Summary
Innate recognition systems, including the Toll-like receptors (TLRs), play a critical role in activating host defenses and proinflammatory pathways in response to infection. Pathogens have developed strategies to subvert TLRs in order to survive and replicate within the host. The model intracellular pathogen, Francisella novicida, modulates host defenses to promote survival and replication in macrophages. TLR2, which recognizes bacterial lipoproteins (BLPs), is critical for activating host defenses and proinflammatory cytokine production in response to Francisella infection. Here we show that the F. novicida protein FTN_0757 acts to repress BLP production, dampening TLR2 activation. The ΔFTN_0757 mutant strain induced robust TLR2-dependent cytokine production in macrophages compared to wild-type bacteria, and produced increased amounts of BLPs. The deletion of one BLP (FTN_1103) from ΔFTN_0757 decreased the total BLP concentration to near wild-type levels and correlated with a decrease in the induction of TLR2 signaling. The overproduction of BLPs also contributed to the in vivo attenuation of the ΔFTN_0757 mutant, which was significantly rescued when FTN_1103 was deleted. Taken together, these data reveal a novel mechanism of immune evasion by the downregulation of BLP expression to subvert TLR2 activation, which is likely used by numerous other intracellular bacterial pathogens.
Introduction
Early detection of microbial pathogens by pattern recognition receptors (PRRs) is an important component in the initiation of an effective immune response aimed at clearing infections (Akira et al., 2006). One group of PRRs, the Toll-like receptors (TLRs), are type I integral membrane proteins present on the surface of a diverse set of host cells, as well as in endosomes. TLRs are responsible for the recognition of a number of different microbial components, or pathogen-associated molecular patterns (PAMPs) (Kumar et al., 2009). For example, lipid A from Gram-negative bacteria is recognized by TLR4, flagellin by TLR5, CpG DNA by TLR9, and bacterial lipoproteins (BLPs) by TLR2 (Lien et al., 2000, Hayashi et al., 2001, Hemmi et al., 2000, Aliprantis et al., 1999, Brightbill et al., 1999). Upon recognition of their cognate PAMP, TLRs signal to activate transcription factors, including NF-κB, which lead to the production of proinflammatory cytokines, chemokines, and antimicrobial peptides (Kumar et al., 2009). Production of these proteins stimulates an array of host defenses including the activation of macrophages and the recruitment of neutrophils, which help to fight infection (Kumar et al., 2009, Sjostedt et al., 1994).
Pathogens have developed a variety of mechanisms to prevent TLR signaling. Some pathogens secrete effector proteins into host cells that block components of the TLR signaling pathways. For example, the enteropathogenic Escherichia coli effector NleE directly blocks NF-κB activation by preventing IκB degradation, and the Brucella spp. effector Btp1 binds the cytosolic TIR domain of TLR2 and TLR4, preventing the recruitment of downstream signaling proteins (Nadler et al., 2010, Radhakrishnan et al., 2009, Salcedo et al., 2008). Pathogens can also prevent TLR signaling by modifying PAMPs. For instance, alterations in the amino acid sequence of the flagellin monomer allow Helicobacter spp. to prevent recognition by TLR5 (Andersen-Nissen et al., 2005). Specific modifications to the structure of lipid A, such as the addition of acyl chains (Salmonella spp.) or the removal of acyl chains (Yersinia pestis), facilitate evasion of TLR4 signaling (Tanamoto et al., 2000, Kawahara et al., 2002). Furthermore, recognition of pathogens by TLRs can be subverted by preventing the release of PAMPs. For example, the masking of flagella by a lipid membrane sheath is used by Vibrio spp. to prevent recognition of flagellin by TLR5 (Follett et al., 1963, Yoon et al., 2008).
The Gram-negative bacterium Francisella novicida, a model intracellular pathogen closely related to highly virulent F. tularensis, has evolved strategies to subvert host defense proteins including TLRs (Rohmer et al., 2007, Telepnev et al., 2003). Similar to other Francisella spp., F. novicida can infect and replicate within host macrophages, which express numerous TLRs (Anthony et al., 1991a, Muzio et al., 2000). Extensive modification of its lipid A, including alterations in the length and number of acyl chains, results in a lack of signaling through TLR4 (Wang et al., 2007, Kanistanon et al., 2008). This is a critical component of Francisella pathogenesis since mutants that cannot modify lipid A are severely attenuated in vivo (Wang et al., 2007). Furthermore, Francisella does not encode flagellin and therefore does not activate TLR5 signaling (Li et al., 2006, Cole et al., 2006). The bacteria are, however, recognized by TLR2, which plays an important role in host defense as indicated by the increased susceptibility to infection of mice lacking TLR2 (Abplanalp et al., 2009, Malik et al., 2006a). Thus, molecular strategies used by Francisella spp. to subvert TLR2 signaling would likely promote pathogenesis.
The specific proteins used by Francisella spp. to suppress host defenses, and their mechanisms of action, are largely unknown. We and others previously used in vivo genetic screens to identify critical Francisella virulence determinants (Weiss et al., 2007, Su et al., 2007, Kadzhaev et al., 2009, Kraemer et al., 2009, Peng et al., 2011). One gene that we identified, FTN_0757 (also termed FTT_0584 before the F. novicida genome was sequenced), is necessary for F. novicida virulence in mice and has been shown to be involved in the suppression of several pro-inflammatory cytokines (Weiss et al., 2007, Peng et al., 2011). However, its mechanism of action is unknown. Therefore, we set out to elucidate how FTN_0757 contributes to the subversion of innate inflammatory responses and better define the breadth of its effect on the host response.
Here, we demonstrate that FTN_0757 action leads to the suppression of a large panel of NF-κB-dependent genes, as well as genes encoding other host defense proteins. We show that the increased production of cytokines and chemokines in response to infection by the FTN_0757 mutant is due to hyperstimulation of TLR2. More specifically, we show that FTN_0757 functions to limit the expression and production of BLPs that induce proinflammatory mediators through TLR2. One BLP, FTN_1103, is highly overproduced in the FTN_0757 mutant and accounts for the majority of the increased BLP content. Deletion of FTN_1103 from the FTN_0757 mutant significantly reduces the activation of TLR2 and rescues the virulence defect of the mutant in vivo. To our knowledge, this is the first demonstration that suppression of BLP content by an intracellular pathogen allows subversion of TLR2-dependent responses and promotes virulence. Furthermore, this work may provide insights into ways by which other pathogens escape recognition by TLR2.
Results
FTN_0757 Suppresses TLR2-dependent Proinflammatory Responses in Macrophages
Previous studies by our laboratory and others showed that a ΔFTN_0757 mutant of F. novicida induced increased macrophage secretion of several proinflammatory cytokines compared to wild-type bacteria (Peng et al., 2011, Weiss et al., 2007). However, the full scope of this hyperinflammatory response and the mechanism underlying this phenotype were unclear. As a first step towards defining the extent of this effect, we sought to measure the breadth of the inflammatory response induced by the ΔFTN_0757 mutant during macrophage infection using microarray analysis. We harvested RNA from murine bone marrow-derived macrophages (BMDM) infected with wild-type F. novicida or the ΔFTN_0757 mutant, as well as uninfected controls. Microarray analysis revealed that there was a broad and robust increase in the number and magnitude of macrophage genes expressed in response to infection by the ΔFTN_0757 mutant compared to wild-type bacteria (Tables S1, S2 and Figures 1A). Furthermore, Ingenuity Pathway Analysis revealed that host defense pathways consisting of genes encoding PRRs that recognize bacteria, and proteins important for proinflammatory cytokine signaling, were induced to higher levels in ΔFTN_0757-infected macrophages than in macrophages infected with wild-type bacteria (Figure S1A). This analysis also identified 53 NF-κB-regulated genes as being induced in macrophages infected with the ΔFTN_0757 mutant, compared to only 25 genes in wild-type-infected macrophages (Figures S1B, C).
Figure 1. The ΔFTN_0757 mutant induces robust TLR2-dependent macrophage activation.
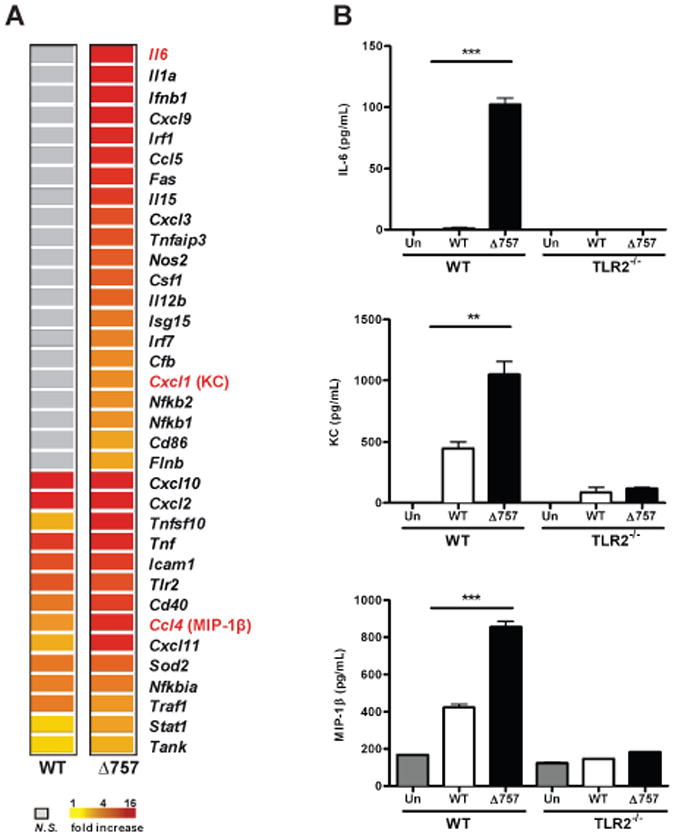
Wild-type and TLR2-/- bone marrow-derived macrophages (BMDM) were either left untreated (Un) or infected with wild-type (WT) or the ΔFTN_0757 mutant (Δ757) of F. novicida for 4 hours at an MOI of 100:1. (A) A heat map of immune genes differentially expressed in infected macrophages compared to uninfected macrophages. (B) The total amount of IL-6, KC and MIP-1β secreted into the culture supernatant by macrophages infected with the indicated strains 4 hours after infection with an MOI of 100:1 was measured by ELISA. Data are representative of three independent experiments. Error bars represent the standard deviation of triplicate samples. ** p = 0.0069; *** p < 0.0005.
To validate the microarray results, we first measured IL-6 production since it was the most differentially expressed gene in macrophages infected with the ΔFTN_0757 mutant compared to those infected with wild-type bacteria (Figure 1A). In agreement with our microarray data, macrophages infected with the ΔFTN_0757 mutant secreted significantly higher levels of IL-6 than wild-type-infected macrophages (Figure 1B). This response was dependent on TLR2 since TLR2-/- cells did not produce detectable levels of IL-6 (Figure 1B). In addition to increased cytokine production, macrophages infected with the ΔFTN_0757 mutant secreted significantly higher amounts of the chemokines KC and MIP-1β compared to those infected with wild-type bacteria, further validating our microarray results (Figure 1B). This response was also TLR2-dependent. To ensure that differences in cytokine and chemokine production were not due to differences in the ability of these strains to replicate within macrophages, we assessed the bacterial burden and found that both wild-type and ΔFTN_0757 bacteria replicated with the same kinetics in wild-type and TLR2-/- macrophages (Figures S2A, B). Taken together, these findings validate our microarray results by demonstrating that TLR2 is required for the hyperinflammatory response elicited in macrophages infected with the ΔFTN_0757 mutant.
FTN_0757 Represses Production of Bacterial Lipoproteins
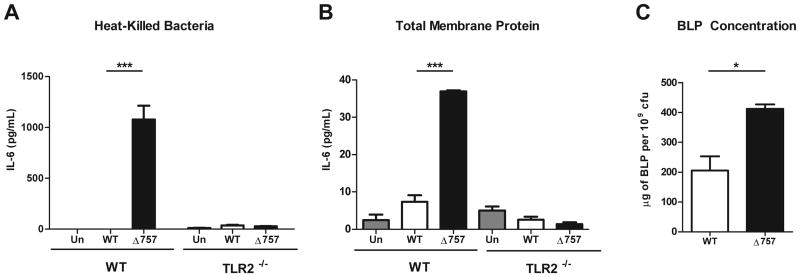
Since we have shown that infection with ΔFTN_0757 elicits a broad and robust increase in TLR2-dependent signaling compared to wild-type bacteria, we sought to explore the mechanism responsible for this phenotype. Bacteria can block TLR signaling through active processes such as secretion of effectors, or dampen host signaling by modulating PAMPs in numerous ways that lessen the immunostimulatory capacity of the bacteria (Nadler et al., 2010, Radhakrishnan et al., 2009, Salcedo et al., 2008, Tanamoto et al., 2000, Kawahara et al., 2002, Andersen-Nissen et al., 2005). To test whether the FTN_0757-dependent subversion of TLR2 signaling was an active process requiring live bacteria, we treated macrophages with heat-killed preparations of wild-type and ΔFTN_0757 mutant bacteria. We used IL-6 as a marker for the proinflammatory response due to its robust induction during macrophage infection with ΔFTN_0757 (Figures 1A, B). We found that heat-killed ΔFTN_0757 induced a significant increase in IL-6 production compared to killed wild-type bacteria, and that this response was completely dependent on TLR2 (Figure 2A). This demonstrated that the suppression of cytokine production by FTN_0757 is not dependent on an active process, but is instead due to a difference in a heat-resistant component(s) of the bacteria. Since the immunostimulatory moiety of BLPs is heat-resistant, BLPs signal through TLR2, and Francisella is known to encode BLPs that activate TLR2 (Vidal et al., 1998, Cole et al., 2006, Li et al., 2006, Abplanalp et al., 2009, Malik et al., 2006a, Thakran et al., 2008), we hypothesized that changes in BLPs were responsible for the hyperinflammatory phenotype of ΔFTN_0757.
Figure 2. The ΔFTN_0757 mutant has increased TLR2-stimulating activity and BLP levels.
Wild-type or TLR2-/- macrophages were stimulated with (A) heat-killed wild-type or ΔFTN_0757 (Δ757) at a ratio of 20:1 bacterial cell equivalents per macrophage or (B) total membrane protein fractions derived from the indicated strains at a 1:1 ratio. At 4 hours, supernatants were collected and IL-6 concentrations were quantified by ELISA. (C) BLPs were extracted from the total membrane protein fraction and their concentrations measured via the BCA assay and normalized to bacterial cfu. Bars represent the mean and standard deviation. Data are representative of at least 2 independent experiments. * p ≤ 0.05; *** p < 0.0001.
BLPs are located in bacterial membranes, so we next isolated the total membrane protein fraction from the wild-type and ΔFTN_0757 strains and tested them for their TLR2-stimulating activity. The membrane fraction from ΔFTN_0757 induced increased IL-6 production compared to the wild-type fraction (Figure 2B). This response was TLR2-dependent, similar to what we observed with the heat-killed preparation (Figure 2A) and infection with live bacteria (Figure 1B). This is consistent with the hypothesis that differences in BLPs are responsible for the hyperinflammatory phenotype of ΔFTN_0757. To further explore this possibility, we fractionated and quantified BLPs from the total membrane protein fraction of each strain. Strikingly, we found that the ΔFTN_0757 strain contained roughly twice as much total BLP as wild-type bacteria (Figure 2C). This data provides a potential explanation for the increased induction of proinflammatory cytokines elicited by ΔFTN_0757, since higher levels of BLPs would likely lead to increased TLR2 activation. To rule out the possibility that BLPs from the ΔFTN_0757 strain had increased activity on a molar basis compared to BLPs from the wild-type strain, we treated macrophages with equal concentrations of the BLP fraction from each strain. Both BLP fractions induced an equivalent amount of IL-6 production in a TLR2-dependent manner (Figure S3), demonstrating that the BLPs from each strain had the same intrinsic TLR2-stimulating activity. Taken together, these data suggest that the hyperinflammatory phenotype of ΔFTN_0757 is due to its increased BLP content leading to more robust TLR2 activation, rather than differences in the ability of its BLPs to act as TLR2 ligands.
FTN_0757 Represses the Expression of the Bacterial Lipoprotein FTN_1103
The significant increase in BLP concentration in the ΔFTN_0757 strain could be due to an increase in the levels of a small number of specific BLPs, or a more global increase in overall BLP production. In order to differentiate between these possibilities, we analyzed the respective protein composition of the BLP fraction from each strain via SDS-PAGE. While most protein bands were present at similar levels, a specific band of approximately 30 kD was highly enriched in the BLP fraction of the ΔFTN_0757 strain compared to wild-type (Figure 3A). Utilizing an LC-MS/MS peptide mass fingerprinting approach, we identified the band to be FTN_1103. Although annotated in the NCBI database as a hypothetical protein, FTN_1103 contains the typical amino acid motifs associated with BLPs, including a positively charged N-terminal region, a hydrophobic H-region, and a conserved lipobox motif (Leu-Gly-Ser) adjacent to the invariant cysteine at residue 29, which would serve as a lipidation site (Figure 3B). Also, consistent with its presence in the BLP fraction, FTN_1103 is predicted to be a BLP by the PRED-LIPO lipoprotein prediction server with a reliability score of 0.996 (Bagos et al., 2008). To further show that the protein overproduced in ΔFTN_0757 is indeed FTN_1103, we generated a ΔFTN_0757/ΔFTN_1103 double deletion strain and analyzed its BLP content. The enriched protein that we previously identified as FTN_1103 was absent in the BLP fraction of the ΔFTN_0757/ΔFTN_1103 mutant, confirming its identity as FTN_1103 (Figure 3A). Together, these data demonstrate that FTN_1103 is a BLP, which we showed is highly overproduced in ΔFTN_0757.
Figure 3. The ΔFTN_0757 mutant produces increased amounts of the BLP FTN_1103.
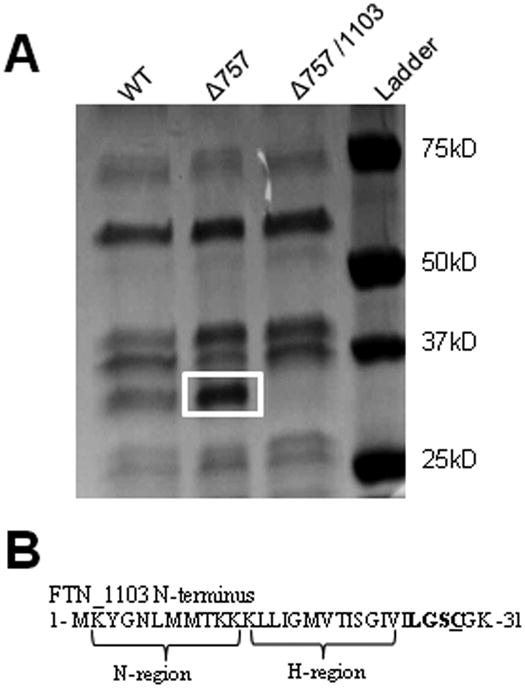
(A) 108 cfu equivalents of the BLP fraction from wild-type or the ΔFTN_0757 mutant (Δ757) were separated by SDS-PAGE and stained with Coomassie G-250. The most enriched band in the ΔFTN_0757 lane (white box) compared to the wild-type lane was subjected to LC-MS/MS analysis and identified as FTN_1103. Deletion of FTN_1103 from the ΔFTN_0757 mutant resulted in loss of the enriched band. (B) The N-terminus of FTN_1103 contains canonical BLP motifs including a positively charged N-region, hydrophobic H-region, a lipobox motif (bold), and conserved cysteine (underline).
We next measured the proportion of the increased BLP pool in ΔFTN_0757 that was due to the increase in production of FTN_1103. Deletion of FTN_1103 in the ΔFTN_0757 strain led to a large reduction in the total BLP concentration, almost to the level present in the wild-type strain (Figure 4A). Altogether these data demonstrate that FTN_0757 is required to repress expression of the BLP FTN_1103, and suggest that overproduction of FTN_1103 may cause the increased TLR2-stimulating activity of the ΔFTN_0757 strain.
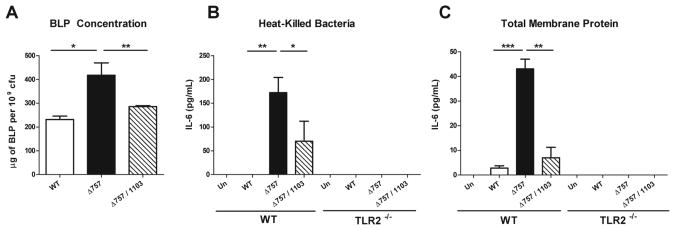
Figure 4. FTN_1103 is responsible for increased TLR2 signaling in the ΔFTN_0757 mutant.
(A) BLPs were extracted from the total membrane protein fraction of wild-type, the ΔFTN_0757 mutant (Δ757), or the ΔFTN_0757/ΔFTN_1103 (Δ757/1103) strains, and their concentrations measured via the BCA assay and normalized to bacterial cfu. Wild-type or TLR2-/- macrophages were stimulated with (B) heat-killed bacteria at a ratio of 20:1 bacterial cell equivalents per macrophage, or (C) total membrane protein fractions at a 1:1 ratio for 4 hours. Supernatants were collected and IL-6 concentrations were quantified by ELISA. Bars represent the mean and standard deviation. Data are representative of at least 2 independent experiments. * p ≤ 0.05; ** p ≤ 0.005; *** p < 0.0001.
To learn more about how FTN_0757 regulates FTN_1103, we tested whether this occurs at the transcriptional level by quantifying the expression level of FTN_1103 mRNA isolated from broth-grown wild-type or ΔFTN_0757 strains (Figure S4A). We observed a large increase in FTN_1103 expression in the ΔFTN_0757 strain compared to wild-type, confirming the observation that lack of FTN_0757 results in an increase in FTN_1103 expression. Furthermore, to investigate whether overexpression occurred in the context of a macrophage infection, we analyzed RNA from wild-type- or ΔFTN_0757-infected macrophages and found that the ΔFTN_0757 strain overexpressed FTN_1103 compared to wild-type (Figure S4B). Additionally, in order to further prove that FTN_0757 is a regulator of FTN_1103, we generated a strain in which the groE promoter drives increased expression of FTN_0757, compared to its natural promoter (Figure S4C) (Maier et al., 2004). During growth in broth, this strain overexpressing FTN_0757 exhibited a significant decrease in FTN_1103 expression compared to wild-type (Figure S4D). This is notable, since this result implies that FTN_1103 expression is controlled by the action of FTN_0757, rather than changes in FTN_1103 expression being an indirect effect of the absence of FTN_0757. Furthermore, the effect of FTN_0757 on FTN_1103 expression is specific since we did not observe a significant difference in gene expression of other predicted BLPs (such as dsbA/ FTN_0771)(Thakran et al., 2008) or genes within the Francisella Pathogenicity Island (FPI) between the wild-type and ΔFTN_0757 strains (data not shown). Altogether, these data suggest that FTN_0757 acts as a negative regulator of FTN_1103, and that FTN_1103 is the primary contributor to the increased BLP content in the ΔFTN_0757 strain.
FTN_0757 Represses FTN_1103 to Evade TLR2 Activation in Macrophages
To determine whether overproduction of FTN_1103 in ΔFTN_0757 was the major basis for the increased TLR2 activation induced by this strain, we treated macrophages with either heat-killed preparations or total membrane protein fractions derived from wild-type, ΔFTN_0757, or ΔFTN_0757/ΔFTN_1103. Similar to our previous observation, macrophages stimulated with preparations from ΔFTN_0757 elicited a significantly increased TLR2-dependent IL-6 response as compared to those treated with preparations from the wild-type strain (Figures 4B, C). However, macrophages treated with equivalent fractions from the ΔFTN_0757/ΔFTN_1103 mutant secreted significantly lower levels of IL-6 compared to those treated with ΔFTN_0757 preparations (Figures 4B, C). Deletion of FTN_1103 did not reduce the TLR2-stimulatory activity of ΔFTN_0757 completely to wild-type levels. This is likely due to smaller increases in the production of other BLPs and correlates with the incomplete reduction of BLP levels in the ΔFTN_0757/ΔFTN_1103 mutant (Figure 4A). These data demonstrate that the major cause of the increased TLR2-stimulating capacity of killed and membrane preparations of ΔFTN_0757 is the overproduction of FTN_1103.
We next wanted to determine whether FTN_1103 overexpression contributed to the increase in TLR2-dependent cytokine production elicited by ΔFTN_0757 during infection of macrophages. First, however, we determined if FTN_0757 and FTN_1103 expression was altered during the course of macrophage infection. We found that FTN_0757 expression was induced early during infection (1 hr) (Figure 5A), when Francisella is located within host phagosomes and co-localizes with TLR2 (Cole et al., 2007). This correlated with a decrease in expression of FTN_1103 (Figure 5B). As the infection progressed past 2 hours, when Francisella has escaped the phagosome and resides in the cytosol (Santic et al., 2005), FTN_0757 expression decreased, correlating with an increase in FTN_1103 expression. These data suggest that differential regulation of both FTN_0757 and FTN_1103 occurs during infection of host cells, and therefore, may contribute to subdued recognition by TLR2 by preventing FTN_1103 overexpression during early phases of macrophage infection.
Figure 5. FTN_0757 and FTN_1103 expression are modulated during macrophage infection and correlate with evasion of TLR2 signaling.
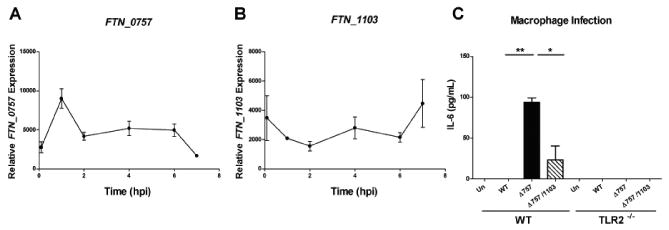
RNA was collected from wild-type F. novicida prior to infection of macrophages, or at 1, 2, 4, 6 and 7 hours after infection with an MOI of 20:1. Quantitative real-time PCR was performed for (A) FTN_0757 or (B) FTN_1103 and ΔΔCT values were normalized to those of the helicase, uvrD (FTN_1594). Points represent the mean and bars the standard deviation. (C) Wild-type or TLR2-/- macrophages were uninfected (Un) or infected with wild-type (WT), ΔFTN_0757 (Δ757), or ΔFTN_0757/ΔFTN_1103 (Δ757/1103) strains at an MOI of 20:1. Supernatants were collected at 4 hpi and IL-6 concentrations were quantified by ELISA. Bars represent the mean and standard deviation. Data are representative of at least 3 independent experiments. * p ≤ 0.05; ** p ≤ 0.005.
We next sought to determine the contribution of FTN_1103 to the increase in TLR2-dependent cytokines observed during ΔFTN_0757 infection. We infected macrophages with the ΔFTN_0757/ΔFTN_1103 mutant and notably, observed the amount of IL-6 induced by this strain was much less than that induced by macrophages infected with ΔFTN_0757 (Figure 5C). This is consistent with our results for stimulation with heat-killed preparations and membrane protein fractions (Figures 4B, C). As a further control, we genetically restored FTN_1103 into the ΔFTN_0757/ΔFTN_1103 strain. Following infection of macrophages with this strain, we observed a restoration of the hyperinflammatory defect of the ΔFTN_0757 strain (Figure S5). Collectively, these data show that the TLR2-dependent cytokine response induced by ΔFTN_0757 is due to the overproduction of FTN_1103, and suggest that FTN_0757 alters FTN_1103 expression during the course of infection to dampen recognition by TLR2.
FTN_0757 Repression of BLP Expression is Critical for F. novicida Virulence in vivo
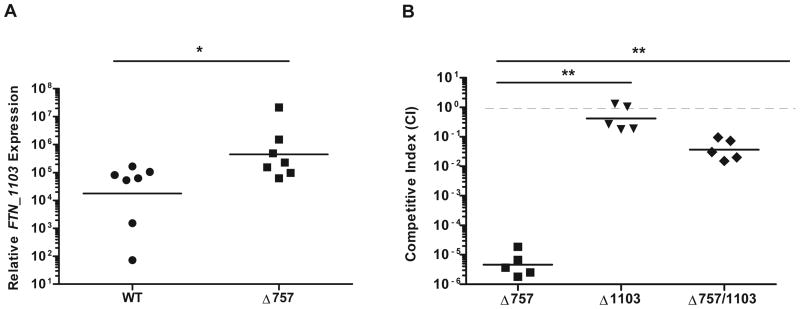
The ΔFTN_0757 mutant is severely attenuated in mice compared to wild-type bacteria (Weiss et al., 2007). Since deletion of FTN_1103 in the ΔFTN_0757 mutant rescued the majority of its hyperinflammatory phenotype during macrophage infection, we tested whether it would also rescue its virulence attenuation in vivo. First, as a control, we tested whether FTN_1103 expression was upregulated in the ΔFTN_0757 mutant during in vivo infection. We measured the level of FTN_1103 expression in the wild-type and ΔFTN_0757 strains at 6 hours after subcutaneous infection of mice, an early timepoint when the loads of each strain were similar (data not shown). We found that the FTN_1103 transcript was indeed present at higher levels in the ΔFTN_0757 mutant compared to wild-type bacteria (Figure 6A). This demonstrates that the regulation of FTN_1103 by FTN_0757 occurs during in vivo infection, similar to our findings with bacteria grown in rich media and during macrophage infection (Figures S4A, B).
Figure 6. Deletion of FTN_1103 significantly rescues the in vivo virulence defect of the ΔFTN_0757 mutant.
(A) Wild-type C57BL/6 mice were infected subcutaneously with 2×106 cfu of wild-type F. novicida or the ΔFTN_0757 mutant. At 6 hpi, the skin was harvested and RNA was extracted. Quantitative real-time PCR was used to determine relative expression of FTN_1103, which was normalized to the constitutively expressed uvrD. (B) Mice were infected subcutaneously with a 1:1 mixture of wild-type F. novicida and either the ΔFTN_0757 (Δ757), ΔFTN_0757/ΔFTN_1103 (Δ757/1103), or ΔFTN_1103 mutant (Δ1103). At 48 hpi, the spleen was harvested and the colony forming units (cfu) for each strain were enumerated after overnight growth. The competitive index (CI) = (mutant cfuoutput / wild-type cfuoutput) / (mutant cfuinput / wild-type cfuinput). Bars represent the geometric mean of CI values from each group of mice. Data shown are representative of three independent experiments. * p ≤ 0.05; ** p ≤ 0.005.
To determine whether overexpression of FTN_1103 contributed to in vivo attenuation of the ΔFTN_0757 mutant, we performed competition experiments. We infected mice subcutaneously with a 1:1 mixture of wild-type and either the ΔFTN_0757 or ΔFTN_0757/ΔFTN_1103 mutant and enumerated bacteria in the spleen at 48 hours post-infection (hpi). The ΔFTN_0757 mutant was >100,000-fold attenuated compared to wild-type bacteria (Figure 6B), in agreement with our previous work (Weiss et al., 2007). In contrast, the ΔFTN_0757/ΔFTN_1103 mutant was present at much higher levels than the ΔFTN_0757 mutant, and was only ∼100-fold attenuated compared to wild-type. This represents 1,000-fold complementation of the ΔFTN_0757 mutant as a result of deleting FTN_1103. The lack of complete complementation correlates with the BLP content, macrophage stimulation and infection experiments using the ΔFTN_0757/ΔFTN_1103 strain, and might be attributed to an increase in the expression of other BLPs in the ΔFTN_0757 mutant (Figures 4, 5). As a control, we tested the phenotype of the ΔFTN_1103 mutant and found that it was present at levels close to those of the wild-type strain, indicating that FTN_1103 alone does not play an important role in virulence under these infection conditions (Figure 6B). Taken together, these data indicate that increased FTN_1103 production in the ΔFTN_0757 mutant significantly contributes to its attenuation in vivo.
Discussion
During in vivo infection, macrophages are among the first cells that Francisella encounters after entering the host (Hall et al., 2008), and TLR2 expressed on the surface of these immune cells recognizes Francisella BLPs and induces a proinflammatory cytokine response aimed at clearing this pathogen. Our findings reveal a novel mechanism of TLR2 evasion: repression of BLP expression. We found that F. novicida FTN_0757 strongly represses the expression of FTN_1103, a BLP whose function is unknown but is not essential for bacterial replication in vitro or virulence in vivo. Furthermore, FTN_1103 is temporally repressed during the first hours of macrophage infection (Figures 5A, B), when the bacteria localize to the phagosome and could be recognized by TLR2 (Cole et al., 2007). Subsequently, FTN_1103 expression is upregulated once the bacteria escape the phagosome and enter the cytosol (Santic et al., 2008), which is devoid of TLR2. Therefore, we propose a model whereby F. novicida uses FTN_0757 to temporally limit BLP expression in order to evade TLR2 signaling in the phagosome.
Since TLR2 is essential for controlling Francisella infection in vivo (Abplanalp et al., 2009, Malik et al., 2006b), evading TLR2 activation may provide this bacterium with precious time to replicate inside the host without triggering an inflammatory response, also enabling it to prevent robust activation of the immune system. Accordingly, the ΔFTN_0757 mutant, which is unable to downregulate BLP (FTN_1103) expression and evade TLR2, was severely attenuated in vivo. This mutant elicited robust production of a broad array of TLR2- and NF-κB-dependent inflammatory chemokines and cytokines during infection of macrophages (Figures 1, S1). Macrophages infected with the ΔFTN_0757 mutant produced significantly higher levels of IL-12, a cytokine whose production in vivo can induce T cells and NK cells to produce IFN-γ (De Pascalis et al., 2008), a potent inducer of macrophage antibacterial defenses including nitric oxide production. Activation of these macrophage defenses by IFN-γ confers these cells with the ability to not only inhibit Francisella replication, but to kill the bacteria (Parsa et al., 2008). In addition, infection with the ΔFTN_0757 mutant induces the robust production of chemokines such as KC (CXCL1) and CXCL2, which promote neutrophil recruitment (Soehnlein et al., 2010). Neutrophils are non-permissive for Francisella replication and essential for host defense against infection; therefore, an influx of these cells in vivo could greatly reduce the bacterial burden (Sjostedt et al., 1994). Together, these findings highlight a few of the ways in which TLR2 activation could lead to the inhibition of Francisella replication in vivo. These data also highlight the importance of evading TLR2 for F. novicida pathogenesis, and the way that bacteria can use the previously unrecognized strategy of downregulating BLP to accomplish this task.
In addition to contributing to the evasion of innate defenses, suppression of BLP expression could also play an important role in evading adaptive immune responses. For example, TLR2 signaling induces the expression of co-stimulatory proteins, such as CD86 (Figure 1A), that promote adaptive responses. Thus, lower BLP levels could result in diminished initiation of adaptive responses. BLPs can also be recognized as antigens by the adaptive immune system, and downregulation of their expression could also serve as a mechanism to lower levels of immunogenic antigens. In fact, the spirochete Borrelia burgdorferi downregulates the expression of one of its most highly immunogenic BLP, OspC, at the onset of the humoral immune response in order to avoid detection by antibodies and subsequent killing (Liang et al., 2002). B. burgdorferi strains that are unable to downregulate OspC expression in vivo are rapidly cleared from mice in an antibody-dependent manner (Embers et al., 2008). By extension, the ΔFTN_0757 mutant may induce a more potent antibody-mediated immune response than wild-type bacteria, and therefore downregulating FTN_1103 may also lead to evasion of antibody responses. In addition, our data suggest that another consequence of OspC downregulation may be the evasion of TLR2 activation. As TLR2 signaling can contribute to antibody responses (Dickinson et al., 2010), downregulation of BLP may therefore represent a two-pronged approach to dampen the TLR2 activation signal as well as limit the expression of major antigens against which the antibody response is directed.
Since downregulation of BLPs can lead to evasion of TLR2, this raises the question of whether the levels of other TLR activators are modulated by pathogens? In fact, our findings are somewhat reminiscent of the repression of flagellin expression by the Salmonella virulence factor TviA when this pathogen invades the intestinal mucosa and can be recognized by TLR5, but not when it is in the lumen which lacks TLR5 (Winter et al., 2010). Furthermore, since adaptive responses are generated against flagellin, its downregulation may also be a means of evading adaptive immune responses. It will be interesting to determine in the future whether the levels of additional TLR ligands such as LPS or lipoteichoic acid (LTA) are modulated by pathogens as a mechanism of host evasion.
Together, our findings reveal a novel mechanism utilized by F. novicida to evade TLR2 activation. To our knowledge, this is the first demonstration of an intracellular bacterial pathogen that downregulates BLP expression to evade innate immune recognition, and this may represent a new paradigm that is used by other intracellular pathogens to evade TLR2.
Experimental procedures
Bacterial strains and growth conditions
Francisella novicida strain U112 was kindly provided by Dr. Denise Monack (Stanford University, Stanford, CA). All bacterial cultures were grown overnight at 37°C with aeration in tryptic soy broth (TSB) supplemented with 0.2% L-cysteine (BD Biosciences, Sparks, MD). When necessary, the media was supplemented with kanamycin (30 μg/ml) or tetracycline (20 μg/ml).
Bacterial mutagenesis
Mutant strains (ΔFTN_0757 and ΔFTN_1103) were constructed by allelic replacement as described previously (Brotcke et al., 2006, Anthony et al., 1991b) using primers in Table 1. To excise the Flippase Recognition Target (FRT)-flanked kanamycin resistance cassette and create unmarked strains, the kanamycin-resistant mutants were transformed with plasmid pLG42 encoding the Flp-recombinase, performed as previously described (Gallagher et al., 2008). The ΔFTN_0757/ΔFTN_1103 deletion strain was generated by transforming an unmarked ΔFTN_0757 strain with genomic DNA from the marked ΔFTN_1103 strain, and selecting for kanamycin resistance. FTN_1103 was complemented in cis into the ΔFTN_0757/ΔFTN_1103 deletion strain through allelic replacement, as described previously (Weiss et al., 2007, Anthony et al., 1991b).
Table 1. Primers used in this study.
| Primer Name | Sequence |
|---|---|
| FTN_0757 Deletion | |
| 757-F1 | tgaatcaaattcaagcacac |
| 757-arm1-rev | tatcgataccgtcgacctcaaaatga |
| 757-FRT-fwd | ccgagtaaaagaggtcattttgaggt |
| 757-FRT-rev | gctaatttcataccaagcatgcatag |
| 757-arm2-fwd | tatcgatcctgcagctatgcatgctt |
| 757-R1 | aactaacgacatctttccag |
| FTN_1103 Deletion | |
| 1103-F1 | ttagctgtaatggaggtatt |
| 1103-arm1-rev | ttatcgataccgtcgacctcatttttc |
| 1103-FRT-fwd | gttgtaggggattattgaaaaatgag |
| 1103-FRT-rev | cttaagcaaaaaatctctgattagcat |
| 1103-arm2-fwd | tatcgatcctgcagctatgctaatca |
| 1103-R1 | cgatagtgatagtagagctg |
| FTN_1103 Complementation | |
| 1103comp-F1 | tagtaatgtgatcggagtgt |
| 1103comp-arm1-rev | atcgataccgtcgacctcgttaagtt |
| 1103comp-FRT-fwd | tgggctgttggccaacttaacgaggt |
| Gro-FTN_0757 Construction | |
| gro757-F1 | atatattctagaatcagctccacttaaagaag |
| gro757-KAN-fwd | gtatgtgaagctagttcctagcggccgcatctctttgggttgtcact |
| gro757-KAN-rev | agtgacaacccaaagagatgcggccgctaggaactagcttcacatac |
| gro757-gro-fwd | cagaattggttaattggttgtgcggccgcttgtatggattagtcgagct |
| gro757-gro-rev | agctcgactaatccatacaagcggccgcacaaccaattaaccaattctg |
| gro757-Nterm-fwd | agtattagttcgtcgtgcaatgaatttcaaaatattgccaa |
| gro757-Nterm-rev | ttggcaatattttgaaattcattgcacgacgaactaatact |
| gro757-R1 | atatatGTCGACtgatgatagtagcttagctc |
| qRT-PCR | |
| uvrD-RT-fwd | gggatgtcgccttttgattttc |
| uvrD-RT-rev | ctcttttgtcccttgtgcttgc |
| 1103-RT-fwd | atggtgggcagtctagcgca |
| 1103-RT-rev | acccaactcaccatcgccaca |
Preparation of bacterial fractions
Overnight cultures of bacteria were subcultured 1:50 into 50ml of TSB with 0.2% cysteine and grown to an OD600 of 0.9 – 1.0. Cultures were centrifuged at 5,000 × g for 10 minutes to pellet the bacterial cells. For cell-free supernatants, the remaining supernatant was passed through a 0.22μm filter (Millipore, Billerica, MA), and stored at -20°C until use. For heat-killed bacteria, the bacterial pellet was resuspended in PBS (Lonza, Walkersville, MD), boiled at 100°C for 1 hour, and then stored at -20°C until use. For isolation of membrane fractions, resuspended cells were lysed via freeze-thawing for three cycles. The cell lysate was then centrifuged at 10,000 × g for 10 minutes to remove unlysed cells. The cleared supernatant was then centrifuged at 120,000 × g for 2 hours to pellet the total membrane fraction. Membrane pellets were resuspended in 1ml PBS and stored at -20°C until use. For enrichment of bacterial lipoproteins, pelleted membrane fractions were resuspended in 200μl PBS and 500μl n-butanol (Sigma-Aldrich, St. Louis, MO) and centrifuged at 27,000 × g for 90 minutes. The aqueous phase, containing an enrichment of bacterial lipoproteins, was collected and stored at -20°C until use (Jones et al., 1995). Protein fractions were normalized either by colony forming units (cfu) or by protein concentration, measured via the bicinchoninic acid assay (Thermo Scientific, Waltham, MA), as indicated. Twenty micrograms or 108 cfu-equivalents of each fraction were separated via 12-20% SDS-PAGE (Bio-Rad, Hercules, CA) and stained with Coomassie Blue G-250 (Teknova, Hollister, CA).
Microarray analysis
All RNA samples were checked for purity using a ND-1000 spectrophotometer (NanoDrop Technologies) and for integrity by electrophoresis on a 2100 BioAnalyzer (Agilent Technologies). The samples were amplified using the Nugen WT Pico Kit (NuGEN Technology) and the target reactions were run with 25 ng of total RNA. The amplification products were processed through the EXON Module (NuGEN Technology), which creates sense-strand cDNA targets. The sense strand cDNA Targets were then fragmented and labeled using NuGEN's FL-Ovation™ cDNA Biotin Module V2 (NuGEN Technology). Labeled targets were hybridized to GeneChip® Mouse Gene 1.0ST arrays (Affymetrix, Inc.), following Standard Nugen Protocols for target hybridization to the Affymetrix Gene Arrays. The hybridizations were run for 16 hours, 45°C, 60 rpm in an Affymetrix Hybridization Oven 640. The Cartridge arrays were washed and stained using the Affyemtrix Fluidics Stations 450, following Affymetrix protocols. Scanning was performed on an Affymetrix GeneChip 3000 7G scanner, and Affymetrix GCOS software was used to perform image analysis and generate raw intensity data. Probe sets of all samples were normalized by RMA, which includes global background adjustment and quantile normalization. Using the gene annotation provided by Affymetrix, we discarded 11,537 probe sets that did not match to known genes. Student's t-test (p < 0.02) and a fold-change filter (mean fold-change > 25%) were used to identify genes differentially expressed in macrophages infected with ΔFTN0757 strain compared to those infected with wild-type strain for 4 hours. The expression levels of NF-κB-regulated genes were visualized using Ingenuity Pathway Analysis (Ingenuity Systems) software.
Protein identification by mass spectrometry
After staining with Coomassie Blue G-250, the band of interest was excised and subjected to in-gel digestion (12.5 μg/ml trypsin). Extracted peptides were loaded onto a C18 column (75μm inner diameter, 15cm long, ∼300nl/min flow rate, 1.9μm resin) (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany) and eluted using a 10-30% gradient (Buffer A: 0.1% formic acid, 1% ACN; Buffer B: 0.1% formic acid, 99.9% ACN). The eluted peptides were detected by Orbitrap (300-1600 m/z; 1,000,000 automatic gain control target; 500-ms maximum ion time; resolution, 30,000 full-width at half-maximum) followed by ten data-dependent MS/MS scans in the linear ion trap quadrupole (2 m/z isolation width, 35% collision energy, 5,000 automatic gain control target, 200-ms maximum ion time) on a hybrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA). The acquired tandem mass spectrometer (MS/MS) spectra were searched against and a decoy-concatenated F. novicida database (3,393 redundant protein targets) from the NCBI RefSeq protein database project (September 2011) using the Sorcerer-SEQUEST Algorithm version 3.11 r11 (Sage-N Research, San Jose, CA). Search results were filtered with a 1% FDR and summarized by in-house programs, as described by Gozal et al (Gozal et al., 2011).
Macrophage experiments and infections
Murine bone marrow-derived macrophages (BMDM) were prepared from 6-8 week old wild-type and TLR2-/- C57BL/6 mice and cultured as described (Weiss et al., 2007). Macrophages were cultured in 96-well plates (5-8×104 cells/well) in high glucose Dulbecco's modified Eagle's medium (DMEM) (Lonza, Walkersville, MD) supplemented with 10% heat-inactivated fetal bovine serum (HyClone, Logan, UT) and 10% L929-conditioned media (conditioned DMEM) containing M-CSF (macrophage colony stimulating factor) overnight. The media was removed and bacteria were added at a multiplicity of infection (MOI) of 20 or 100 bacteria per macrophage. Plates were centrifuged for 15 minutes at 335 × g at room temperature to promote uptake of bacteria. Macrophages were incubated for 30 minutes at 37°C and washed two times before adding warm conditioned DMEM. The concentrations of IL-6, KC, and MIP-1β in culture supernatants at the indicated timepoints after infection were quantified by ELISA (BD Biosciences, Sparks, MD). For treatment with bacterial components, cells were washed gently and media containing heat-killed bacteria, membrane fractions, or bacterial lipoprotein fractions at the given concentrations was added. Macrophages were stimulated for the indicated duration of time, before the cell-culture supernatant was collected.
RNA extraction and quantitative real-time PCR
Overnight cultures of the indicated bacteria were subcultured 1:50 into 50mL TSB with 0.2% L-cysteine and grown to an OD600 of 0.9 to 1.0. RNA was isolated using TRI Reagent (Molecular Research Center, Cincinnati, OH) and purified using the RNeasy Mini Kit (Qiagen, Germantown, MD) according to the manufacturers' instructions. Quantitative real-time RT-PCR (qRT-PCR) was performed using the Power Sybr Green RNA to CT 1-Step Kit (Applied Biosystems, Carlsbad, CA) and gene-specific primers (Table 1) using an Applied Biosystems StepOne Cycler. Relative transcript levels were calculated by normalizing CT values to DNA helicase II (uvrD, FTN_1594).
Mouse infections
For competition experiments, groups of five wild-type C57BL/6 mice were infected subcutaneously with a 1:1 ratio of wild-type and the indicated mutant strain of F. novicida (total of 1 × 105 cfu) in sterile PBS. At 48 hpi, the skin, spleen and liver of infected mice were harvested and homogenized in PBS. Serial dilutions were plated on Mueller-Hinton agar supplemented with 0.1% L-cysteine with or without kanamycin for enumeration of bacterial burden in each organ. The competitive index (CI) was calculated using the formula, CI = (mutant cfu in output/wild-type cfu in output)/(mutant cfu in input/wild-type cfu in input). All experimental procedures were approved by the Emory University Institutional Animal Care and Use Committee (protocol #069-2008Y).
Statistics
All experiments were analyzed using the unpaired Student's t test except Figure 6A, which was analyzed by a Mann-Whitney test. The CI values from competition experiments were analyzed using the one-sample Student's t test and CI values of the ΔFTN_0757 and ΔFTN_0757/ ΔFTN 1103 strains were both significantly different from 1 (***, p < 0.0001).
Supplementary Material
(A) Ingenuity Pathway Analysis was used to identify innate immune pathways whose genes were differentially expressed in macrophages infected with wild-type F. novicida (WT) or the ΔFTN_0757 mutant (Δ757) at 4 hpi. The red dashed line represents a p-value (right-tailed Fisher Exact Test) cutoff of 0.001. (B, C) A network map of NF-κB-related genes differentially expressed in macrophages infected with (B) WT or (C) Δ757 at 4 hpi. Solid and dashed lines represent direct and indirect interactions reported for the genes, respectively. The colors represent the mean fold-change in gene expression at 4 hpi compared to control in two biological replicates.
Wild-type or TLR2-/- macrophages were infected with wild-type (WT) or the ΔFTN_0757 mutant (Δ757) bacteria at an MOI of (A) 20:1 or (B) 100:1. At 4 hours post infection, macrophage lysates were plated, and colony forming units (cfu) were counted. Data are representative of three independent experiments. Error bars represent the standard deviation of triplicate samples.
Wild-type or TLR2-/- macrophages were unstimulated (Un) or stimulated with 1μg of total BLP preparations from wild-type (WT) or ΔFTN_0757 (Δ757) strains. At 4 hours post-stimulation, supernatants were collected and IL-6 concentrations were quantified by ELISA. Bars represent the mean and standard deviation.
RNA was harvested from wild-type or ΔFTN_0757 (Δ757) grown in (A) broth or (B) macrophages at an MOI of 20:1. Quantitative real-time PCR was performed for FTN_1103 and ΔΔCT values were normalized to those of the helicase, uvrD (FTN_1594). (C, D) RNA was harvested from wild-type or a strain overexpressing FTN_0757 (gro-Δ757) grown in broth culture, and qRT-PCR was performed for (C) FTN_0757 and (D) FTN_1103. Bars represent the mean and standard deviation. * p ≤ 0.05; ** p ≤ 0.005; *** p < 0.0001.
Macrophages were untreated (Un) or infected with wild-type (WT) F. novicida, ΔFTN_0757 (Δ757), ΔFTN_0757/ΔFTN_1103 (Δ757/1103), or the ΔFTN_0757/ΔFTN_1103 mutant complemented with FTN_1103 (Δ757/1103 + 1103), at an MOI of 20:1. At 4 hpi, the concentration of IL-6 secreted into the culture supernatant was measured by ELISA. Data are representative of three independent experiments. Error bars represent the standard deviation of triplicate samples. * p < 0.05.
Acknowledgments
We thank Brooke Napier and Dr. Thomas Henry for critical reading of this manuscript. The project described was supported by NIH grant #U54 AI057157 from the Southeastern Regional Center of Excellence for Emerging Infections and Biodefense. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. This research was also supported by the National Science Foundation Graduate Research Fellowship (C.L.J. and T.R.S.), as well as the ARCS Foundation (T.R.S.). We are grateful to the Vanderbilt Genome Sciences Resource which is supported by the Vanderbilt Ingram Cancer Center (P30 CA68485), the Vanderbilt Digestive Disease Center (P30 DK58404), the Vanderbilt Vision Center (P30 EY08126), and NIH/NCRR (G20 RR030956), for microarray processing. The Resource collaborates with the Computational Genomics Core of the Center Human Genetic Research and the Center for Quantitative Sciences. We thank the Emory Proteomics Core, which is supported in part by the Proteomics Core of the Emory Neuroscience NINDS Core Facilities grant, P30NS055077. The authors declare no conflicts of interest.
References
- Abplanalp AL, Morris IR, Parida BK, Teale JM, Berton MT. TLR-dependent control of Francisella tularensis infection and host inflammatory responses. PLoS One. 2009;4:e7920. doi: 10.1371/journal.pone.0007920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Aliprantis AO, Yang RB, Mark MR, Suggett S, Devaux B, Radolf JD, et al. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science. 1999;285:736–739. doi: 10.1126/science.285.5428.736. [DOI] [PubMed] [Google Scholar]
- Andersen-Nissen E, Smith KD, Strobe KL, Barrett SL, Cookson BT, Logan SM, Aderem A. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc Natl Acad Sci U S A. 2005;102:9247–9252. doi: 10.1073/pnas.0502040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony LD, Burke RD, Nano FE. Growth of Francisella spp. in rodent macrophages. Infect Immun. 1991a;59:3291–3296. doi: 10.1128/iai.59.9.3291-3296.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony LS, Gu MZ, Cowley SC, Leung WW, Nano FE. Transformation and allelic replacement in Francisella spp. J Gen Microbiol. 1991b;137:2697–2703. doi: 10.1099/00221287-137-12-2697. [DOI] [PubMed] [Google Scholar]
- Bagos PG, Tsirigos KD, Liakopoulos TD, Hamodrakas SJ. Prediction of lipoprotein signal peptides in Gram-positive bacteria with a Hidden Markov Model. J Proteome Res. 2008;7:5082–5093. doi: 10.1021/pr800162c. [DOI] [PubMed] [Google Scholar]
- Brightbill HD, Libraty DH, Krutzik SR, Yang RB, Belisle JT, Bleharski JR, et al. Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science. 1999;285:732–736. doi: 10.1126/science.285.5428.732. [DOI] [PubMed] [Google Scholar]
- Brotcke A, Weiss DS, Kim CC, Chain P, Malfatti S, Garcia E, Monack DM. Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis. Infection and immunity. 2006;74:6642–6655. doi: 10.1128/IAI.01250-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole LE, Elkins KL, Michalek SM, Qureshi N, Eaton LJ, Rallabhandi P, et al. Immunologic consequences of Francisella tularensis live vaccine strain infection: role of the innate immune response in infection and immunity. J Immunol. 2006;176:6888–6899. doi: 10.4049/jimmunol.176.11.6888. [DOI] [PubMed] [Google Scholar]
- Cole LE, Shirey KA, Barry E, Santiago A, Rallabhandi P, Elkins KL, et al. Toll-like receptor 2-mediated signaling requirements for Francisella tularensis live vaccine strain infection of murine macrophages. Infection and immunity. 2007;75:4127–4137. doi: 10.1128/IAI.01868-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pascalis R, Taylor BC, Elkins KL. Diverse myeloid and lymphoid cell subpopulations produce gamma interferon during early innate immune responses to Francisella tularensis live vaccine strain. Infect Immun. 2008;76:4311–4321. doi: 10.1128/IAI.00514-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson GS, Piccone H, Sun G, Lien E, Gatto L, Alugupalli KR. Toll-like receptor 2 deficiency results in impaired antibody responses and septic shock during Borrelia hermsii infection. Infect Immun. 2010;78:4579–4588. doi: 10.1128/IAI.00438-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embers ME, Alvarez X, Ooms T, Philipp MT. The failure of immune response evasion by linear plasmid 28-1-deficient Borrelia burgdorferi is attributable to persistent expression of an outer surface protein. Infect Immun. 2008;76:3984–3991. doi: 10.1128/IAI.00387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follett EA, Gordon J. An Electron Microscope Study of Vibrio Flagella. J Gen Microbiol. 1963;32:235–239. doi: 10.1099/00221287-32-2-235. [DOI] [PubMed] [Google Scholar]
- Gallagher LA, McKevitt M, Ramage ER, Manoil C. Genetic dissection of the Francisella novicida restriction barrier. J Bacteriol. 2008;190:7830–7837. doi: 10.1128/JB.01188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozal YM, Seyfried NT, Gearing M, Glass JD, Heilman CJ, Wuu J, et al. Aberrant septin 11 is associated with sporadic frontotemporal lobar degeneration. Mol Neurodegener. 2011;6:82. doi: 10.1186/1750-1326-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JD, Woolard MD, Gunn BM, Craven RR, Taft-Benz S, Frelinger JA, Kawula TH. Infected-host-cell repertoire and cellular response in the lung following inhalation of Francisella tularensis Schu S4, LVS, or U112. Infection and immunity. 2008;76:5843–5852. doi: 10.1128/IAI.01176-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- Jones JD, Bourell KW, Norgard MV, Radolf JD. Membrane topology of Borrelia burgdorferi and Treponema pallidum lipoproteins. Infect Immun. 1995;63:2424–2434. doi: 10.1128/iai.63.7.2424-2434.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadzhaev K, Zingmark C, Golovliov I, Bolanowski M, Shen H, Conlan W, Sjostedt A. Identification of genes contributing to the virulence of Francisella tularensis SCHU S4 in a mouse intradermal infection model. PLoS One. 2009;4:e5463. doi: 10.1371/journal.pone.0005463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanistanon D, Hajjar AM, Pelletier MR, Gallagher LA, Kalhorn T, Shaffer SA, et al. A Francisella mutant in lipid A carbohydrate modification elicits protective immunity. PLoS Pathog. 2008;4:e24. doi: 10.1371/journal.ppat.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara K, Tsukano H, Watanabe H, Lindner B, Matsuura M. Modification of the structure and activity of lipid A in Yersinia pestis lipopolysaccharide by growth temperature. Infect Immun. 2002;70:4092–4098. doi: 10.1128/IAI.70.8.4092-4098.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer PS, Mitchell A, Pelletier MR, Gallagher LA, Wasnick M, Rohmer L, et al. Genome-wide screen in Francisella novicida for genes required for pulmonary and systemic infection in mice. Infect Immun. 2009;77:232–244. doi: 10.1128/IAI.00978-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochem Biophys Res Commun. 2009;388:621–625. doi: 10.1016/j.bbrc.2009.08.062. [DOI] [PubMed] [Google Scholar]
- Li H, Nookala S, Bina XR, Bina JE, Re F. Innate immune response to Francisella tularensis is mediated by TLR2 and caspase-1 activation. J Leukoc Biol. 2006;80:766–773. doi: 10.1189/jlb.0406294. [DOI] [PubMed] [Google Scholar]
- Liang FT, Jacobs MB, Bowers LC, Philipp MT. An immune evasion mechanism for spirochetal persistence in Lyme borreliosis. J Exp Med. 2002;195:415–422. doi: 10.1084/jem.20011870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien E, Means TK, Heine H, Yoshimura A, Kusumoto S, Fukase K, et al. Toll-like receptor 4 imparts ligand-specific recognition of bacterial lipopolysaccharide. J Clin Invest. 2000;105:497–504. doi: 10.1172/JCI8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier TM, Havig A, Casey M, Nano FE, Frank DW, Zahrt TC. Construction and characterization of a highly efficient Francisella shuttle plasmid. Appl Environ Microbiol. 2004;70:7511–7519. doi: 10.1128/AEM.70.12.7511-7519.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik M, Bakshi CS, Sahay B, Shah A, Lotz SA, Sellati TJ. Toll-like receptor 2 is required for control of pulmonary infection with Francisella tularensis. Infection and immunity. 2006a;74:3657–3662. doi: 10.1128/IAI.02030-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik M, Bakshi CS, Sahay B, Shah A, Lotz SA, Sellati TJ. Toll-like receptor 2 is required for control of pulmonary infection with Francisella tularensis. Infect Immun. 2006b;74:3657–3662. doi: 10.1128/IAI.02030-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzio M, Bosisio D, Polentarutti N, D'Amico G, Stoppacciaro A, Mancinelli R, et al. Differential expression and regulation of toll-like receptors (TLR) in human leukocytes: selective expression of TLR3 in dendritic cells. J Immunol. 2000;164:5998–6004. doi: 10.4049/jimmunol.164.11.5998. [DOI] [PubMed] [Google Scholar]
- Nadler C, Baruch K, Kobi S, Mills E, Haviv G, Farago M, et al. The type III secretion effector NleE inhibits NF-kappaB activation. PLoS Pathog. 2010;6:e1000743. doi: 10.1371/journal.ppat.1000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsa KV, Butchar JP, Rajaram MV, Cremer TJ, Gunn JS, Schlesinger LS, Tridandapani S. Francisella gains a survival advantage within mononuclear phagocytes by suppressing the host IFNgamma response. Molecular immunology. 2008;45:3428–3437. doi: 10.1016/j.molimm.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K, Broz P, Jones J, Joubert LM, Monack D. Elevated AIM2-mediated pyroptosis triggered by hypercytotoxic Francisella mutant strains is attributed to increased intracellular bacteriolysis. Cell Microbiol. 2011;13:1586–1600. doi: 10.1111/j.1462-5822.2011.01643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan GK, Yu Q, Harms JS, Splitter GA. Brucella TIR Domain-containing Protein Mimics Properties of the Toll-like Receptor Adaptor Protein TIRAP. J Biol Chem. 2009;284:9892–9898. doi: 10.1074/jbc.M805458200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer L, Fong C, Abmayr S, Wasnick M, Larson Freeman TJ, Radey M, et al. Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 2007;8:R102. doi: 10.1186/gb-2007-8-6-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo SP, Marchesini MI, Lelouard H, Fugier E, Jolly G, Balor S, et al. Brucella control of dendritic cell maturation is dependent on the TIR-containing protein Btp1. PLoS Pathog. 2008;4:e21. doi: 10.1371/journal.ppat.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santic M, Asare R, Skrobonja I, Jones S, Abu Kwaik Y. Acquisition of the vacuolar ATPase proton pump and phagosome acidification are essential for escape of Francisella tularensis into the macrophage cytosol. Infect Immun. 2008;76:2671–2677. doi: 10.1128/IAI.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santic M, Molmeret M, Abu Kwaik Y. Modulation of biogenesis of the Francisella tularensis subsp. novicida-containing phagosome in quiescent human macrophages and its maturation into a phagolysosome upon activation by IFN-gamma. Cell Microbiol. 2005;7:957–967. doi: 10.1111/j.1462-5822.2005.00529.x. [DOI] [PubMed] [Google Scholar]
- Sjostedt A, Conlan JW, North RJ. Neutrophils are critical for host defense against primary infection with the facultative intracellular bacterium Francisella tularensis in mice and participate in defense against reinfection. Infect Immun. 1994;62:2779–2783. doi: 10.1128/iai.62.7.2779-2783.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soehnlein O, Lindbom L. Phagocyte partnership during the onset and resolution of inflammation. Nature reviews Immunology. 2010;10:427–439. doi: 10.1038/nri2779. [DOI] [PubMed] [Google Scholar]
- Su J, Yang J, Zhao D, Kawula TH, Banas JA, Zhang JR. Genome-wide identification of Francisella tularensis virulence determinants. Infect Immun. 2007;75:3089–3101. doi: 10.1128/IAI.01865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanamoto K, Azumi S. Salmonella-type heptaacylated lipid A is inactive and acts as an antagonist of lipopolysaccharide action on human line cells. J Immunol. 2000;164:3149–3156. doi: 10.4049/jimmunol.164.6.3149. [DOI] [PubMed] [Google Scholar]
- Telepnev M, Golovliov I, Grundstrom T, Tarnvik A, Sjostedt A. Francisella tularensis inhibits Toll-like receptor-mediated activation of intracellular signalling and secretion of TNF-alpha and IL-1 from murine macrophages. Cell Microbiol. 2003;5:41–51. doi: 10.1046/j.1462-5822.2003.00251.x. [DOI] [PubMed] [Google Scholar]
- Thakran S, Li H, Lavine CL, Miller MA, Bina JE, Bina XR, Re F. Identification of Francisella tularensis lipoproteins that stimulate the toll-like receptor (TLR) 2/TLR1 heterodimer. J Biol Chem. 2008;283:3751–3760. doi: 10.1074/jbc.M706854200. [DOI] [PubMed] [Google Scholar]
- Vidal V, Scragg IG, Cutler SJ, Rockett KA, Fekade D, Warrell DA, et al. Variable major lipoprotein is a principal TNF-inducing factor of louse-borne relapsing fever. Nat Med. 1998;4:1416–1420. doi: 10.1038/4007. [DOI] [PubMed] [Google Scholar]
- Wang X, Ribeiro AA, Guan Z, Abraham SN, Raetz CR. Attenuated virulence of a Francisella mutant lacking the lipid A 4′-phosphatase. Proc Natl Acad Sci U S A. 2007;104:4136–4141. doi: 10.1073/pnas.0611606104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss DS, Brotcke A, Henry T, Margolis JJ, Chan K, Monack DM. In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci U S A. 2007;104:6037–6042. doi: 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Godinez I, Yang HJ, Russmann H, Andrews-Polymenis HL, Baumler AJ. A rapid change in virulence gene expression during the transition from the intestinal lumen into tissue promotes systemic dissemination of Salmonella. PLoS Pathog. 2010;6:e1001060. doi: 10.1371/journal.ppat.1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SS, Mekalanos JJ. Decreased potency of the Vibrio cholerae sheathed flagellum to trigger host innate immunity. Infect Immun. 2008;76:1282–1288. doi: 10.1128/IAI.00736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Ingenuity Pathway Analysis was used to identify innate immune pathways whose genes were differentially expressed in macrophages infected with wild-type F. novicida (WT) or the ΔFTN_0757 mutant (Δ757) at 4 hpi. The red dashed line represents a p-value (right-tailed Fisher Exact Test) cutoff of 0.001. (B, C) A network map of NF-κB-related genes differentially expressed in macrophages infected with (B) WT or (C) Δ757 at 4 hpi. Solid and dashed lines represent direct and indirect interactions reported for the genes, respectively. The colors represent the mean fold-change in gene expression at 4 hpi compared to control in two biological replicates.
Wild-type or TLR2-/- macrophages were infected with wild-type (WT) or the ΔFTN_0757 mutant (Δ757) bacteria at an MOI of (A) 20:1 or (B) 100:1. At 4 hours post infection, macrophage lysates were plated, and colony forming units (cfu) were counted. Data are representative of three independent experiments. Error bars represent the standard deviation of triplicate samples.
Wild-type or TLR2-/- macrophages were unstimulated (Un) or stimulated with 1μg of total BLP preparations from wild-type (WT) or ΔFTN_0757 (Δ757) strains. At 4 hours post-stimulation, supernatants were collected and IL-6 concentrations were quantified by ELISA. Bars represent the mean and standard deviation.
RNA was harvested from wild-type or ΔFTN_0757 (Δ757) grown in (A) broth or (B) macrophages at an MOI of 20:1. Quantitative real-time PCR was performed for FTN_1103 and ΔΔCT values were normalized to those of the helicase, uvrD (FTN_1594). (C, D) RNA was harvested from wild-type or a strain overexpressing FTN_0757 (gro-Δ757) grown in broth culture, and qRT-PCR was performed for (C) FTN_0757 and (D) FTN_1103. Bars represent the mean and standard deviation. * p ≤ 0.05; ** p ≤ 0.005; *** p < 0.0001.
Macrophages were untreated (Un) or infected with wild-type (WT) F. novicida, ΔFTN_0757 (Δ757), ΔFTN_0757/ΔFTN_1103 (Δ757/1103), or the ΔFTN_0757/ΔFTN_1103 mutant complemented with FTN_1103 (Δ757/1103 + 1103), at an MOI of 20:1. At 4 hpi, the concentration of IL-6 secreted into the culture supernatant was measured by ELISA. Data are representative of three independent experiments. Error bars represent the standard deviation of triplicate samples. * p < 0.05.