Abstract
In Drosophila embryonic development, the Bicoid (Bcd) protein establishes positional information of downstream developmental genes like hunchback (hb), which has a strong anterior expression and a sharp on-off boundary in the mid-embryo. The role of Bcd cooperative binding in the positioning of the Hb pattern has been previously demonstrated. However, there are discrepancies in the reported results about the role of this mechanism in the sharp Hb border. Here, we determined the Hill coefficient (nH) required for Bcd to generate the sharp border of Hb in wild-type (WT) embryos. We found that an nH of approximately 6.3 (s.d. 1.4) and 10.8 (s.d. 4.0) is required to account for Hb sharpness at early and late cycle 14A, respectively. Additional mechanisms are possibly required because the high nH is likely unachievable for Bcd binding to the hb promoter. To test this idea, we determined the nH required to pattern the Hb profile of 15 embryos expressing an hb14F allele that is defective in self-activation and found nH to be 3.0 (s.d. 1.0). This result indicates that in WT embryos, the hb self-activation is important for Hb sharpness. Corroborating our results, we also found a progressive increase in the required value of nH spanning from 4.0 to 9.2 by determining this coefficient from averaged profiles of eight temporal classes at cycle 14A (T1 to T8). Our results indicate that there is a transition in the mechanisms responsible for the sharp Hb border during cycle 14A: in early stages of this cycle, Bcd cooperative binding is primarily responsible for Hb sharpness; in late cycle 14A, hb self-activation becomes the dominant mechanism.
Keywords: Drosophila melanogaster, Bicoid, hunchback, cooperative binding, Hill coefficient, self-regulation
INTRODUCTION
It has been shown that cooperative binding plays a central role in pattern formation and in the interpretation of morphogenetic positional information during embryonic development. One classical example is the Bicoid (Bcd) protein that establishes the anterior–posterior (AP) developmental axis during embryonic development in Drosophila melanogaster. This morphogenetic protein determines positional information for downstream developmental genes such as hunchback (hb), Krüppel (Kr) and knirps (kni), which also show cross-regulation (Berleth et al., 1988; Crauk and Dostatni, 2005; de Lachapelle and Bergmann, 2010; Driever and Nusslein-Volhard, 1988; He et al., 2010a; Jäckle et al., 1986; Jaeger et al., 2004; Manu et al., 2009; Nusslein-Volhard et al., 1987; Papatsenko and Levine, 2011). It has been shown that cooperative binding is critical for Bcd transcriptional activity (Burz and Hanes, 2001; Burz et al., 1998; Driever et al., 1989; Lebrecht et al., 2005; Lopes et al., 2005; Lopes et al., 2008; Ma et al., 1996) and plays a central role in reducing transcriptional noise during Drosophila development (Holloway et al., 2011).
The hb gene encodes a morphogenetic protein (Lehmann and Nusslein-Volhard, 1987; Papatsenko and Levine, 2008; Tautz et al., 1987) and exhibits anterior and posterior expression patterns (Fig. 1). The hb regulatory region has two distinct promoters, P1 (distal) and P2 (proximal); each controls the expression of specific transcripts that encode the same protein (Margolis et al., 1995; Spirov et al., 2002; Spirov et al., 2000; Tautz et al., 1987)1. In the anterior region of the embryo, the regulation of hb by Bcd and Hb proteins (Driever and Nusslein-Volhard, 1989; Lukowitz et al., 1994; Margolis et al., 1995; Simpson-Brose et al., 1994; Struhl et al., 1989; Treisman and Desplan, 1989) produces a uniform expression pattern with a sharp on–off boundary at mid-embryo (Fig. 1). It was shown that an ~300 bp region upstream of the hb coding region, the core part of the proximal promoter, that contains 6 main Bcd sites is sufficient to confer full regulation of hb by Bcd (Driever et al., 1989; Schroder et al., 1988; Struhl et al., 1989). Cooperative binding of Bcd to these six sites has also been previously demonstrated (Burz et al., 1998; Lopes et al., 2005; Ma et al., 1996). Moreover, to show that Bcd cooperative binding could determine the positioning of the hb pattern, Driever et al. used a series of lacZ artificial constructs that were driven by fragments of the native hb promoter and contained different numbers of Bcd binding sites (Driever et al., 1989). It was then suggested that this mechanism could account for the sharpness of the hb pattern because the sharpness of the lacZ patterns increased with the number and strength of Bcd sites. However, none of the lacZ constructs analyzed achieved wild-type (WT) Hb sharpness. This result was found even when an artificial construct was driven by 6 strong and 6 weak Bcd sites, which showed the strongest level of expression. Since these experiments, many efforts have been dedicated to characterizing the role of Bcd cooperative binding in hb pattern formation (Burz et al., 1998; Crauk and Dostatni, 2005; Gregor et al., 2007a; Lebrecht et al., 2005; Lopes et al., 2005; Lopes et al., 2008; Ma et al., 1996).
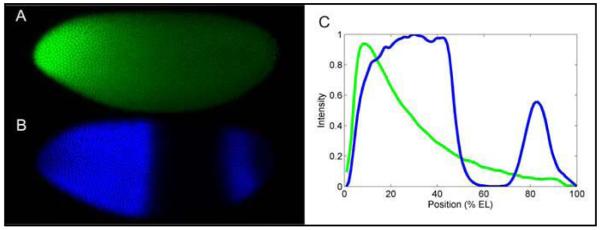
Fig. 1. Hb and Bcd protein profiles (Lopes et al., 2008).
An embryo at mid-nuclear cleavage cycle 14A immunostained for Bcd (A) and Hb (B). (C) Fluorescence intensities for A and B are shown in green and blue, respectively, as a function of position along the AP axis. The diffusion of Bcd protein, translated from its mRNA localized at the anterior end of the egg, forms an exponential concentration gradient. In A and B, the anterior pole is on left and the dorsal side is on top.
He et al. contributed to the above discussion with immunofluorescence to determine Bcd and Hb protein profiles from 28 WT embryos at early cycle 14A and found that a Hill coefficient (nH) of approximately 5.1 ± 2.7 in the dorsal and 4.9 ± 2.7 in the ventral side was sufficient to account for Hb sharpness (He et al., 2010b). Using the same technique with 9 early embryos at cycle 14A and visual inspection, Gregor et al. found an nH of approximately 5.0 (Gregor et al., 2007a). This result indicates that Bcd cooperative binding is sufficient to account for hb regulation. However, the coefficients required are higher than those that are observed experimentally. Using DNAse footprint assays in vitro, an estimated nH of 3.6 was found for Bcd binding to a 250 bp fragment of the native hb promoter containing the six Bcd strong sites (Ma et al., 1996). Using gel-shift assays, the nH was estimated as 3.0 (standard deviation, s.d. 0.031) for the binding of a homeodomain-containing fragment of Bcd (called Bcd89–154) to a 230 bp hb element also containing the six Bcd strong sites (Burz et al., 1998). Using immunofluorescence and fluorescent in situ hybridization (FISH), early embryos at cycle 14A (He et al., 2011) were used to show that the nH required for Bcd to pattern the hb transcriptional profile (6.1±2.6, calculated from 14 embryos) was higher than the coefficient required to pattern the Hb protein profile (5.2±0.4, calculated from 5 embryos). In addition, Houchmandzadeh et al. have suggested that Bcd alone cannot account for Hb sharpness because an nH of more than 10 is required based on their estimations (Houchmandzadeh et al., 2002). Finally, the idea that Hb sharpness is caused by hb self-activation through bistable kinetics was proposed based on work using a systems biology approach that combined immunofluorescence and a reaction-network model (Lopes et al., 2008).
During cycle 14A, the hb expression pattern exhibits a significant variation of approximately 30% (Lopes et al., 2008; Surkova et al., 2008) and reaches a maximum level of expression around mid-cycle. Bcd protein concentration, in turn, decreases continuously from its maximum level at the beginning of the cycle (Gregor et al., 2007b; Surkova et al., 2008). These patterns of temporal variation indicate that the contribution of both Bcd and Hb proteins must be temporally modulated and that a precise characterization of Bcd cooperative binding and hb self-activation in Hb sharpness must account for the variations in different stages of cycle 14A.
Here, we analyzed Bcd cooperativity levels using 30 WT Bcd and Hb profiles. We found that an nH of 6.3 (s.d. 1.4) is required to account for Hb sharpness at early cycle 14A. However, at late stages of this cycle, an nH of approximately 10.8 (s.d. 4.0) is required. Bcd binding is not likely to reach this level of cooperativity. Thus, we investigated additional regulation that could be taking place by determining the nH required to pattern the Hb profile of 15 embryos expressing an hb14F allele that is defective in self-regulation. We found an nH of 3.0 (s.d. 1.0), which is in agreement with previous in vitro results (Burz et al., 1998; Ma et al., 1996). This result indicates that hb self-activation contributes to Hb sharpness in WT embryos, which was suggested in earlier studies (Lopes et al., 2008; Simpson-Brose et al., 1994). We verified our results using an independent set of data (Poustelnikova et al., 2004; Surkova et al., 2008) to follow the progressive increase in the nH required to account for Hb sharpness from early to late cycle 14. Taken together, our results indicate that there is a transition in the mechanisms responsible for hb sharpness during cycle 14A: in early stages of this cycle, Bcd cooperative binding is mainly responsible for hb sharpness; in late cycle 14A, hb self-activation becomes the dominant mechanism.
MATERIALS AND METHODS
Dataset
WT Oregon-R and hb mutant hb14F (Hülskamp et al., 1994) embryos were stained for Bcd and Hb proteins. For the pThb5 construct (driven by a fragment of the hb promoter (Driever et al., 1989)), lacZ expression was visualized by FISH. These data were previously used in different analysis (Holloway et al., 2011; Lopes et al., 2008).
Embryo fixation and staining
Embryos were incubated with guinea pig and rat primary antibodies against Hb and Bcd, respectively, followed by secondary antibodies labeled with Alexa Fluor 647 (to Hb) and 488 (to Bcd; Molecular Probes). For lacZ embryos, simultaneous immunostaining was used to detect Hb and Bcd, and FISH was used to detect lacZ transcripts using the Janssens et al. method (Janssens et al., 2005). Immunostaining was performed as above to detect Hb (Alexa Fluor 647) and Bcd (Alexa Fluor 555). After hybridization, lacZ mRNA was visualized by sequential incubation with a rabbit antibody to fluorescein (Molecular Probes) and then an antibody to rabbit labeled with Alexa Fluor 488 (Molecular Probes).
Confocal Microscopy
Whole-embryo images were taken using a laser confocal scanning microscope (Leica TCS SP2) and the Janssens et al. method (Janssens et al., 2005). Fluorophores were excited by a laser at different wavelengths (488, 555, and 647 nm) and detected via a filterless spectral separation system. To reduce image noise from the photomultiplier tubes, each embryo was scanned sequentially 16 times, and the results were averaged.
Image Processing
Protein and RNA profiles were determined by averaging intensities in each dorsoventral pixel column from the central 10% strip of the AP axis. The quality of this direct method was checked by nuclear masks that were manually made based on several costained lacZ embryos using the multiple ROI feature in ImageJ software (Moodley and Murrell, 2004). Each circular ROI with radii comparable to the nuclear radii was manually positioned to outline a given nucleus. Nuclear-resolution AP profiles from this method were of comparable quality to pixel-resolution profiles from our direct extraction method.
Temporal Classification
In addition to confocal scanning, all embryos were observed along the dorsal edge using Differential Interference Contrast (DIC) optics. Distances were measured from the egg surface to the invaginating membrane and from the surface to the cortex. The ratio of the membrane depth to the cortex depth was used to estimate the embryo age in minutes using a published standard curve (Merrill et al., 1988).
The straight line fitting of the Hill equation
The original form of the Hill equation can be written as (Hill, 1910):
| (1) |
In this equation, used to generate the plots of Fig. 3, Fr is the fraction of site occupancy, i.e. the number of bound sites by the total number of sites, nH is the Hill coefficient, [L] is the (total) ligand concentration and K the equilibrium constant. This constant is related to [L]1/2, the ligand concentration that produces half of promoter occupancy, by .
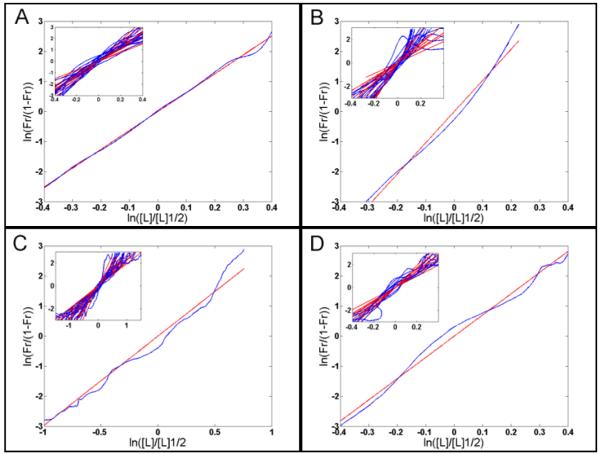
Fig. 3. Estimating the required Hill coefficient to account for Hb sharpness using an alternative plot.
(A) The same data from the 15 WT early embryos used for Fig. 2A but plotted similarly to Fig. 4A by Gregor et al. (Gregor et al., 2007a). (B) The same 15 WT late embryos as in Fig. 2B but plotted as in A. In these graphics, the red, green, cyan and black lines show the plots for an nH of 5, 6, 7 and 10, respectively. It is easy to see that in (A), the nH choice of 6 fits the data better, while in (B), the best choice is 12. This method (see equation (1) in Material and Methods) does not allow a precise determination of nH because visual inspection is used. The straight-line fitting, used for Figs. 2 and 4, is more precise and accurate.
For practical applications, it is useful to write the above equation as
| (2) |
This equation can be easily used in a linear fitting to determine nH, which is the inclination of the straight line. This procedure was used to generate the plots of Figs. 2 and 4.
Fig. 2. Estimating the required Hill coefficient to pattern Hb profiles in embryos of different stages or backgrounds (Lopes et al., 2008).
The Hill coefficient is given by the inclination of the straight line (see equation (2) in Material and Methods). (A): Plot from a WT embryo at early cycle 14A exhibiting an nH of 6.3 that is similar to the mean nH (6.3, s.d. 1.4) measured from 15 embryos at this stage (inset). (B): The same plot as in (A) but from a WT embryo at late cycle 14A with an nH of approximately 11.6 that is similar to the mean nH (10.8, s.d. 4.0) measured from 15 embryos at this stage (inset). (C): Same plot as in (A) but from an hb14F embryo at cycle 14A with an nH of approximately 3.0. Inset: 15 embryos at this stage with the mean nH (3.0, s.d. 1.0). (D): The same plot as above from an embryo expressing the pThb5 lacZ artificial construct at cycle 14A and exhibiting an nH of 7.0. Inset: 9 embryos with mean nH (7.0, s.d. 1.5). We assumed early and late cycle 14A embryos corresponded to stages T1-T4 and T5-T6, respectively, according to previous classification (Surkova et al., 2008).
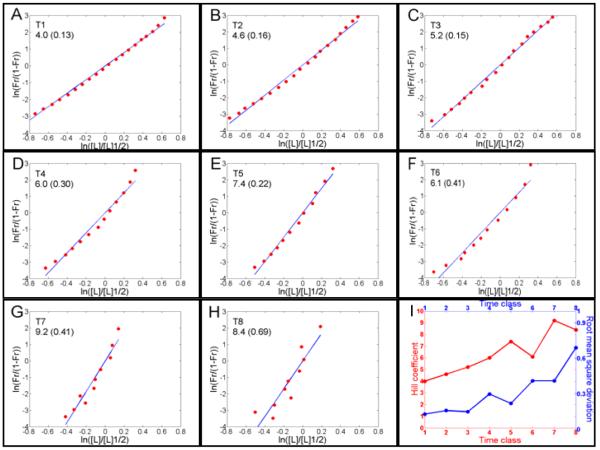
Fig. 4. Required Hill coefficients for each temporal class in cycle 14A necessary to account for Hb sharpness as determined from averaged data.
Poustelnikova et al., 2004; Surkova et al., 2008)(A) – (H): Plots for time classes T1-T8, which last for approximately 6 minutes each. The Hill coefficients, shown in the insets followed by the r.m.s.d., were determined by the straight-line inclination of these plots. (I): Evolution of the Hill coefficient (red line) from A to H and r.m.s.d. (blue line) as a function of time classes. The increase in r.m.s.d. indicates a reduction in how suitable the Hill approach is for describing this system. The increase in nH indicates that Bcd cooperative binding cannot account for the sharpness of the Hb pattern after T4. To generate these plots, we used the equation (2) in Material and Methods.
A recent result has shown that the protein signal obtained from immunostaining experiments is proportional to the real protein concentration (Gregor et al., 2007a). Using this result, we determined [L]/[L]1/2 as the quotient [Bcdsignal]/[Bcdsignal]1/2. In this relation, [Bcdsignal] is the signal intensity of the Bcd protein at each position in the AP axis of the Drosophila embryo and [Bcdsignal]1/2 is the [Bcdsignal] observed at the position where half of the Hb protein signal is detected. To determine Fr we assume that the level of protein signal is proportional to the fraction of site occupancy (Gregor et al., 2007a; He et al., 2010b). This assumption allowed us to determine Fr as the quotient [Hbsignal]/[Hbsignal]max. In this relation, [Hbsignal] is the signal intensity of the Hb protein at each position in the AP axis and [Hbsignal]max is the maximum level of Hb protein signal. To fit the equations (1) and (2), a region of interest (ROI) in the posterior border of the anterior Hb pattern was defined. We used a ROI in the range of 5% to 95% of the maximum Hb profile. This ROI was based on Gregor et al results about the 10% accuracy for the detection of protein concentrations (Gregor et al., 2007a). In addition, this ROI accounts for the estimated error level introduced by confocal images (Myasnikova et al., 2009).
Fitting procedures
Fitting was performed using the Matlab platform with the respective equations for root mean square deviation (r.m.s.d.) and standard deviation (s.d.):
| (3) |
and
| (4) |
where “exp” is the experimental data and “the” is the corresponding theoretical values for each position i along the AP axis in the embryo. m and μ are the number of experimental points and its mean, respectively.
RESULTS
Bcd cooperative binding cannot account for late Hb sharpness
Here we are not estimating nH or analyzing the bcd/hb relationship. In our analysis, we are estimating the Hill coefficient required for generating the sharpness of the hb expression pattern in WT and hb14 mutant embryos. In order to analyze the ability of the Bcd gradient to generate each hb pattern, we compared this estimated nH with the experimentally determined values.
To determine what value of nH is necessary for Bcd cooperative binding to produce the sharp Hb border, we used 15 embryos at early cycle 14A and found an nH of 6.3 (s.d., 1.4, Fig. 2A; Table 1). This is in agreement with similar results for early embryos found by He et al. (He et al., 2011; He et al., 2010b) and Gregor et al. (Gregor et al., 2007a). To test what nH is necessary to account for mature Hb sharpness, we used 15 embryos at late cycle 14A and found an nH of 10.8 (s.d. 4.0, Fig. 2B; Table 1). This coefficient value is approximately three times that of the coefficient estimated for human hemoglobin (nH = 2.8 (Stryer, 1995)) and is not likely to ever be reached by Bcd binding to the six sites shown to be sufficient to reproduce hb regulation by Bcd (Driever et al., 1989; Schroder et al., 1988; Struhl et al., 1989).
Table 1.
Estimating the required Hill coefficient to account for Hb sharpness in embryos at different stages of cycle 14 or with different backgrounds.
|
|
Background | MeannH (s.d./mean r.m.s.d.) |
Number of embryos |
Age (minutes) |
| hb14F | 3.0 (1.0/0.25) | 15 | 32-39 | |
| early WT | 6.3 (1.4/0.20) |
15 | 4-23 | |
| late WT | 10.8 (4.0/0.35) | 15 | 26-53 | |
| lacZ | 7.0 ( 1.5/0.36) |
9 | 20-37 | |
Gregor et al. based their results on early embryos at cycle 14 to reduce the influence of other transcriptional regulators on Hb profiles as much as possible (Gregor et al., 2007a). The authors found that nH = 5.0 and suggested that Bcd cooperative binding accounts for hb regulation in early embryos. To verify the effect of embryo age using the Gregor et al. approach, we plotted our Bcd/Hb data from early and late embryos in the same semi-log plot that they used. We found that the ratios between the axis for early (Fig. 3A, very similar to Fig. 4A in Gregor et al. (Gregor et al., 2007a)) and late embryos (Fig. 3B) are different and result in different coefficient estimations: 6 for early and 10 for late embryos. Although similar values were found using both of the above approaches, the latter is less appropriate because a visual inspection was used and the differences between the plots for early and late embryos were not as evident as for the straight lines (Fig. 2A,B).
These results indicate that Bcd plays a critical role in sharpening early Hb profiles but cannot fully account for the sharpness of the mature Hb pattern; thus, other regulators must be performing a role in sharpening the Hb pattern. Below, we estimated nH values for Bcd binding to the endogenous hb regulatory region in WT embryos and tested the role of additional regulators.
hb self-activation is critical for Hb sharpness
The role of hb self-activation in the sharpness of the Hb pattern has previously been demonstrated (Lopes et al., 2008). This effect is more evident in late embryos due to Hb protein accumulation. Therefore, to perform a confident estimation for the effect of additional regulation, it is necessary to determine hb self-regulatory effects.
Performing this estimation is possible using hb14F embryos, which harbor an hb allele that is deficient for self-activation (Hülskamp et al., 1994; Lopes et al., 2008). Using 15 embryos, we found that an nH of approximately 3.0 (s.d. 1.0, Fig. 2C) is required to account for Hb pattern in this allele. This coefficient value is in good agreement with the 3.6 and 3.0 (s.d. 0.031) found by Ma et al. and Burz et al. (Burz et al., 1998), respectively. However, these values are significantly lower than our estimations for the nH required to pattern the Hb profile at early (6.3 s.d. 1.4) or late (10.8 s.d. 4.0) cycle 14A. This difference suggests that in WT embryos the nH value required to pattern the Hb profile is affected by other mechanisms, most likely hb self-activation.
To check the effect of additional regulation on Hill coefficients, we used the patterns of beta-galactosidase transcripts from 9 early embryos expressing a lacZ construct driven by a fragment of the native hb promoter (pThb5 (Driever et al., 1989)) that contained six Bcd, two Hb and two Kr sites (Treisman and Desplan, 1989) (Fig. 2D). We reasoned that visualizing lacZ transcripts instead of β-galactosidase protein would give results that are more accurate because transcripts can be degraded normally, as other endogenous transcripts are. Using this visualization approach, we found that an nH of 7.0 (s.d. 1.5) is required to pattern the profile of the lacZ transcripts. This nH value is higher than the nH estimated to pattern the Hb profile in early (6.3, s.d. 1.4) cycle 14A embryos but lower than the estimation for late (10.8, s.d. 4.0) cycle 14A embryos. Our result quantitatively shows that additional regulation of the target gene can affect the determination of the Hill coefficient.
The cooperative-binding and self-regulating stages of Hb pattern formation
To verify the above results using an independent set of experimental data, we used quantitative data from the FlyEx (Poustelnikova et al., 2004; Surkova et al., 2008) database (http://urchin.spbcas.ru/flyex or http://flyex.uchicago.edu/flyex/index.jsp), which divides cycle 14A into 8 time classes, T1 to T8. We used the averaged (integrated) data for Bcd and Hb protein profiles, which are very close to the profiles of individual embryos. This approach allowed us to estimate the required Hill coefficient necessary for Bcd to produce the Hb pattern at each temporal class (Fig. 4). We found that the required nH varies from 4.0 (r.m.s.d., 0.13; at T1) to 9.2 (r.m.s.d. 0.41; at T7; Fig. 4I) (see Materials and Methods for definition of r.m.s.d.). The required nH at T1 (4.0, r.m.s.d. 0.13) is also in agreement with our estimation using hb14F (3.0, s.d. 1.0), the estimation of Burz et al. (Burz et al., 1998) (3.0, s.d. 0.031) and the estimation of Ma et al. (Ma et al., 1996) (3.6). These data also indicate that the bcd/hb14F pair and the WT bcd/hb (at T1) are the only cases where Hb sharpness is fully produced by Bcd cooperative binding. Moreover, even though Bcd cooperative binding is performing a critical role for Hb sharpness during time classes T2 (nH 4.6, r.m.s.d. 0.16) and T3 (nH = 5.2, r.m.s.d. 0.15), it cannot account for the overall sharpness of Hb expression. Furthermore, the required nH ranges from 6.0 to 9.2 for the T5 to T8 stages (Fig. 4I) and makes it even less feasible that Bcd cooperativity alone accounts for the Hb sharpness. Even in the T4 stage, where the required nH is 6.0, Bcd could not solely account for the Hb sharpness because only three of the six main Bcd binding sites are strong (Driever and Nusslein-Volhard, 1989); thus, the maximum nH of 6 is not feasible. The effect of hb self-activation is more pronounced after Hb protein accumulation because hb self-activation plays a critical role in Hb sharpness (Lopes et al., 2008). The above results suggest that cycle 14A occurs in two distinct stages: a stage that relies predominantly on Bcd cooperative binding from T1 until T4 (the first 24 minutes of the cycle) and a stage that relies predominantly on self-regulation by Hb from T4 until T8 (the remaining 24 minutes).
The early stages of the bcd/hb system are best described by the Hill approach
To check the applicability of the Hill approach to the bcd/hb system, we determined the r.m.s.d. for the coefficient fitting of each time class in the averaged data (Fig. 4I). We observed a five-fold increase in the r.m.s.d. from 0.13 for T1 to 0.69 for T8. This increase indicates that T1 is the best time class for the application of the Hill approach. This finding is expected because the nH of 4.0 for T1 is similar to the nH found for hb14F, the self-activation mutant where the hb pattern exhibits only the effect of Bcd cooperative binding. The progressive increase in r.m.s.d. after T1 (r.m.s.d. in T4, 0.30, is more than two times higher than that of T1, 0.13) also indicates that the bcd/hb system is progressively less accurately described by this approach. This loss of appropriateness results from the progressive accumulation of Hb protein, which shows a maximum concentration at mid-cycle 14A (Lopes et al., 2008; Surkova et al., 2008). The observed increase in r.m.s.d. is in agreement with the division of cycle 14A into cooperative (T1 until T4) and self-regulating (T4 until T8) stages.
To perform a similar analysis of Hill equation fitting for the unaveraged data shown in Fig. 2, it was unsuitable to use r.m.s.d. because this measurement would be indistinguishable from the effects of intrinsic transcriptional noise (He et al., 2010a; Holloway et al., 2011) and from fitting by the Hill approach. To circumvent this limitation, we used the standard deviation (s.d.) because we used 15 embryos for each assay except for the 9 embryos used for the pThb5 lacZ construct. From the mean s.d. for hb14F (1.02), early WT (1.45), late WT (6.08) and the lacZ construct (1.52), we note that the bcd/hb14F system is the one better described by the Hill approach, as describe above. The early WT, lacZ, and late WT embryos each show a progressive increase in the mean s.d., which indicates that these systems are progressively more poorly described by the Hill approach. This result is consistent with the increased effect of hb self-activation in these systems. Taken together, these results indicate that the r.m.s.d. and s.d. of the straight-line fitting of the Hill equation can be used to verify the applicability of the Hill approach.
DISCUSSION
Driever et al. first suggested that Bcd cooperative binding could account for Hb sharpness (Driever et al., 1989). Since then, there have been studies that demonstrate that Bcd cooperative DNA binding occurs; however, discrepant results exist about whether the resulting effects are sufficient to account for the sharp borders of expression observed for Hb and other Bcd target genes. In particular, the importance of Hb self-activation has been the subject of debate. To help resolve these discrepancies, we estimated the nH required to pattern Hb profile in the hb14F self-activation mutant allele, which does not exhibit hb self-regulation. Analysis of this mutant allowed us to distinguish contributions to the overall nH by both Bcd cooperativity and by Hb self-activation. The nH of 3.0 (s.d. 1.0) that we found from hb14F embryos agrees with the nH values for Bcd binding to fragments of the hb promoter containing the six main Bcd binding sites of 3.6 and 3.0 (s.d. 0.031) from Ma et al. (Ma et al., 1996) and Burz et al. (Burz et al., 1998), respectively. Although these values seem contradictory to others in the literature (Gregor et al., 2007a; He et al., 2011; He et al., 2010b), we detail below how these values are in fact explainable.
A time resolved analysis was needed to understand the role of Bcd cooperative binding in hb patterning because Bcd and Hb patterns show significant variation during cycle 14A. By performing this analysis, we found that Bcd cooperative binding does account for Hb sharpness at very early stages of cycle 14A because the required nH for time class T1 is similar to that estimated for hb14F (Fig. 4I, Table 1). This result indicates that Hb protein (from zygotic or embryonic origin) must have little or no role in the sharpness of very early Hb patterning. For time classes T2 to T4, however, the effect of hb self-activation increases because the required nH increases progressively (Fig. 4I). This increase in nH agrees with the nH we found for cycle 14A (Table 1). Taken together with the progressive decrease in Bcd and increase in Hb concentrations (Gregor et al., 2007b; Surkova et al., 2008), these results indicate that a combination of well-established Bcd cooperative binding and nascent self-regulatory effects are important for the establishment of Hb sharpness for time classes T1 until T4. Self-regulation becomes predominant as reflected in the nH for time classes T5 to T8 and in late WT embryos (Fig. 4I, Table 1). This driven change is in agreement with the ability of Bcd cooperative binding to determine the positioning of the hb pattern (Driever et al., 1989; Lopes et al., 2008) because positioning is determined in the early stages of cycle 14 (Fig. 2C in Lopes et al. (Lopes et al., 2008) and Fig. 2G in Surkova et al. (Surkova et al., 2008)). This behavior also agrees with the previous result that the control of Hb sharpness and positioning by hb self-activation and Bcd cooperative binding, respectively, are separate processes that can be altered independently (Lopes et al., 2008). The results also agree with an nH estimation that indicates that a coefficient greater than 10 is required for Hb sharpness (Houchmandzadeh et al., 2002).
Taken together, our results indicate that the apparent discrepancy between the various nH can be accounted for by two time-dependent components. Early Bcd cooperative DNA binding sets up a later Hb self-regulation step, and together these are sufficient for establishing a sharp border of expression. Driver et al. noted that “One would not expect that such a mechanism immediately generates a sharp border … Subsequent regulatory interactions between hb and other zygotic genes might be involved in the establishment and maintenance of a border as sharp as that observed in later blastoderm stages” (Driever and Nusslein-Volhard, 1989). Lewis et al. used theoretical arguments to specifically propose that cooperative binding cannot account for sharp borders and that a self-activation mechanism is sufficient for sharp borders (Lewis et al., 1977). This last argument is also in agreement with the systems biology approach and proposal of Lopes et al. that hb self-activation via bistable kinetics is responsible for the sharp border of the Hb pattern (Lopes et al., 2008).
Crauk and Dostatni used lacZ constructs with three Bcd binding sites, and their results led them to propose that Bicoid determines sharp and precise hb expression (Crauk and Dostatni, 2005). However, three binding sites, even if they are strong, are not expected to provide full regulation of hb by Bcd as suggested by the authors. In addition, the experimental technique used, (non-fluorescent) in situ hybridization, can be affected by artificial saturation. Artificial saturation can occur depending on the reaction exposure time and can increase the measurement of sharpness (see Fig. S3 in Lopes et al. (Lopes et al., 2008)). Other than hb itself, the two best candidates for having a role in Hb sharpening are Kr and kni. These candidates are best because of the relative position of their expression domains (hb domain is adjacent to kni and overlaps the Kr domain (Surkova et al., 2008; Tomancak et al., 2007)) and regulation of hb by Kr has previously been demonstrated (Jäckle et al., 1986; Treisman and Desplan, 1989). However, no effect on Hb sharpness was reported, even though Kr;kni double mutants showed disruptions in Hb pattern precision (Manu et al., 2009). In addition, there are discrepancies in the described effects of single mutations in Kr or kni on Hb positioning. Jäckle and collaborators reported an anterior shift of the Hb pattern in Kr mutant embryos (Jäckle et al., 1986), whereas Houchmandzadeh et al. reported no significant shift of the Hb pattern in Kr or kni single mutant embryos (Houchmandzadeh et al., 2002). However, the strong decrease in sharpness measured for the hb14F allele (21.8% reduction, from 80.2° in WT to 62.8° in hb14F (Lopes et al., 2008)) indicates that Kr or kni must have no effect on Hb sharpness because both of these genes are not affected in hb14F. Finally, another explanation for the sharp border of the Hb pattern could be transcriptional synergy. It has been show that a greater than additive transcriptional response can be achieved by more than one activator acting simultaneously (Carey et al., 1990). However, experimental data that could indicate that this mechanism is responsible for the sharp Hb border are still needed. This mechanism could explain the enhanced nH of pThb5 construct, however the effect of hb activation on this construct is dictated by the sharp border of the hb expression pattern. Because of this influence, it is difficult to distinguish between the contribution of transcriptional synergy and the sharp border of Hb pattern to the enhanced nH of pThb5.
Highlights.
Bcd cooperative binding to the native hunchback promoter exhibits a Hill coefficient of approximately 3.0.
A Hill coefficient of approximately 10.0 is necessary for sharp, mature Hunchback patterning.
Bicoid cooperative binding cannot account for sharpness in the mature Hunchback pattern.
Self-activation of hunchback is necessary to produce the sharp border of the mature Hunchback pattern.
ACKNOWLEDGMENTS
We thank John Reinitz for fruitful discussions on the model and for sharing lab facilities; David Holloway for discussions and exchanging of ideas; Steve Hanes and Ricardo Fabbri for manuscript revisions and comments; Ah-Ram Kim and the Bloomington Stock Center for providing lacZ probes and hb14F flies, respectively; Carlos Alonso, Ah-Ram Kim, and Lisa Prazak for instructions on experiments; and Svetlana Surkova for helping with embryo temporal classification.
Acknowledgments
ROLE OF THE FUNDING SOURCE FJPL was supported by the Brazilian National Research Council (CNPq)/Ministry of Science and Technology (MCT, Brazil), the Research Foundation of the State of Rio de Janeiro (FAPERJ, Brazil), grant 1-R01-GM072022 of the joint National Science Foundation (NSF)/National Institute of General Medical Sciences (NIGMS) BioMath program, and grant 2-ROI-RR07801 of the National Institutes of Health. PMB was supported by the CNPq/MCT, Brazil and the Research Foundation of the State of Rio de Janeiro (FAPERJ, Brazil). AVS was supported by grant 1-R01-GM072022 of the joint NSF/NIGMS BioMath program.
The study sponsors had no role in the study design or in the collection, analysis, or interpretation of data. The study sponsors were not involved in writing the report or in the decision to submit the paper for publication.
Footnotes
ACCESSION NUMBERS Bicoid (Gene ID: 40830) Hunchback (ID: 41032)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
For details http://www.evol.nw.ru/spirov/hox_pro/hunchback.html
REFERENCES
- Berleth T, et al. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. Embo Journal. 1988;7:1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burz DS, Hanes SD. Isolation of mutations that disrupt cooperative DNA binding by the Drosophila Bicoid protein. Journal of Molecular Biology. 2001;305:219–230. doi: 10.1006/jmbi.2000.4287. [DOI] [PubMed] [Google Scholar]
- Burz DS, et al. Cooperative DNA-binding by Bicoid provides a mechanism for threshold-dependent gene activation in the Drosophila embryo. Embo Journal. 1998;17:5998–6009. doi: 10.1093/emboj/17.20.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M, et al. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature. 1990;345:361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- Crauk O, Dostatni N. Bicoid determines sharp and precise target gene expression on the Drosophila embryo. Current Biology. 2005;15:1888–1898. doi: 10.1016/j.cub.2005.09.046. [DOI] [PubMed] [Google Scholar]
- de Lachapelle AM, Bergmann S. Precision and scaling in morphogen gradient read-out. Molecular Systems Biology. 2010;6 doi: 10.1038/msb.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W, Nusslein-Volhard C. A gradient of bicoid protein in Drosophila embryos. Cell. 1988;54:83–93. doi: 10.1016/0092-8674(88)90182-1. [DOI] [PubMed] [Google Scholar]
- Driever W, Nusslein-Volhard C. The Bicoid protein Is a Positive regulator of Hunchback transcription in the early Drosophila embryo. Nature. 1989;337:138–143. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- Driever W, et al. Determination of spatial domains of zygotic gene-expression in the Drosophila embryo by the affinity of binding-sites for the Bicoid morphogen. Nature. 1989;340:363–367. doi: 10.1038/340363a0. [DOI] [PubMed] [Google Scholar]
- Gregor T, et al. Probing the limits to positional information. Cell. 2007a;130:153–164. doi: 10.1016/j.cell.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T, et al. Stability and nuclear dynamics of the bicoid morphogen gradient. Cell. 2007b;130:141–152. doi: 10.1016/j.cell.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, et al. A Multiscale Investigation of Bicoid-Dependent Transcriptional Events in Drosophila Embryos. Plos One. 2011;6 doi: 10.1371/journal.pone.0019122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, et al. Shaping a Morphogen Gradient for Positional Precision. Biophysical Journal. 2010a;99:697–707. doi: 10.1016/j.bpj.2010.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, et al. Distance measurements via the morphogen gradient of Bicoid in Drosophila embryos. Bmc Developmental Biology. 2010b;10 doi: 10.1186/1471-213X-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AV. Proceedings of the Physiological Society. Journal of Physiology. 1910;40 [Google Scholar]
- Holloway DM, et al. Gene Expression Noise in Spatial Patterning: hunchback Promoter Structure Affects Noise Amplitude and Distribution in Drosophila Segmentation. PLoS Comput Biol. 2011;7:e1001069. doi: 10.1371/journal.pcbi.1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houchmandzadeh B, et al. Establishment of developmental precision and proportions in the early Drosophila embryo. Nature. 2002;415:798–802. doi: 10.1038/415798a. [DOI] [PubMed] [Google Scholar]
- Hülskamp M, et al. Differential regulation of target genes by different alleles of the segmentation gene Hunchback in Drosophila. Genetics. 1994;138:125–134. doi: 10.1093/genetics/138.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäckle H, et al. Cross-regulatory interactions among the gap genes of Drosophila. Nature. 1986;324:668–670. [Google Scholar]
- Jaeger J, et al. Dynamic control of positional information in the early Drosophila embryo. Nature. 2004;430:368–371. doi: 10.1038/nature02678. [DOI] [PubMed] [Google Scholar]
- Janssens H, et al. A high-throughput method for quantifying gene expression data from early Drosophila embryos. Development Genes and Evolution. 2005;215:374–381. doi: 10.1007/s00427-005-0484-y. [DOI] [PubMed] [Google Scholar]
- Lebrecht D, et al. Bicoid cooperative DNA binding is critical for embryonic patterning in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13176–13181. doi: 10.1073/pnas.0506462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R, Nussleinvolhard C. Hunchback, a gene required for segmentation of an anterior and posterior region of the Drosophila embryo. Developmental Biology. 1987;119:402–417. doi: 10.1016/0012-1606(87)90045-5. [DOI] [PubMed] [Google Scholar]
- Lewis J, et al. Thresholds in Development. Journal of Theoretical Biology. 1977;65:579–590. doi: 10.1016/0022-5193(77)90216-8. [DOI] [PubMed] [Google Scholar]
- Lopes FJP, et al. A kinetic mechanism for Drosophila Bicoid cooperative binding. Journal of Theoretical Biology. 2005;235:185–198. doi: 10.1016/j.jtbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Lopes FJP, et al. Spatial Bistability Generates hunchback Expression Sharpness in the Drosophila Embryo. PLoS computational Biology. 2008;4 doi: 10.1371/journal.pcbi.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, et al. Regulatory and coding regions of the segmentation gene Hunchback are functionally conserved between Drosophila-virilis and Drosophila-melanogaster. Mechanisms of Development. 1994;45:105–115. doi: 10.1016/0925-4773(94)90024-8. [DOI] [PubMed] [Google Scholar]
- Ma XG, et al. The Drosophila morphogenetic protein Bicoid binds DNA cooperatively. Development. 1996;122:1195–1206. doi: 10.1242/dev.122.4.1195. [DOI] [PubMed] [Google Scholar]
- Manu, et al. Canalization of Gene Expression in the Drosophila Blastoderm by Gap Gene Cross Regulation. Plos Biology. 2009;7:591–603. doi: 10.1371/journal.pbio.1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis JS, et al. Posterior stripe expression of Hunchback Is driven from 2 promoters by a common enhancer element. Development. 1995;121:3067–3077. doi: 10.1242/dev.121.9.3067. [DOI] [PubMed] [Google Scholar]
- Merrill PT, et al. Requirements for autosomal gene activity during precellular stages of Drosophila-melanogaster. Development. 1988;104:495–509. doi: 10.1242/dev.104.3.495. [DOI] [PubMed] [Google Scholar]
- Moodley K, Murrell H. A colour-map plugin for the open source, Java based, image processing package, Image. J. Computers & Geosciences. 2004;30:609–618. [Google Scholar]
- Myasnikova E, et al. Estimation of errors introduced by confocal imaging into the data on segmentation gene expression in Drosophila. Bioinformatics. 2009;25:346–352. doi: 10.1093/bioinformatics/btn620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslein-Volhard C, et al. Determination of anteroposterior polarity in Drosophila. Science. 1987;238:1675–1681. doi: 10.1126/science.3686007. [DOI] [PubMed] [Google Scholar]
- Papatsenko D, Levine M. The Drosophila Gap Gene Network Is Composed of Two Parallel Toggle Switches. PLoS ONE. 2011;6:e21145. doi: 10.1371/journal.pone.0021145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papatsenko D, Levine MS. Dual regulation by the Hunchback gradient in the Drosophila embryo. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2901–2906. doi: 10.1073/pnas.0711941105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poustelnikova E, et al. A database for management of gene expression data in situ. Bioinformatics. 2004;20:2212–2221. doi: 10.1093/bioinformatics/bth222. [DOI] [PubMed] [Google Scholar]
- Schroder C, et al. Differential regulation of the 2 transcripts from the Drosophila gap segmentation gene Hunchback. Embo Journal. 1988;7:2881–2887. doi: 10.1002/j.1460-2075.1988.tb03145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson-Brose M, et al. Synergy between the Hunchback and Bicoid morphogens is required for anterior patterning in Drosophila. Cell. 1994;78:855–865. doi: 10.1016/s0092-8674(94)90622-x. [DOI] [PubMed] [Google Scholar]
- Spirov AV, et al. HOX Pro DB: the functional genomics of hox ensembles. Nucleic Acids Research. 2002;30:2. doi: 10.1093/nar/30.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirov AV, et al. HOX Pro: a specialized database for clusters and networks of homeobox genes. Nucleic Acids Research. 2000;28:3. doi: 10.1093/nar/28.1.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, et al. The gradient morphogen Bicoid is a concentration-dependent transcriptional activator. Cell. 1989;57:1259–1273. doi: 10.1016/0092-8674(89)90062-7. [DOI] [PubMed] [Google Scholar]
- Stryer L. Biochemistry. Freeman; New York: 1995. [Google Scholar]
- Surkova S, et al. Characterization of the Drosophila segment determination morphome. Developmental Biology. 2008;313:844–862. doi: 10.1016/j.ydbio.2007.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, et al. Finger protein of novel structure encoded by Hunchback, a 2nd member of the gap class of Drosophila segmentation genes. Nature. 1987;327:383–389. [Google Scholar]
- Tomancak P, et al. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biology. 2007;8 doi: 10.1186/gb-2007-8-7-r145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman J, Desplan C. The products of the Drosophila gap genes Hunchback and krüppel bind to the Hunchback promoters. Nature. 1989;341:335–337. doi: 10.1038/341335a0. [DOI] [PubMed] [Google Scholar]