SUMMARY
Chlamydia trachomatis is an obligate intracellular bacterial pathogen that is the most common cause of sexually transmitted bacterial infections and is the etiological agent of trachoma, the leading cause of preventable blindness. The organism infects epithelial cells of the genital tract and eyelid resulting in a damaging inflammatory response. C. trachomatis grows within a vacuole termed the inclusion, and its growth depends on numerous host factors, including lipids. Although a variety of mechanisms are involved in the acquisition of host cell cholesterol and glycosphingolipids by C. trachomatis, none of the previously documented pathways for lipid acquisition are absolutely required for growth. Here we demonstrate that multiple components of the host high density lipoprotein (HDL) biogenesis machinery including the lipid effluxers, ABCA1 and CLA 1, and their extracellular lipid acceptor, apoA-1, are recruited to the inclusion of C. trachomatis-infected cells. Furthermore, the apoA-1 that accumulates within the inclusion co-localizes with pools of phosphatidylcholine. Knockdown of ABCA1, which mediates the cellular efflux of cholesterol and phospholipids to initiate the formation of HDL in the serum, prevents the growth of C. trachomatis in infected HeLa cells. In addition, drugs that inhibit the lipid transport activities of ABCA1 and CLA 1 also inhibit the recruitment of phospholipids to the inclusion and prevent chlamydial growth. These results strongly suggest that C. trachomatis co-opts the host cell lipid transport system involved in the formation of HDL to acquire lipids, such as phosphatidylcholine, that are necessary for growth.
INTRODUCTION
During replication within the inclusion, Chlamydia acquires essential nutrients including nucleotides (Hatch, 1975b, McClarty et al., 1991) and amino acids (Hatch, 1975a, McClarty, 1994) from the host. In addition, C. trachomatis acquires host cell lipids through multiple mechanisms. Host-derived sphingomyelin (Hackstadt et al., 1995, Hackstadt et al., 1996) and cholesterol (Carabeo et al., 2003) are derived from the Golgi and delivered to the inclusion via a vesicular transport pathway that is sensitive to the Arf1 inhibitor, brefeldin A (BFA) (Hackstadt et al., 1996, Carabeo et al., 2003). The delivery of sphingomyelin to the inclusion via this vesicular transport pathway is dependent upon the Arf1 guanine nucleotide exchange factor, GBF1 (Elwell et al., 2011). Non-vesicular transport pathways are also involved in the delivery of host sphingolipids to the inclusion of infected cells. The lipid carrier protein, CERT, transports ceramide from the ER to the inclusion (Elwell et al., 2011, Derre et al., 2011) where ceramide is converted into sphingomyelin by host sphingomyelin synthases (Elwell et al., 2011). This CERT-dependent transport pathway for the delivery of lipids to the inclusion appears to primarily contribute to chlamydial growth regulation (Elwell et al., 2011, Derre et al., 2011), whereas lipids delivered to the inclusion via the vesicular transport pathway contribute to the maintenance of inclusion membrane integrity (Elwell et al., 2011). Finally, recent studies have indicated that cytoplasmic lipid droplets, which are made up of a core of neutral lipids surrounded by a phospholipid monolayer (Martin et al., 2006), translocate across the chlamydial inclusion membrane (Cocchiaro et al., 2008) where the lipids associated with the droplet may be made available for chlamydial growth.
The acquisition of host sphingolipids by C. trachomatis is facilitated by the fragmentation of the Golgi, which is triggered by chlamydial infection (Heuer et al., 2009). Furthermore, studies using inhibitors that block multivesicular body function suggest that host-derived sphingolipids traffic through the multivesicular body prior to undergoing delivery to the inclusion (Beatty, 2006). While each of the lipid transport pathways described above contribute to the acquisition of host lipids by Chlamydia, inhibitors of the individual pathways only partially block chlamydial growth (Carabeo et al., 2003, Beatty, 2006, Heuer et al., 2009, Elwell et al., 2011, Derre et al., 2011). These results suggest that Chlamydia co-opts multiple, redundant pathways to acquire host lipids, such as sphingomyelin and cholesterol, that are essential for growth.
The import of host-derived glycerophospholipids (hereafter referred to as phospholipids) into the inclusion requires their deacylation by the host calcium-dependent phospholipase A2 releasing lysophospholipid, which is reacylated by a bacterial branched chain fatty acid prior to its incorporation into bacterial cell membranes (Wylie et al., 1997). Although previous studies indicated that the acquisition of host phospholipids by C. trachomatis is BFA-insensitive (Wylie et al., 1997), the precise mechanism involved in phospholipid acquisition by C. trachomatis is unclear. In the studies described here, we investigated whether host proteins involved in phospholipid and cholesterol efflux may be involved in lipid acquisition by C. trachomatis with the ultimate goal of defining host cell pathways that are critical for the growth of Chlamydia in infected cells. Specifically, we examined whether the host machinery involved in the biogenesis of high density lipoprotein (HDL) is involved in regulating the growth of C. trachomatis in infected cells. The formation of HDL in the plasma is mediated by the sequential transport of lipids to extracellular apoA-1 by the transporters ABCA1, ABCG1, and the SR-B1 scavenger receptor, respectively. ABCA1, the initial transporter in the HDL biogenesis pathway, mediates the efflux of cellular cholesterol and phospholipids to extracellular lipid-free apoA-1 in the serum (Zannis et al., 2006, Oram et al., 2000, Wang et al., 2000). Lipidated apoA-1 then serves as an acceptor for additional cholesterol efflux by ABCG1 (Gelissen et al., 2006) and subsequently by SR-B1 (Rhainds et al., 2004, Van Eck et al., 2005). Loss of function mutations in ABCA1 (Zannis et al., 2006, Rust et al., 1999), ABCG1 (Yvan-Charvet et al., 2007), SR-B1 (Zannis et al., 2006, Braun et al., 2002, Van Eck et al., 2004), and apoA-1 (Zannis et al., 2006, Matsunaga et al., 1991) can enhance atherosclerotic lesion development.
The studies presented here indicate that C. trachomatis infection alters the intracellular trafficking of several components of the host HDL biogenesis machinery, inducing ABCA1, CLA 1, the human homologue of rodent SR-B1 scavenger receptor (Calvo et al., 1993), and apoA-1 to accumulate in the inclusion of infected HeLa cells. The inclusion-associated population of apoA-1 co-localizes with pools of phosphatidylcholine in infected cells. In addition, glyburide, a drug that inhibits the ability of ABCA1 and B1 type scavenger receptor members to transport lipids to apoA-1 (Nieland et al., 2004), inhibits the accumulation of phosphatidylcholine within the inclusion of infected cells and prevents chlamydial growth. Finally, knockdown of ABCA1 prevents the growth of C. trachomatis in infected HeLa cells. These data indicate that multiple elements of the host HDL biogenesis machinery are recruited to the inclusion of C. trachomatis-infected cells where they play a critical role in chlamydial growth regulation most likely by providing a source of phospholipids that are essential for growth.
RESULTS
ABCA1 accumulates in the inclusion membrane of C. trachomatis-infected HeLa cells
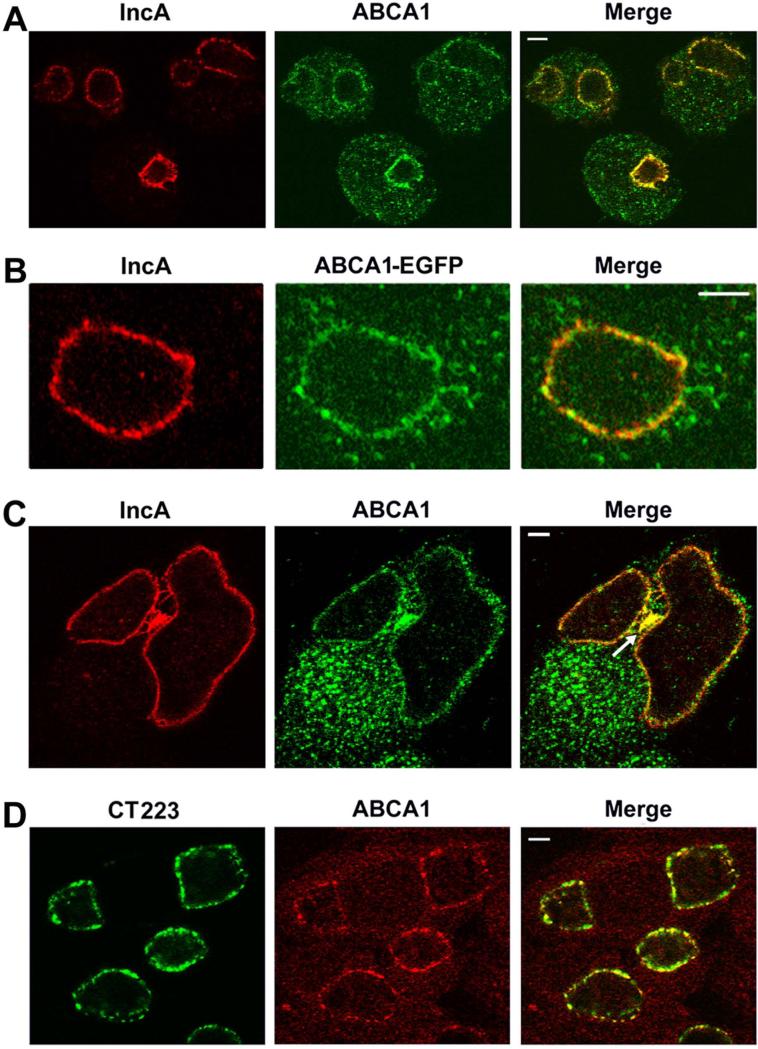
Immunolocalization studies investigated whether host ABCA1 transporters are recruited to the inclusion of Chlamydia-infected cells. For these studies we employed an ABCA1 antibody that detected a ~240kD protein in a HeLa cell lysate (data not shown), which is consistent with previous reports for the size of ABCA1 (Arakawa et al., 2002). Localization analysis with this antibody revealed that ABCA1 primarily accumulates in a punctate, intracellular membrane compartment in HeLa cells (data not shown). To determine the effect of chlamydial infection on ABCA1 localization, HeLa cells infected with C. trachomatis serovar D were fixed 24 hours post-infection (PI) and stained with antibodies directed against ABCA1 and IncA, an inclusion membrane protein (Bannantine et al., 1998). Confocal analysis of these cells revealed that ABCA1 still primarily resided in intracellular membrane compartments and a substantial percentage of the intracellular pool of ABCA1 overlapped the distribution of IncA in the inclusion membrane of infected cells (Fig. 1A). To confirm the results obtained with the ABCA1-specific antibodies, HeLa cells transfected with an ABCA1-EGFP fusion (Tanaka et al., 2003) were infected with C. trachomatis. The cells were fixed 48 hours PI and confocal analysis revealed that ABCA1-EGFP also accumulated in the inclusion membrane where it overlapped the localization of IncA (Fig. 1B). The stained cells in Fig. 1A were infected at a multiplicity of infection (MOI=2) and cells containing multiple inclusions were observed. The higher magnification image in Fig. 1C illustrates that IncA and ABCA1 accumulate in the inclusion membrane of two adjacent inclusions. In addition, both IncA and ABCA1 accumulate in a membrane aggregate (marked with an arrow in Fig. 1C) that lies between the two inclusions. Further analyses compared the localization of ABCA1 in infected cells to an additional C. trachomatis inclusion membrane protein, CT223 (Bannantine et al., 2000, Alzhanov et al., 2009). Confocal analysis revealed that ABCA1 localization was very similar to the localization profile of CT223 at 24 hours PI (Fig. 1D). Collectively, these data demonstrate that ABCA1 is recruited to the inclusion membrane of Chlamydia-infected cells and to additional intracellular membrane compartments that contain inclusion membrane marker proteins.
Figure 1.
Non-transfected HeLa cells (A, C, and D) or HeLa cells transfected with the plasmid encoding ABCA1-EGFP (B) were infected with Chlamydia trachomatis serovar D. The cells were fixed at 24 hours (A, C, and D) or 48 hours (B) PI with 4% paraformaldehyde in PBS (A - C) or with methanol (D). The cells in A - C were then permeabilized in PBS containing 0.1% Triton X-100 and incubated with a mouse monoclonal antibody directed against IncA (A - C) and a rabbit polyclonal antibody directed against ABCA1 (A and C). Alternatively, the cells in D were directly incubated with a monoclonal antibody directed against CT223 and a rabbit polyclonal antibody directed against ABCA1. Following washing, the cells were incubated with the appropriate secondary antibodies prior to confocal analysis. The arrow in C points to a membrane aggregate between adjacent inclusions that is positive for IncA and ABCA1. The white bars in A and D are 5μm, the bar in B is 10μm, and the bar in C is 2.5μm.
Multiple components of the host HDL biogenesis machinery are recruited to the inclusion of C. trachomatis-infected HeLa cells
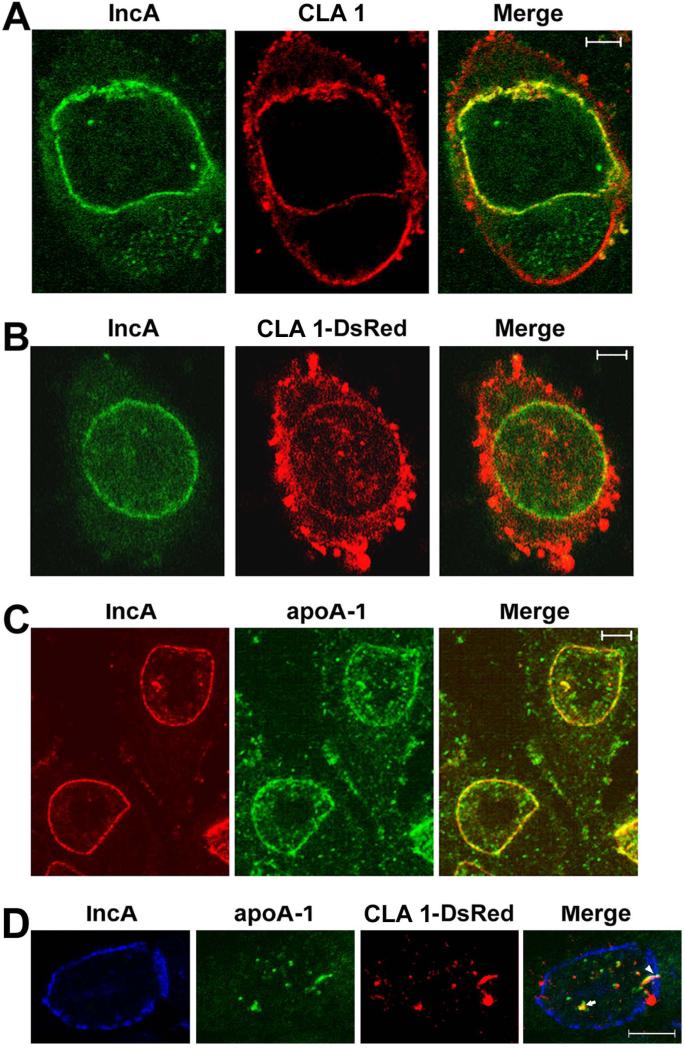
Localization studies investigated whether CLA 1 (Calvo et al., 1993, Calvo et al., 1997), and the extracellular lipid acceptor involved in HDL biogenesis, apoA-1 (Zannis et al., 2006, Wang et al., 2000, Van Eck et al., 2005), are also recruited to the inclusion of Chlamydia-infected cells. Immunoblotting analyses using antibodies that recognize CLA 1 and apoA-1 detected single proteins of ~84kD and ~29kD, respectively, in a lysate prepared from C. trachomatis-infected HeLa cells at 32 hours PI (Supp. Fig. S1A). The sizes of these proteins were again consistent with previously reported sizes of CLA 1 and apoA-1 (Calvo et al., 1997, Ritter et al., 2002). Confocal analysis of C. trachomatis-infected HeLa cells that were fixed at 48 hours PI revealed that CLA 1 was present both on the cell surface and in the inclusion membrane where it overlapped the distribution of IncA (Fig. 2A). This localization profile was confirmed in experiments using a CLA 1-DsRed monomer fusion (Fig. 2B). Foci containing CLA 1-DsRed were also observed within the lumen of the inclusion of the infected cell (Fig. 2B).
Figure 2.
Non-transfected HeLa cells (A and C) or HeLa cells transfected with the plasmid encoding the CLA 1-DsRed monomer fusion (B and D) were infected with C. trachomatis serovar D and fixed forty-eight hours PI with 4% paraformaldehyde in PBS. The cells were permeabilized in PBS containing 0.1% saponin (A) or 0.1% TX-100 (B - D) and incubated with a monoclonal antibody directed against IncA (A - D), a rabbit polyclonal antibody that recognizes CLA 1 (A), or a rabbit polyclonal antibody directed against apoA-1 (C and D). Following washing, the cells were incubated with the appropriate secondary antibodies prior to confocal analysis. The arrow in D points to adjacent foci of apoA-1 and CLA 1-DsRed within the lumen of the inclusion. The arrowhead in D points to an elongated tubular structure containing apoA-1 and CLA 1-DsRed that appears to span the inclusion membrane. The white bars in A and B are 5μm, and the bars in C and D are 10μm.
Additional analyses revealed that apoA-1 accumulated both in the inclusion membrane and in discrete foci in the lumen of the inclusion of infected cells at 48 hours PI (Fig. 2C). In some infected cells, the CLA 1-DsRed fusion protein and apoA-1 primarily accumulated in the lumen of the inclusion (Fig. 2D). The lumenal foci containing CLA 1-DsRed were often immediately adjacent to lumenal clusters of apoA-1 (arrow in Fig. 2D). In addition, elongated tubular elements containing CLA 1-DsRed and apoA-1 were observed that appeared to traverse the inclusion membrane of the infected cell (arrowhead in Fig. 2D). Whether these elongated tubular structures represent intermediates in the transport of CLA 1 and apoA-1 from the cytoplasm to the lumen of the inclusion remains to be determined. To further demonstrate that the foci of apoA-1 observed in Figs. 2C and 2D were indeed present within the lumen of the inclusion, infected cells were fixed at 32 hours PI and stained with IncA and apoA-1 antibodies. The cells were then imaged by confocal microscopy. As shown in Supp. Fig. 1B, foci of apoA-1 were detected within the lumen of the inclusion in consecutive 0.5μm confocal slices. This lumenal population of apoA-1 is also seen in the xz-slice from a Z-stack of an infected cell (Supp. Fig. S1C). Since it was possible that antibodies in the apoA-1 antisera recognized antigens associated with Chlamydia in fixed cells, we also compared the localization profiles obtained with apoA-1 and chlamydial Hsp60 antibodies. This analysis revealed that the apoA-1 antibody does not stain Chlamydia within the lumen of the inclusion (Supp. Fig. S1D). At this time, the factors that determine whether apoA-1 accumulates in the inclusion membrane or within the inclusion of infected cells are not understood. Furthermore, although the data in Figs. 1 and 2 suggest that a subset of ABCA1, CLA1, and apoA-1 accumulate in the inclusion membrane of infected cells, it is possible that these proteins are present in cytoplasmic membranes immediately adjacent to the inclusion. Future immuno-electron microscopic analyses will distinguish between these possibilities.
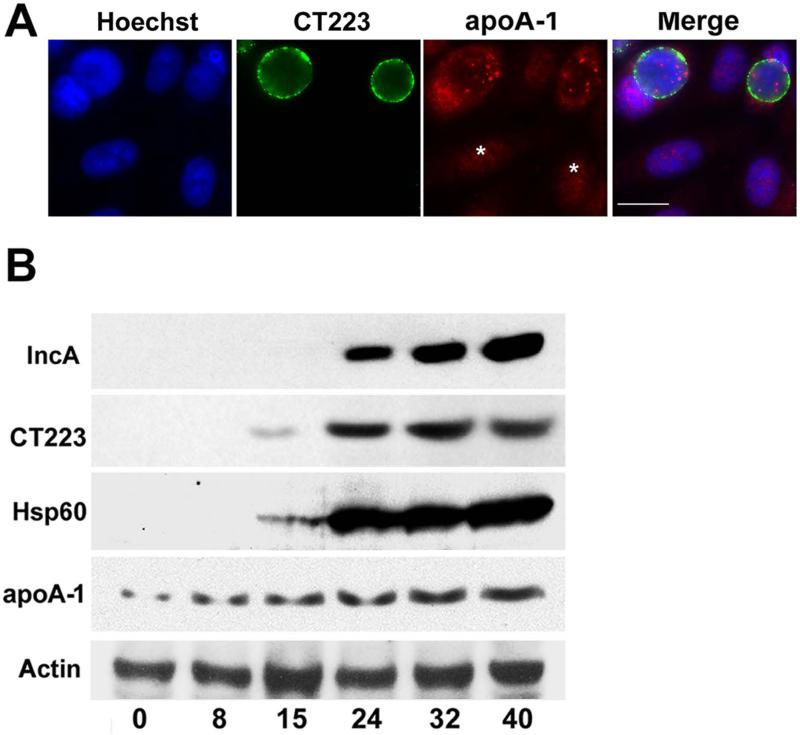
Immunofluorescence analyses suggested that the level of cellular associated apoA-1 in Chlamydia-infected cells was higher than that observed in uninfected cells (Fig. 3A). To more carefully assess the effect of chlamydial infection on intracellular apoA-1 levels, lysates prepared from uninfected HeLa cells and from cells that were harvested at various times PI were probed with antibodies directed against IncA, CT223, chlamydial Hsp60, and apoA-1. Hsp60 and CT223 were present in these lysates in detectable amounts at 15 hours PI, while IncA was first detected in the lysate from 24 hours PI (Fig. 3B). Blotting analysis of the lysates with apoA-1 specific antibodies revealed that the level of cellular associated apoA-1 was approximately 4-fold higher in cells at 40 hours PI when compared to uninfected cells (Fig. 3B). In addition, there was approximately a 2-fold increase in apoA-1 levels by 8 hours PI, which is prior to the time that the chlamydial proteins were detected in this blotting assay (Fig. 3B). To investigate the origin of this observed increase in cellular-associated apoA-1, qRT-PCR was used to measure apoA-1 transcript levels in infected HeLa cells during the chlamydial developmental cycle. These studies revealed that apoA-1 transcript levels did not change significantly during the course of the infection (data not shown). This result suggested that the increased levels of cellular-associated apoA-1 in Chlamydia-infected cells resulted from the enhanced translation and/or stabilization of the endogenous pool of apoA-1, or the enhanced uptake of exogenous apoA-1.
Figure 3.
HeLa cells infected with C. trachomatis serovar D were methanol fixed 24 hours PI (A). The cells were then stained with a monoclonal antibody directed against CT223 and a rabbit polyclonal antibody directed against apoA-1. Following washing, the cells were incubated with the appropriate secondary antibodies. DNA was visualized by staining with Hoechst. The images in A were collected on a Zeiss Axioplan II microscope. Alternatively, immunoblotting analyses characterized the expression of chlamydial and host proteins during the developmental cycle (B). Lysates were prepared from uninfected HeLa cells (0 time) or from cells that were infected with C. trachomatis at various times PI. The samples were subjected to blotting analysis using the indicated antibodies. Times PI in B are indicated at the bottom of the figure. Asterisks in A denote uninfected cells. The white bar in A is 20μm.
Phosphatidylcholine is recruited to apoA-1 containing foci in the lumen of the inclusion of infected cells
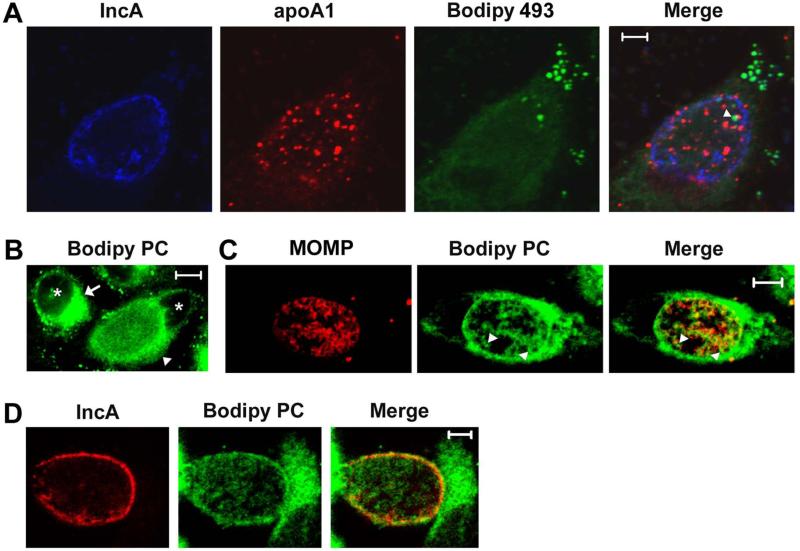
The demonstration that ABCA1, CLA 1, and apoA-1 accumulate in the inclusion of Chlamydia-infected cells suggested that these proteins may be involved in the delivery of lipids to the vacuole where Chlamydia grow. To address this possibility, we initially investigated whether the foci of apoA-1 that accumulate in the lumen of the inclusion of infected cells are associated with specific lipids. A previous study demonstrated that neutral lipids in the form of lipid droplets translocate from the host cell cytoplasm to the lumen of the inclusion during the course of a chlamydial infection (Cocchiaro et al., 2008). To determine whether apoA-1 associates with these neutral lipid containing droplets within the inclusion, Chlamydia-infected cells were fixed at 48 hours PI and incubated with antibodies directed against apoA-1 and IncA and with the dye Bodipy 493, which specifically stains lipid droplets (Cocchiaro et al., 2008). As shown in Fig. 4A, lipid droplets accumulate in the cytoplasm and within the inclusion of infected cells. While the inclusion-associated droplets were occasionally adjacent to the lumenal foci of apoA-1 (arrowhead in Fig. 4A), we never observed co-localization of lipid droplets with apoA-1. Similar results were obtained with cells that were fixed at 32 hours PI (data not shown). These results indicate that apoA-1 does not serve as a site of neutral lipid accumulation within the lumen of the inclusion of infected cells.
Figure 4.
HeLa cells infected with Chlamydia trachomatis serovar D were fixed with 4% paraformaldehyde in PBS at 48 hours PI. The cells were then permeabilized in PBS containing 0.1% saponin and incubated with antibodies directed against Inc A and apoA-1. The cells were then washed and incubated with the appropriate secondary antibodies. Following washing, the cells were stained with the dye Bodipy 493 and imaged by confocal microscopy (A). Alternatively, infected cells were incubated in the presence of β-BODIPY FL C5-HPC in fatty acid-free BSA at a final concentration of 1.5μM for 1 hour prior to fixation in 4% paraformaldehyde in PBS at 48 hours PI. These labeled cells were either directly imaged by confocal microscopy (B) or permeabilized by a brief incubation in methanol. Permeabilized cells were incubated with monoclonal antibodies directed against MOMP (C) or IncA (D) followed by the appropriate secondary antibody and imaged by confocal microscopy. The arrowhead in A denotes a lipid droplet that lies adjacent to a foci of apoA-1 within an inclusion of an infected cell. The arrow in B indicates the perinuclear pool of β-BODIPY FL C5-HPC that accumulates in uninfected cells, while the arrowhead in B points to an infected cell where β-BODIPY FL C5-HPC accumulates in membranes that surround the inclusion and within the inclusion. The arrowheads in C indicate β-BODIPY FL C5-HPC that accumulates at sites within the inclusion that do not stain with MOMP antibodies. The asterisks in B indicate nuclei. The white bars are 5μm.
Since ABCA1 has the capacity to transport phospholipids to apoA-1 (Zannis et al., 2006, Oram et al., 2000, Wang et al., 2000), we next examined whether inclusion-associated apoA-1 may specifically associate with phospholipids in infected cells. For these studies, HeLa cells infected with C. trachomatis Serovar D were incubated with a fluorescent analogue of phosphatidylcholine, β-BODIPY FL C5-HPC, for 1 hour at 37°C. Following fixation in paraformaldehyde at 48 hours PI, the cellular distribution of β-BODIPY FL C5-HPC was analyzed by confocal microscopy. This analysis revealed that β-BODIPY FL C5-HPC primarily accumulated in a perinuclear, Golgi-like compartment in uninfected HeLa cells (marked by arrow in Fig. 4B). This pattern of localization was altered in Chlamydia-infected cells where the majority of the β-BODIPY FL C5-HPC accumulated in membranes that surrounded the inclusion (marked by arrowhead in Fig. 4B). In addition, this phosphatidylcholine analogue appeared to accumulate in the membrane of individual bacteria within the inclusion (Fig. 4B). Similar results were obtained with live cells that were imaged following a 1 hour incubation with β-BODIPY FL C5-HPC (data not shown). The redistribution of β-BODIPY FL C5-HPC observed in Chlamydia-infected cells is very similar to the redistribution of BODIPY FL C5-ceramide that occurs upon the fragmentation of the Golgi that is induced by C. trachomatis infection of epithelial cells (Hackstadt et al., 1995, Hackstadt et al., 1996, Heuer et al., 2009). To determine whether the β-BODIPY FL C5-HPC that accumulates within the inclusion of Chlamydia-infected cells indeed accumulates in bacterial cell membranes, paraformaldehyde fixed cells were briefly permeabilized with methanol and stained with antibodies against the C. trachomatis major outer membrane protein, MOMP. While this permeabilization step extracted β-BODIPY FL C5-HPC from fixed cells to a variable extent, it is clear that a subset of the remaining lipid within the inclusion co-localizes with MOMP in bacterial cell membranes (Fig. 4C). Furthermore, some of the β-BODIPY FL C5-HPC accumulates in regions of the inclusion devoid of MOMP (marked by arrowheads in Fig. 4C). Staining of these permeabilized cells with IncA antibodies further revealed that this phosphatidylcholine analogue accumulates in the inclusion membrane of infected cells (Fig. 4D). Similar analyses using HeLa cells infected with C. trachomatis Serovar L2 yielded identical results (data not shown). A previous report showed that another fluorescent analogue of phosphatidylcholine, 6:0-N-NBD-phosphatidylcholine, accumulated in the inclusion membrane but was not incorporated into the bacterial cell membrane of C. trachomatis Serovar L2 in infected HeLa cells (Hackstadt et al., 1996). Whether the differing abilities of 6:0-N-NBD-phosphatidylcholine and β-BODIPY FL C5-HPC to undergo incorporation into bacterial cell membranes reflects the effect of the fluorescent NBD and BODIPY moieties on phosphatidylcholine trafficking within cells is not known.
To determine whether the non-bacterial associated population of β-BODIPY FL C5-HPC in the inclusion is associated with apoA-1, infected cells were fixed at 48 hours PI, permeabilized with methanol, and stained with apoA-1 and IncA antibodies. The image in Fig. 5A illustrates that a large percentage of the β-BODIPY FL C5-HPC remaining within this permeabilized cell co-localizes with the apoA-1-containing foci within the lumen of the inclusion (Fig. 5A). A similar pattern of co-localization between β-BODIPY FL C5-HPC and apoA-1 within the inclusion was observed in cells that were fixed and permeabilized at 24 hours PI (Fig. 5B). In the cell shown in Fig. 5B, a pool of perinuclear β-BODIPY FL C5-HPC that was not completely dispersed as a result of chlamydial infection is marked by an arrow. Although the mechanisms that result in the accumulation of β-BODIPY FL C5-HPC in apoA-1-containing foci in the lumen of the inclusion are not known, these results suggest a role for apoA-1 in directing the accumulation of phospholipids within the inclusion of Chlamydia-infected cells. A wide variety of lipid transport proteins associate with and transport lipids between different membrane compartments within cells (D'Angelo et al., 2008), and recent studies have indicated a role for CERT in the delivery of ceramide to the inclusion of Chlamydia-infected cells (Elwell et al., 2011, Derre et al., 2011). However, to our knowledge our data represents the first example where the restricted localization of a substantial intracellular pool of a specific lipid is potentially dependent upon its association with a carrier protein, such as apoA-1.
Figure 5.
HeLa cells infected with Chlamydia trachomatis serovar D were incubated in the presence of 1.5μM β-BODIPY FL C5-HPC in fatty acid-free BSA for 1 hour prior to fixation in 4% paraformaldehyde in PBS at 48 (A) or 24 (B) hours PI. The labeled cells were then permeabilized by a brief incubation in methanol and incubated with a monoclonal antibody directed against IncA and a rabbit polyclonal antibody directed against apoA-1. Following incubation with appropriate secondary antibodies, the cells were analyzed by confocal microscopy. The arrow in B indicates the perinuclear pool of β-BODIPY FL C5-HPC in an infected cell. The white bars are 5μm.
Inhibition of the lipid transport activities of ABCA1 and/or CLA 1 inhibits chlamydial growth
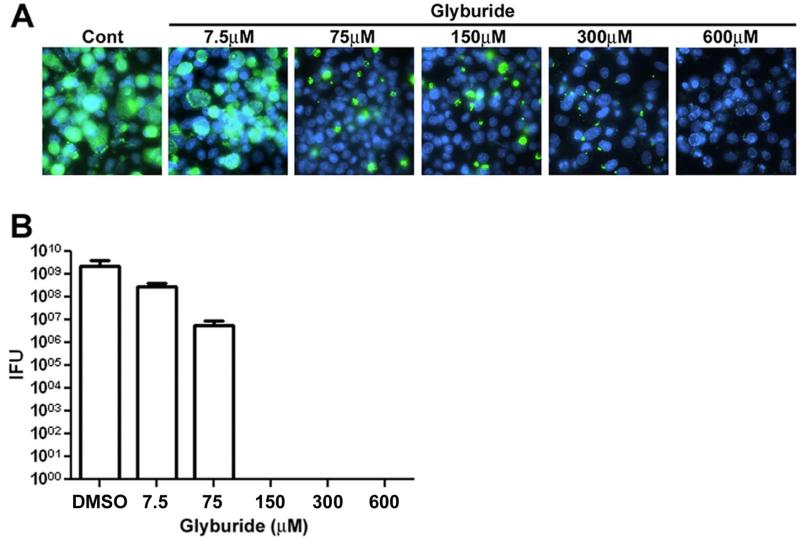
The localization data described above suggests a role for the HDL biogenesis machinery in delivering phospholipids to the inclusion of Chlamydia-infected cells. To test whether the lipid transport activities associated with ABCA1 and/or CLA 1 are required for chlamydial growth, we investigated whether glyburide, a drug that inhibits the ability of both ABCA1 and B1 type scavenger receptor members to efflux lipids to extracellular apoA-1, affects chlamydial growth. Previous studies have shown that glyburide concentrations in excess of 100μM are necessary to partially inhibit the lipid efflux activities of both ABCA1 and B1 type scavenger receptor members, such as CLA 1 (Smith et al., 2004, Nieland et al., 2004). In our analyses, HeLa cells were infected with C. trachomatis in media containing glyburide concentrations ranging from 7.5μM to 600μM. The drug was added to the infected cells 30 minutes PI to avoid possible effects of the drug on the entry of C. trachomatis into cells. At 48 hours PI, control and drug treated cells were fixed and stained with chlamydial Hsp60 antibodies (Fig. 6A). These analyses revealed that glyburide had a dose-dependent effect on chlamydial growth, and a concentration of 300μM was sufficient to completely inhibit chlamydial growth (Fig. 6A). To further evaluate the effect of glyburide on chlamydial growth we determined the number of infection-forming units (IFUs) of C. trachomatis present in infected HeLa cells treated with various drug concentrations. These studies indicated that 150μM glyburide was sufficient to completely block the production of infectious C. trachomatis (Fig. 6B) suggesting that the lipid transport activities associated with ABCA1 and/or CLA 1 are required for chlamydial growth in HeLa cells.
Figure 6.
HeLa cells were infected with C. trachomatis serovar D. At 30 minutes PI, the indicated amounts of glyburide in DMSO or DMSO alone (Cont) were added to the cells and the infection was allowed to proceed for 48 hours at which time the cells were fixed with methanol and incubated with a monoclonal antibody directed against Hsp60. Following washing, the cells were incubated with secondary antibody and random fields of cells were imaged on a Zeiss Axioplan II microscope (A). DNA was visualized by staining with Hoechst. The number of IFUs in infected cells at 48 hours PI was determined for each experimental condition (B). The values shown in B are the average of three independent experiments. Standard deviations in B are shown.
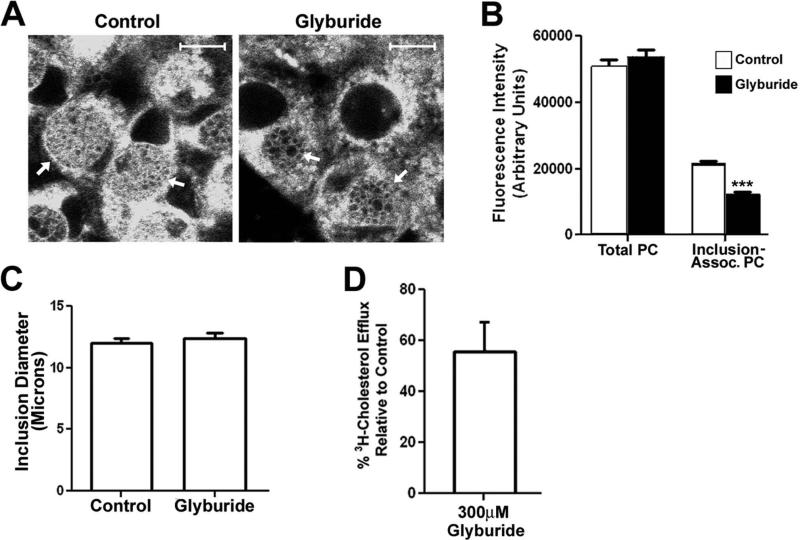
Additional experiments investigated whether the inhibitory effect of glyburide on chlamydial growth was potentially due to the ability of this drug to inhibit the recruitment of phospholipids to the inclusion of Chlamydia-infected cells. For these analyses, HeLa cells infected with C. trachomatis Serovar D were incubated for 28 hours to allow for inclusion development and bacterial growth. At this time, the cells were incubated in the absence or presence of 300μM glyburide for 3 hours. β-BODIPY FL C5-HPC was then added to the media of control and drug-treated cells, and the cells were incubated an additional hour at 37°C. The cells were then fixed and imaged by confocal microscopy. Although 300μM glyburide did not completely prevent the accumulation of β-BODIPY FL C5-HPC in the inclusion of infected cells (arrows in Fig. 7A), the fluorescence associated with the inclusions of the drug treated cells appeared reduced compared to control cells. Quantification of this experiment revealed that the glyburide treatment resulted in a 43% reduction in β-BODIPY FL C5-HPC levels within the inclusions of infected cells (Fig. 7B). This reduced level of inclusion-associated phospholipid in drug treated cells was not due to an effect of glyburide treatment on the total cellular uptake of β-BODIPY FL C5-HPC, which was very similar in control and drug-treated cells (Fig. 7B). In addition, the reduced inclusion-associated fluorescence was not due to an effect of the drug on inclusion size (Fig. 7C). Previous investigators have shown that glyburide has virtually an identical effect on the cholesterol and phospholipid transport activities of ABCA1 (Smith et al., 2004). To determine whether the reduced levels of phospholipid within the inclusion of drug-treated cells correlated with the inhibitory effect of glyburide on the lipid transport activity of ABCA1, we measured the effect of 300μM glyburide on the ability of ABCA1 to efflux 3H-cholesterol to extracellular lipid-free apoA-1. These studies revealed that treatment of cells with this concentration of drug resulted in a 45% reduction in ABCA1-dependent cholesterol efflux to extracellular lipid-free apoA-1 (Fig. 7D). The very similar inhibitory effect of glyburide on phospholipid accumulation within the inclusion of infected cells (Fig. 7B) and on the lipid transport activity of ABCA1 (Fig. 7D), is consistent with a role for ABCA1 and apoA-1 in directing the accumulation of phospholipids within the inclusion of Chlamydia-infected cells.
Figure 7.
HeLa cells infected with C. trachomatis serovar D were incubated at 37°C for 28 hours. At this time, the cells were incubated in the absence or presence of 300μM glyburide for 3 hours at 37°C. β-BODIPY FL C5-HPC in fatty acid-free BSA was then added to the media of control and drug-treated cells at a final concentration of 1.5μM and the cells were incubated an additional hour at 37°C. The cells were then fixed in 4% paraformaldehyde in PBS and imaged by confocal microscopy (A). The levels of total and inclusion-associated β-BODIPY FL C5-HPC fluorescence in control and glyburide-treated cells were quantified using the imaging software for the Zeiss LSM 510 confocal microscope as described in the Methods (B). The average diameter of the inclusions in control and glyburide-treated cells was also quantified using the Zeiss imaging software (C). The values shown in the histograms in B and C represent the average values obtained from the analysis of at least 75 cells from two independent experiments. *** p<0.0001 (two-tailed t-test). The effect of 300μM glyburide on the ability of ABCA1 to efflux 3H-cholesterol to extracellular lipid-free apoA-1 (D) was also determined as described in the Materials and Methods. The value shown in D, which is the average of two independent experiments, is expressed as the % 3H-cholesterol efflux to apoA-1 relative to that observed in control cells. Standard deviations are shown in B-D. Arrows in A point to inclusions in control and drug-treated cells. White bars in A are 10μm.
ABCA1 knockdown inhibits chlamydial growth in HeLa cells
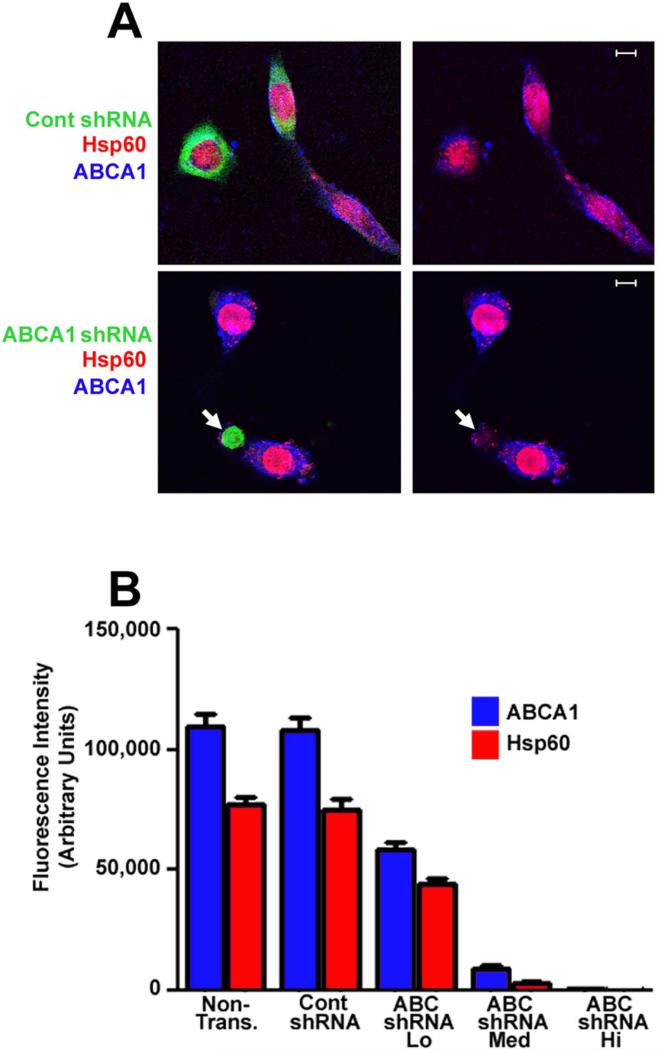
The data described above indicate that glyburide inhibits both chlamydial growth and the trafficking of phospholipids to the inclusion of Chlamydia-infected cells. However, in addition to ABCA1 and BI type scavenger receptors, glyburide inhibits the activity of several other cellular proteins (Vila-Carriles et al., 2007, Lehtihet et al., 2003, White et al., 1988) and its ability to block chlamydial growth may be due to its inhibitory effect on one or more of these other cellular targets. To more directly address the role of ABCA1 in regulating chlamydial growth, we knocked down the expression of this transporter using a shRNA approach. HeLa cells were transfected with plasmids encoding 2 shRNAs directed against ABCA1 or with a control shRNA. The shRNA-encoding plasmids also encoded a GFP reporter. In our initial assays, we attempted to establish stable cell lines that were stably transfected with the plasmids encoding the ABCA1 shRNAs. However, all of the clonal lines we established expressed low levels of the ABCA1 shRNAs using the GFP reporter as a measure of expression, and these clones exhibited minimal knockdown of ABCA1 (data not shown). For this reason, we opted to develop a transient transfection and fluorescence-based assay to measure the effect of the ABCA1 shRNAs on ABCA1 protein levels and chlamydial infection. This approach allowed us to determine how the growth of C. trachomatis in HeLa cells was affected over a broad range of expression of ABCA1. HeLa cells were transfected with the ABCA1 shRNAs or the control shRNA, and 24 hours post-transfection the cells were infected and allowed to grow an additional 48 hours. At this time, the cells were fixed and stained with ABCA1 and chlamydial Hsp60-specific antibodies. As shown in Fig. 8A, the control shRNA had no apparent effect on ABCA1 or Hsp60 levels in infected cells. However, ABCA1 and Hsp60 staining was very low in cells expressing high levels of ABCA1-specific shRNAs (arrows in Fig. 8A). Control experiments indicated that many of the Hsp60-positive bacteria associated with cells expressing high levels of ABCA1-specific shRNAs are intracellular and could only be stained when the cells were permeabilized prior to incubation with primary and secondary antibodies (data not shown). This result indicates that ABCA1 knockdown does not prevent chlamydial entry into cells but rather prevented growth following entry. These knockdown studies were quantified by determining the fluorescence intensity resulting from ABCA1 and chlamydial Hsp60 staining in non-transfected and shRNA-expressing cells. This analysis demonstrated that the control shRNA had no effect on ABCA1 or Hsp60 levels in infected cells (Fig. 8B). In contrast, ABCA1 and Hsp60 were dramatically reduced in cells expressing high levels of the ABCA1-specific shRNAs (Fig. 8B). Cells expressing low or medium amounts of the ABCA1-specific shRNAs exhibited a reduction in ABCA1 protein levels proportional to the level of the ABCA1 shRNA (Fig. 8B). In addition, the observed reduction in ABCA1 levels correlated with a corresponding decrease in Hsp60 levels in infected cells. These data illustrate the power of this fluorescence based assay, which suggests that the extent to which Chlamydia grow in these cells is dependent upon the cellular level of ABCA1 and complete knockdown of ABCA1 appears to be sufficient to completely block chlamydial growth. Taken together, the localization, knockdown, and drug studies presented here strongly suggest that the delivery of phospholipids to the inclusion by components of the host HDL biogenesis machinery is required for the growth of C. trachomatis within infected HeLa cells.
Figure 8.
HeLa cells were transfected with GFP-V-RS plasmids encoding two ABCA1-specific shRNAs or a control shRNA (A). Twenty-four hours post-transfection the cells were infected with C. trachomatis serovar D and the cells were subsequently fixed with 4% paraformaldehyde in PBS at 48 hours PI. The cells were permeabilized and incubated with rabbit ABCA1-specific and mouse Hsp60-specific antibodies. Following washing, the cells were incubated with the appropriate secondary antibodies and imaged by confocal microscopy. The arrows in A indicate a cell expressing high levels of the ABCA1-specific shRNAs. Z-stacks were acquired for each transfected cell and non-transfected control cells and the fluorescence intensity of the GFP reporter encoded by the pGFP-V-RS plasmid was used as a measure of shRNA levels within an individual transfected cell. The cells were arbitrarily grouped into high, medium, and low shRNA expressers based on the detector gain on the confocal microscope required to reach a saturating level of fluorescence for the GFP reporter. In each transfected and non-transfected cell, the fluorescence intensity through the entire Z-stack that resulted from staining with the ABCA1 and Hsp60 antibodies was quantified using the imaging software for the Zeiss LSM 510 confocal microscope. The values shown in the histogram in B represent the average value obtained from Z-stacks of at least 25 different cells. The average Hsp60 fluorescence intensity in cells expressing high levels of ABCA1-specific shRNAs was 225. This value is too small to be seen in the histogram in B. White bars in A are 5μm.
DISCUSSION
During growth and replication within infected cells, Chlamydia acquires cholesterol and sphingolipids from the host via vesicular (Hackstadt et al., 1995, Hackstadt et al., 1996, Carabeo et al., 2003, Elwell et al., 2011, Heuer et al., 2009, Beatty, 2006) and non-vesicular transport (Elwell et al., 2011, Derre et al., 2011) pathways. The fact that inhibitors of these pathways (Hackstadt et al., 1996, Carabeo et al., 2003, Elwell et al., 2011, Heuer et al., 2009, Beatty, 2006, Derre et al., 2011) do not completely block chlamydial growth has led to the suggestion that the pathways Chlamydia utilizes for acquiring cholesterol and sphingolipids are redundant. In contrast to cholesterol and sphingolipid acquisition, the mechanism(s) involved in the acquisition of host cell phospholipids by Chlamydia were not known prior to this study. We hypothesized that host lipid transporters may be involved in both phospholipid and cholesterol acquisition by Chlamydia. Our analyses have revealed that the host ABCA1 transporter, whose normal cellular function is to mediate the efflux of cellular cholesterol and phospholipids to lipid-free apoA-1 to initiate the formation of HDL (Zannis et al., 2006, Oram et al., 2000, Wang et al., 2000), is recruited to the inclusion membrane of C. trachomatis-infected HeLa cells. Localization analyses further indicated that additional components of the HDL biogenesis machinery including apoA-1 and CLA 1 are recruited to the inclusion of Chlamydia-infected cells. Several lines of evidence suggest that these elements of the HDL biogenesis machinery play a critical role in the delivery of essential phospholipids to the inclusion of infected cells. 1) The fluorescent phosphatidylcholine analogue, β-BODIPY FL C5-HPC, co-localizes with foci of apoA-1 within the inclusion of infected cells. 2) Glyburide, a drug that inhibits the ability of both ABCA1 (Smith et al., 2004, Nieland et al., 2004) and B1 type scavenger receptors (Smith et al., 2004), such as CLA 1, to transport lipids to extracellular apoA-1, inhibits the accumulation of phosphatidylcholine within the inclusion of infected cells and prevents chlamydial growth. 3) Knockdown of ABCA1 prevents the growth of C. trachomatis in infected HeLa cells. These data suggest that ABCA1 and perhaps CLA 1 in conjunction with their lipid acceptor apoA-1 provide a source of phospholipids that is required for the growth of Chlamydia within infected cells.
The concentration of glyburide used in our studies exhibits virtually an identical inhibitory effect on the apoA-1-dependent lipid transport activity of ABCA1 and on the accumulation of the phosphatidylcholine analogue, β-BODIPY FL C5-HPC, in the inclusion of Chlamydia-infected cells (Fig. 7). These data are consistent with a role for ABCA1 and apoA-1 in directing phosphatidylcholine accumulation within the inclusion. This hypothesis is further strengthened by the fact that apoA-1 co-localizes with pools of β-BODIPY FL C5-HPC within the lumen of the inclusion of infected cells. While other pathways may contribute to the transport of phospholipids to the inclusion, interfering with the ABCA1/apoA-1-dependent pathway with drugs or by reducing ABCA1 protein levels is sufficient to prevent chlamydial growth.
The accumulation of ABCA1, CLA 1 and apoA-1 in the inclusion of infected cells may reflect a global redistribution of host proteins of the endosomal/lysosomal pathway to the organelle where Chlamydia grow. Alternatively, the accumulation of these proteins in the inclusion could result from the modification of specific sorting events that are altered as a consequence of Chlamydia infection. A candidate protein that could be involved in this process is Rab4, one of several small GTP-binding proteins recruited to the inclusion membrane of C. trachomatis-infected cells (Rzomp et al., 2003, Rzomp et al., 2006, Moorhead et al., 2010). ABCA1 normally undergoes internalization from the plasma membrane and recycles from endosomes back to the cell surface in a Rab4-dependent fashion (Neufeld et al., 2001, Azuma et al., 2009). Whether C. trachomatis infection alters Rab4 function resulting in the recruitment of ABCA1 and apo-A1, which associates with ABCA1 (Wang et al., 2000), to the inclusion of infected cells is currently under investigation.
The localization profiles described here for ABCA1, apoA-1, and the inclusion membrane-associated population of CLA 1 could arise from the fusion of intracellular vesicles containing these proteins with the inclusion membrane of Chlamydia-infected cells. However, alternative mechanisms must be involved in directing CLA 1 to foci within the inclusion of infected cells. The elongated tubular structure containing CLA 1 and apoA-1 (arrowhead in Fig. 2D) appears to span the inclusion membrane of this infected cell. This image suggests that cytoplasmic membrane vesicles containing CLA 1 and apoA-1 may be engulfed by the inclusion by an autophagic-type mechanism (Glick et al., 2010). Alternatively, CLA 1-containing vesicles may initially fuse with the inclusion membrane. Subsequently, regions of the inclusion membrane containing CLA 1 could bud into the lumen of the inclusion analogous to the intra-lumenal budding of vesicles that occurs during multivesicular body biogenesis (Hurley et al., 2010). Whether these mechanisms or others are involved in the recruitment of CLA 1 to foci within the lumen of the inclusion of infected cells will be investigated in future studies.
In conclusion our studies have demonstrated that several of the components of the host cell HDL biogenesis machinery are recruited to the inclusion of C. trachomatis-infected cells. Knockdown and inhibitor studies strongly suggest that the lipid transport activities of ABCA1 and perhaps CLA 1 are required for the growth of C. trachomatis in HeLa cells. Our analyses have indicated that pools of phosphatidylcholine accumulate in the inclusion of infected cells in close proximity to apoA-1, the lipid acceptor of both ABCA1 and CLA 1. How the pools of phospholipid associated with apoA-1 are made available for chlamydial growth is currently unknown. However, it is interesting to note that CLA 1, which lies adjacent to apoA-1 within the inclusion of infected cells (Fig. 2D), can both interact with apoA-1 (Zannis et al., 2006) and transfer lipids from apoA-1-containing HDL complexes (Acton et al., 1996, Ji et al., 1997). Whether CLA 1 or other host or chlamydial encoded proteins are required for making the pools of phospholipid delivered to the inclusion available for chlamydial growth will be defined in future studies.
EXPERIMENTAL PROCEDURES
Antibodies, Plasmids, and Reagents
ABCA1 rabbit polyclonal antibodies were obtained from Novus. Rabbit polyclonal antibodies against a peptide corresponding to amino acids 450-509 of the murine scavenger receptor, SR-B1, were obtained from Novus. This region of SR-B1 is 82% identical to CLA 1, the human homologue of SR-B1 (Calvo et al., 1993, Acton et al., 1994). Rabbit polyclonal antibodies directed against apo-A1 were obtained from BioVision. Mouse monoclonal antibodies directed against MOMP were obtained from Argene, and mouse monoclonal antibodies directed against the chlamydial inclusion membrane proteins, IncA and CT223, were provided by Dr. D. D. Rockey. Mouse monoclonal antibodies directed against the Chlamydia Hsp60 protein were provided by Dr. R. P. Morrison. Mouse monoclonal antibodies directed against actin were obtained from Sigma Chemicals. Various AlexaFluor conjugated secondary antibodies, Hoechst, and β-BODIPY FL C5-HPC were obtained from Invitrogen, while horseradish peroxidase (HRP) conjugated secondary antibodies were obtained from BioRad. Glyburide and fatty acid-free BSA were purchased from Sigma Chemicals, lipid-free apoA-1 was purchased from Millipore, and 3H-cholesterol was purchased from New England Nuclear.
A scrambled control shRNA and 2 ABCA1-specific shRNAs in the pGFP-V-RS vector were purchased from Origene. The pGFP-V-RS vector also encodes GFP under the control of a separate promoter. The ABCA1-specific shRNAs target the sequences: CTCTGCTATCTCCAACCTCATCAGGAAGC (nucleotides 3853-3881) and ATCTGCTTCCAGCAGAAGTCCTATGTGTC (nucleotides 5477-5505) in the human ABCA1 mRNA. ABCA1-EGFP (Tanaka et al., 2003) in pcDNA3 was provided by Dr. K. Ueda. A CLA 1 cDNA was amplified by RT-PCR from RNA isolated from HeLa cells. The CLA 1 cDNA was then fused at its C-terminus to DsRed monomer in the pDsRedN1 vector from Clontech. The resulting fusion, CLA 1-DsRed monomer, was sequenced prior to its use in transfection studies.
Cell Culture and Chlamydial Infections
HeLa cells grown in DMEM containing 10% fetal calf serum were infected with C. trachomatis serovar D (strain UW-3/Cx) or C. trachomatis serovar L2 (strain 434/Bu). At various times PI, the cells were fixed and processed for immunostaining, or the cells were harvested and the number of IFUs was determined as described previously (Caldwell et al., 1981). In some instances, glyburide was added to infected cells at 30 minutes PI and maintained in the media for the duration of the infection.
HeLa cells were transfected with the ABCA1-EGFP or CLA 1-DsRed monomer expressing plasmids or the pGFP-V-RS plasmids encoding the ABCA1-specific or control shRNAs using the Effectene transfection reagent (Qiagen). Cells were fixed at various times post-transfection and processed for microscopy as described in the text.
Immunoblotting Assays
Lysates were prepared from uninfected HeLa cells or HeLa cells infected with C. trachomatis at various times PI by direct lysis in SDS sample buffer. The lysates were then subjected to immunoblotting analysis with various antibodies. Immunoreactive species in blotting analyses were detected using HRP-conjugated secondary antibodies and chemiluminescence detection systems. Quantification of immunoblots was carried out using Scion Image.
Localization and Knockdown Analyses
HeLa cells infected with C. trachomatis were fixed at various times PI by incubation in 4% paraformaldehyde in phosphate buffered-saline (PBS) for 10 minutes. Following rinsing in PBS, the cells were permeabilized in PBS containing 0.1% Triton X-100 or 0.1% saponin prior to primary antibody incubation. Alternatively, cells were fixed by incubation in ice-cold methanol for 30 seconds. The cells were then rinsed with PBS and incubated with primary antibodies. In each instance, the cells were rinsed, incubated with the appropriate AlexaFluor conjugated secondary antibody prior to microscopic analysis.
HeLa cells were transfected with pGFP-V-RS plasmids encoding the ABCA1-specific and control shRNAs. Twenty-four hours post-transfection the cells were infected with C. trachomatis (MOI=2) and then fixed and processed for staining with ABCA1 and Hsp60 antibodies 48 hours PI. Z-stacks were acquired for each transfected cell and non-transfected control cells using a Zeiss LSM 510 confocal microscope. The fluorescence intensity of the GFP reporter encoded by the pGFP-V-RS plasmid was used as a measure of shRNA levels within an individual transfected cell. The cells were arbitrarily grouped into high, medium, and low shRNA expressers based on the setting of the detector gain on the confocal microscope required to reach a saturating level of fluorescence for the GFP reporter. The setting for high cells was <800; the setting for medium cells was 800 – 900; and the setting for low cells was >900. In each transfected and non-transfected cell, the fluorescence intensity through the entire Z-stack that resulted from staining with the ABCA1 and Hsp60 antibodies was quantified using the imaging software for the Zeiss LSM 510 confocal microscope. The values presented in the text represent the average value obtained from Z-stacks of at least 25 different cells.
Quantitative RT-PCR
Total RNA was isolated using Masterpure (Epicentre) and used as a template to synthesize first strand cDNA. 3μg RNA was reverse-transcribed using Transcriptor reverse transcriptase (Roche) and random hexamer primers. Subsequent qRT-PCR analysis was carried out using a LightCycler 480 System (Roche) using primers specific for apoA-1 and probes from the Universal Probe Library system (Roche). The expression of apoA-1 and the control gene (GAPDH) were quantified based on their threshold cycle values and apoA-1 levels were normalized using the control gene value.
β-BODIPY FL C5-HPC Labeling of C. trachomatis-Infected HeLa Cells
HeLa cells infected with C. trachomatis serovar D were incubated in the presence of 1.5μM β-BODIPY FL C5-HPC in fatty acid-free BSA (Wustner et al., 2001) for 1 hour at 37°C at various times PI. The cells were then rinsed in PBS and fixed in 4% paraformaldehyde in PBS. The cells were then directly imaged by confocal microscopy or permeabilized by extraction in methanol. Permeabilized cells were stained with various antibodies prior to confocal analysis. In some experiments, HeLa cells were back-extracted for 1 hour in media containing fatty acid-free BSA for 1 hour prior to microscopic analysis and identical results were observed.
HeLa cells infected with C. trachomatis serovar D were incubated at 37°C for 28 hours. At this time, the cells were incubated in the absence or presence of 300μM glyburide for 3 hours at 37°C. β-BODIPY FL C5-HPC in fatty acid-free BSA was then added to the media of control and drug-treated cells at a final concentration of 1.5μM and the cells were incubated an additional hour at 37°C. The cells were then fixed in 4% paraformaldehyde in PBS and imaged by confocal microscopy. Z-stacks were acquired for control and glyburide-treated cells using identical acquisition parameters and the levels of total and inclusion-associated β-BODIPY FL C5-HPC fluorescence in control and glyburide-treated cells were quantified using the imaging software for the Zeiss LSM 510 confocal microscope. In this analysis, the lines demarcating the boundaries of inclusions were drawn inside the inclusion membrane to insure that cytoplasmic fluorescence was excluded from the inclusion-associated fluorescence value. The values shown in the text represent the average values obtained from the analysis of at least 75 cells from two independent experiments.
Cholesterol Efflux Assays
Cholesterol efflux assays were carried out as described previously (Smith et al., 2004). Briefly, HeLa cells were incubated with 3H-cholesterol at a final concentration 0.5μCi/ml overnight in DMEM containing 10% fetal calf serum and 0.5% BSA. After washing once in DMEM, the cells were incubated for 4 hours at 37°C in the absence or presence of 300μM glyburide in DMEM containing 10% fetal calf serum, 0.5% fatty acid-free BSA, and 10μg/ml of lipid-free apoA-1. At this time, the media was collected and saved. The cells were rinsed once in DMEM and then scraped from the dish and collected. The resulting cell pellet was dissolved by overnight incubation in 0.1M NaOH, and the amount of 3H-cholesterol in the media and associated with the cells was determined by scintillation counting. The percentage of 3H-cholesterol that was effluxed from the cells was determined and the values shown in the text represent the average from two independent experiments and are expressed as the percent 3H-cholesterol efflux in the presence of glyburide relative to that observed in the non-glyburide treated control.
Supplementary Material
Supp. Fig. S1. A lysate prepared from HeLa cells at 32 hours PI with C. trachomatis serovar D was subjected to immunoblotting analyses using antibodies directed against CLA 1 and apoA-1 (A). Alternatively, HeLa cells infected with C. trachomatis were fixed at 32 hours PI and stained with antibodies directed against apoA-1 (red) and IncA (green) (B and C). The numbered images in B correspond to consecutive 0.5μm confocal slices through an infected cell. The upper panel in C is a xy confocal slice through an infected cell and the lower panel is a xz confocal slice through the same cell. HeLa cells infected with C. trachomatis were also fixed at 32 hours PI and stained with antibodies directed against apoA-1 (red) and chlamydial Hsp60 (green) (D). The numbered images in D correspond to consecutive 0.5μM confocal slices through an infected cell. Molecular weight markers are indicated in A and the white bars in B-D are 10μm.
ACKNOWLEDGEMENTS
This work was supported in part by a University of Tennessee Health Science Center Pathogenesis Center of Excellence Grant (JVC), and a grant from the National Institutes of Health, AI 070693 (RJB). Antibodies against IncA and CT223 were kindly provided by Dr. D. D. Rockey, and antibodies against chlamydial Hsp60 were kindly provided by Dr. R. P. Morrison. The ABCA1-EGFP cDNA was kindly provided by Dr. K. Ueda.
REFERENCES
- Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science. 1996;271:518–520. doi: 10.1126/science.271.5248.518. [DOI] [PubMed] [Google Scholar]
- Acton SL, Scherer PE, Lodish HF, Krieger M. Expression cloning of SR-BI, a CD36-related class B scavenger receptor. J Biol Chem. 1994;269:21003–21009. [PubMed] [Google Scholar]
- Alzhanov DT, Weeks SK, Burnett JR, Rockey DD. Cytokinesis is blocked in mammalian cells transfected with Chlamydia trachomatis gene CT223. BMC Microbiol. 2009;9:2. doi: 10.1186/1471-2180-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa R, Yokoyama S. Helical apolipoproteins stabilize ATP-binding cassette transporter A1 by protecting it from thiol protease-mediated degradation. J Biol Chem. 2002;277:22426–22429. doi: 10.1074/jbc.M202996200. [DOI] [PubMed] [Google Scholar]
- Azuma Y, Takada M, Shin HW, Kioka N, Nakayama K, Ueda K. Retroendocytosis pathway of ABCA1/apoA-I contributes to HDL formation. Genes Cells. 2009;14:191–204. doi: 10.1111/j.1365-2443.2008.01261.x. [DOI] [PubMed] [Google Scholar]
- Bannantine JP, Griffiths RS, Viratyosin W, Brown WJ, Rockey DD. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell Microbiol. 2000;2:35–47. doi: 10.1046/j.1462-5822.2000.00029.x. [DOI] [PubMed] [Google Scholar]
- Bannantine JP, Stamm WE, Suchland RJ, Rockey DD. Chlamydia trachomatis IncA is localized to the inclusion membrane and is recognized by antisera from infected humans and primates. Infect Immun. 1998;66:6017–6021. doi: 10.1128/iai.66.12.6017-6021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty WL. Trafficking from CD63-positive late endocytic multivesicular bodies is essential for intracellular development of Chlamydia trachomatis. J Cell Sci. 2006;119:350–359. doi: 10.1242/jcs.02733. [DOI] [PubMed] [Google Scholar]
- Braun A, Trigatti BL, Post MJ, Sato K, Simons M, Edelberg JM, et al. Loss of SR-BI expression leads to the early onset of occlusive atherosclerotic coronary artery disease, spontaneous myocardial infarctions, severe cardiac dysfunction, and premature death in apolipoprotein E-deficient mice. Circ Res. 2002;90:270–276. doi: 10.1161/hh0302.104462. [DOI] [PubMed] [Google Scholar]
- Caldwell HD, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31:1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo D, Gomez-Coronado D, Lasuncion MA, Vega MA. CLA-1 is an 85-kD plasma membrane glycoprotein that acts as a high-affinity receptor for both native (HDL, LDL, and VLDL) and modified (OxLDL and AcLDL) lipoproteins. Arterioscler Thromb Vasc Biol. 1997;17:2341–2349. doi: 10.1161/01.atv.17.11.2341. [DOI] [PubMed] [Google Scholar]
- Calvo D, Vega MA. Identification, primary structure, and distribution of CLA-1, a novel member of the CD36/LIMPII gene family. J Biol Chem. 1993;268:18929–18935. [PubMed] [Google Scholar]
- Carabeo RA, Mead DJ, Hackstadt T. Golgi-dependent transport of cholesterol to the Chlamydia trachomatis inclusion. Proc Natl Acad Sci U S A. 2003;100:6771–6776. doi: 10.1073/pnas.1131289100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocchiaro JL, Kumar Y, Fischer ER, Hackstadt T, Valdivia RH. Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc Natl Acad Sci U S A. 2008;105:9379–9384. doi: 10.1073/pnas.0712241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo G, Vicinanza M, De Matteis MA. Lipid-transfer proteins in biosynthetic pathways. Curr Opin Cell Biol. 2008;20:360–370. doi: 10.1016/j.ceb.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Derre I, Swiss R, Agaisse H. The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites. PLoS Pathog. 2011;7:e1002092. doi: 10.1371/journal.ppat.1002092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwell CA, Jiang S, Kim JH, Lee A, Wittmann T, Hanada K, et al. Chlamydia trachomatis co-opts GBF1 and CERT to acquire host sphingomyelin for distinct roles during intracellular development. PLoS Pathog. 2011;7:e1002198. doi: 10.1371/journal.ppat.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelissen IC, Harris M, Rye KA, Quinn C, Brown AJ, Kockx M, et al. ABCA1 and ABCG1 synergize to mediate cholesterol export to apoA-I. Arterioscler Thromb Vasc Biol. 2006;26:534–540. doi: 10.1161/01.ATV.0000200082.58536.e1. [DOI] [PubMed] [Google Scholar]
- Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T, Rockey DD, Heinzen RA, Scidmore MA. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 1996;15:964–977. [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T, Scidmore MA, Rockey DD. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc Natl Acad Sci U S A. 1995;92:4877–4881. doi: 10.1073/pnas.92.11.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch TP. Competition between Chlamydia psittaci and L cells for host isoleucine pools: a limiting factor in chlamydial multiplication. Infect Immun. 1975a;12:211–220. doi: 10.1128/iai.12.1.211-220.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch TP. Utilization of L-cell nucleoside triphosphates by Chlamydia psittaci for ribonucleic acid synthesis. J Bacteriol. 1975b;122:393–400. doi: 10.1128/jb.122.2.393-400.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer D, Rejman Lipinski A, Machuy N, Karlas A, Wehrens A, Siedler F, et al. Chlamydia causes fragmentation of the Golgi compartment to ensure reproduction. Nature. 2009;457:731–735. doi: 10.1038/nature07578. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Jian B, Wang N, Sun Y, Moya ML, Phillips MC, et al. Scavenger receptor BI promotes high density lipoprotein-mediated cellular cholesterol efflux. J Biol Chem. 1997;272:20982–20985. doi: 10.1074/jbc.272.34.20982. [DOI] [PubMed] [Google Scholar]
- Lehtihet M, Welsh N, Berggren PO, Cook GA, Sjoholm A. Glibenclamide inhibits islet carnitine palmitoyltransferase 1 activity, leading to PKC-dependent insulin exocytosis. Am J Physiol Endocrinol Metab. 2003;285:E438–446. doi: 10.1152/ajpendo.00057.2003. [DOI] [PubMed] [Google Scholar]
- Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- Matsunaga T, Hiasa Y, Yanagi H, Maeda T, Hattori N, Yamakawa K, et al. Apolipoprotein A-I deficiency due to a codon 84 nonsense mutation of the apolipoprotein A-I gene. Proc Natl Acad Sci U S A. 1991;88:2793–2797. doi: 10.1073/pnas.88.7.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClarty G. Chlamydiae and the biochemistry of intracellular parasitism. Trends Microbiol. 1994;2:157–164. doi: 10.1016/0966-842x(94)90665-3. [DOI] [PubMed] [Google Scholar]
- McClarty G, Tipples G. In situ studies on incorporation of nucleic acid precursors into Chlamydia trachomatis DNA. J Bacteriol. 1991;173:4922–4931. doi: 10.1128/jb.173.16.4922-4931.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead AM, Jung JY, Smirnov A, Kaufer S, Scidmore MA. Multiple host proteins that function in phosphatidylinositol-4-phosphate metabolism are recruited to the chlamydial inclusion. Infect Immun. 2010;78:1990–2007. doi: 10.1128/IAI.01340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld EB, Remaley AT, Demosky SJ, Stonik JA, Cooney AM, Comly M, et al. Cellular localization and trafficking of the human ABCA1 transporter. J Biol Chem. 2001;276:27584–27590. doi: 10.1074/jbc.M103264200. [DOI] [PubMed] [Google Scholar]
- Nieland TJ, Chroni A, Fitzgerald ML, Maliga Z, Zannis VI, Kirchhausen T, Krieger M. Cross-inhibition of SR-BI- and ABCA1-mediated cholesterol transport by the small molecules BLT-4 and glyburide. J Lipid Res. 2004;45:1256–1265. doi: 10.1194/jlr.M300358-JLR200. [DOI] [PubMed] [Google Scholar]
- Oram JF, Vaughan AM. ABCA1-mediated transport of cellular cholesterol and phospholipids to HDL apolipoproteins. Curr Opin Lipidol. 2000;11:253–260. doi: 10.1097/00041433-200006000-00005. [DOI] [PubMed] [Google Scholar]
- Rhainds D, Brissette L. The role of scavenger receptor class B type I (SR-BI) in lipid trafficking. defining the rules for lipid traders. Int J Biochem Cell Biol. 2004;36:39–77. doi: 10.1016/s1357-2725(03)00173-0. [DOI] [PubMed] [Google Scholar]
- Ritter M, Buechler C, Boettcher A, Barlage S, Schmitz-Madry A, Orso E, et al. Cloning and characterization of a novel apolipoprotein A-I binding protein, AI-BP, secreted by cells of the kidney proximal tubules in response to HDL or ApoA-I. Genomics. 2002;79:693–702. doi: 10.1006/geno.2002.6761. [DOI] [PubMed] [Google Scholar]
- Rust S, Rosier M, Funke H, Real J, Amoura Z, Piette JC, et al. Tangier disease is caused by mutations in the gene encoding ATP-binding cassette transporter 1. Nat Genet. 1999;22:352–355. doi: 10.1038/11921. [DOI] [PubMed] [Google Scholar]
- Rzomp KA, Moorhead AR, Scidmore MA. The GTPase Rab4 interacts with Chlamydia trachomatis inclusion membrane protein CT229. Infect Immun. 2006;74:5362–5373. doi: 10.1128/IAI.00539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzomp KA, Scholtes LD, Briggs BJ, Whittaker GR, Scidmore MA. Rab GTPases are recruited to chlamydial inclusions in both a species-dependent and species-independent manner. Infect Immun. 2003;71:5855–5870. doi: 10.1128/IAI.71.10.5855-5870.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JD, Le Goff W, Settle M, Brubaker G, Waelde C, Horwitz A, Oda MN. ABCA1 mediates concurrent cholesterol and phospholipid efflux to apolipoprotein A-I. J Lipid Res. 2004;45:635–644. doi: 10.1194/jlr.M300336-JLR200. [DOI] [PubMed] [Google Scholar]
- Tanaka AR, Abe-Dohmae S, Ohnishi T, Aoki R, Morinaga G, Okuhira K, et al. Effects of mutations of ABCA1 in the first extracellular domain on subcellular trafficking and ATP binding/hydrolysis. J Biol Chem. 2003;278:8815–8819. doi: 10.1074/jbc.M206885200. [DOI] [PubMed] [Google Scholar]
- Van Eck M, Bos IS, Hildebrand RB, Van Rij BT, Van Berkel TJ. Dual role for scavenger receptor class B, type I on bone marrow-derived cells in atherosclerotic lesion development. Am J Pathol. 2004;165:785–794. doi: 10.1016/S0002-9440(10)63341-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eck M, Pennings M, Hoekstra M, Out R, Van Berkel TJ. Scavenger receptor BI and ATP-binding cassette transporter A1 in reverse cholesterol transport and atherosclerosis. Curr Opin Lipidol. 2005;16:307–315. doi: 10.1097/01.mol.0000169351.28019.04. [DOI] [PubMed] [Google Scholar]
- Vila-Carriles WH, Zhao G, Bryan J. Defining a binding pocket for sulfonylureas in ATP-sensitive potassium channels. FASEB J. 2007;21:18–25. doi: 10.1096/fj.06-6730hyp. [DOI] [PubMed] [Google Scholar]
- Wang N, Silver DL, Costet P, Tall AR. Specific binding of ApoA-I, enhanced cholesterol efflux, and altered plasma membrane morphology in cells expressing ABC1. J Biol Chem. 2000;275:33053–33058. doi: 10.1074/jbc.M005438200. [DOI] [PubMed] [Google Scholar]
- White CW, Rashed HM, Patel TB. Sulfonylureas inhibit metabolic flux through rat liver pyruvate carboxylase reaction. J Pharmacol Exp Ther. 1988;246:971–974. [PubMed] [Google Scholar]
- Wustner D, Mukherjee S, Maxfield FR, Muller P, Herrmann A. Vesicular and nonvesicular transport of phosphatidylcholine in polarized HepG2 cells. Traffic. 2001;2:277–296. doi: 10.1034/j.1600-0854.2001.9o135.x. [DOI] [PubMed] [Google Scholar]
- Wylie JL, Hatch GM, McClarty G. Host cell phospholipids are trafficked to and then modified by Chlamydia trachomatis. J Bacteriol. 1997;179:7233–7242. doi: 10.1128/jb.179.23.7233-7242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvan-Charvet L, Ranalletta M, Wang N, Han S, Terasaka N, Li R, et al. Combined deficiency of ABCA1 and ABCG1 promotes foam cell accumulation and accelerates atherosclerosis in mice. J Clin Invest. 2007;117:3900–3908. doi: 10.1172/JCI33372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannis VI, Chroni A, Krieger M. Role of apoA-I, ABCA1, LCAT, and SR-BI in the biogenesis of HDL. J Mol Med (Berl) 2006;84:276–294. doi: 10.1007/s00109-005-0030-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp. Fig. S1. A lysate prepared from HeLa cells at 32 hours PI with C. trachomatis serovar D was subjected to immunoblotting analyses using antibodies directed against CLA 1 and apoA-1 (A). Alternatively, HeLa cells infected with C. trachomatis were fixed at 32 hours PI and stained with antibodies directed against apoA-1 (red) and IncA (green) (B and C). The numbered images in B correspond to consecutive 0.5μm confocal slices through an infected cell. The upper panel in C is a xy confocal slice through an infected cell and the lower panel is a xz confocal slice through the same cell. HeLa cells infected with C. trachomatis were also fixed at 32 hours PI and stained with antibodies directed against apoA-1 (red) and chlamydial Hsp60 (green) (D). The numbered images in D correspond to consecutive 0.5μM confocal slices through an infected cell. Molecular weight markers are indicated in A and the white bars in B-D are 10μm.