Abstract
The alloreactive human T cell clone MBM15 was found to exhibit dual specificity recognizing both an antigen in the context of the HLA class I A2 molecule and an antigen in the context of the HLA class II DR1. We demonstrated that the dual reactivity that was mediated via a single clonal T cell population depended on specific peptide binding. For complete recognition of the HLA-A2-restricted specificity the interaction of CD8 with HLA class I is essential. Interestingly, interaction of the CD8 molecule with HLA class I contributed to the HLA-DR1-restricted specificity. T cell clone MBM15 expressed two in-frame T cell receptor (TCR) Vα transcripts (Vα1 and Vα2) and one TCR Vβ transcript (Vβ13). To elucidate whether two TCR complexes were responsible for the dual recognition or one complex, cytotoxic T cells were transduced with retroviral vectors encoding the different TCR chains. Only T cells transduced with the TCR Vα1Vβ13 combination specifically recognized both the HLA-A2+ and HLA-DR1+ target cells, whereas the Vα2Vβ13 combination did not result in a TCR on the cell surface. Thus a single TCRαβ complex can have dual specificity, recognizing both a peptide in the context of HLA class I as well as a peptide in the context of HLA class II. Transactivation of T cells by an unrelated antigen in the context of HLA class II may evoke an HLA class I-specific T cell response. We propose that this finding may have major implications for immunotherapeutic interventions and insight into the development of autoimmune diseases.
The function of the major histocompatibility complex (MHC) molecules is to bind and display peptides to T lymphocytes. Allogeneic MHC molecules can induce strong T cell responses, which is reflected by the mixed lymphocyte reaction in vitro and the high incidence of graft rejection and graft versus host disease after transplantation of organs or hematopoietic cells over MHC barriers. T cell recognition of allogeneic MHC is often peptide specific, resembling self-MHC-restricted T cell recognition of foreign antigens (1–3). However, alloreactive T cells are heterogeneous in their degree of peptide specificity (4), and a minor population might be peptide independent or recognize motifs shared by many peptides (5). Several alloreactive T cell clones have been described to be crossreactive, recognizing two unrelated peptides in the context of two different allogeneic MHC class I molecules (6, 7). In addition, alloreactive cytotoxic T lymphocytes recognizing an endogenously processed peptide binding to allogeneic MHC molecules and recognizing a different peptide in the context of self-MHC class I have been described (8). In one instance, crossreactive cytotoxic T lymphocytes showing dual recognition for both HLA class I and class II molecules also have been reported (9, 10). Those authors postulated that based on the shared structural motif between the HLA-B27 and the DR2 B5*0101 chain, the reactivity pattern reflected presentation of identical or structurally related peptides by HLA-B27 and HLA-DR2. In several of these previously mentioned studies, cold-target inhibition experiments were performed to confirm that one clonal T cell population was mediating the crossreactivity. However, whether the crossreactivity of these alloreactive T cell clones was mediated via one or two T cell receptor (TCR) αβ complexes was not investigated. This hypothesis may be possible because 20% of peripheral human T cells and 10% of mouse T cells express two different TCRαβ complexes at the cell surface (11).
In this study alloreactive T cells against HLA-A2 were generated by using haploidentical siblings. One alloreactive CD8+ T cell clone demonstrated dual specificity recognizing both the HLA class I molecule A2 and the HLA class II molecule DR1. We demonstrated that the lytic activity restricted by HLA-A2 and HLA-DR1 depended on specific peptide binding. In addition, for both the HLA-A2- and HLA-DR1-restricted lysis the interaction of the CD8 molecule on the T cells with the HLA class I molecules on the target cells was contributing to the lytic activity of the T cell clone. By retroviral gene transfer of individual TCR α chains in combination with the TCR β chain into cytotoxic T cells we demonstrated that the alloreactive lytic activity of MBM15 restricted by HLA-A2 as well as by HLA-DR1 was mediated by one TCRαβ complex.
Materials and Methods
T Cell Clones.
T cell clones MBM15 and MBM13 were derived from a mixed lymphocyte reaction with peripheral blood mononuclear cells from haploidentical siblings by limiting dilution analysis. The serologically determined HLA type of the responder (M) was A1, B7, B44, Cw7, Cw16, DR7, DR13, DQ1, and DQ2. The HLA type of the stimulator (BM) was A1, A2 (A*0201), B7, B57, Cw6, Cw7, DR7, DR13, DQ1, DQ2. The T cell clones were cultured in Iscove's modified Dulbecco's medium (BioWhittaker) containing 10% pooled normal human serum and 100 units/ml IL-2 (Chiron). T cells were stimulated every 2 weeks with a mixture of irradiated allogeneic peripheral blood mononuclear cells, irradiated HLA-A2+ Epstein–Barr virus (EBV)-transformed B cells (EBV-LCL), 800 ng/ml phytohemagglutinin (PHA, Murex Diagnostics, Dartford, U.K.), and 100 units/ml IL-2.
Target Cells.
As target cells, HLA-typed PHA-stimulated T cells, the HLA class I-deficient Burkitt lymphoma cell line Daudi (12), and EBV-LCLs were used. Part of the panel of EBV-LCLs was kindly provided by T. J. H. Pool and J. Anholts (Leiden University Medical Center). HPLC fractions of HLA-A*0201 eluted peptides were loaded on the HLA-A2+ lymphoblastoid processing defective cell line T2 and used as target cells in cytotoxicity assays (13).
Cytotoxicity Assay.
Target cells were harvested and labeled with 50 μCi Na251CrO4 for 60 min at 37°C, washed, and added to various numbers of effector cells, in a final volume of 150 μl Iscove's modified Dulbecco's medium supplemented with 10% FBS in 96-well U-bottomed microtiter plates. For blocking experiments using mAbs RIV6 (anti-CD4) or FK18 (anti-CD8), the effector cells were preincubated with saturating concentrations of mAb for 30 min at 37°C before addition to the target cells. For blocking experiments of HLA class I or class II molecules, the target cells were preincubated with saturating concentrations of mAb for 30 min at 37°C before addition to the effector cells. The mAbs used for the blocking of HLA class I and class II were BB7.2 (anti-HLA-A2), W6/32 (anti-class I α2 and α3), TP25 (anti-class I α3) (14), B8.11.2 (anti-HLA-DR), and SPV-L3 (anti-HLA-DQ). Cold-target inhibition assays were performed by addition of various ratios of unlabeled (cold) target cells to the 51Cr-labeled (hot) target cells before the addition of the effector cells. After 5 h of incubation of target and effector cells at 37°C and 5% CO2, 25 μl of supernatant was harvested and measured in a luminescence counter (Topcount-NXT, Packard). The mean percentage of specific lysis of triplicate wells was calculated as follows: specific lysis = [(experimental release − spontaneous release)/(maximal release − spontaneous release)] × 100.
Isolation of HLA-A*0201 Binding Peptides.
Isolation of HLA-A*0201 binding peptides was performed as described (13), with minor modifications. Briefly, 5 × 109 HLA-A*0201+ EBV-LCLs (EBV-PH) were lysed at 4°C in 20 mM Tris⋅HCl buffer, 150 mM NaCl, and 1% Nonidet P-40 (pH 7.5) with a mixture of protease inhibitors. Cell lysates were subjected to affinity chromatography using the BB7.2 mAb (IgG2a, anti-HLA-A2). HLA-A2-bound peptides were eluted from the column with 10% acidic acid, filtered through Centricon 3 (Amicon), and concentrated to 500 μl for HPLC fractionation. HPLC fractionation was conducted with a C18 reverse-phase column using 0.1% trifluoroacetic acid as organic modifier and a gradient of 1% acetonitril/min. The individual fractions were incubated with the HLA-A2+ processing-defective cell line T2 to analyze for sensitization of the cytolytic activity of MBM13 and MBM15 in a 4-h 51Cr release assay.
Construction of Retroviral Vectors and Generation of Retroviral Supernatant.
By reverse transcriptase–PCR, using primers that cover the total repertoire of known TCR chains, the TCR α and β usage of the T cell clone MBM15 was determined and cloned into retroviral vectors. The Moloney murine leukemia virus-based retroviral vector LZRS and packaging cells ϕ-NX-A used for this purpose were kindly provided by G. Nolan, Stanford University, Palo Alto, CA (15). Two bicistronic retroviral vectors were constructed in which the multiple cloning site is linked to the downstream internal ribosome entry sequence and the marker gene green fluorescent protein (GFP) (16) or truncated form of the nerve growth factor (ΔNGF-R) (17). The TCR Vα chains of MBM15 were cloned into the retroviral vectors in combination with GFP and the TCR Vβ chain in combination with the ΔNGF-R. Retroviral vectors encoding GFP or ΔNGF-R alone were used as control vectors in the experiments. Furthermore, retroviral vectors were constructed encoding the HLA-A2 (A*0201) or the HLA-DR1 (DRA/DRB1*0101) molecule. The constructs were transfected into ϕ-NX-A cells by using calcium phosphate (Life Technologies, Gaithersburg, MD) and 2 days later 2 μg/ml puromycin (CLONTECH) was added. Ten to 14 days after transfection 6 × 106 cells were plated per 10-cm Petri dish (Becton Dickinson) in 10 ml Iscove's modified Dulbecco's medium supplemented with 10% FBS without puromycin. The next day the medium was refreshed and on the following day retroviral supernatant was harvested, centrifuged, and frozen in aliquots at −70°C.
Isolation and Retroviral Transduction of αβ T Cells Derived from Peripheral Blood.
Unselected αβ+ T cells derived from peripheral blood were stimulated with PHA (800 ng/ml) and IL-2 (120 units/ml) at a concentration of 0.5 × 106 cells/ml. After 2 days of culture the T cells were transduced with retroviral supernatant. The transduction procedure used for the peripheral blood T cells was based on the method developed by Hanenberg et al. (18) using recombinant human fibronectin fragments CH-296 (19). Briefly, 5 × 106 T cells were cultured on CH-296-coated Petri dishes together with 1 ml of thawed retroviral supernatant for 6 h or overnight at 37°C, washed, and transferred to 24-well culture plates.
Flow Cytometric Analysis and Fluorescence-Activated Cell Sorting.
The transduction efficiency, measured by the expression of the markers GFP and ΔNGF-R, was analyzed by flow cytometry, 3–5 days after transduction. ΔNGF-R expression was detected by using the mAb 20.4 (anti-human NGF-R) obtained from American Type Culture Collection. In addition, flow cytometric analyses were performed by using mAbs specific for TCR Vβ13.1 (ImmunoTech, Marseille, France), CD3, CD4, and CD8 (BD). Transduced T cells were sorted by fluorescence-activated cell sorting on the basis of GFP+ ΔNGF-R+ in combination with CD8 positivity.
Results
Dual HLA-A2- and HLA-DR1-Specific Recognition of T Cell Clone MBM15.
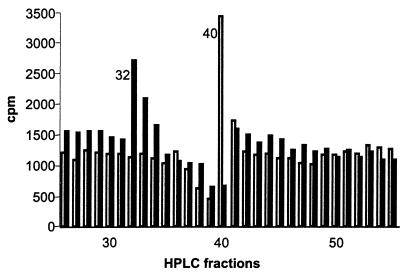
HLA-A2 alloreactive T clones were generated by performing a mixed lymphocyte reaction with peripheral blood mononuclear cells from haploidentical siblings. Several different T cell clones were isolated from which two T cell clones MBM13 and MBM15 were selected. Panel studies using HLA-typed EBV-LCLs and blocking experiments with anti-HLA-A2 antibodies demonstrated that both T cell clones were HLA-A2 restricted (data not shown). To test the HLA-A2 subtype specificity of the two T cell clones, cytotoxicity assays were performed with a panel of EBV-LCLs with known HLA-A2 subtypes. As shown in Table 1, HLA-A2-specific recognition of MBM15 is restricted by HLA-A*0201/A*0202/A*0204/A*0207 and A*0209. In contrast, the recognition of MBM13 is restricted by HLA-A*0201/A*0204 and A*0209 subtypes, illustrating that the recognition pattern of the two T cell clones is different. In addition, experiments performed with HPLC fractions of peptides eluted from HLA-A*0201 molecules and loaded on T2 cells also demonstrated a difference in fine specificity of the two T cell clones, because the alloreactive T cell clones MBM15 and MBM13 lysed T2 cells loaded with different HPLC fractions (Fig. 1). The HLA-A2 binding epitopes recognized by the two allogeneic T cell clones have to be derived from ubiquitous antigenic proteins, because many different HLA-A*0201+ target cells including EBV-LCLs, PHA-stimulated T cells, leukemic cells, fibroblasts, and melanoma cell lines were recognized (data not shown). In addition to the HLA-A2-restricted alloreactivity, T cell clone MBM15 exhibited a second specificity. Using a large panel of different HLA-typed EBV-LCLs as target cells we demonstrated that all HLA-DR1+ target cells were recognized by T cell clone MBM15, independent of the HLA class I haplotype whereas MBM13 did not recognize these DR1+ EBV-LCLs. A selection of this panel is shown in Table 2.
Table 1.
Cytolytic activity of the MBM13 and MBM15 T cell clones against a panel of HLA-typed EBV-LCL
| Name | Cell type | HLA-A2 subtype | MBM15 | MBM13 |
|---|---|---|---|---|
| % specific
lysis (3:1)
| ||||
| PH | EBV-LCL | 0201 | 51 | 64 |
| M7 | EBV-LCL | 0202 | 47 | < |
| DK6 | EBV-LCL | 0203 | < | < |
| HO | PHA blast | 0204 | 31 | 59 |
| AMW | EBV-LCL | 0205 | < | < |
| SO | EBV-LCL | 0206 | < | < |
| MA | EBV-LCL | 0207 | 62 | < |
| KLO | EBV-LCL | 0208 | < | < |
| OZB | EBV-LCL | 0209 | 50 | 81 |
Cytotoxic activity was marked with < when the percentage-specific lysis of the target cells was <10% at an effector-to-target cell ratio (E/T) of 3:1.
Figure 1.
Cytotoxic activity of the T cell clones MBM15 and MBM13 against T2 cells loaded with HPLC fractions of HLA-A2-eluted peptides. The closed bars represent T cell clone MBM15, the open bars represent T cell clone MBM13. The cytotoxic activity is shown as cpm. Background lysis of T2 by MBM15 and MBM13 in the absence of HPLC fractions was 1,400 cpm and 1,100 cpm, respectively. The maximum lysis of T2 by 0.5% Triton X-100 was 3,600 cpm.
Table 2.
Cytolytic activity of the MBM13 and MBM15 T cell clones against a panel of HLA-typed EBV-LCL
| Name | HLA class I | HLA class II* | MBM15 | MBM13 |
|---|---|---|---|---|
| % specific lysis
(3:1)†
| ||||
| BM | A1 A2 B7 B57 Cw6 Cw7 | DR7 DR13 DQ2 DQ1 | ++ | ++ |
| M | A1 B7 B44 Cw7 Cw16 | DR7 DR13 DQ2 DQ1 | < | < |
| PH | A2 B62 Cw2 Cw3 | DR11 DR7 DQ2 DQ7 | ++ | ++ |
| JY | A2 B7 Cw7 | DR4 DR13 DQ1 DQ3 | ++ | ++ |
| WO | A1 A3 B35 B57 Cw4 Cw6 | DR101 DR4 DQ5 DQ8 | ++ | < |
| LE | A1 A3 B7 B44 Cw7 | DR15 DR11 DQ1 DQ7 | < | < |
| TS | A3 B7 B35 Cw4 Cw7 | DR11 DQ6 | < | < |
| BA | A1 A3 B7 B8 Cw7 | DR15 DR3 DQ1 DQ2 | < | < |
| GU | A1 A24 B13 B57 Cw6 | DR3 DR7 DQ2 DQ3 | < | < |
| SC | A3 A24 B35 Cw4 | DR101 DR13 DQ5 DQ6 | ++ | < |
| FO | A3 A24 B44 B61 Cw2 | DR101 DR4 DQ1 DQ7 | ++ | < |
| WT | A11 B35 Cw4 | DR101 DQ5 | ++ | < |
| LW | A33 B14 Cw8 | DR102 DQ5 | < | < |
| MZ | A24 B14 Cw2 Cw8 | DR102 DQ5 | < | < |
The HLA-DR1 subtyping was molecular defined.
Cytotoxic activity was marked with < when the percentage-specific lysis of the target cells was <10% at an E/T of 3:1. Cytotoxic activity was marked with ++ when the percentage-specific lysis of the target cells was >20% at an E/T of 3:1.
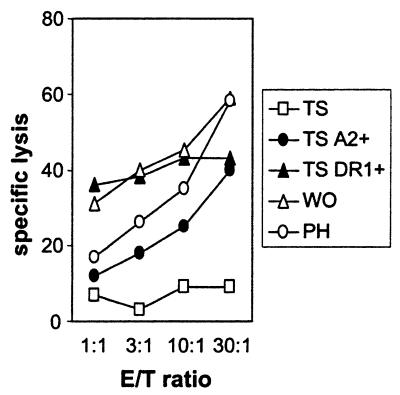
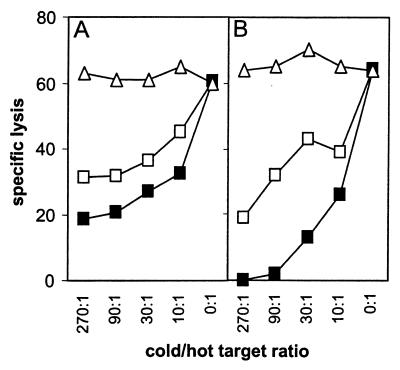
The experiments using different HLA-DR1-subtyped EBV-LCLs demonstrated that all HLA-DRB1*0101+ EBV-LCLs were recognized by MBM15, whereas EBV-LCLs positive for HLA-DRB1*0102 were not recognized. The HLA-DRB1*0102 molecule has 1-aa difference in the peptide binding domain, resulting in a difference in peptides binding to HLA-DRB1*0102 compared with HLA-DRB1*0101 (20). These results indicate that the HLA-DR1-restricted recognition is not a conformation similarity between HLA-A2 and HLA-DR1 but is peptide specific. Experiments performed with T2 cells transduced with HLA-DR1 (DRA/DRB1*0101) supported these results. HLA-DR1+ T2 cells were not recognized by MBM15, even after stabilization of the HLA-A2 molecule by HLA-A2 binding peptides (data not shown). Because the HLA-DR1 molecules present on T2 cells were filled mainly with the invariant chain-derived peptide (CLIP), the T2-DR1+ cells were loaded with the DR1 binding peptide of human influenza hemagglutinin (HA). Although the HA-loaded DR1+ T2 cells were recognized by a HA-specific T cell clone, MBM15 was not able to recognize these cells, demonstrating the peptide specificity of the HLA-DR1-restricted recognition of MBM15. EBV-TS cells negative for both HLA-A2 and HLA-DR1, and therefore not recognized by the MBM15 T cell clone, were transduced with either HLA-A2 (A*0201) or HLA-DR1 (DRB1*0101). Cytotoxic experiments performed with these transduced target cells confirmed the dual specificity restricted by HLA-A2 and HLA-DR1 of MBM15 (Fig. 2). To prove that MBM15 was a clonal population and that the class I- and class II-specific recognition was mediated by the same clonal T cell, cold-target inhibition studies were performed, using HLA-A2+ (EBV-PH), HLA-DR1+ (EBV-WO), and as control HLA-A2−/DR1− EBV-LCL (EBV-BA). The results demonstrated that the HLA-A2-restricted lysis was specifically blocked by HLA-DR1+ cold target cells and vice versa, but not by HLA-A2/DR1− cold target cells (Fig. 3).
Figure 2.
Cytotoxic activity of T cell clone MBM15 against different target cells. Cytotoxic activity was measured against the target cells EBV-TS (TS, HLA-A2/HLA-DR1−), transduced with HLA-A2 (TS A2+) or transduced with HLA-DR1 (TS DR1+). As positive controls EBV-LCLs positive for HLA-A2 (PH) or positive for HLA-DR1 (WO) were used. Cytotoxicity was measured in a 5-h 51Cr release assay.
Figure 3.
Cold-target inhibition analysis of the cytotoxicity of T cell clone MBM15. 51Cr-labeled target cells were (A) EBV-PH (HLA-A2+) and (B) EBV-WO (HLA-DR1+). Effector to labeled (hot) target cell ratio was 10:1. The following unlabeled (cold) target cells were added in different ratios to the labeled target cells: EBV-PH (□), EBV-WO (■), and the HLA-A2/DR1− EBV-BA (▵).
One TCR Complex Mediates Both the HLA Class I- and HLA Class II-Restricted Recognition.
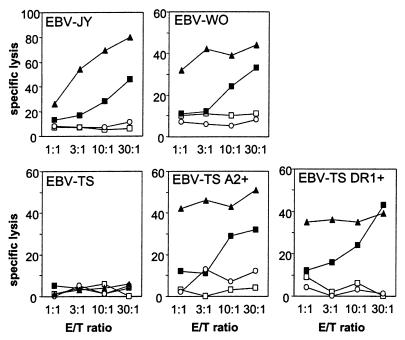
By reverse transcriptase–PCR, we established that MBM15 expressed two in-frame TCRα transcripts, Vα1 and Vα2, and one TCRβ gene transcript, Vβ13.1. Flow cytometry confirmed that 100% of the T cells expressed TCR Vβ13.1 at the cell surface (data not shown). No antibodies were available for analysis of the specific TCRα chain expression at the cell surface. To investigate whether both TCR complexes were essential for the dual recognition or one of the two TCR complexes, three retroviral vectors were constructed by expressing TCR Vα1 or Vα2 in combination with GFP and TCR Vβ13 in combination with the marker gene ΔNGF-R. PHA-stimulated unselected T cells derived from freshly isolated unrelated peripheral blood and negative for HLA-A2 and HLA-DR1 were transduced with the combinations of TCR Vα1 and Vβ13 or TCR Vα2 and Vβ13. CD8+ T cells double positive for GFP and ΔNGF-R of both transductions and control GFP/ΔNGF-R transduction were sorted by fluorescence-activated cell sorting and tested for functionality in cytotoxicity assays. HLA-A2+ EBV-LCL (EBV-JY), HLA-DR1+ EBV-LCL (EBV-WO), and control HLA-A2/DR1− EBV-LCL (EBV-TS) were used as target cells in the 51Cr release assays (Fig. 4). As positive control the parental T cell clone MBM15 was included in these experiments. The results demonstrated that the TCR Vα1Vβ13-transduced CD8+ T cell population specifically recognized the HLA-A2 and HLA-DR1 expressing target cells almost as effectively as the parental T cell clone MBM15. In contrast, no specific lysis was observed with the TCR Vα2Vβ13 and GFP/ΔNGF-R control transduced CD8+ T cells. In addition, EBV-LCL (EBV-TS) negative for HLA-A2 and HLA-DR1, transduced with either HLA-A2 or HLA-DR1, were used as target cells. The results shown in Fig. 4 demonstrate that both MBM15 and TCR Vα1Vβ13-transduced CD8+ T cells lyse the HLA-A2- and HLA-DR1-transduced EBV-TS, confirming that the dual specificity restricted by HLA-A2 and HLA-DR1 was mediated only via the TCR Vα1Vβ13 complex. Jurkat cells deprived of a TCR α chain, transduced with Vα2, were not able to express a TCRαβ complex at the cell surface (data not shown). Transduction of the same Jurkat cell line with Vα1 or other cloned TCR α chains was able to form a TCRαβ complex at the cell surface, with or without the presence of the corresponding TCR β chain. These results indicate that the Vα2 chain derived from MBM15 is not expressed at the cell surface of Vα2Vβ13-transduced CD8+ T cells.
Figure 4.
Cytotoxic activity of TCR Vα1Vβ13-transduced (■), TCR Vα2Vβ13-transduced (○), and control GFP/ΔNGF-R-transduced CD8+ T cells (□) derived from peripheral blood, and the parental T cell clone MBM15 (▴) against HLA-A2+ EBV-LCL (JY), HLA-DR1+ EBV-LCL (WO), and HLA-A2/DR1− EBV-LCL (TS) transduced with HLA-A2 or HLA-DR1, in a 5-h 51Cr release assay.
Role of the Coreceptor CD8 in Dual Recognition.
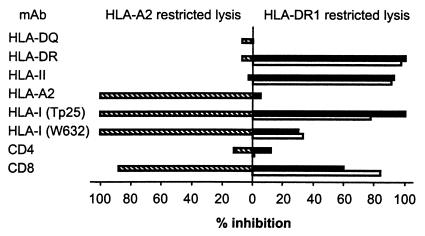
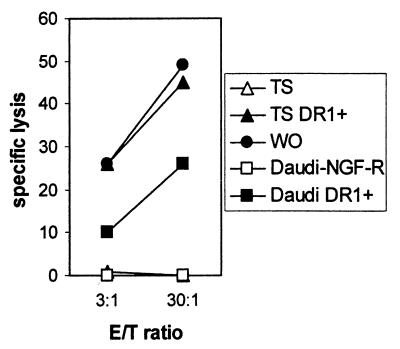
Antibody blocking experiments confirmed the HLA-A2 and HLA-DR1 restricted reactivities of MBM15, because the HLA-DR1-restricted lysis was blocked by the addition of mAb directed against HLA-DR (B8.11.2), whereas the HLA-A2 lytic activity was not blocked with this mAb (Fig. 5). Conversely, the anti-HLA-A2 mAb only blocked the HLA-A2-restricted lysis. In these experiments blocking of the CD8 molecule with saturating concentrations of mAb resulted in an almost complete inhibition of the lytic activity of MBM15 against both HLA-A2+ target cells (88% inhibition) and the two HLA-DR1+ target cells (84% and 60%). Using an antibody directed against the α3 domain of the HLA-class I molecules (TP25) (14) we confirmed that the HLA-A2- and the HLA-DR1-restricted recognition was decreased when the CD8-HLA class I interaction was blocked (Fig. 5). In addition, transduction of the β2 microglobulin/HLA class I-deficient Burkitt lymphoma cell line Daudi with HLA-DR1 (DRB1*0101) demonstrated that the HLA-DR1-restricted lysis of MBM15 was lower against DR1-transduced Daudi cells than against endogenously DR1 expressing EBV-LCLs (EBV-WO) and DR1-transduced EBV-LCLs (EBV-TS) (Fig. 6). Thus, the interaction of the CD8 molecule with non-TCR-bound HLA class I molecules on the target cell contributed to the antigen-specific recognition of the HLA class II-restricted specificity of MBM15. These results demonstrate that the CD8 molecule expressed by the T cell clone MBM15 served as a coreceptor for both the HLA-A2- and the HLA-DR1-restricted recognition.
Figure 5.
Percentage inhibition of the HLA-A2- and HLA-DR1-restricted lytic activity of T cell clone MBM15, using saturating concentrations of different mAbs. The cytotoxicity of MBM15 against the HLA-A2+ EBV-PH (hatched bars), the HLA-DR1+ EBV-WO (open bars), and the HLA-DR1+ EBV-WT (filled bars) were, respectively, 54%, 31%, and 40% at an E/T ratio of 3:1.
Figure 6.
Cytotoxic activity of T cell clone MBM15 against HLA-DR1-transduced Daudi cells. The cytotoxic activity of MBM15 was measured against Daudi cells transduced with HLA-DR1 or ΔNGF-R control vector. EBV-LCLs positive for HLA-DR1 (WO) or transduced with HLA-DR1 (TS DR1+) were used to determine the HLA-DR1-restricted lysis in the presence of HLA class I. To exclude NK or LAK-mediated lysis of MBM15, 25-fold excess of unlabeled K562 and Daudi was added. Cytotoxicity was measured in a 4-h 51Cr release assay.
Discussion
In this report we demonstrate that a single TCRαβ complex can exhibit dual specificity, recognizing both an antigenic peptide in the context of HLA class I and an antigenic peptide in the context of HLA class II. The dual cytotoxic activity restricted by HLA-A2 and HLA-DR1 was confirmed by panel studies with HLA-typed EBV-LCL and by blocking experiments with mAb. Anti-HLA-A2 antibodies could inhibit HLA-A2-specific lysis but not HLA-DR1-specific lysis, and conversely, anti-HLA-DR-specific mAb only blocked the HLA-DR1-specific lysis. In addition, EBV-LCL negative for HLA-A2 and HLA-DR1, and therefore not recognized by MBM15, were lysed by MBM15 when the cells were transduced with either HLA-A2 or HLA-DR1. HPLC fractionation of the HLA-A2 eluted peptides demonstrated the peptide-specific HLA-A2-restricted recognition. Furthermore, the difference in the recognition pattern of HLA-DRB1*0101 and DRB1*0102 and the inability to recognize HLA-DR1-transduced T2 cells indicated the peptide-specific recognition of HLA-DRB1*0101.
Cold target competition experiments demonstrated that the same clonal T cell population mediated these two specificities. The HLA-A2-specific lysis could be blocked efficiently with unlabeled target cells expressing HLA-DR1 and vice versa.
Because, 20% of human T cells and 10% of peripheral mouse T cells can coexpress two different TCR Vα chains on the cell surface as part of two functional and independent TCRαβ complexes, the dual specificity might be caused by two independent TCRαβ complexes recognizing two different HLA/peptide complexes. By retroviral gene transfer of the two TCRαβ complexes into cytotoxic T cells, we demonstrated that the dual specificity was not mediated via two different TCRαβ complexes, but that a single TCRαβ complex exhibited both specificities.
Thus far, T cell crossreactivity often has been explained by the molecular mimicry model, which proposes that different MHC/peptide complexes may form antigenic surfaces that are similar in shape, charge, or both (21–23) and therefore can be recognized by one TCR complex. The HLA class I and class II molecules possess a very similar overall antigen-presenting groove consisting of α1 plus α2 domains and α1 plus β1 domains for HLA class I and class II, respectively. However, unique structural features of the two HLA classes dictate the binding of peptides differing in length and composition (24). Therefore, the antigenic surfaces of the HLA-A2/peptide and the HLA-DR1/peptide complex recognized by the dual-specific TCR of MBM15 would very likely be significantly different in shape and charge. In agreement with this finding is a recent report showing that TCR footprint regions of two complexes recognized by a single TCR are significantly different in shape and charge (25). Crystal structure analysis of four distinct class I-restricted TCRs in complex with peptide and MHC class I have demonstrated a diagonal orientation of the TCRs, regardless of TCR specificity or species origin (26). In contrast, recent elucidation of the crystal structure of a MHC class II-restricted TCR in complex with peptide and MHC class II revealed that the class II-restricted TCR is oriented in a orthogonal mode relative to its peptide-MHC II ligand, necessitated by the amino-terminal extension of peptide residues projecting from the MHC class II antigen-binding groove (27). These results indicate that the antigenic surface of HLA-A2/peptide and HLA-DR1/peptide do not necessarily require the same configuration, but that the TCR will be able to bind the complex in different orientations. It therefore will be of interest to study the crystal structure of this particular TCR in complex with the two different peptide-HLA ligands, to unravel the conformational similarities and differences of the two complexes. For that purpose, the identification of the two peptides recognized by MBM15 in the context of HLA-A2 and HLA-DR1 is needed. Using tetrameric complexes composed of the HLA molecules and these specific peptides, tetramer-positive T cells can be selected and tested for dual specificity to determine the frequency of similar T cells in different individuals.
Crossreactive cytotoxic T lymphocytes showing dual recognition for both HLA class I and class II previously have been reported (9, 28, 29). In addition, several cytotoxic T lymphocyte lines and clones of the CD8 phenotype exhibiting class II restricted reactivity (30–32) and alloreactive CD4+ T cells exhibiting a class I-restricted proliferative capacity have been described (33). Based on the assumption that during T cell development CD8+ T cells are selected on HLA class I and CD4+ T cells on HLA class II, we hypothesize that these previously described T cells also have a specificity recognizing a peptide in the MHC for which they are selected. However, determination of the second specificity is not a trivial experiment and therefore not many dual specificities have been described. Therefore we hypothesize that dual recognition for both HLA class I and HLA class II may represent a more general phenomenon.
The alloreactive cytotoxicity of the class II-restricted CD8+ T cell clones described was independent of CD8, implying that the affinity of the TCRs for class II was high enough to be independent of the additional avidity provided by the CD8 coreceptor (9, 10, 29). For alloreactive CD4+ T cells that exhibit a class I-restricted proliferative capacity, the interaction of CD4 with non-TCR-bound MHC class II molecules did contribute to antigen-specific activation of these class I-restricted CD4+ T cells (33). We demonstrated by using blocking mAbs directed against CD8, HLA class I, and in particular against the α3 domain of the HLA class I molecule that the interaction between the CD8 coreceptor expressed on the alloreactive T cells and the non-TCR-bound HLA class I present on the target cells supported the interaction of the TCR complex with HLA-DR1. In addition, we demonstrated that the recognition of HLA class I-deficient Daudi cells transduced with HLA-DR1 is clearly less than of other DR1-transduced target cells that coexpress HLA class I. We postulate a model in which the TCR binds to the MHC class II/peptide complex, whereas the CD8 molecule expressed by the same T cell will bind to any HLA class I molecule present on the target cell, to increase the total avidity of the interaction between T cell and target cell.
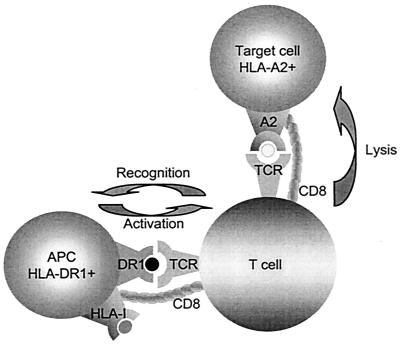
The results shown in this manuscript suggest that certain concepts of crossreactivity or promiscuity need to be revised. Autoimmune reactions have been proposed to be caused by mimicry of different peptides presented by the same HLA molecule. This assumption might be too limited, because we demonstrated that dissimilar conformations composed of HLA class I/peptide or HLA class II/peptide complexes can trigger “transactivation.” Based on our observations discussed in this manuscript we propose a hypothesis of HLA class II-associated transactivation that may trigger HLA class I-specific effector functions of T cells, leading to unexpected and undesirable reactions (Fig. 7). In this model CD8+ T cells, capable of recognizing an endogenous autoantigen but anergized due to the inappropriate context, in which the autoantigens are presented to the T cells, may be capable of dual recognizing an unrelated class II-associated antigen derived, for instance, from an infectious agent. If these dual-recognizing T cells would be activated by the class II-associated antigen presented by a professional antigen-presenting cell, the anergy of the T cell may be abrogated, leading to the autoimmune response. This hypothesis could in part explain a possible correlation between a certain HLA class II polymorphism associated with an autoimmune disease and an apparently unrelated HLA class I-specific effector mechanism, leading to destruction of the tissue involved. Similarly, class II-associated activation of CD8+ T cells may lead to class I-specific organ destruction by alloreactive T cells, leading to rejection or graft versus host reactions after transplantation. We suggest that dual HLA class I- and class II-specific recognition by single T cells may have major implications for immunotherapeutic interventions and insight into the development of autoimmune diseases.
Figure 7.
A single TCR from a CD8+ T cell can interact both with peptides presented in HLA class I and unrelated peptides presented in HLA class II. The CD8+ T cell may not be directly activated by the class I/peptide complex presented on the target cells in the absence of appropriate expression of costimulatory molecules. Transactivation by the specific but unrelated peptides presented in class II-expressing professional antigen-presenting cells (APC) may lead to an immune response against the class I-associated peptide. In this model transactivation by e.g., viral peptides presented by these professional APC may result in undesired autoimmune or alloreactive effector T cell responses.
Acknowledgments
We thank T. J. H. Pool and J. Anholts (Department of Immunohematology and Blood Transfusion, Leiden University Medical Center) for providing HLA-typed EBV-LCLs. M.H.M.H. was supported by a Gisela Thier fellowship. This study was supported in part by a grant from the J. A. Cohen Institute for Radiopathology and Radiation Protection.
Abbreviations
- ΔNGF-R
truncated form of the nerve growth factor receptor
- GFP
green fluorescent protein
- EBV
Epstein–Barr virus
- EBV-LCL
EBV-transformed B cells
- TCR
T cell receptor
- PHA
phytohemagglutinin
- HA
hemagglutinin
- E/T
effector-to-target cell ratio
References
- 1.Panina-Bordignon P, Corradin G, Roosnek E, Sette A, Lanzavecchia A. Science. 1991;252:1548–1550. doi: 10.1126/science.1710827. [DOI] [PubMed] [Google Scholar]
- 2.Wang W, Man S, Gulden P H, Hunt D F, Engelhard V H. J Immunol. 1998;160:1091–1097. [PubMed] [Google Scholar]
- 3.Man S, Salter R D, Engelhard V H. Int Immunol. 1992;4:367–375. doi: 10.1093/intimm/4.3.367. [DOI] [PubMed] [Google Scholar]
- 4.Rotzschke O, Falk K, Faath S, Rammensee H G. J Exp Med. 1991;174:1059–1071. doi: 10.1084/jem.174.5.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elliott T J, Eisen H N. Proc Natl Acad Sci USA. 1990;87:5213–5217. doi: 10.1073/pnas.87.13.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith K D, Huczko E, Engelhard V H, Li Y Y, Lutz C T. Transplantation. 1997;64:351–359. doi: 10.1097/00007890-199707270-00030. [DOI] [PubMed] [Google Scholar]
- 7.Tallquist M D, Yun T J, Pease L R. J Exp Med. 1996;184:1017–1026. doi: 10.1084/jem.184.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Udaka K, Wiesmuller K H, Kienle S, Jung G, Walden P. J Immunol. 1996;157:670–678. [PubMed] [Google Scholar]
- 9.Aparicio P, Jaraquemada D, Lopez de Castro J A. J Exp Med. 1987;165:428–443. doi: 10.1084/jem.165.2.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez D, Barber D F, Villadangos J A, Lopez de Castro J A. J Immunol. 1993;150:2675–2686. [PubMed] [Google Scholar]
- 11.Padovan E, Casorati G, Dellabona P, Meyer S, Brockhaus M, Lanzavecchia A. Science. 1993;262:422–424. doi: 10.1126/science.8211163. [DOI] [PubMed] [Google Scholar]
- 12.Klein E, Klein G, Nadkarni J S, Nadkarni J J, Wigzell H, Clifford P. Cancer Res. 1968;28:1300–1310. [PubMed] [Google Scholar]
- 13.den Haan J M, Sherman N E, Blokland E, Huczko E, Koning F, Drijfhout J W, Skipper J, Shabanowitz J, Hunt D F, Engelhard V H, et al. Science. 1995;268:1476–1480. doi: 10.1126/science.7539551. [DOI] [PubMed] [Google Scholar]
- 14.Tanabe M, Sekimata M, Ferrone S, Takiguchi M. J Immunol. 1992;148:3202–3209. [PubMed] [Google Scholar]
- 15.Kinsella T M, Nolan G P. Hum Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- 16.Heemskerk M H, Blom B, Nolan G, Stegmann A P, Bakker A Q, Weijer K, Res P C, Spits H. J Exp Med. 1997;186:1597–1602. doi: 10.1084/jem.186.9.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruggieri L, Aiuti A, Salomoni M, Zappone E, Ferrari G, Bordignon C. Hum Gene Ther. 1997;8:1611–1623. doi: 10.1089/hum.1997.8.13-1611. [DOI] [PubMed] [Google Scholar]
- 18.Hanenberg H, Xiao X L, Dilloo D, Hashino K, Kato I, Williams D A. Nat Med. 1996;2:876–882. doi: 10.1038/nm0896-876. [DOI] [PubMed] [Google Scholar]
- 19.Heemskerk M H, Hooijberg E, Ruizendaal J J, van der Weide M M, Kueter E, Bakker A Q, Schumacher T N, Spits H. Cell Immunol. 1999;195:10–17. doi: 10.1006/cimm.1999.1520. [DOI] [PubMed] [Google Scholar]
- 20.Verreck F A, van de, P. A, Drijfhout J W, Amons R, Coligan J E, Konig F. Immunogenetics. 1996;43:392–397. doi: 10.1007/BF02199809. [DOI] [PubMed] [Google Scholar]
- 21.Oldstone M B. Cell. 1987;50:819–820. doi: 10.1016/0092-8674(87)90507-1. [DOI] [PubMed] [Google Scholar]
- 22.Wucherpfennig K W, Strominger J L. Cell. 1995;80:695–705. doi: 10.1016/0092-8674(95)90348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quaratino S, Thorpe C J, Travers P J, Londei M. Proc Natl Acad Sci USA. 1995;92:10398–10402. doi: 10.1073/pnas.92.22.10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madden D R. Annu Rev Immunol. 1995;13:587–622. doi: 10.1146/annurev.iy.13.040195.003103. [DOI] [PubMed] [Google Scholar]
- 25.Zhao R, Loftus D J, Appella E, Collins E J. J Exp Med. 1999;189:359–370. doi: 10.1084/jem.189.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teng M K, Smolyar A, Tse A G, Liu J H, Liu J, Hussey R E, Nathenson S G, Chang H C, Reinherz E L, Wang J H. Curr Biol. 1998;8:409–412. doi: 10.1016/s0960-9822(98)70160-5. [DOI] [PubMed] [Google Scholar]
- 27.Reinherz E L, Tan K, Tang L, Kern P, Liu J, Xiong Y, Hussey R E, Smolyar A, Hare B, Zhang R, et al. Science. 1999;286:1913–1921. doi: 10.1126/science.286.5446.1913. [DOI] [PubMed] [Google Scholar]
- 28.Aparicio P, Lopez de Castro J A. Hum Immunol. 1989;24:295–299. doi: 10.1016/0198-8859(89)90022-0. [DOI] [PubMed] [Google Scholar]
- 29.Schilham M W, Lang R, Benner R, Zinkernagel R M, Hengartner H. J Immunol. 1986;137:2748–2754. [PubMed] [Google Scholar]
- 30.Kirberg J, Baron A, Jakob S, Rolink A, Karjalainen K, von Boehmer H. J Exp Med. 1994;180:25–34. doi: 10.1084/jem.180.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heemskerk M H, Schilham M W, Schoemaker H M, Spierenburg G, Spaan W J, Boog C J. Eur J Immunol. 1995;25:1109–1112. doi: 10.1002/eji.1830250438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spits H, Ijssel H, Thompson A, de Vries J E. J Immunol. 1983;131:678–683. [PubMed] [Google Scholar]
- 33.de Bueger M, Bakker A, Goulmy E. Eur J Immunol. 1992;22:875–878. doi: 10.1002/eji.1830220338. [DOI] [PubMed] [Google Scholar]