Abstract
The trimeric human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) spike is a molecular machine that mediates virus entry into host cells and is the sole target for virus-neutralizing antibodies. The mature Env spike results from cleavage of a trimeric gp160 precursor into three gp120 and three gp41 subunits. Here we describe an ~11-Å cryo-EM structure of the trimeric HIV-1 Env precursor in its unliganded state. The three gp120 and three gp41 subunits form a cage-like structure with an interior void surrounding the trimer axis. Interprotomer contacts are limited to the gp41 transmembrane region, the torus-like gp41 ectodomain, and a gp120 trimer association domain composed of the V1/V2 and V3 variable regions. The cage-like architecture, which is unique among characterized viral envelope proteins, restricts antibody access, reflecting requirements imposed by HIV-1 persistence in the host.
Human immunodeficiency virus type (HIV-1) establishes persistent infections in humans that often lead to depletion of CD4+ T lymphocytes and acquired immunodeficiency syndrome (AIDS). To infect human T lymphocytes, HIV-1 utilizes a trimeric envelope glycoprotein (Env) spike on the virion surface to engage the host receptors, CD4 and a chemokine receptor (either CCR5 or CXCR4), and to fuse the viral and target cell membranes1,2. During synthesis and folding in virus-producing cells, the Env precursors trimerize and are heavily modified by N-linked glycosylation3,4. HIV-1 Env is one of the most heavily glycosylated proteins in nature; each Env protomer usually contains 27 or more glycosylation sites, three to five times more than the number of glycosylation sites found in other viral envelope glycoproteins. In the Golgi complex, the gp160 Env precursor is cleaved into a gp120 exterior subunit and a gp41 transmembrane subunit by a host furin protease3,4. Proteolytic cleavage renders the Env trimer competent for membrane fusion, and occurs immediately prior to cell surface expression and incorporation into virus particles. The Env spike on the virion surface is the only virus-specific component potentially accessible to neutralizing antibodies5. A typical HIV-1 virion contains only 10-14 Env spikes, so each spike must defend itself against antibody binding. The dense ‘glycan shield’ and a high degree of interstrain structural variability help the Env trimer to evade the humoral immune response4.
The sequential binding of gp120 to the target cell receptors, CD4 and the chemokine receptor, triggers Env transitions from the unliganded state to ‘fusion-ready’ conformations4. CD4 binding results in the formation and exposure of two Env elements: the gp120 chemokine receptor-binding site and a trimeric gp41 coiled coil. The resulting ‘pre-hairpin intermediate’ binds the chemokine receptor, and the hydrophobic ‘fusion peptide’ at the amino terminus of gp41 is thought to insert into the target cell membrane. The collapse of the pre-hairpin intermediate into a stable six-helix bundle results in the fusion of viral and cell membranes. Previous studies using xray crystallography have revealed the structure of the monomeric gp120 core, in most cases with the V1/V2 and V3 variable regions deleted, in the CD4-bound state6-8. The gp41 ectodomain, with truncations of the fusion peptide and disulfide-bonded loop, crystallized as a six-helix bundle, which likely represents the end-stage, post-fusion conformation9-11. The energetically stable six-helix bundle is composed of a trimeric coiled coil derived from the gp41 heptad repeat (HR1) region, with helices derived from the gp41 HR2 region packed against the coiled coil in an antiparallel fashion. The structure of the HIV-1 Env trimer complex in its unliganded state, which represents the major target for most neutralizing antibodies, is poorly defined. Despite extensive efforts, heavy glycosylation, structural plasticity and lability have frustrated attempts to obtain a high-resolution structure of the HIV-1 Env trimer. Cryo-electron tomography of virion Env spikes and cryo-electron microscopy (cryo-EM) analysis of purified Env variants have yielded structural models at 20-30 Å resolution12-18. At this resolution, the organization of the assembled gp120 and gp41 subunits in the trimer could not be assessed. We applied single-particle cryo-EM analysis19 to reconstruct an ~11-Å structure of a membrane-bound HIV-1 Env trimer, including the complete exterior and transmembrane regions, in an unliganded state. The improved details in our cryo-EM structure reveal the architectural organization of gp120 and gp41 subunits in an unliganded trimeric Env precursor. The new HIV-1 Env trimer structure allows a better understanding of the function of this molecular machine and provides insights into the mechanism of HIV-1 entry and immune evasion.
RESULTS
Structure determination
The Env glycoprotein derived from a primary, neutralization-resistant (Tier 2) clade B isolate, HIV-1JR-FL, was chosen for this study. The gp120-gp41 proteolytic cleavage site in the HIV-1JR-FL Env was eliminated by two single-residue changes (R508S and R511S in standard HXB2 numbering). To improve the expression level on the cell surface, the gp41 cytoplasmic tail was truncated starting from Tyr712. The modified Env, designated Env(−)ΔCT, thus contains the complete ectodomain and transmembrane regions, and was purified from the plasma membrane of Env-expressing cells after solubilization in Cymal-5 detergent (see Methods). This procedure ensures that the purified Env(−)ΔCT trimers derive from membrane-bound Env complexes that are glycosylated and have passed the quality-control checkpoints of the secretory pathway3,4. Importantly, HIV-1 Env(−)ΔCT complexes purified in this manner retain epitopes that are dependent upon conformation, glycosylation and quaternary structure (Supplementary Fig. 1). The detergent Cymal-5 was exchanged to Cymal-6 before preparation for cryo-EM imaging. The membrane glycoprotein, under the protection of Cymal-6, was flash-frozen on holey carbon film-coated EM grids and cryo-EM image data were collected at liquid nitrogen temperature. The imaging quality was found to be critically affected by the choice of the detergent and its concentration in the vitrified cryo-EM samples. A dataset of 90,306 single-particle images was assembled and subjected to multivariate data analysis, maximum-likelihood alignment and classification19-21. An initial model was generated by angular reconstitution from two-dimensional class averages refined by a maximum-likelihood approach20,21. The model was then further refined by a projection-matching algorithm to a final resolution of 10.8 Å, measured by Fourier shell correlation (FSC) with a 0.5-cutoff criterion22 (see Supplementary Figs. 2-5 for more technical details).
Overall architecture
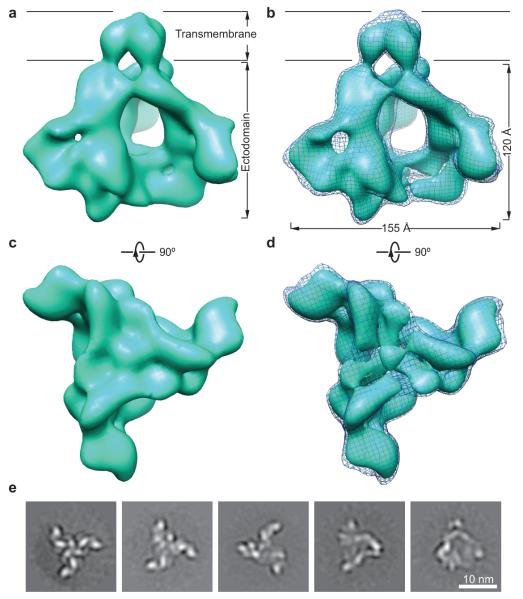
The HIV-1JR-FL Env(−)ΔCT complex resembles a triangular pyramid, with an overall tetrahedral shape and a remarkable central empty space (Fig. 1 and Supplementary Movie 1). The transmembrane domain of gp41 shows three lobes that associate at the trimer axis. Extending from the viral membrane, each protomer projects approximately 30° away from the trimer axis. Narrow interprotomer joints at the base of each projection suggest interactions among the gp41 ectodomains. At the membrane-distal end of the Env complex, each protomeric ‘leaf’ turns approximately 90° towards the trimer axis and there participates in a six-way junction at the gp120-gp120 interfaces.
Figure 1.
The cryo-EM structure of the membrane-bound HIV-1 Env trimer at ~11-Å resolution. (a) The reconstruction of the HIV-1JR-FL Env(−)ΔCT trimer is shown as a solid surface viewed from a perspective parallel to the viral membrane. The approximate boundaries of the transmembrane region and ectodomain are indicated. (b) The Env trimer reconstruction is visualized at two different levels of contour. The lower and higher levels of contour are illustrated as a meshwork and a solid surface representation, respectively. (c) The Env trimer reconstruction in a solid surface representation is viewed from the perspective of the target cell, at the same contour level as that shown in a. (d) The Env trimer reconstruction is shown at two different levels of contour in the same way as in b, viewed from the perspective of the target cell. (e) The images show typical reference-free class averages produced by maximum-likelihood alignment with no C3 symmetry imposed. Scale bar, 10 nm. See Supplementary Figures 3 and 4 and Supplementary Movies 2 and 3 for more details.
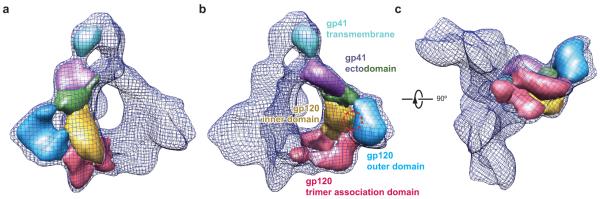
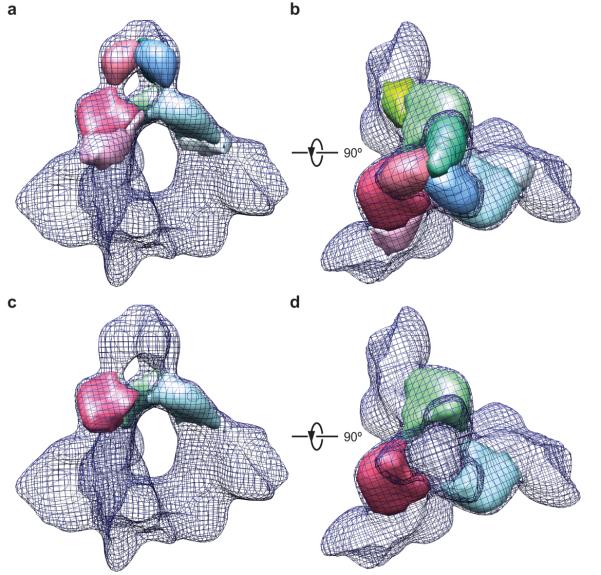
The cryo-EM reconstruction appears to be of sufficient quality to permit map segmentations that make biological sense and are compatible with available structural information. Each Env protomer can be segmented into eight sub-structures (Fig. 2). Limited by the resolution of the reconstruction itself, the map segmentation may not precisely define the interfaces between structural subunits and domains. Nonetheless, the overall subunit definitions conveyed by the segments are instructive. The gp41 transmembrane domain and the gp41 ectodomain clearly partitioned into discrete segments, and two gp41 ectodomain segments were defined. The inner domain and outer domain of gp120 are also visible in two segmented sub-structures adjacent to the more distal gp41 ectodomain segment. The rest of the gp120 segmentation appears as two arms extending from the gp120 inner and outer domains toward a central triangular junction at the gp120 trimer axis (Figs. 1d and 2c).
Figure 2.
The map segmentation of an Env protomer. (a, b) The map segmentation of an Env protomer in solid surface representation is wrapped in a meshwork representation of the Env timer, viewed from a perspective parallel to the viral membrane. Two different Env protomers are segmented in a and b. The segments approximately correspond to the indicated gp120 and gp41 domains. The red dashed circle indicates the approximate position of the CD4-binding site on the gp120 subunit. (c) The segmentation is viewed from the perspective of the target cell.
gp120 inner and outer domains
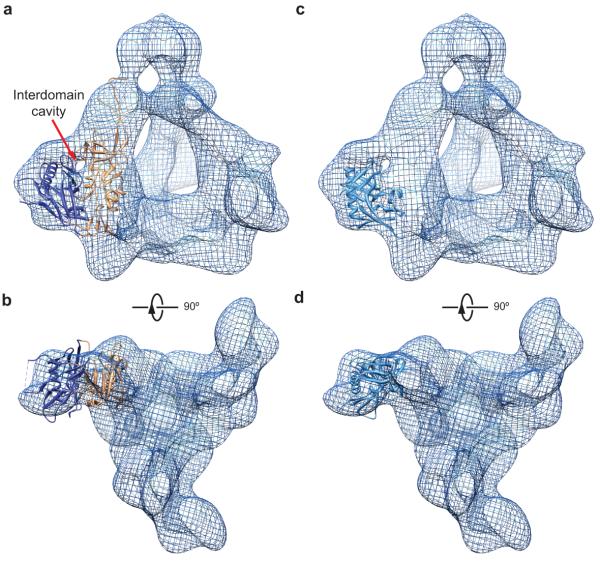
The spatial relationship between the gp120 outer domain and inner domain is apparent in the density map, although at this resolution it is not possible to distinguish any secondary structures within the domains. The size and shape of the gp120 segments corresponding to the inner domain and outer domain are consistent with those seen in xray crystal structures (compare Fig. 2a-c with Fig. 3a,b). Interestingly, the cryo-EM reconstruction clearly shows a cavity at the interface between the gp120 outer domain and inner domain. This prominent cavity is reminiscent of a similar feature observed in the crystal structure of the CD4-bound gp120 core monomer8, and also provides a structural constraint for fitting the crystal structure of the gp120 core into the cryo-EM map. Indeed, the crystal structure of the CD4-bound gp120 core8 was fitted as a rigid body into the cryo-EM map, with a good agreement on the position of the inter-domain cavity (Fig. 3a). In this fitting configuration, the gp120 inner domain in its CD4-bound state fit the density well (Fig. 3a,b); however, the gp120 outer domain fit less well, as can be best appreciated when viewed from the perspective of the target cell (Fig. 3b). The crystal structure of the gp120 outer domain (in the CD4-bound state) alone fit well as a rigid body into the cryo-EM map in a slightly different fitting configuration (Fig. 3c,d). However, in this case, the associated CD4-bound inner domain structure was out of fit (not shown). Thus, the crystal structures of the inner domain and outer domain cannot simultaneously fit the cryo-EM map while the CD4-bound state of gp120 is maintained. These observations support the existence of conformational differences between the unliganded and CD4-bound states in the Env trimer. In fact, there is a substantial body of literature documenting a major conformational change in the gp120 subunit upon CD4 binding14-17,23-28. Our analysis is consistent with this notion and suggests that CD4 engagement modifies the spatial relationship between the gp120 inner domain and outer domain.
Figure 3.
Fit of the crystal structure of CD4-bound gp120 core into the cryo-EM density of the unliganded Env trimer in two different configurations. (a, b) The CD4-bound gp120 core structure (PDB ID: 3JWD)8 can fit into the local Env trimer cryo-EM density identified by segmentation as the gp120 inner domain and outer domain (See Fig. 2). This fitting configuration represents the best fit of the gp120 core crystal structure, taking into account both the inner and outer domains. The cryo-EM density is shown as a meshwork representation. The red arrow in a marks the interdomain cavity in the gp120 subunit. The gp120 core is missing the V1/V2 and V3 variable regions8. The inner domain and outer domain of the gp120 core crystal structure are coloured orange and dark blue, respectively. A fraction of the residues of the gp120 outer domain does not fit in the cryo-EM density in this fitting configuration, suggesting conformational differences between the unliganded and CD4-bound states. (c, d) The gp120 outer domain from the CD4-bound gp120 core structure8 was fitted into the cryo-EM density of the Env trimer, without consideration of the fit of the gp120 inner domain. This allows a much better fitting of the gp120 outer domain than that seen in a and b; only the LV5 variable loop has a few residues out of the cryo-EM density. Under the same fitting configuration, the inner domain of the CD4-bound gp120 core is largely outside the Env trimer cryo-EM density (not shown). This observation suggests that the gp120 inner domain rotates with respect to the outer domain upon binding CD4.
The outer domain in the crystal structure of an unliganded gp120 core monomer from simian immunodeficiency virus (SIV)29 also fit very well into our cryo-EM map in the same fitting configuration as that used for the CD4-bound HIV-1 gp120 core. However, in this configuration, the inner domain of the SIV gp120 core did not fit the cryo-EM map. Apparently, the unliganded gp120 structure differs between SIV in the monomeric form and HIV-1 in the trimeric form.
At a higher level of contour in the cryo-EM map, the weak density linking the gp120 inner domain and outer domain at the midpoint of the interdomain interface agrees with the location of interdomain strands in the CD4-bound gp120 core structure6-8. Taken together, the structural consistency between the cryo-EM density map and previously reported x-ray crystallography data provides an independent check on the reconstruction accuracy and resolution estimation of the cryo-EM model.
gp120 trimer association
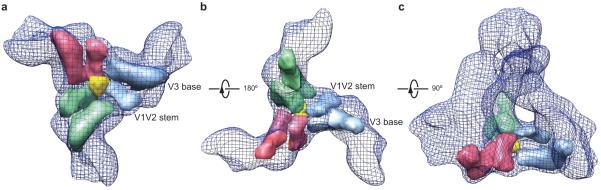
The cryo-EM map exhibits rich features around the gp120-gp120 interfaces, revealing that the interprotomer contacts among the gp120 subunits involve a six-way junction (Fig. 4a). Two elongated arm-like elements extend from the core of each gp120 subunit toward the trimer axis. One arm originates in the gp120 outer domain at the base of the V3 variable region and terminates about 1.7 nm away from the trimer center. The V3 variable region, which is flanked by disulfide-bonded cysteine residues at its base, likely contributes to this arm. The other arm extends from the gp120 inner domain and points directly at the trimer axis (Fig. 4b, c). This arm likely contains the V1/V2 stem, which is composed of two antiparallel β-strands stabilized at both ends by disulfide bonds6. The V1/V2 stem projects ~ 2.7 nm from the inner domain and serves as a point of origin and return for the V1 and V2 variable regions. At the gp120 trimer center, a triangular junction about 1.5 nm across points its apex toward the target cell (Fig. 4a). The interprotomer contacts among the three gp120 subunits involve this central triangular junction and the six arms; together, these structures can account for the expected mass of the gp120 V1/V2 and V3 regions. These results agree with recent electron tomography studies of wild-type and V1/V2-truncated HIV-1 Env spikes18, which suggested that the V1/V2 regions reside in the membrane-distal apex of the spike.
Figure 4.
The gp120 trimer association domains. (a) The segmented local densities of gp120 that associate near the trimer axis are shown as solid surfaces wrapped in a meshwork representing the overall Env trimer, viewed from the perspective of the target cell. Note the central triangular junction (yellow) with a diameter of around 1.5 nm. (b) The gp120 trimer association domain segments are viewed from the perspective of the viral membrane. Three arms extend from the V3 base of each gp120 subunit toward the trimer axis and appear to support the central triangular junction. The approximate positions of the V1/V2 stem and V3 base are labelled in a and b. (c) The gp120 trimer association domain segments are viewed from a perspective parallel to the viral membrane.
The V1/V2 and V3 variable regions have been speculated to be highly disordered in the unliganded state of HIV-1 Env. This speculation is not supported by our cryo-EM map. If the structure of these variable regions is mostly disordered in the unliganded state, the averaging of cryo-EM single-particle projections should eliminate or substantially weaken their corresponding density. On the contrary, our cryo-EM map exhibits a very stable reconstruction of the central triangle and six-arm junction that is very unlikely to be accounted for by disordered conformations. Our analysis instead favors the possibility that the V1/V2 and V3 regions of gp120 fold into a relatively stable structure. Such a model likely explains the observation that, in primary HIV-1 strains, changes in the V3 variable region can result in a decreased association of gp120 with the Env trimer26.
A recent crystal structure of a neutralizing antibody, PG9, bound to the gp120 V1/V2 region expressed as a fusion with a heterologous scaffold protein suggested that V1/V2 can form secondary structures30. Perhaps because the V1/V2-scaffold protein is missing the gp120 core and V3 region and was analyzed in a monomeric state, the V1/V2 segment of this structure does not fit into our cryo-EM density and therefore may not exhibit the V1/V2 conformation in the unliganded trimer. Given the limited resolution of the present cryo-EM structure, any conclusion regarding the difference in V1/V2 structure between the unliganded trimer and the artificially engineered scaffold protein would be premature. However, both studies suggest some potentially common hypotheses that deserve further investigation. First, the V1/V2 and V3 regions are able to form secondary structures despite their variable nature. Second, the secondary structure(s) formed from V1/V2 and V3 might be metastable and might inter-convert between different forms, depending on the environment and structural context. Third, the overall architecture of the V1/V2 and V3 structures should be conserved across different isolates to maintain the integrity of a functional Env trimer.
gp41 trimer association
The transmembrane region of the gp41 trimer segregates as an entity distinct from the rest of the cryo-EM structure. The transmembrane segment of each gp41 protomer forms a rod-like shape with an approximate length of 3 nm (Fig. 5a,b). The diameter of the trimeric Env transmembrane region can accommodate a three-helix bundle. The extra density surrounding the three-helix bundle might be a mixture of detergents, glycans and protein components, and is not separable at this resolution.
Figure 5.
Architecture of gp41 trimer association. (a, b) The gp41 segments are shown as solid surfaces wrapped in a meshwork representing the overall Env trimer, viewed from a perspective parallel to the viral membrane (a) and from the perspective of the viral membrane (b). (c, d) The gp41 ectodomain segments that form the torus and contribute to interprotomer interactions are shown. The views are from a perspective parallel to the viral membrane (c) and from the perspective of the viral membrane (d).
In contrast to the tightly packed transmembrane regions, the three leaf-like shapes of the gp41 ectodomain spread out from the tripod base formed by the distal ends of three transmembrane rods. Approximately 2 nm from the viral membrane, two lobes of each ‘leaf’ transversely join the cognate elements of the neighbouring subunits to form a torus-like structure (Fig. 5c,d). The gp41 ectodomain extends obliquely to form an interface with the gp120 inner domain approximately 3 nm away from the base of the torus. The direct contacts between the gp120 and gp41 subunits within each protomer are apparently centered on a region approximately halfway along the height of the pyramidal ‘tetrahedron’.
In addition to the gp120 trimer association domain, the gp41 transmembrane region and ectodomain torus make interprotomer contacts that likely stabilize the trimeric Env cage, which bounds a central void surrounding the trimer axis. This central void in the unliganded Env complex contrasts with the densely packed trimeric coiled coil and six-helix bundle that surround the trimer axis of the pre-hairpin intermediate and post-fusion state, respectively. Neither the crystal structure of the post-fusion gp41 core trimer in a six-helix bundle conformation9-11 nor the deduced structure of the gp41 HR1 coiled coil fit into the gp41 component of the cryo-EM density. The torus-like architecture instead indicates a different fold of the gp41 ectodomain in the unliganded Env precursor, and presumably in the proteolytically processed Env trimer as well. Such a model is consistent with studies of HIV-1 variants suggesting that the gp41 coiled coil is not formed in the Env trimer prior to CD4 binding31. Moreover, the gp41 ectodomain, and the HR1 region in particular, play roles in maintaining the non-covalent interaction with gp120 and sequestering the hydrophobic fusion peptide32-35. The contacts that the gp41 ectodomain makes in the unliganded Env trimer presumably keep the gp41 trimer from folding into downstream conformations.
Comparison with tomographic map of the virion Env spike
Comparison of our cryo-EM structure with the 20-Å electron tomographic map14,15 of the proteolytically processed36 and functional37 Env spike on HIV-1 virions indicates a remarkable agreement in overall architecture (Fig. 6). The general agreement of the density maps suggests that our methods to extract and purify the membrane-bound Env trimer preserve a conformation similar to that of the unliganded, pre-fusion conformation of the Env spike on the native virions. Previous studies found that uncleaved Env trimers bind poorly neutralizing antibodies better than proteolytically cleaved Env trimers38,39. However, at a resolution of 10-20 Å, any structural differences resulting from proteolytic cleavage are not apparent. Thus, consistent with studies suggesting that proteolytic processing occurs very late during HIV-1 Env movement through the secretory pathway3, the major quaternary interactions that maintain trimer integrity are established before gp120-gp41 cleavage. Furthermore, any conformational changes in the Env ectodomain resulting from association with the viral membrane or the presence of the gp41 cytoplasmic tail40 are also too subtle to be detected at this level of resolution.
Figure 6.
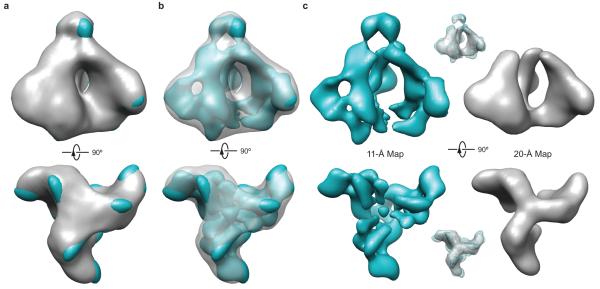
Comparison of the 11-Å cryo-EM structure of the trimeric HIV-1 Env precursor with the 20-Å electron tomographic model of the native HIV-1 Env trimer on virions. (a) The 20-Å model of the HIV-1 Env trimer on virions (grey surface)14 and the 11-Å model of the HIV-1 Env precursor (cyan surface) are superimposed, using the trimer axis as a common reference. Both models are shown at a comparable level of contour. (b) The same models and levels of contour as in a are shown, but the 20-Å tomographic model of the virion Env trimer is shown as a transparent grey surface. (c) The 11-Å cryo-EM model (left) and the 20-Å tomographic model (right) are visualized side by side as solid surface representations at higher levels of contour than those shown in a and b. Upper and lower inserts show the superposition of the two models, with the 11-Å cryo-EM model shown as a meshwork representation and the 20-Å tomographic model as a transparent surface representation.
Comparing our cryo-EM structure to earlier tomographic maps14,15,17, one immediately senses the considerable enrichment of structural features resulting from an improved resolution. The cage-like architecture and the striking central void in the HIV-1 Env trimer become unambiguously manifest. This topology is unusual among viral envelope glycoproteins41 and likely contributes to HIV-1 immune evasion and persistence. By positioning conserved surfaces like the CD4-binding site on the inward-facing side of the protomeric leaves, the accessibility of antibodies to these sites is sterically restricted by the adjacent protomer. Other structural features evident in our cryo-EM map are not apparent in the tomographic map. Whether the 20-Å tomographic map is visualized at a low level (Fig. 6a, b) or high level (Fig. 6c) of contour, the inter-domain cavity between the gp120 inner domain and outer domain is not visible. The six-way junction at the gp120 trimerization interface evident in our cryo-EM map is simplified into a three-way joint in the tomographic map. At a high level of contour (Fig. 6c), the density of the gp41 transmembrane region disappears in the 20-Å tomographic map, but is retained in our cryo-EM map at an amplitude comparable to that of the rest of the density. The 11-Å density map allows a clear-cut visualization of the gp41 torus-like architecture over a wide range of contour levels. These differences in the resolvability of Env features reflect the expected improvement in the density map due to higher resolution, suggesting that the FSC at a 0.5-cutoff provides an appropriate estimation of resolution in this case.
DISCUSSION
In the absence of constraints imposed by the V1/V2 and V3 variable regions, the HIV-1 gp120 core assumes the CD4-bound conformation42. Peptides corresponding to the heptad repeat regions (HR1 and HR2) of the gp41 ectodomain form a stable six-helix bundle in the absence of interactions with gp120 (refs. 9-11). Thus, outside of their structural context in the unliganded HIV-1 Env trimer, both gp120 and gp41 exhibit a propensity to assume conformations that are associated with downstream states on the virus entry pathway. Our cryo-EM map allows us to conclude that gp120 is not in the CD4-bound state in the unliganded Env precursor. The proposed location of the gp120 V1/V2 and V3 variable regions in the trimer association domain of the unliganded HIV-1 Env trimer is consistent with these variable regions restraining movement into the CD4-bound conformation by virtue of their interactions at the gp120 trimer interface. These interactions are likely disrupted upon CD4 binding as the Env trimer ‘opens’14. Changes in V1/V2 and V3 have been shown to render the HIV-1 Env more prone to assume the CD4-bound conformation spontaneously, allowing virus entry into cells expressing no CD4 or low levels of CD4 (refs. 43-45).
The visualization of the HIV-1 gp41 ectodomain in our cryo-EM map allows us to rule out the presence of the gp41 six-helix bundle or the HR1 trimeric coiled coil at the trimer axis of the unliganded Env precursor. This conclusion is consistent with those of several previous studies indicating that the formation of the HR1 trimeric coiled coil and the six-helix bundle are steps in virus entry that occur after CD4 binding31-34.
All approaches to higher-resolution structure determination require enrichment and preferably purification of the molecular complex of interest. The metastability of the HIV-1 Env trimer creates significant challenges to the purification of Env complexes that retain a native conformation. Soluble gp140 glycoproteins that lack the gp41 transmembrane region and cytoplasmic tail are conveniently expressed and purified; however, the epitopes for the PG9/PG16 neutralizing antibodies, which are dependent upon Env quaternary structure, are disrupted in the soluble gp140 glycoproteins, suggesting that these proteins do not completely retain the native conformation of the unliganded Env trimer46.
As an alternative to the soluble gp140 glycoproteins, we explored approaches to solubilize and purify the membrane-bound HIV-1 Env trimer in a native conformation. We screened a wide spectrum of detergent candidates such as CHAPS, DDM (ndodecyl-β-D-maltoside), OG (β-octyl-D-glucoside), FOS-CHOLINE-12, etc. The detergent Cymal-5 was found to be best suited for Env trimer purification. Importantly, the membrane-bound HIV-1JR-FL E168K Env trimers purified by this approach are recognized by the PG16 antibody equivalently to cell-surface-expressed HIV-1JR-FL E168K Env trimers (Supplementary Fig. 1b,c)49. The PG16 neutralizing antibody recognizes a gp120 epitope that is very sensitive to changes in the V1/V2 and V3 variable regions, in the associated glycans, and in the quaternary structure of the Env trimer30,46. Other conformation-dependent neutralizing antibodies, including VRC01 (refs. 47,48), b12 (ref. 7) and 2G12 (ref. 49), recognized the original HIV-1JR-FL Env(−)ΔCT glycoprotein and its E168K variant following purification (Supplementary Fig. 1). Thus, the approach that we developed to purify the membrane-bound Env trimer largely preserves its conformational and structural integrity.
Although single-particle cryo-EM has achieved near-atomic resolution for highly symmetric icosahedral particles50, only a few cases have been reported for membrane protein structures at a resolution better than ~10 Å by single-particle cryo-EM51,52. One formidable barrier impeding single-particle cryo-EM reconstruction of membrane proteins at subnanometer resolution is the requirement for detergents. Detergents form nanometer-sized micelles in the protein solutions that may increase the background noise and further reduce the cryo-EM image contrast. In this study, we screened a number of combinations of detergent and solution configuration to maximize the image contrast prior to 3D reconstruction. The best imaging results were obtained by switching from Cymal-5 to Cymal-6 after Env(−)ΔCT purification and prior to imaging.
Our results demonstrate that the purified Env(−)ΔCT trimers are sufficiently homogeneous in conformation to allow an ~11-Å reconstruction by single-particle cryo-EM. In addition to the choice of detergents that maintain inter-protomer interactions and quaternary epitopes, our use of an uncleaved form of the Env trimer may also have contributed to conformational homogeneity. Indeed, it has been difficult to obtain homogeneous preparations of proteolytically processed wild-type HIV-1 Env trimers due to the lability of these complexes. Cleaved HIV-1 Env trimers have been stabilized by the introduction of an artificial disulfide bond between gp120 and gp41 (SOSgp160ΔCT)16,53. In some cases, the alteration of a gp41 isoleucine residue to proline and deletion of the transmembrane region have been applied to SOSgp160ΔCT to create SOSIP soluble gp140 glycoproteins53. Both SOSgp160ΔCT and SOSIP soluble gp140 trimers have been studied by single-particle cryo-EM or cryoelectron tomography approaches16,17,54. The 20-Å cryo-EM structure of the SOSgp160ΔCT trimer exhibits a tripod conformation, with an unclear assignment of the transmembrane region16. These dissimilarities between the SOSgp160ΔCT structure and our Env(−)ΔCT structure might result from differences, including proteolytic cleavage, between the Env constructs, their choice of Triton X-100 as a detergent, and/or their use of glutaraldehyde crosslinking16. The SOSIP soluble gp140 structure has been analyzed in two studies17,54. A 14-Å negative staining EM structure of the SOSIP gp140 trimer complexed with PGT antibodies demonstrated missing or weakened density around the gp120 trimer axis compared with our 11-Å Env(−)ΔCT structure54. The six-way junction in the Env(−)ΔCT structure has apparently collapsed and partially disappears in the 14-Å negative-staining EM reconstruction of the SOSIP soluble gp140 trimer54. Similarly, a 20-Å resolution cryo-electron tomographic structure of the unliganded SOSIP soluble gp140 trimer17 also exhibited rather weak density in the gp120 trimer association domain. The basis for the differences in the gp120 trimer association domain between the SOSIP soluble gp140 and the Env(−)ΔCT trimers will require further investigation; however, the poor recognition of the SOSIP soluble gp140 trimers by quaternary structure-dependent antibodies like PG16 (ref. 46) could potentially be explained by the metastability of the six-way junction at the gp120-gp120 interfaces. The future availability of structural data at higher resolution for these different forms of Env should clarify these issues and expedite the improvement of Env trimer mimics as immunogens.
METHODS
Methods and any associated references are available in the online version of the paper.
ONLINE METHODS
Purification of the membrane-bound Env trimer
Then env cDNA was codonoptimised and subcloned into the pcDNA3.1(−) expression plasmid (Invitrogen). The Env(−)ΔCT glycoprotein contains a heterologous signal sequence from CD5 in place of the wild-type HIV-1 Env signal peptide. Site-directed mutagenesis was used to change the proteolytic cleavage site between gp120 and gp41, substituting Ser for Arg508 and Arg511. The Env cytoplasmic tail was truncated by introduction of a stop codon at Tyr712; a sequence encoding a (Gly)2(His)6 tag was inserted immediately before the stop codon. The plasmid expressing the Env(−)ΔCT glycoprotein was transfected into the 293F cells. After 36 h, cells expressing the envelope glycoproteins were harvested and washed with phosphate-buffered saline (PBS) at 4 °C. The cell pellets were homogenized in a homogenization buffer (250 mM sucrose, 10 mM Tris-HCl [pH 7.4]) and a cocktail of protease inhibitors [Roche Complete tablets]). The plasma membranes were then extracted from the homogenates by ultracentrifugation and sucrose gradient separation. The extracted crude plasma membrane pellet was collected and solubilized in a solubilization buffer containing 100 mM (NH4)2SO4, 20 mM Tris-HCl (pH 8), 300 mM NaCl, 20 mM imidazole, 1% (wt/vol) Cymal-5 (Affymatrix) and a cocktail of protease inhibitors (Roche Complete tablets). The membranes were solubilized by incubation at 4 °C for 30 min on a rocking platform. The suspension was ultracentrifuged for 30 min at 200,000 × g at 4 °C. The supernatant was collected and mixed with a small volume of pre-equilibrated Ni-NTA beads (QIAGEN) for 8-12 h on a rocking platform at 4 °C. The mixture was then injected into a small column and washed with a buffer containing 100 mM (NH4)2SO4, 20 mM Tris-HCl (pH 8), 1 M NaCl, 30 mM imidazole and 0.5% Cymal-5. The bead-filled column was eluted with a buffer containing 100 mM (NH4)2SO4, 20 mM Tris-HCl (pH 7.4), 250 mM NaCl, 250 mM imidazole and 0.5% Cymal-5. The eluted Env glycoprotein solution was concentrated, diluted in a buffer containing 20 mM Tris-HCl, pH 7.4, 300 mM NaCl and 0.01% Cymal-6, and reconcentrated to ~2.5 mg ml−1 prior to cryo-sample preparation. The recognition of the purified Env glycoproteins by a number of conformation-dependent antibodies, including VRC01, b12 and 2G12, as well as CD4-Ig, was measured in an enzyme-linked immunosorbent assay (ELISA) (see below). The VRC01 and b12 antibodies recognize conformation-dependent epitopes near the CD4-binding site of gp120 (refs. 7, 47, 48). The 2G12 antibody recognizes a high-mannose glycan array on the gp120 outer domain49. CD4-Ig consists of the two N-terminal domains of CD4 fused to the Fc portion of the immunoglobulin heavy chain27. We also examined whether our Env solubilization and purification approach affected the integrity of an epitope that is recognized by the PG16 antibody and that is sensitive to changes in the quaternary structure of the HIV-1 Env trimer46. To this end, we studied PG16 binding to the Env(−)ΔCT E168K glycoprotein. The wild-type HIV-1JR-FL isolate is highly resistant to neutralization by the PG16 antibody, but the E168K change renders the HIV-1JR-FL Env sensitive to PG16 (ref. 46). PG16 binding to the purified Env(−)ΔCT E168K glycoprotein was tested in the ELISA.
Enzyme-linked immunosorbent assay
A white, high-binding microtiter plate (Corning) was coated by incubating 0.5 μg of mouse anti-polyhistidine antibody (sc-53073, Santa Cruz Biotechnology) in 100 μl PBS in each well overnight. Wells were blocked with blocking buffer (5% non-fat dry milk [Bio-Rad] in 20 mM Tris-HCl, pH 7.4 and 300 mM NaCl) for 2 h and then washed twice with wash buffer (20 mM Tris-HCl, pH 7.4 and 300 mM NaCl). Approximately 0.5 μg of purified Env trimer in blocking buffer was added to each well, the plate was incubated for 60 min and washed thrice with wash buffer. Different concentrations of specific Env ligands (conformation-dependent antibodies and the CD4-Ig fusion protein) in blocking buffer were added to the wells and the plate was incubated for another 45 min. After three washes, peroxidase-conjugated F(ab’)2 fragment donkey anti-human IgG (1:3600 dilution; Jackson ImmunoResearch Laboratories) in blocking buffer was added to each well. The plate was incubated for 30 min, washed six times, and 80 μl of SuperSignal chemiluminescent substrate (Pierce) was added to each well. The relative light units in each well were measured for two seconds with a Centro LB 960 luminometer (Berthold Technologies, TN). All procedures were performed at room temperature.
Flow cytometry
293T cells were transfected, by either calcium phosphate co-precipitation or by using the Effectene transfection reagent (Qiagen), with a plasmid encoding the Env(−)ΔCT E168K glycoproteins. After 48 h, approximately half a million cells were analyzed by flow cytometry as previously described55, but with primary antibody incubation for 30 min, and secondary antibody (Allophycocyaninconjugated F(ab’)2 fragment donkey anti-human IgG antibody, Jackson ImmunoResearch Laboratories) incubation for 15 min, both at room temperature. Cells were analyzed with a BD FACSCanto II flow cytometer (BD Biosciences).
Cryo-EM reconstruction and model analysis
To prepare the cryo-sample for single-particle imaging, 2.5 μl of the 2.5 mg ml−1 Env(−)ΔCT solution was spread on a C-flat holey carbon grid (Electron Microscopy Sciences) in a chamber of 100% humidity, held for 2 s, blotted by filter papers for 2 s at 4 °C, and then flash-plunged into liquid ethane by Vitrobot (FEI). The prepared cryo-grids were transferred into the CT3500 cryo-transfer system (Gatan) in liquid nitrogen and were used for single-particle image data collection at −183 °C. Focus pairs of micrographs were recorded on a Tecnai F20 TEM (FEI) with a field-emission gun at 200 kV and a calibrated magnification of 200,835× on a 4k × 4k slow-scan CCD camera (Gatan). The electron dose of each exposure was 10.0 electrons Å−2. The defocus of the second set of micrographs differed from that of the first set by 1.0 μm. Micrographs were screened for drift, astigmatism and visibility of Thon rings in the power spectra. Parameters of the contrast transfer function (CTF) of each micrograph were determined with the CTFFind3 program56,57. A total of 90,306 single-particle images (in a dimension of 320 × 320 pixels and a pixel size of 0.747 Å) selected from the closer-to-focus micrographs were used for reconstruction. Each single-particle image was decimated by 4 times to a dimension of 80 × 80 pixels prior to further image analysis, resulting in a pixel size of 2.99 Å. The images were CTF-corrected by the phase flipping method. These single-particle images were then subjected to multivariate data analysis and classification. Images in each class were aligned in a reference-free manner and the class averages were refined by a maximum-likelihood approach20,21. These class averages were used to perform angular reconstitution to yield an initial model. The initial model was further refined by the projection-matching algorithm with C3 symmetry imposed. The angular increment was progressively decreased from 10° to 1° in the refinement. For the last round of refinement, the new classes of images were re-aligned by a maximum-likelihood approach20,21. The above image analysis was implemented in customized computational procedures and workflows, combining the functions of SPIDER, XMIPP and homemade FORTRAN programs58,59. The final reconstruction at 10.8 Å, measured by FSC-0.5 cutoff, was not corrected for its temperature factor. Segmentation of the cryo-EM density, fitting of crystal structures into the map and figure illustrations were done in UCSF Chimera60.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank D. R. Burton for helpful discussion and encouragement; M. Ericsson, L. Trakimas and E. Benecchi for help in initial sample screening by conventional electron microscopy; D. Bell, E. Hodges and D. Wei for assistance and coordination in data collection; J. Faltskog, S. Doktor and B. Battle for laboratory coordination and assistance in building a high-performance computing system; Y. McLaughlin and E. Carpelan for assistance in manuscript preparation. The experiments and data processing were performed in part at the Center for Nanoscale Systems at Harvard University, a member of National Nanotechnology Infrastructure Network (NNIN), which is supported by the National Science Foundation under NSF award no. ECS-0335765. This work was funded by the National Institutes of Health (NIH) (AI93256, AI67854 and AI24755 to J.S.), by an Innovation Award (J.S.) and a Fellowship Award (Y.M.) from the Ragon Institute of MGH, MIT and Harvard, by the International AIDS Vaccine Initiative, and by a gift from the late William F. McCarty-Cooper (J.S.).
Footnotes
Accession codes
The cryo-EM map is deposited into the EM Databank under accession code EMD-5418.
AUTHOR CONTRIBUTIONS
Y.M. and J.S. conceived this study. Y.M. designed experimental protocols; Y.M., L.W., C.G., A.H., S.H.X., H.H., X.Y. and J.S. performed the research; Y.M. and J.S. analyzed the results and wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no conflicts of interest.
REFERENCES
- 1.Barre-Sinoussi F, et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 2.Allan JS, et al. Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science. 1985;228:1091–1094. doi: 10.1126/science.2986290. [DOI] [PubMed] [Google Scholar]
- 3.Moulard M, Decroly E. Maturation of HIV envelope glycoprotein precursors by cellular endoproteases. Biochim. Biophys. Acta. 2000;1469:121–132. doi: 10.1016/s0304-4157(00)00014-9. [DOI] [PubMed] [Google Scholar]
- 4.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 5.Barin F, McLane MF, Allan JS, Lee TH, Groopman JE, Essex M. Virus envelope protein of HTLV-III represents major target antigen for antibodies in AIDS patients. Science. 1985;228:1094–1096. doi: 10.1126/science.2986291. [DOI] [PubMed] [Google Scholar]
- 6.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou T, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pancera M, et al. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl. Acad. Sci. USA. 2010;107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 10.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 11.Buzon V, Natrajan G, Schibli D, Campelo F, Kozlov MM, Weissenhorn W. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Pathog. 2010;6:e1000880. doi: 10.1371/journal.ppat.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu P, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 13.Zanetti G, Briggs JAG, Grunewald K, Sattentau QJ, Fuller SD. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2006;2:e83. doi: 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramanian S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White TA, et al. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 2010;6:e1001249. doi: 10.1371/journal.ppat.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu SR, et al. Single-particle cryoelectron microscopy analysis reveals the HIV-1 spike as a tripod structure. Proc. Natl. Acad. Sci. USA. 2010;107:18844–18849. doi: 10.1073/pnas.1007227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris A, et al. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc. Natl. Acad. Sci. USA. 2011;108:11440–11445. doi: 10.1073/pnas.1101414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu G, Liu J, Taylor KA, Roux KH. Structural comparison of HIV-1 envelope spikes with and without the V1/V2 loop. J. Virol. 2011;85:2741–2750. doi: 10.1128/JVI.01612-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frank J. Three-dimensional electron microscopy of macromolecular assemblies: visualization of biological molecules in their native state. Oxford Univ. Press; 2006. [Google Scholar]
- 20.Sigworth FJ. A maximum-likelihood approach to single-particle image refinement. J. Struct. Biol. 1998;122:328–339. doi: 10.1006/jsbi.1998.4014. [DOI] [PubMed] [Google Scholar]
- 21.Scheres SHW, et al. Maximum-likelihood multi-reference refinement for electron microscopy images. J. Mol. Biol. 2005;348:139–149. doi: 10.1016/j.jmb.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 22.Liao HY, Frank J. Definition and estimation of resolution in single-particle reconstructions. Structure. 2010;18:768–775. doi: 10.1016/j.str.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sattentau QJ, Moore JP. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J. Exp. Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myszka DG, et al. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. USA. 2000;97:9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan W, Bazick J, Sodroski J. Characterization of the multiple conformational states of free monomeric and trimeric human immunodeficiency virus envelope glycoproteins after fixation by cross-linker. J. Virol. 2006;80:6725–6737. doi: 10.1128/JVI.00118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiang SH, et al. A V3 loop-dependent gp120 element disrupted by CD4 binding stabilizes the human immunodeficiency virus envelope glycoprotein trimer. J. Virol. 2010;84:3147–3161. doi: 10.1128/JVI.02587-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finzi A, et al. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Mol. Cell. 2010;37:656–667. doi: 10.1016/j.molcel.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang SH, et al. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J. Virol. 2002;76:9888–9899. doi: 10.1128/JVI.76.19.9888-9899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen B, et al. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433:834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 30.McLellan JS, et al. Structure of HIV-1 gp120 V1V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mische CC, Yuan W, Strack B, Craig S, Farzan M, Sodroski J. An alternative conformation of the gp41 heptad repeat 1 region coiled coil exists in the human immunodeficiency virus (HIV-1) envelope glycoprotein precursor. Virology. 2005;338:133–143. doi: 10.1016/j.virol.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 32.Helseth E, Olshevsky U, Furman C, Sodroski J. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J. Virol. 1991;65:2119–2123. doi: 10.1128/jvi.65.4.2119-2123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang S, York J, Shu W, Stoller MO, Nunberg JH, Lu M. Interhelical interactions in the gp41 core: implications for activation of HIV-1 membrane fusion. Biochemistry. 2002;41:7283–7292. doi: 10.1021/bi025648y. [DOI] [PubMed] [Google Scholar]
- 34.Dimitrov AS, Louis JM, Bewley CA, Clore GM, Blumenthal R. Conformational changes in HIV-1 gp41 in the course of HIV-1 envelope glycoprotein-mediated fusion and inactivation. Biochemistry. 2005;44:12471–12479. doi: 10.1021/bi051092d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sen J, et al. Alanine scanning mutagenesis of HIV-1 gp41 heptad repeat 1: insight into the gp120-gp41 interaction. Biochemistry. 2010;49:5057–5065. doi: 10.1021/bi1005267. [DOI] [PubMed] [Google Scholar]
- 36.Chertova E, et al. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 2002;76:5315–5325. doi: 10.1128/JVI.76.11.5315-5325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossio JL, et al. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J. Virol. 1998;72:7992–8001. doi: 10.1128/jvi.72.10.7992-8001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chakrabarti BK, et al. HIV type 1 Env precursor cleavage state affects recognition by both neutralizing and nonneutralizing gp41 antibodies. AIDS Res. Hum. Retroviruses. 2011;27:877–887. doi: 10.1089/aid.2010.0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pancera M, Wyatt R. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology. 2005;332:145–156. doi: 10.1016/j.virol.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 40.Edwards TG, et al. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J. Virol. 2002;76:2683–2691. doi: 10.1128/JVI.76.6.2683-2691.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrison SC. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwon YD, et al. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc. Natl. Acad. Sci. U.S.A. 2012;109:5663–5668. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Musich T, et al. A conserved determinant in the V1 loop of HIV-1 modulates the V3 loop to prime low CD4 use and macrophage infection. J. Virol. 2011;85:2397–2405. doi: 10.1128/JVI.02187-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kolchinsky P, Kiprilov E, Bartley P, Rubenstein R, Sodroski J. Loss of a single N-linked glycan allows CD4-independent HIV-1 infection by altering the position of the gp120 V1/V2 variable loops. J. Virol. 2001;75:3435–3443. doi: 10.1128/JVI.75.7.3435-3443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang PF, et al. A variable region 3 (V3) mutation determines a global neutralization phenotype and CD4-independent infectivity of a human immunodeficiency virus type 1 envelope associated with a broadly cross-reactive, primary virus-neutralizing antibody response. J. Virol. 2002;76:644–655. doi: 10.1128/JVI.76.2.644-655.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou T, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trkola A, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH. 3.3 Å cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell. 2010;141:472–482. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yau WCY, Rubinstein JL. Subnanometre-resolution structure of the intact Thermus thermophilus H+-driven ATP synthase. Nature. 2012;481:214–218. doi: 10.1038/nature10699. [DOI] [PubMed] [Google Scholar]
- 52.Becker T, et al. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science. 2009;326:1369–1373. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Binley JM, et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 2000;74:627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pejchal R, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herschhorn A, Marasco WA, Hizi A. Antibodies and lentiviruses that specifically recognize a T cell epitope derived from HIV-1 Nef protein and presented by HLA-C. J. Immunol. 2010;185:7623–7632. doi: 10.4049/jimmunol.1001561. [DOI] [PubMed] [Google Scholar]
- 56.Huang Z, Baldwin PR, Mullapudi S, Penczek PA. Automated determination of parameters describing power spectra of micrograph images in electron microscopy. J. Struct. Biol. 2003;144:79–94. doi: 10.1016/j.jsb.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 57.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 58.Shaikh TR, et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc. 2008;3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scheres SHW, Nunez-Ramirez R, Sorzano COS, Carazo JM. Image processing for electron microscopy single-particle analysis using XMIPP. Nat. Protoc. 2008;3:977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pettersen EF, et al. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.