CsrA can also repress translation without contacting the SD sequence of the target transcript.
Repression of quorum sensing by binding exclusively to the sdiA mRNA coding region
Bacteria have the ability to regulate gene expression in response to population density by a process known as quorum sensing. A common quorum sensing system in Gram-negative bacterial species involves the synthesis and detection of N-acyl-L-homoserine lactones (AHL). Since AHLs can freely diffuse across cell membranes, as the bacterial population increases, the concentration of AHLs in the cellular environment increases. Once a critical level of AHL is reached, AHL reenters the cell and is detected by a response regulator, thereby altering expression of target genes. Although E. coli does not synthesize AHLs, it contains an AHL receptor homolog (SdiA), which appears to sense and respond to AHLs produced by other bacterial species (reviewed in Ng et al., 2009; Smith et al., 2011; Soares and Ahmer, 2011). CsrA represses translation of sdiA by binding two sites in the sdiA transcript. In contrast to all other known CsrA-regulated genes, CsrA inhibits translation by interacting solely within the early coding region of the sdiA mRNA, without binding to or otherwise occluding the sdiA SD sequence (Yakhnin et al., 2011a).
Repression of Pseudomonas aeruginosa biofilm formation by modifying RNA structure
An important microbial survival strategy is to form surface-associated multicellular communities known as biofilms (Costerton et al., 1999). Biofilms provide protection against stresses, predation, the immune system, and antimicrobial compounds (Wang et al., 2005 and references therein). Biofilm formation is regulated in response to environmental conditions and cues, although the specifics vary considerably among different species (Goller and Romeo, 2008). This complex process requires the establishment of cell-cell and cell-surface attachments, which are mediated by proteins, polysaccharides and nucleic acids (Petrova and Sauer, 2012; Karatan and Watnick, 2009). Following the demonstration that E. coli CsrA inhibits biofilm formation in E. coli (Jackson et al, 2002; Wang et al., 2005), RsmA of Pseudomonas aeruginosa was found to bind to psl mRNA and thereby represses genes necessary for synthesis of the biofilm polysaccharide, Psl (Irie et al., 2010). All previously known translational repression mechanisms involved CsrA binding directly to the SD sequence. However, in this case, RsmA repressed translation by stabilizing a hairpin structure that sequesters the pslA SD sequence. RsmA binds to the loop of this structure, reflecting the fact that the highest affinity CsrA binding sites are in the loops of RNA hairpins (Liu et al., 1997; Dubey et al., 2005), and that RNA binding affects secondary structure, causing RNA bending (Schubert et al., 2007).
CsrA-mediated activation mechanisms
For several years, CsrA has been known to activate gene expression (Romeo, 1998). For example, CsrA activates expression of the E. coli flhDC operon by binding to and stabilizing the mRNA (Wei et al., 2001; unpublished results). FlhD4C2 is a hexameric DNA binding protein that plays a critical role in activating flagellum biosynthesis and chemotaxis (Wang et al., 2006; reviewed in Smith and Hoover, 2009). CsrA binds to two sites centered at +5 and +50 of the 198-nt untranslated flhDC leader transcript. Bound CsrA protected the flhDC mRNA from in vitro cleavage by RNase E, an essential endoribonuclease that exhibits partial 5′ end dependence. The major RNase E cleavage sites were ≥ 50 nt downstream from the distal CsrA binding site. Thus, bound CsrA appears to stabilize flhDC mRNA by inhibiting RNase E interaction with the 5′ end of the transcript, rather than by directly shielding the RNA (unpublished results).
The molybdenum cofactor, MOCO, serves as a redox center for enzymes of anaerobic metabolism. moaA is one of several genes needed for MOCO synthesis. The untranslated leader of moaA mRNA is an apparent MOCO-sensing riboswitch, in which bound MOCO is thought to stabilize an RNA structure that inhibits translation (Regulski et al., 2008). moaA was identified as a CsrA target by RNA-seq studies (Edwards et al., 2011). CsrA binds to the highly structured moaA leader and causes RNA structural changes that apparently result in activation of moaA translation (Patterson-Fortin, Vakulskas, Baker, Bhardwaj, Babitzke and Romeo, unpublished results). To our knowledge, this is the only known example of a riboswitch aptamer that interacts with two different regulatory factors.
Autoregulatory circuitry of the E. coli Csr system
The discovery that CsrB and CsrC sRNAs sequester and antagonize CsrA was central to understanding how the Csr system functions (Liu et al., 1997; Weilbacher et al., 2003). This kind of regulation is possible because the affinity of CsrA for CsrB/C is higher than for most mRNA targets and the concentration of CsrA in the cell (6 to 17 μM) is much greater than its dissociation constant (Kd) for target RNAs (Gudapaty et al., 2001). Thus, the relative amount of CsrA that is not bound to RNA should be low, and the level of CsrA that is available for binding mRNA targets is determined by the amount of CsrB/C in the cell. Studies of the expression of the Csr components and their genetic interactions in E. coli Csr revealed that CsrA indirectly activates the transcription of its antagonistic sRNAs, CsrB/C, an effect that is directly mediated via the BarA-UvrY two component signal transduction system (TCS) (Gudapaty et al., 2001; Suzuki et al., 2002; Weilbacher et al., 2003). This TCS regulates transcription of Csr sRNAs in many other γ-proteobacterial species, where it is known by a variety of other names (reviewed in Lapouge et al., 2007; Heroven et al., 2012; Babitzke and Romeo, 2007; Molofsky and Swanson, 2004; Mikkelsen et al.,2011). Signaling by the membrane-bound BarA sensor-kinase and its homologues is somehow triggered by products of carbon metabolism such as acetate, short chain carboxylic acids, and perhaps TCA cycle intermediates in some species (Chavez et al., 2010; Takeuchi et al., 2011). These findings suggest that the Csr system responds to the metabolic state of the bacterium and to products that are common in the lumen of the mammalian intestine (discussed in Chavez et al., 2010; Edwards et al., 2011). The central negative feedback loop of the Csr system suggests a homeostatic circuit for control of CsrA activity (Fig. 2). An interesting consequence of this circuitry is that disruption of the gene for either of the regulatory RNAs causes a compensatory increase in the other RNA (Weilbacher et al., 2003). Compensatory regulation was later documented in other Csr systems (reviewed in Heroven et al., 2012), as well as distinct bacterial circuits that make use of redundant regulatory sRNAs, e.g. the Qrr 1–4 RNAs of the Vibrio cholerae quorum sensing circuitry (Svenningsen et al., 2009).
Fig. 2.
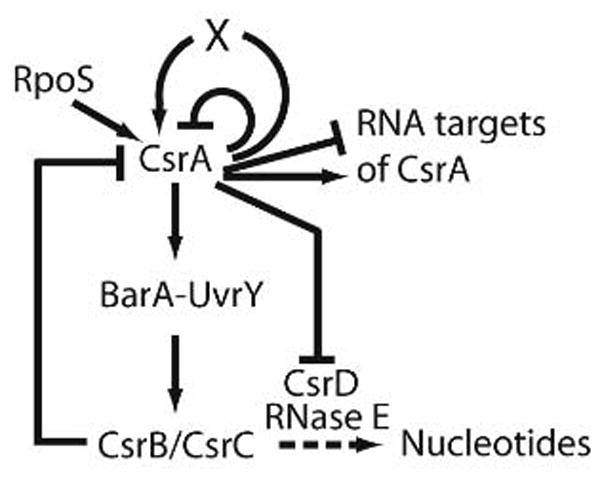
Autoregulatory loops of the Csr system. CsrA binds to its own mRNA and inhibits translation initiation. It also indirectly activates its own transcription at P3, an RpoS-dependent promoter. CsrA also activates the expression of its two RNA antagonists, CsrB and CsrC, via the BarA-UvrY two-component signal transduction system. Phosphorylated UvrY directly activates transcription of csrB and csrC. The sRNAs CsrB and CsrC contain several CsrA binding sites such that each sRNA is capable of sequestering several CsrA dimers. CsrA modestly represses expression of CsrD, a protein that is required for degradation of CsrB and CsrC by RNase E and PNPase (from Yakhnin et al., 2011).
The Csr system also contains additional autoregulatory loops. CsrA directly represses its own translation, while at the same time it indirectly activates its transcription (Yakhnin et al., 2011). Repression involves binding of CsrA to four sites in the csrA mRNA leader, including the SD sequence, which prevents ribosome loading and translation. Thus, CsrA-mediated autorepression provides a rapid mechanism to reduce CsrA synthesis when the concentration of free CsrA reaches a critical level in the cell. Five promoters, two of which use the alternative sigma factor σs, drive transcription of csrA. Transcription of P3 is CsrA activated, σs-dependent, and is primarily responsible for the increase in CsrA that occurs when cells enter the stationary phase of growth. Furthermore, CsrA activates the gene for σs when cells are at low temperature and in the exponential phase of growth (unpublished observations). This should create a positive feedback circuit for signal amplification, i.e. csrA induction, under σs-responsive stresses.
Turnover of CsrB/CsrC sRNAs requires a novel protein, CsrD
RNA synthesis and turnover determine transcript levels and both processes are important in the regulation of gene expression. RNA decay is highly coordinated, and may be affected by several factors, including a variety of endo- and exonucleases, RNA binding proteins, noncoding complementary RNAs, and the translational status of a message (reviewed in Arraiano et al., 2010; Belasco, 2010; Kaberdin and Bläsi, 2006; Kushner, 2002). In E. coli, RNA decay is often initiated by RNase E, an endoribonuclease that preferentially binds to the 5′ monophosphorylated terminus of transcripts, with cleavage occurring in A/U-rich regions adjacent to stem-loop structures. The resulting cleavage products serve as substrates for rapid degradation by the processive 3′ to 5′ exoribonucleases polynucleotide phosphorylase (PNPase) and RNase II. RNase E also provides a scaffold for assembly of the degradosome, a protein complex consisting of RNase E, PNPase, the RNA helicase RhlB, and enolase, which couples endonucleolytic cleavage with exonucleolytic decay (Carpousis et al., 2007).
Recently, a novel regulator of RNA turnover, CsrD, was found to be essential for decay of CsrB and CsrC in E. coli (Suzuki et al., 2006). CsrD destabilizes CsrB/C sRNAs in an RNase E-dependent fashion, increasing CsrA availability in the cell and affecting the expression of all CsrA-regulated genes that were tested. Though a member of the GGDEF-EAL domain family of proteins, which typically synthesize (GGDEF) or degrade (EAL) the secondary signaling molecule c-di-GMP, CsrD does not produce, degrade or respond to this nucleotide. CsrD contains two predicted N-terminal spanning domains followed by coiled-coil, GGDEF and EAL domains. The latter domains are essential for in vivo activity, while the membrane spanning regions are dispensable if the protein is overexpressed. Although the molecular mechanism of CsrD-dependent RNA cleavage has not been defined, CsrD does not appear to be a ribonuclease. CsrD binds to CsrB and CsrC RNAs, albeit without apparent specificity. A working model is that CsrD binding imparts structural changes in CsrB/C, resulting in sRNAs that are no longer refractory to RNase E turnover. Whether RNA binding specificity requires additional factors or conditions is unknown. Preliminary microarray experiments suggest that although CsrD is quite specific, a small number of novel sRNA targets may exist (Vakulskas and Romeo unpublished). CsrD homologues are apparent in Enterobacteriaceae and closely allied families, but are not characteristic of all γ-proteobacteria. CsrA represses csrD expression in E. coli as well as Salmonella, indicative of an additional negative feedback loop of the Csr system (Jonas et al., 2008; Suzuki et al., 2006). However, CsrA effects on csrD expression are weak (2-fold) and CsrA has modest or no effect on CsrB and CsrC turnover in vivo (Gudapaty et al., 2001; Suzuki et al., 2006). Thus, the biological significance of this autoregulatory loop is not clear.
Approaches to determine the complete Csr regulon
The initial studies of CsrA (RsmA) suggested that it is global repressor of genes that are expressed in the stationary phase of growth (Romeo et al. 1993; Yang et al. 1996; Chatterjee et al., 1995). Soon it became apparent that CsrA also activates expression of genes expressed during exponential growth such as glycolytic genes and flhDC (Sabnis et al., 1995; Wei et al., 2001). Furthermore, candidate gene studies and other approaches revealed regulatory roles for CsrA in a variety of proteobacterial processes, which were of special interest to the investigators, e.g. biofilm formation, motility, metabolism, pathogenesis, quorum sensing (reviewed in Romeo 1996; 1998; Molofsky and Swanson 2004; Babitzke and Romeo 2007; Lapouge et al., 2007; Lucchetti-Miganeh et al., 2008; Timmermans and Van Melderen, 1010; Heroven et al., 2012). With the advent of microarray studies, comparison of csrA mutant vs. wild-type strains provided for the systematic examination of the global effects of CsrA in S. Typhimurium and Pseudomonas aeruginosa, which suggested that CsrA affects hundreds of different RNAs, amounting to >10% of the detectable transcripts (Lawhon et al., 2003, Brencic and Lory, 2009; Burrows et al., 2006). Because CsrA is a regulator of regulators, recent studies have used “timed arrays”, aimed at distinguishing direct vs. indirect targets in E. coli (Jonas et al., 2008). For this approach, the csrA gene was induced in a csrA deficient strain background and response of the transcriptome was monitored with respect to the time of induction. As expected, known direct targets of CsrA regulation responded rapidly to csrA induction, while indirect targets responded in delayed fashion. This study revealed that mRNAs for GGDEF domain proteins, which synthesize the secondary messenger c-di-GMP, are direct targets of CsrA.
Although the timed array approach represented an improvement for discovering CsrA targets, it has limitations. For example, RNA binding and translational repression by CsrA does not invariably cause decay of target mRNAs (Baker et al. 2007; Pannuri et al., 2012) and analysis of low abundance transcripts is problematic. To address these limitations, RNA-seq experiments were used to identify the direct RNA targets of CsrA (Edwards et al., 2011). In this study, RNA that copurified with a His6-tagged CsrA protein was analyzed by high throughput sequencing. Altogether, 721 probable direct targets of CsrA binding were identified, representing >15% of E. coli genes (Edwards et al., 2011). The transcripts reside within 20 E. coli COGs (clusters of orthologous groups of proteins), representing virtually all of the major physiological and macromolecular processes of this bacterium (Edwards et al. 2011). Importantly, numerous regulatory genes are represented among the bound RNAs, suggestive of widespread indirect effects of CsrA on the expression of genes that may or may not also be direct regulatory targets. While confirmatory studies are still needed to establish the biological relevance of most of these binding interactions, this study provided groundwork for elucidating new molecular mechanisms of CsrA action and new global regulatory circuits that interact with the Csr system, necessary for defining the systems biology of E. coli.
Interaction of Csr with other regulatory systems generates diverse regulatory circuits
CsrA (RsmA) was reported to affect Erwinia carotovora N-acyl-homoserine lactone production as early as 1995 (Chatterjee et al., 1995) and was shown to directly activate expression of the E. coli flhDC genes (Wei et al., 2001), which encode a DNA binding protein that initiates a regulatory cascade for the expression of genes required for motility and chemotaxis (Smith and Hoover, 2009). Subsequently, CsrA was found to regulate a variety of regulators in several species. A limited number of biological response pathways that have been dissected in detail reveal that CsrA is involved in diverse regulatory circuitry patterns, described below. In view of the many regulatory mRNAs that are likely direct targets of CsrA binding in E. coli (Edwards et al., 2011), other circuitry patterns involving the Csr system are likely to emerge.
Multitier regulation of biofilm formation and PGA synthesis
CsrA represses biofilm formation in a variety of bacteria (Jackson et al., 2002; Irie et al. 2010). In E. coli and other species, the polysaccharide Poly-β-1,6-GlcNAc (PGA) promotes attachment to solid surfaces, cell-cell adherence, and stabilizes the biofilm structure (Wang et al., 2004; Itoh et al., 2005). The pgaABCD operon is required for PGA synthesis (PgaC and PgaD) and secretion (PgaA and PgaB) (Itoh et al., 2008). CsrA inhibits PGA synthesis and biofilm formation by cooperatively binding to six sites in the pgaABCD mRNA leader, competing with the ribosome for binding, and repressing translation of pgaA (Wang et al., 2005). RNA decay studies showed that CsrA also destabilizes the pgaABCD transcript, which is probably a consequence of translational inhibition (Fig. 3A).
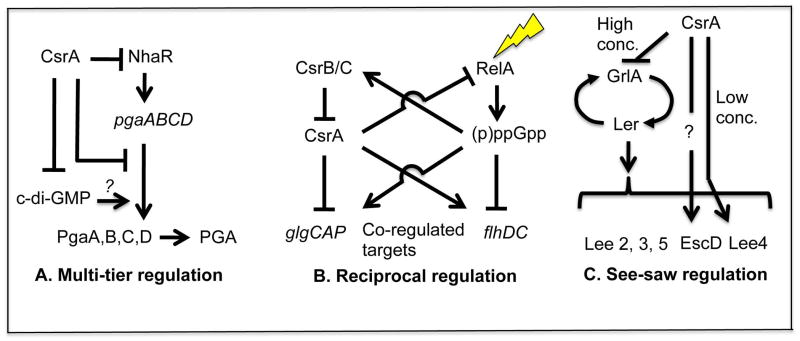
Fig 3.
Interactions of the Csr system with other E. coli regulatory systems shape diverse circuitry.
A. CsrA posttranscriptionally represses several steps in the synthesis of the biofilm polysaccharide β-1,6-N-acetylglucosamine, tightening the regulation of biofilm formation. (from Pannuri et al. 2011).
B. The Csr and stringent response systems interact to reinforce transcriptional control by ppGpp, as well as relieve repression of relA and ppGpp synthesis by CsrA during the stringent response (from Edwards et al. 2011).
C. Expression of pathogenicity genes in the Lee locus is compromised by both low and high activity of CsrA due to its opposing effects on structural genes and the GrlA regulator (Bhatt et al. 2009). The effect of CsrA on escD (?) is indirect.
In addition to its direct interaction with pgaABCD mRNA, there are at least two other ways in which CsrA represses PGA production and biofilm formation by E. coli (Fig. 3A). The LysR family DNA binding protein NhaR activates transcription of the pgaABCD promoter in response to elevated pH or high [Na+] (Goller et al., 2006). CsrA binds to two sites within the 59 nt intervening RNA segment separating nhaA, encoding a sodium-proton antiporter, and nhaR of the nhaAR operon, and represses translation of nhaR by competing with ribosome binding. Interestingly, CsrA does not bind to the nhaA leader or directly affect nhaA expression (Pannuri et al., 2012). The nucleotide c-di-GMP is a widespread bacterial secondary messenger that activates the production of surface adhesins and biofilm formation, and inhibits motility by binding to and regulating transcription factors, glycosyltransferases, and/or riboswitches (Hengge, 2009; Römling et al., 2005). Biofilm formation and synthesis of PGA by E. coli is also activated by c-di-GMP, although the precise mechanism(s) responsible for this effect remain unresolved (Suzuki et al., 2006; Tagliabue et al., 2010; Boehm et al. 2009). CsrA regulates the expression of several genes involved in c-di-GMP synthesis or turnover, repressing genes for c-di-GMP synthesis and reducing the intracellular concentration of c-di-GMP (Jonas et al., 2008; 2010). Thus, CsrA represses biofilm formation and PGA synthesis at multiple levels by (i) binding to the pgaABCD leader, inhibiting pgaA translation and causing pgaABCD mRNA destabilization, (ii) repressing c-di-GMP production, and (iii) repressing translation of nhaR, an activator of pgaABCD transcription. Since CsrA copurified with mRNAs representing both the structural and regulatory genes for a variety of processes (Edwards et al., 2011), we propose that multi-level regulation by CsrA is a pervasive feature of E. coli genetic circuitry, which helps to coordinate multiple influences on a given pathway or process.
Reciprocal regulatory interactions between the Csr and stringent response regulatory systems
The results of RNA-seq studies indicated that E. coli CsrA is a regulator of many other regulatory genes, including relA, dksA and spoT of the stringent response system (Edwards et al., 2011). This system mediates the stress response to amino acid starvation and depletion of other nutrients by the production of a nucleotide secondary messenger, ppGpp, which binds to RNA polymerase and positively or negatively regulates transcription (Potrykus and Cashel, 2008; Barker et al., 2001). The RelA protein synthesizes ppGpp, SpoT both synthesizes and hydrolyzes ppGpp, while DksA binds to RNA polymerase and often, though not invariably, potentiates effects of ppGpp on transcription. A comparison of the transcripts that responded to ppGpp and/or DksA in microarray analyses to the RNAs that copurified with CsrA in the RNA-seq studies revealed extensive overlap, indicating that numerous genes are regulated by both systems (Edwards et al., 2011). By tracing the regulatory interactions between the genes of the Csr and stringent response systems the latter study revealed that during the stringent response, ppGpp accumulation induces synthesis of CsrB and CsrC RNAs, which bind to CsrA and inhibit its activity. Because CsrA and ppGpp are known to exhibit opposite effects on the expression of specific genes, this regulation allows the Csr system to posttranscriptionally reinforce the direct transcriptional effects of ppGpp at a posttranscriptional level (Fig. 3B). The strong positive effect of ppGpp on csrB and csrC expression explains the earlier observation that addition of amino acids to minimal media causes a drastic decrease in CsrB/C RNA levels (Jonas and Melefors, 2009). In addition, CsrA posttranscriptionally represses relA expression. Thus, a positive feedback loop exists for enhancing relA expression and ppGpp levels during the stringent response (Fig. 3B). We predict that similar reciprocal interactions between the Csr system and other stress responsive transcriptional regulatory systems may serve to link the expression of CsrA regulated genes to a variety of stress responses. Thus, while CsrA and σs often have opposite effects on gene expression, the role of the Csr system may be similar that of σs in governing the expression of its regulon in response to numerous distinct stress conditions (Klauck et al., 2007).
See-saw regulation of enteric pathogenicity
While there are now numerous examples in which the Csr system affects expression of bacterial virulence factors, the first evidence of its involvement in mammalian infection was from studies of S. Typhimurium (Altier et al., 2000). A notable observation from this work was that both a CsrA deficiency and CsrA excess disrupted the expression of virulence factors involved in invasion. Recently, this phenomenon, “see-saw” expression, was observed in enteropathogenic E. coli (EPEC), and the circuitry responsible for this pattern was identified (Bhatt et al., 2009). EPEC causes infantile diarrhea by attaching to intestinal microvilli and forming actin-filled protrusions known as pedestals (Chen and Frankel, 2005). The Lee virulence locus of this bacterium contains several operons required for expression of a type III secretion system (TTSS) that is necessary for actin pedestal formation and virulence (McDaniel and Kaper, 1997; Bhatt et al., 2011). CsrA binds to and activates expression of mRNAs encoding secreted proteins and components of the TTSS. Thus, CsrA is required for expression of TTSS structural genes and pedestal formation. CsrA also bound to mRNA of a regulatory operon, grlRA, which encodes an activator of Lee locus gene expression, GrlA. Overexpression of CsrA globally repressed Lee locus gene expression via effects on grlRA, suggesting a two-tier model for see-saw regulation in this bacterium (Fig. 3C). Although its biological significance is yet to be established, the see-saw pattern of regulation in enteric pathogens raises the possibility that the Csr system acts as both an activator and repressor of virulence gene expression, perhaps to coordinate virulence gene expression in the microenvironmental conditions of the mammalian host.
A novel model for Csr circuitry: FliW is an anti-CsrA protein in Bacillus subtilis that creates a checkpoint in flagellum biosynthesis
While the Csr system has been studied extensively in Gram-negative organisms, comparatively little is known about the Csr circuitry and the function of CsrA in Gram-positive bacteria. In the one Gram-positive organism in which CsrA has been studied, it was shown that CsrA represses expression of hag, the gene encoding flagellin of B. subtilis, by binding to two sites in the hag leader RNA (Yakhnin et al., 2007). As one of these sites overlaps the hag SD sequence, CsrA represses translation by essentially the same mechanism as for E. coli and other Gram-negative species (Yakhnin et al., 2007). Of particular interest, FliW of B. subtilis was shown to function as the first known protein antagonist of CsrA activity in any species. FliW, CsrA and Hag proteins participate in a protein partner switching mechanism to control Hag synthesis (Mukherjee et al., 2011; Fig. 4). Following completion of the flagellar hook, secretion of Hag releases FliW from a FliW-Hag complex. FliW then binds to CsrA, which relieves CsrA-mediated repression of hag translation such that Hag synthesis is highest when it is needed for flagellar assembly. Thus, Hag homeostatically restricts its own translation (Mukherjee et al., 2011). Phylogenetic analyses of the genetic neighborhood of CsrA and FliW suggest this model for CsrA regulation may apply to many other bacterial groups, and might even be a more ancient regulatory system than the γ-proteobacterial sRNA model (Mukherjee et al., 2011). Despite the presence of a protein antagonist of CsrA activity in B. subtilis, a gene predicted to encode a CsrB-like sRNA has been identified (Kulkarni et al., 2006); however, it is not yet known whether this gene encodes an sRNA antagonist of CsrA activity.
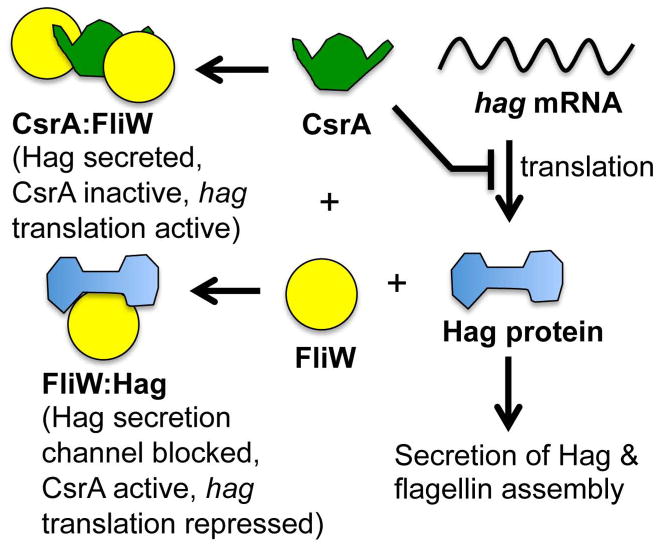
Fig 4.
A partner-switching mechanism involving B. subtilis CsrA, FliW and Hag proteins create a morphogenetic checkpoint for flagellum biosynthesis. CsrA protein represses translation of hag mRNA encoding flagellin. FliW competitively binds to Hag or CsrA, depending on the intracellular concentration of Hag. Prior to the completion of the flagellum secretion channel or upon capping of the pore, high intracellular levels of Hag occupy FliW and CsrA is free to block Hag translation. When the flagellum channel is open and Hag protein is being actively secreted, intracellular Hag levels drop and FliW is free to bind to CsrA, derepressing the synthesis of Hag protein (Mukherjee et al., 2011).
Concluding Remarks
The Csr systems of γ-proteobacteria make use of noncoding RNAs that sequester multiple copies of CsrA and thus inhibit the activity of this global regulatory mRNA binding protein. This rapid and efficient strategy for genetic regulation surfaced at least twice during eubacterial evolution, but is yet unknown in eukaryotic organisms or Achaea. While the basic components of the Csr system and the structural biology of CsrA-RNA binding have been described, new molecular mechanisms of CsrA action are still being uncovered. We expect a variety of different CsrA-dependent activation and repression mechanisms to be described in the near future.
Microarray technology and RNA-seq approaches have proven to be useful for gaining a broad overview of the CsrA regulon. Results from such studies suggest that the Csr system not only directly regulates the expression of hundreds of structural genes, but also dozens of genes encoding regulatory factors. The findings from these approaches have provided clues for deciphering novel patterns of regulatory circuitry in E. coli. The finding that the Csr and stringent response systems exhibit reciprocal regulatory effects on each other, which posttranscriptionally reinforce transcriptional regulation by the stringent response, was an unanticipated discovery. Do similar reciprocal interactions occur between Csr and other regulatory systems whose genes are part of the csrA regulon? This would imply that the Csr system serves as a global, posttranscriptional regulator of stress responses.
The Csr system of E. coli, and perhaps other γ-proteobacteria, contains multiple autoregulatory loops, designed to provide precise control of CsrA levels and activity. In contrast, little is known about the workings of the Csr systems of other bacteria. Bacillus subtilis, the only Gram-positive bacterium in which CsrA has been studied to date, uses an anti-CsrA protein factor, FliW, to control flagellum synthesis. Does this species also express one or more CsrA-antagonistic RNAs? Do many other species express anti-CsrA FliW proteins, as suggested by phylogenetic analyses? Additional studies will be required to determine the phylogenetic distribution of the sRNA-based model for controlling CsrA activity, the FliW model, or perhaps undiscovered regulatory mechanisms.
The recent finding that the rapid turnover of CsrB/C by RNase E and PNPase requires a specificity factor (CsrD), which is not involved in most other RNase E decay pathways, suggests a novel point of regulation in the Csr system. This protein is not a nuclease, and although it contains GGDEF and EAL domains, does not produce, degrade, or mediate a response to c-di-GMP. The precise molecular mechanism of CsrD in E. coli is an important open question. Whether species that express Csr RNAs, but lack apparent CsrD homologues, employ related strategies for triggering decay of these RNAs also remains to be determined.
Acknowledgments
Studies in the authors’ laboratories that contributed to this body of literature were supported by NIH grants GM059969 (T.R. and P.B.) and GM066794 (T.R.) and University of Florida CRIS project, FLA-MCS-004949 (T.R.). We gratefully acknowledge the important contributions of the students and postdoctoral researchers who have worked in our laboratories to elucidate the Csr system.
References
- Altier C, Suyemoto M, Lawhon SD. Regulation of enterica serovar typhimurium invasion genes by csrA. Infect Immun. 2000;68:6790–6797. doi: 10.1128/iai.68.12.6790-6797.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M, Silva IJ, Viegas SC. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev. 2010;34:883–923. doi: 10.1111/j.1574-6976.2010.00242.x. [DOI] [PubMed] [Google Scholar]
- Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Baker CS, Eöry LA, Yakhnin H, Mercante J, Romeo T, Babitzke P. CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. J Bacteriol. 2007;189:5472–5481. doi: 10.1128/JB.00529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CS, Morozov I, Suzuki K, Romeo T, Babitzke P. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol Microbiol. 2002;44:1599–1610. doi: 10.1046/j.1365-2958.2002.02982.x. [DOI] [PubMed] [Google Scholar]
- Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco JG. All things must pass: contrasts and commonalities in eukaryotic and bacterial mRNA decay. Nat Rev Mol Cell Biol. 2010;11:467–478. doi: 10.1038/nrm2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S, Edwards AN, Nguyen HT, Merlin D, Romeo T, Kalman D. The RNA binding protein CsrA is a pleiotropic regulator of the locus of enterocyte effacement pathogenicity island of enteropathogenic Escherichia coli. Infect Immun. 2009;77:3552–3568. doi: 10.1128/IAI.00418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S, Romeo T, Kalman D. Honing the message: post-transcriptional and post-translational control in attaching and effacing pathogens. Trends Microbiol. 2011;19:217–224. doi: 10.1016/j.tim.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaya D, Davisons M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: Versatile small RNAs for adaptive defense and regulation. Ann Rev Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- Boehm A, Steiner S, Zaehringer F, Casanova A, Hamburger F, Ritz D, Keck W, Ackermann M, Schirmer T, Jenal U. Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress. Mol Microbiol. 2009;72:1500–1516. doi: 10.1111/j.1365-2958.2009.06739.x. [DOI] [PubMed] [Google Scholar]
- Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrowes E, Baysse C, Adams C, O’Gara F. Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology. 2006;152:405–418. doi: 10.1099/mic.0.28324-0. [DOI] [PubMed] [Google Scholar]
- Carpousis AJ. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu Rev Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- Chatterjee A, Cui Y, Liu Y, Dumenyo CK, Chatterjee AK. Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density-sensing signal, N-(3-oxohexanoyl)-L-homoserine lactone. Appl Environ Microbiol. 1995;61:1959–1967. doi: 10.1128/aem.61.5.1959-1967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez RG, Alvarez AF, Romeo T, Georgellis D. The physiological stimulus for the BarA sensor kinase. J Bacteriol. 2010;192:2009–2012. doi: 10.1128/JB.01685-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HD, Frankel G. Enteropathogenic Escherichia coli: unravelling pathogenesis. FEMS Microbiol Rev. 2005;29:83–98. doi: 10.1016/j.femsre.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Dubey AK, Baker CS, Romeo T, Babitzke P. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA. 2005;11:1579–1587. doi: 10.1261/rna.2990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey AK, Baker CS, Suzuki K, Jones DA, Romeo T, Babitzke P. CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J Bacteriol. 2003;185:4450–4460. doi: 10.1128/JB.185.15.4450-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AN, Patterson-Fortin LM, Vakulskas CA, Mercante JW, Potrykus K, Camacho MI, Fields JA, Thompson SA, Georgellis D, Cashel M, Babitzke P, Romeo T. Circuitry linking the Csr and stringent response global regulatory systems. Mol Microbiol. 2011;80:1561–1580. doi: 10.1111/j.1365-2958.2011.07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goller C, Wang X, Itoh Y, Romeo T. The cation-responsive protein NhaR of Escherichia coli activates pgaABCD transcription, required for production of the biofilm adhesin poly-β-1,6-N-acetyl-D-glucosamine. J Bacteriol. 2006;23:8022–8032. doi: 10.1128/JB.01106-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollnick P, Babitzke P. Transcription attenuation. Biochim Biophys Acta. 2002;1577:240–250. doi: 10.1016/s0167-4781(02)00455-4. [DOI] [PubMed] [Google Scholar]
- Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu Rev Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2011;3:1–16. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudapaty S, Suzuki K, Wang X, Babitzke P, Romeo T. Regulatory interactions of Csr components: the RNA binding protein CsrA activates csrB transcription in Escherichia coli. J Bacteriol. 2001;183:6017–6027. doi: 10.1128/JB.183.20.6017-6027.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez P, Li Y, Osborne MJ, Pomerantseva E, Liu Q, Gehring K. Solution structure of the carbon storage regulator protein CsrA from Escherichia coli. J Bacteriol. 2005;187:3496–3501. doi: 10.1128/JB.187.10.3496-3501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- Heroven AK, Böhme K, Dersch P. The Csr/Rsm system of Yersinia and related pathogens: A post-transcriptional strategy for managing virulence. RNA Biol. 2012;9 doi: 10.4161/rna.19333. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Irie Y, Starke M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and posttranscriptionally by RsmA. Mol Microbiol. 2010;78:158–172. doi: 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama A. Prokaryotic genome regulation: multifactor promoters, multitarget regulators and hierarchic networks. FEMS Microbiol Rev. 2010;34:628–645. doi: 10.1111/j.1574-6976.2010.00227.x. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Rice JD, Goller C, Pannuri A, Taylor J, Meisner J, Beveridge TJ, Preston JF, III, Romeo T. Roles of pgaABCD genes in synthesis, modification and export of the Escherichia coli biofilm adhesin poly-β-1,6-N-acetyl-D-glucosamine. J Bacteriol. 2008;190:3670–80. doi: 10.1128/JB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Wang X, Hinnebusch BJ, Preston JF, III, Romeo T. Depolymerization of ®-1,6-N-acetyl-D-glucosamine disrupts the integrity of diverse bacterial biofilms. J Bacteriol. 2005;187:382–387. doi: 10.1128/JB.187.1.382-387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DW, Suzuki K, Oakford L, Simecka JW, Hart ME, Romeo T. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J Bacteriol. 2002;184:290–301. doi: 10.1128/JB.184.1.290-301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas K, Edwards AN, Ahmad I, Römeo T, Romling U, Melefors O. Complex regulatory network encompassing the Csr, c-di-GMP and motility systems of Salmonella Typhimurium. Environ Microbiol. 2010;12:524–540. doi: 10.1111/j.1462-2920.2009.02097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas K, Edwards AN, Simm R, Römeo T, Romling U, Melefors O. The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol Microbiol. 2008;70:236–257. doi: 10.1111/j.1365-2958.2008.06411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas K, Melefors O. The Escherichia coli CsrB and CsrC small RNAs are strongly induced during growth in nutrient-poor medium. FEMS Microbiol Lett. 2009;297:80–86. doi: 10.1111/j.1574-6968.2009.01661.x. [DOI] [PubMed] [Google Scholar]
- Kaberdin VR, Bläsi U. Translation initiation and the fate of bacterial mRNAs. FEMS Microbiol Rev. 2006;30:967–979. doi: 10.1111/j.1574-6976.2006.00043.x. [DOI] [PubMed] [Google Scholar]
- Karatan E, Watnick P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev. 2009;73:310–347. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauck E, Typas A, Hengge R. The sigmaS subunit of RNA polymerase as a signal integrator and network master regulator in the general stress response in Escherichia coli. Sci Prog. 2007;90:103–127. doi: 10.3184/003685007X215922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter R, Siegele DA, Tormo A. The stationary phase of the bacterial life cycle. Annu Rev Microbiol. 1993;47:855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- Kulkarni PR, Cui X, Williams JW, Stevens AM, Kulkarni RV. Prediction of CsrA-regulating small RNAs in bacteria and their experimental verification in Vibrio fischeri. Nucleic Acids Res. 2006;34:3361–3369. doi: 10.1093/nar/gkl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner SR. mRNA decay in Escherichia coli comes of age. J Bacteriol. 2002;184:4658–4665. doi: 10.1128/JB.184.17.4658-4665.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawhon SD, Frye JG, Suyemoto M, Porwollik S, McClelland M, Altier C. Global regulation by CsrA in Salmonella typhimurium. Mol Microbiol. 2003;48:1633–1645. doi: 10.1046/j.1365-2958.2003.03535.x. [DOI] [PubMed] [Google Scholar]
- Lapouge K, Sineva E, Lindell M, Starke K, Baker CS, Babitzke P, Haas D. Mechanism of hcnA mRNA recognition in the Gac/Rsm signal transduction pathway of Pseudomonas fluorescens. Mol Microbiol. 2007;66:341–356. doi: 10.1111/j.1365-2958.2007.05909.x. [DOI] [PubMed] [Google Scholar]
- Lapouge K, Schubert M, Allain FH, Haas D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol. 2008;67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- Liu MY, Gui G, Wei B, Preston JF, III, Oakford L, Yuksel U, Giedroc DP, Romeo T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem. 1997;272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- Liu MY, Romeo T. The global regulator CsrA of Escherichia coli is a specific mRNA-binding protein. J Bacteriol. 1997;179:4639–4642. doi: 10.1128/jb.179.14.4639-4642.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MY, Yang H, Romeo T. The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. J Bacteriol. 1995;177:2663–2672. doi: 10.1128/jb.177.10.2663-2672.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchetti-Miganeh C, Burrowes E, Baysse C, Ermel G. The post-transcriptional regulator CsrA plays a central role in the adaptation of bacterial pathogens to different stages of infection in animal hosts. Microbiology. 2008;154:16–29. doi: 10.1099/mic.0.2007/012286-0. [DOI] [PubMed] [Google Scholar]
- Martínez LC, Yakhnin H, Camacho MI, Georgellis D, Babitzke P, Puente JL, Bustamante VH. Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol Microbiol. 2011;80:1637–1656. doi: 10.1111/j.1365-2958.2011.07674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matin A, Auger EA, Blum PH, Schultz JE. Genetic basis of starvation survival in nondifferentiating bacteria. Annu Rev Microbiol. 1989;43:293–316. doi: 10.1146/annurev.mi.43.100189.001453. [DOI] [PubMed] [Google Scholar]
- McDaniel TK, Kaper JB. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol Microbiol. 1997;23:399–407. doi: 10.1046/j.1365-2958.1997.2311591.x. [DOI] [PubMed] [Google Scholar]
- Mercante J, Edwards AN, Dubey AK, Babitzke P, Romeo T. Molecular Geometry of CsrA (RsmA) RNA Binding and its Implications for Regulated Expression. J Mol Biol. 2009;392:511–528. doi: 10.1016/j.jmb.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercante J, Suzuki K, Cheng X, Babitzke P, Romeo T. Comprehensive alanine-scanning mutagenesis of Escherichia coli CsrA defines two subdomains of critical functional importance. J Biol Chem. 2006;281:31832–31842. doi: 10.1074/jbc.M606057200. [DOI] [PubMed] [Google Scholar]
- Mikkelsen H, Sivaneson M, Filloux A. Key two-component regulatory systems that control biofilm formation in Pseudomonas aeruginosa. Environ Microbiol. 2011;13:1666–1681. doi: 10.1111/j.1462-2920.2011.02495.x. [DOI] [PubMed] [Google Scholar]
- Molofsky AB, Swanson MS. Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol Microbiol. 2004;53:29–40. doi: 10.1111/j.1365-2958.2004.04129.x. [DOI] [PubMed] [Google Scholar]
- Moreno R, Fonseca P, Rojo F. Two small RNAs, CrcY and CrcZ, act in concert to sequester the Crc global regulator in Pseudomonas putida, modulating catabolite repression. Mol Microbiol. 2012;83:24–40. doi: 10.1111/j.1365-2958.2011.07912.x. [DOI] [PubMed] [Google Scholar]
- Moreno R, Ruiz-Manzano A, Yuste L, Rojo F. The Pseudomonas putida Crc global regulator is an RNA binding protein that inhibits translation of the AlkS transcriptional regulator. Mol Microbiol. 2007;64:665–675. doi: 10.1111/j.1365-2958.2007.05685.x. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Cui Y, Liu Y, Dumenyo CK, Chatterjee AK. Global regulation in Erwinia species by Erwinia carotovora rsmA, a homologue of Escherichia coli csrA: repression of secondary metabolites, pathogenicity and hypersensitive reaction. Microbiology. 1996;142:427–434. doi: 10.1099/13500872-142-2-427. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Yakhnin H, Kysela D, Sokoloski J, Babitzke P, Kearns DB. CsrA-FliW interaction governs flagellin homeostasis and a checkpoint on flagellar morphogenesis in Bacillus subtilis. Mol Microbiol. 2011;82:447–461. doi: 10.1111/j.1365-2958.2011.07822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannuri A, Yakhnin H, Vakulskas CA, Edwards AN, Babitzke P, Romeo T. Translational Repression of NhaR, a Novel Pathway for Multi-Tier Regulation of Biofilm Circuitry by CsrA. J Bacteriol. 2012;194:79–89. doi: 10.1128/JB.06209-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. Sticky situations – Key components that control bacterial surface attachment. J Bacteriol. 2012 doi: 10.1128/JB.00003-12. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Kulkarni PR, Cui X, Williams JW, Stevens AM, Kulkarni RV. Prediction of CsrA-regulating small RNAs in bacteria and their experimental verification in Vibrio fischeri. Nucleic Acids Res. 2006;34:3361–3369. doi: 10.1093/nar/gkl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulski EE, Moy RH, Weinberg Z, Barrick JE, Yao Z, Ruzzo WL, Breaker RR. A widespread riboswitch candidate that controls bacterial genes involved in molybdenum cofactor and tungsten cofactor metabolism. Mol Microbiol. 2008;68:918–932. doi: 10.1111/j.1365-2958.2008.06208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rife C, Schwarzenbacher R, McMullan D, Abdubek P, Ambing E, Axelrod H, et al. Crystal structure of the global regulatory protein CsrA from Pseudomonas putida at 2.05 A resolution reveals a new fold. Proteins. 2005;61:449–453. doi: 10.1002/prot.20502. [DOI] [PubMed] [Google Scholar]
- Romeo T, Gong M, Liu MY, Brun-Zinkernagel AM. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J Bacteriol. 1993;175:4744–4755. doi: 10.1128/jb.175.15.4744-4755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol. 1998;29:1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x. [DOI] [PubMed] [Google Scholar]
- Römling U, Gomelsky M, Galperin MY. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- Sabnis NA, Yang H, Romeo T. Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA. J Biol Chem. 1995;270:29096–29104. doi: 10.1074/jbc.270.49.29096. [DOI] [PubMed] [Google Scholar]
- Schubert M, Lapouge K, Duss O, Oberstrass FC, Jelesarov I, Haas D, Allain FHT. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat Struct Molec Biol. 2007;14:807–813. doi: 10.1038/nsmb1285. [DOI] [PubMed] [Google Scholar]
- Smith JL, Fratamico PM, Yan X. Eavesdropping by bacteria: the role of SdiA in Escherichia coli and Salmonella enterica serovar Typhimurium quorum sensing. Foodborne Pathog Dis. 2011;8:169–178. doi: 10.1089/fpd.2010.0651. [DOI] [PubMed] [Google Scholar]
- Smith TG, Hoover TR. Deciphering bacterial flagellar gene regulatory networks in the genomic era. Adv Appl Microbiol. 2009;67:257–295. doi: 10.1016/S0065-2164(08)01008-3. [DOI] [PubMed] [Google Scholar]
- Soares JA, Ahmer BM. Detection of acyl-homoserine lactones by Escherichia and Salmonella. Curr Opin Microbiol. 2011;14:188–193. doi: 10.1016/j.mib.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner E, Abdou L, Haas D. Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2009;106:21866–21871. doi: 10.1073/pnas.pnas.0910308106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Babitzke P, Kushner SR, Romeo T. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 2006;20:2605–2617. doi: 10.1101/gad.1461606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Wang X, Weilbacher T, Pernestig AK, Melefors O, Georgellis D, Babitzke P, Romeo T. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J Bacteriol. 2002;184:5130–5140. doi: 10.1128/JB.184.18.5130-5140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsen SL, Tu KC, Bassler BL. Gene dosage compensation calibrates four regulatory RNAs to control Vibrio cholerae quorum sensing. EMBO J. 2009;28:429–439. doi: 10.1038/emboj.2008.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K, Kiefer P, Reimmann C, Keel C, Dubuis C, Rolli J, Vorholt JA, Haas D. Small RNA-dependent expression of secondary metabolism is controlled by Krebs cycle function in Pseudomonas fluorescens. J Biol Chem. 2009;284:34976–85. doi: 10.1074/jbc.M109.052571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliabue L, Antoniani D, Maciag A, Bocci P, Raffaelli N, Landini P. The diguanylate cyclase YddV controls production of the exopolysaccharide poly-N-acetylglucosamine (PNAG) through regulation of the PNAG biosynthetic pgaABCD operon. Microbiology. 2010;156:2901–2911. doi: 10.1099/mic.0.041350-0. [DOI] [PubMed] [Google Scholar]
- Timmermans J, Van Melderen L. Post-transcriptional global regulation by CsrA in bacteria. Cell Mol Life Sci. 2010;67:2897–2908. doi: 10.1007/s00018-010-0381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–89. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. CsrA post-transcriptionally represses pgaABCD, responsible for the synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol. 2005;56:1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- Wang S, Fleming RT, Westbrook EM, Matsumura P, McKay DB. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J Mol Biol. 2006;355:798–808. doi: 10.1016/j.jmb.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Wang X, Preston JF, III, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei BL, Brun-Zinkernagel AM, Simecka JW, Prüß BM, Babitzke P, Romeo T. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol. 2001;40:245–256. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, Morozov I, Baker CS, Georgellis D, Babitzke P, Romeo T. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol Microbiol. 2003;48:657–670. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- Yakhnin H, Baker CS, Berezin I, Evangelista MA, Rassin A, Romeo T, Babitzke P. CsrA represses translation of sdiA, which encodes the N-acylhomoserine-L-lactone receptor of Escherichia coli, by binding exclusively within the coding region of sdiA mRNA. J Bacteriol. 2011a;193:6162–6170. doi: 10.1128/JB.05975-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin H, Pandit P, Petty TJ, Baker CS, Romeo T, Babitzke P. CsrA of Bacillus subtilis regulates translation initiation of the gene encoding the flagellin protein (hag) by blocking ribosome binding. Mol Microbiol. 2007;64:1605–1620. doi: 10.1111/j.1365-2958.2007.05765.x. [DOI] [PubMed] [Google Scholar]
- Yakhnin H, Yakhnin AV, Baker CS, Sineva E, Berezin I, Romeo T, Babitzke P. Complex regulation of the global regulatory gene csrA: CsrA-mediated translational repression, transcription from five promoters by Eσ70 and EσS, and indirect transcriptional activation by CsrA. Mol Microbiol. 2011b;81:689–704. doi: 10.1111/j.1365-2958.2011.07723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Liu MY, Romeo T. Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the CsrA gene product. J Bacteriol. 1996;178:1012–1017. doi: 10.1128/jb.178.4.1012-1017.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]