SUMMARY
Protein ubiquitination is an essential posttranslational modification regulating neurodevelopment, synaptic plasticity, learning and memory, and its dysregulation contributes to the pathogenesis of neurological diseases. Here we report a systematic analysis of ubiquitinated proteome (ubiquitome) in rat brain using a newly developed monoclonal antibody that recognizes the diglycine tag on lysine residues in trypsinized peptides (K-GG peptides). Initial antibody specificity analysis showed that the antibody can distinguish K-GG peptides from linear GG peptides or pseudo K-GG peptides derived from iodoacetamide. To evaluate the false discovery rate of K-GG peptide matches during database search, we introduced a null experiment using bacterial lysate that contains no such peptides. The brain ubiquitome was then analyzed by this antibody enrichment with or without strong cation exchange (SCX) prefractionation. During SCX chromatography, although the vast majority of K-GG peptides were detected in the fractions containing at least three positive charged peptides, specific K-GG peptides with two positive charges (e.g. protein N-terminal acetylated and C-terminal non-K/R peptides) were also identified in early fractions. The reliability of C-terminal K-GG peptides was also extensively investigated. Finally, we collected a dataset of 1786 K-GG sites on 2064 peptides in 921 proteins and estimated their abundance by spectral counting. The study reveals a wide range of ubiquitination events on key components in presynaptic region (e.g. Bassoon, NSF, SNAP25, synapsin, synaptotagmin, and syntaxin) and postsynaptic density (e.g. PSD-95, GKAP, CaMKII, as well as receptors for NMDA, AMPA, GABA, serotonin, and acetylcholine). We also determined ubiquitination sites on amyloid precursor protein and alpha synuclein that are thought to be causative agents in Alzhermer’s and Parkinson’s disorders, respectively. As K-GG peptides can also be produced from Nedd8 or ISG15 modified proteins, we quantified these proteins in the brain and found that their levels are less than 2% of ubiquitin. Together, this study demonstrates that a large number of neuronal proteins are modified by ubiquitination, and provides a feasible method for profiling the ubiquitome in the brain.
Keywords: ubiquitin, synapse, antibody, proteomics, mass spectrometry
INTRODUCTION
Ubiquitin (Ub) is a small protein of 76 amino acids conserved in eukaryotic cells and functions as a posttranslational modifier to regulate nearly all cellular events1,2. The modification reaction is mediated by E1 activating, E2 conjugating, and E3 ligating enzymes, resulting in an isopeptide bond between the Ub C-terminal carboxyl group and the amine group on a lysine side chain or N-terminus of affected substrates3. Polyubiquitination of the substrates occurs by extending Ub chains through any of eight amino groups in Ub itself (M1, K6, K11, K27, K29, K33, K48 or K63)4–6. Alternatively, cysteine, serine and threonine residues are implicated as Ub conjugating sites but direct biochemical evidence (e.g. validation by mass spectrometry) is still missing7–9. Moreover, ubiquitination is reversible by the action of a large family of deubiquitinating enzymes (DUBs)10. Although initially discovered as a principle signal for proteasomal degradation, Ub plays fundamental roles in a variety of proteasome-independent processes, such as chromatin remodeling11, DNA repair12, endocytotic trafficking13, autophagy14, and immune defense15. The human genome encodes two Ub E1s, ~40 E2s, ~600 E3s and 95 DUBs16,17, generating the ubiquitinated proteome (ubiquitome) including thousands of protein substrates in different cell types and tissues. In addition, Ub like modifications have been found in eukaryotes (e.g. SUMO, Nedd8, and ISG15)18 and even in prokaryotes (e.g. Pup)19.
The role of ubiquitin in neuronal development, synaptic plasticity, learning and memory is well recognized and its dysfunction may contribute to disease development20–23. For example, a number of E3 enzymes (e.g. Highwire, APC, SCF, Mindbomb, and TRIM3), DUBs (Fat facets/FAM, UCH-L1, UCH-L3, USP14), and proteasome play important functions in neuronal morphogenesis and synaptic formation23. Genetic mutations in Parkin E3 ligase and UCH-L1 are linked to Parkinson’s disease; UBE3A and ATXN3 are associated with Angelman syndrome and spinal cerebral ataxia, respectively23. Recently, mutations in UBQLN2, a Ub receptor regulating proteasomal degradation, have been shown to cause a rare type of familial amyotrophic lateral sclerosis24. Furthermore, Ub-positive inclusion staining is a pathological hallmark in numerous neurodegenerative diseases, suggesting a critical role for Ub in disease development25. However, the investigation of ubiquitin pathways in neuronal regulation is limited by the lack of systematic tools to map the ubiquitome in the brain.
With rapid development of mass spectrometry-based proteomics technologies26, large-scale analyses of the ubiquitome have become possible5. As trypsin digestion of ubiquitinated substrates generates signature peptides carrying a Gly-Gly (GG) tag on modified lysine residues (K-GG peptides), the resulting monoisotopic mass shift (+114.0429) is detectible by mass spectrometry27,28. However, because of low stoichiometry of protein ubiquitination in cells, pre-enrichment of the ubiquitome is essential for successful analysis. The ubiquitome has been purified by various approaches including Ub-binding domains, Ub antibodies, and epitope-tagged Ub (e.g., FLAG, HA, myc, Strep, His, and biotin tag)29–36, but the analysis is still complicated by co-purified contaminants and difficulty in detecting K-GG peptides for modification site determination. Recently, a new strategy has been developed to directly enrich K-GG peptides instead of ubiquitinated proteins using monoclonal antibodies37–39. This strategy leads to the detection of up to 19,000 ubiquitinated sites in mammalian cell lines after accumulating ubiquitinated species by proteasomal inhibition. But its application to complex tissue samples has not been reported yet.
In this study, we utilized this antibody enrichment strategy for analyzing the ubiquitome from rat brain. We also fully characterized the K-GG peptide antibody by pure proteins/peptides, and evaluated the target-decoy strategy for processing K-GG peptide matches during database search. Our study demonstrated that the antibody strategy provided a feasible method to detect many key ubiquitination events during neuronal regulation, and revealed a large number of unknown ubiquitinated targets for subsequent functional investigation.
EXPERIMENTAL PROCEDURES
Generation of various GG peptides for antibody characterization
Iodoacetamide modification of ubiquitin was performed based on the protocol previously reported40. Purified mono-Ub was denatured in 8 M urea, and incubated with 50 mM IAA at 90°C for 20 min. The sample was cooled down to 4°C, diluted with 50 mM ammonium bicarbonate to reduce urea to 2 M, and digested with trypsin (30 ng/µl) at 37°C overnight. The resulting peptides were acidified by TFA (1%), desalted with Zip-Tip C18 (Millipore), and analyzed by LC-MS/MS. Genuine Ub K48-GG peptide was generated by digestion of K48 di-Ub (BostonBiochem), while M1-GG peptide and K48 peptide without GG tag were chemically synthesized. Finally, these samples were mixed to ensure similar levels (~5 picomoles) of pseudo-K48-GG, K48-GG, M1-GG peptides and K48 peptide without GG tag. The mixture was desalted, dried and subjected to enrichment by K-GG antibody coupled to protein A agarose beads (2 µl, Cell Signaling Technology). The flowthrough and elution were desalted for MS analysis (Q-Exactive, Thermo Scientific).
Escherichia coli peptide preparation
E. coli DH5α strain was harvested and lysed in lysis buffer A (0.1 M Tris, pH 8.5, 8 M urea, 0.15% sodium deoxycholate). Approximately 80 µg of protein was digested with Lys-C (100:1 substrate-to-enzyme ratio) at 21°C for 1 h, diluted to 2 M Urea by a dilution buffer (0.1 M Tris-HCl, pH 8.5, 0.15% sodium deoxycholate) and further digested by trypsin (100:1 ratio at 37°C overnight).
Rat brain peptide preparation and fractionation by strong cation exchange chromatography
Adult Rat brain (Pel Freez Biologicals) was dissected to obtain cerebral cortex, excised with a razor blade and vortexed in the presence of a mixture of three different sizes of glass beads (0.5 mm, 1 mm and 5 mm in diameter, Sigma) in the lysis buffer A (above) until all the tissue was visibly lysed. The lysate was clarified at 15,000 × g at 4°C for 10 min, and quantified by BCA (Thermo Scientific). Brain protein (40 mg) was digested with Lys-C (1:200, 21°C, 30 min), diluted to 2 M urea and digested by trypsin (200:1 ratio at 37°C overnight). The peptide sample was acidified, desalted with Sep-Pak C18 (Millipore), and eluted by 40% acetonitrile (ACN) plus 0.1% TFA. While half of the eluent was dried for direct K-GG antibody enrichment, the other half were dried and resuspended in SCX binding buffer (5 mM KH2PO4, pH 3, 25% ACN), loaded onto an SCX column (250mm×94 mm, polyLC), and eluted with a gradient from 18 to 38% of SCX elution buffer (5 mM KH2PO4, pH 3, 1 M KCl, 25% ACN) over 40 min at a flow rate of 1.5 ml/min. Peptide eluents were collected every minute and the solution charge state was determined by analyzing a fraction (10 µl) of each fraction. Based on the charge state analysis, fractions 5–15, 23–29, and 30–55 were pooled together, and the remaining fractions were analyzed separately.
Enrichment of K-GG peptides by immunoaffinity purification
The enrichment was performed largely based on the manufacturer's protocol with some modifications. The SCX fractions were desalted, dried and dissolved in IAP buffer (50 mM MOPS/NaOH, pH 7.2, 10 mM Na2HPO4, 50 mM NaCl). The K-GG antibody beads (2 µg Ab per mg starting protein, 1 µg Ab per µl beads, Cell Signaling Technology) were incubated with the peptide solution at 4°C with gentle rotation for 30 min. The beads were then washed at 4°C with the IAP buffer plus 0.15% sodium deoxycholate three times, followed by 5 mM ammonium bicarbonate wash once. Gentle centrifugation (1500 × g for 15 sec) was used to separate beads from the solution. Captured peptides were eluted by 0.15% TFA at 21°C for 5 min, desalted with Zip-Tip C18 (Millipore) for LC-MS/MS analysis. In some cases, the unbound peptide sample was subjected to a second round of IAP and subjected to analysis as indicated.
Mass spectrometry analysis
The peptide samples were analyzed either on LTQ-Velos Orbitrap (Thermo Scientific) coupled with Nano Acquity UPLC (Waters) or on Q-Exactive (Thermo Scientific) coupled with Easy-nLC 1000 (Thermo Scientific), and separated on a C18 reversed phase column (100 mm, 75 µm ID, 2.7 µm HALO beads, Michrom Biosources; buffer A: 0.1% formic acid; buffer B: 0.1% formic acid plus 70% AcN; 10–40% gradient in ~1 h for SCX samples or ~3 h for directly enriched K-GG peptide samples; 0.3 µl/min flow rate). The LTQ-Velos Orbitrap settings included one survey scan (60,000 resolution in Orbitrap, 106 automatic gain control (AGC), and 500 ms maximal ion time), and top 20 low resolution MS/MS scans (5000 AGC, 250 ms maximal ion time, 3 m/z isolation window, default collision-induced dissociation, and 15 sec dynamic exclusion). The Q-Exactive program was one survey scan (70,000 resolution, 106 AGC, 30 ms maximal ion time), followed by top 10 MS/MS scans (17,500 resolution, 105 AGC, 500 ms maximal ion time, 3 m/z isolation window, default higher-energy C-trap dissociation (HCD), and 15 sec dynamic exclusion). Charge state screening was enabled to exclude precursor ions with single charge or unassigned charge state.
During the quantification of Ub, Nedd8 and ISG15, a targeted MS/MS method on Q-Exactive was used to assess protein abundance based on multiple peptides, similar to a previous analysis41. HCD collision energy was optimized using authentic standards. Heavy and light forms of each target peptide were multiplexed (n = 2) with an isolation window of 0.4 m/z and maximal ion time of 1000 ms. The relative isotopic abundance of heavy peptides to light standards was determined by measuring co-eluted peak area of the matching peptides (10 ppm tolerance) in high resolution (70,000) survey scans, and confirmed by the MS/MS spectra with known standard spectra.
Data processing and analysis
Acquired MS/MS raw files were converted into mzXML format and searched by Sequest algorithm (version 28 revision 13) against a composite target/decoy database to estimate false discovery rate (FDR)42,43. The target protein database was generated by merging the NCBI reference and Uniprot rat databases (48,255 protein entries). The decoy protein database was generated by reversing all target protein sequences. Spectra were searched with ±5 ppm for precursor ion mass tolerance, ±0.02 (Q-Exactive data) or ±0.5 (LTQ-Velos data) for product ion mass tolerance, fully tryptic restriction, dynamic mass shift for oxidized Met (+15.9949) and GG-tagged Lys (+114.0429), three maximal missed cleavages, and four maximal modification sites. Only b and y ions were considered during the search. For combined modification analysis, additional dynamic mass shifts were +42.0106 on N-terminus for N-terminal acetylation, −89.0299 on N-terminus for N-terminal acetylation after Met removal, +0.9840 on Gln/Asn with partial tryptic restriction for deamidation, or +79.9663 on Ser/Thr/Tyr for phosphorylation.
Assigned peptide spectra matches were first filtered by MS mass accuracy (±4 standard deviations, ~±1.4 ppm on Q-Exactive, which was determined by all empirical good matches of doubly charged peptides with Xcorr at least 2.5). These good matches were also used for global mass re-calibration prior to the filtering. The survived matches were grouped by precursor ion charge state and further filtered by Xcorr and ΔCn value. The cutoff values for XCorr and ΔCn were adjusted until a peptide FDR lower than 1% was achieved. In addition, all spectra of GG peptides with multiple modifications were manually validated. If one GG peptide was matched to multiple proteins, the peptide was represented by the protein with the highest spectral counts according to the rule of parsimony.
Quantification of Ub, Nedd8 and ISG15 in metabolically labeled mouse brain
Mouse brain was labeled by stable isotope (K +6.0201 Da) using the metabolic labeling strategy in three generations41,44. Brain tissue was lysed as above and quantified by BCA (Thermo Scientific) and Coomassie staining on a short SDS gel45. Recombinant purified Ub, Nedd8 and ISG15 (Boston Biochem) were subjected to amino acid analysis (AAA Service Laboratory) to determine exact protein concentration. The free (<12 kDa) and conjugated forms (>12 KDa) of brain proteins were separated by loading onto a 9% Tris-glycine SDS gel. The standard proteins were mixed, run on a short SDS gel, excised and mixed with the gel bands corresponding to the Ub/Nedd8 free and conjugated forms, respectively. The samples were in-gel trypsin digested (20 ng/µl trypsin, 37°C overnight), and analyzed by Q-Exactive MS (Thermo Scientific). The targeted data analysis was performed with Xcalibur software (Thermo Scientific).
RESULTS
Neither M1-GG peptides nor iodoacetamide pseudo-GG peptides bind to the K-GG antibody
Among the two K-GG monoclonal Abs commercially available, the Ab from Lucerna was generated against GG-modified histone and was shown to distinguish K-GG peptides from M1-GG peptides (linear peptide modified on the N-terminal amine group)37, the other from Cell Signaling Technology was produced against the sequence CXXXXXXKGGXXXXXX (X = any amino acid except Cys, Trp) but is not fully characterized38. As the latter Ab was made against more randomized antigens, we decided to use the Ab for our analysis. In addition, iodoacetamide (IAA), a common Cys alkylation reagent to inhibit DUB activity during purification, may react with Lys residue under high temperature to derive a mass-indistinguishable, pseudo-GG tag40 (Fig. 1A). Therefore, we compared the binding of various peptides to this Ab (Fig. 1B, 1C). A mixture of comparable level of four types of peptides (M1-GG, pseudo-GG, Ub K48 GG, and Ub K48 without GG tag) was incubated with the Ab beads, followed by MS analysis of the flow-through and eluent samples. Whereas the majority of Ub K48 GG peptide was bound to the beads, all other peptides showed trace amount of binding (less than 0.1%), indicating that the Ab has high specificity only to K-GG peptides.
Fig. 1. Specificity of the K-GG antibody.
(A). Lys residues modified by GG tag or IAA (two IAA molecules alkylating the amine group).
(B). The detection of K48-GG (LIFAGK(GG)QLEDGR), K48 pseudo-GG, and another negative control peptide (K48 without GG, LIFAGKQLEDGR). These peptides were produced by either IAA reaction or chemical synthesis, and mixed as input for the Ab affinity purification. The equal percentage (3%) of the flow through and the eluent was run by LC-MS/MS for quantitative comparison. The signal of each peptide was extracted from ion current and plotted after normalization according to the strongest peak. Retention times are also indicated.
(C). The analysis of M1-GG (GGMQIFVK) and other pseudo-GG in ubiquitin in the same samples. The K33 pseudo-GG peptide was eluted in two peaks that may be due to different configuration by proline isomerization.
Removal of false positive K-GG peptides by the target-decoy strategy
Although the target/decoy strategy43 is a widely used method to filter false positive peptides, its use to control false discovery rate (FDR) for GG peptides has not been validated and some concerns has been raised, because the addition of dynamic modification dramatically increases the database search space46. Thus, we used E. coli total lysate as a negative control to evaluate if the target/decoy strategy is suitable for data filtering, since the prokaryotic cells contain no Ub and the digested lysate has no GG peptides. All potential peptide matches were grouped by charge state, and then filtered stepwise by mass accuracy and matching scores (Xcorr and ΔCn) (Fig. 2A). As expected, in the dataset before filtering, we observed equal numbers of GG peptides matched to the target and decoy databases (n = 562), while more total peptides were matched to the target than the decoy databases, suggesting that about 3953 (n = 5613 − 1660) peptides were true positive non-GG peptides (Fig. 2B).
Fig. 2. Filtering of false positive GG peptides by the target-decoy method.
(A). Multiple steps to filter false positive GG matches during data processing.
(B). The total and GG peptides matched to the target-decoy database before filtering. E. coli peptide mixture containing no GG peptides was used in the analysis.
(C). The effect of mass accuracy cutoffs on the acceptance of various peptides. The LC-MS/MS systematic mass deviation was evaluated by standard deviation (SD) of empirical good peptide matches. The false discovery rate at 4 SD is 25.2%.
(D). The peptides matched to the target-decoy database after mass accuracy filtering at 4 SD.
(E). Distribution of XCorr and ΔCn values of either target or decoy GG peptides after mass accuracy filtering.
(F). The peptides after final XCorr and ΔCn based filtering, resulting in ~1% peptide FDR.
(G). The LC-MS/MS runs of an E. coli sample and a rat K-GG fraction were merged into one raw dataset and filtered by mass accuracy. The distribution of target and decoy GG peptides was shown.
(H). The accepted total and GG peptides in the merged dataset with ~1% peptide FDR.
We then tested the effect of precursor mass tolerance on FDR and accepted peptide numbers (Fig. 2C). With increased stringency for mass tolerance, the FDR was decreased but the number of accepted peptides was lower. We found that 1.4 ppm mass tolerance showed a good compromise between minimizing FDR and maximizing true positives. After filtering, more than half of the false positive GG peptides were removed. The remaining target and decoy GG peptides still showed almost equal numbers (209 versus 216) (Fig. 2D), and similar score distributions (Fig. 2E), suggesting that the mass accuracy filter showed no bias in removing false GG peptides in either the target or decoy databases. Moreover, we manually accepted highly reliable peptides (doubly charged peptides, XCorr at least 2.5)42 to analyze mass error of the LC-MS/MS platform. All data points were fitted to a Gaussian distribution with a standard deviation of ~0.35 ppm. Therefore, our selected mass tolerance was about 4 times the standard deviation, which theoretically included > 99.9% of true positives. Indeed, the 1.4 ppm threshold accepted 3863 (n = 4515 − 652) potential true matches (Fig. 2D) with a recovery rate of approximately 97.7% (3863/3953). Finally, the peptide matches were further filtered by dynamically adjusting XCorr and ΔCn until peptide FDR reached ~1%. While the vast majority of GG peptides were removed, leaving only 7 target and 5 decoy GG peptides (Fig. 2F), the true positive number was estimated to be 3621 (n = 3636 − 15), indicating a recovery rate of 91.6% (3621/3953).
To further examine if the target-decoy strategy has a systematic bias in analyzing null experiment data (i.e. from E. coli) versus GG peptide containing samples, we merged raw data from a E. coli sample and a rat GG peptide enriched fraction, and filtered the data by mass accuracy. It was clear that the target GG peptides showed a different score distribution from the decoy GG peptides (Fig. 2G), indicating the presence of true positives in this mixed dataset. After XCorr and ΔCn filtering to ~1% peptide FDR, the decoy GG peptides from E. coli or Rat samples were almost eliminated (with only one remaining, Fig. 2H). Overall, these testing experiments demonstrated the effectiveness of the target-decoy strategy in data processing for GG peptides.
Analysis of rat brain ubiquitome with or without implementation of SCX fractionation
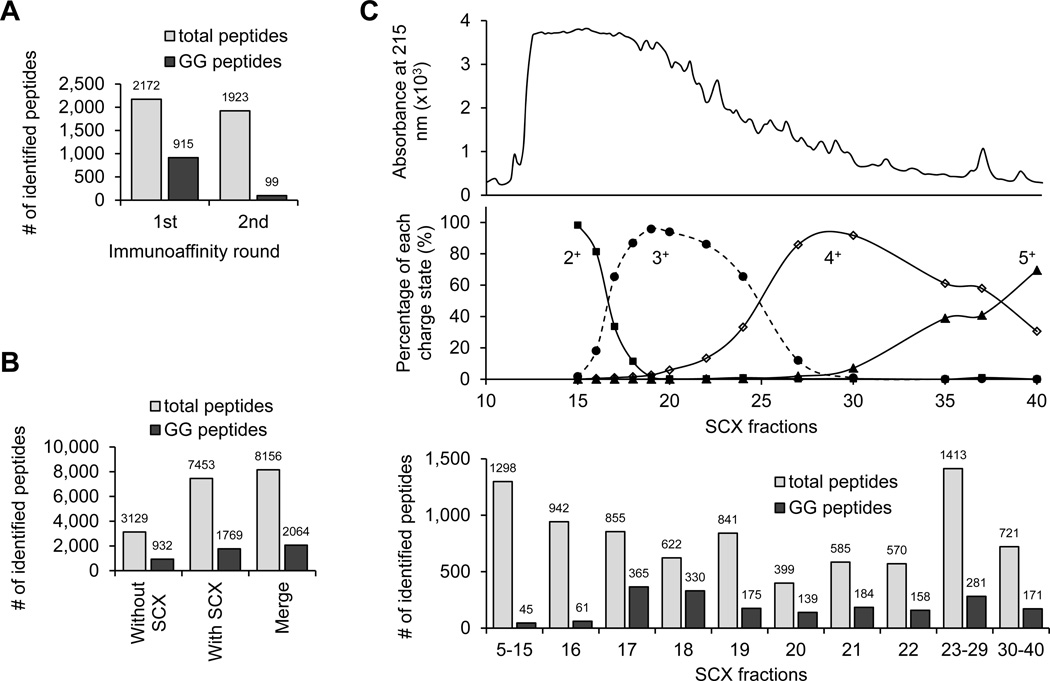
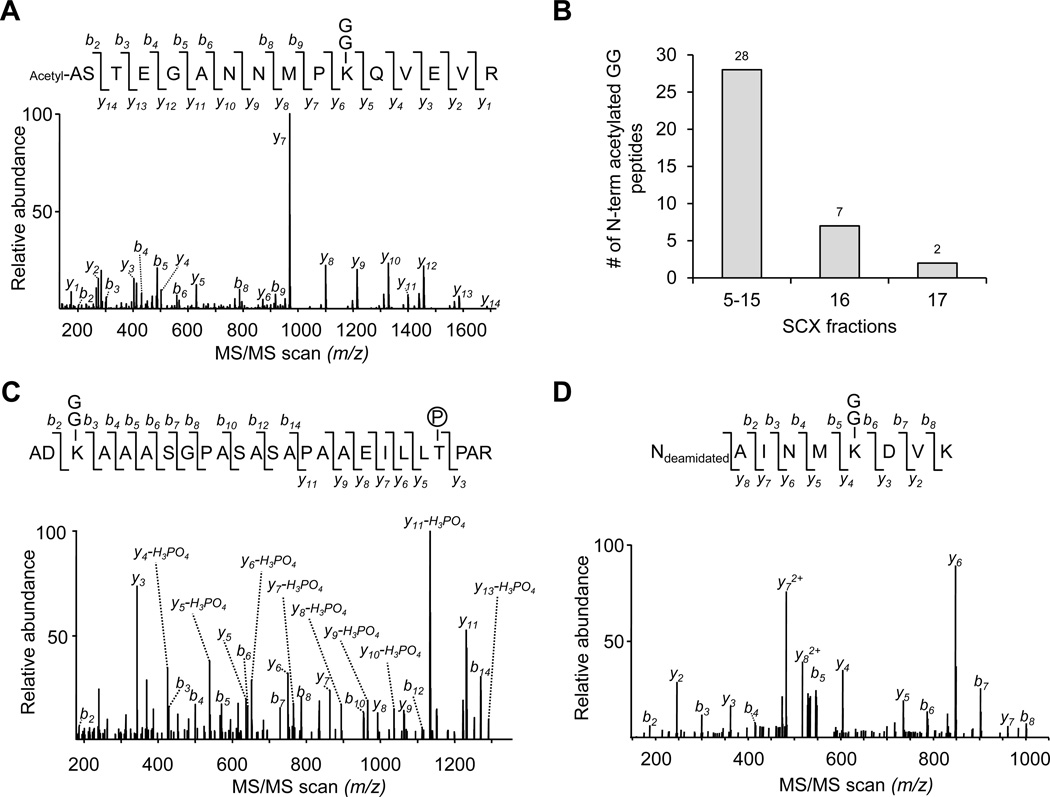
Since SCX fractionation is able to decrease the complexity of complex peptide samples and is commonly used for complex protein mixture analysis, we attempted to implement SCX fractionation prior to the Ab enrichment (Fig. 3A). In addition, most tryptic peptides are doubly charged, but typical GG peptides are at least triply charged in SCX buffer, so the SCX column may allow for enrichment of GG peptides (Fig. 3B). Compared with the previously reported method, in which GG peptides were isolated directly from digested total cell lysate by multiple cycles of affinity pulldown38, we digested 40 mg of rat cerebral cortex protein and equally split the sample for testing both approaches. During the multiple cycles of Ab enrichment analysis, 915 GG peptides were identified out of 2172 total peptides in the 1st cycle, and only 99 GG peptides were identified out of 1923 total peptides in the 2nd cycle, indicating that the majority of GG peptides were recovered in the first round (Fig. 4A). In the SCX-Ab enrichment study, a total of 1769 GG peptides were identified out of 7453 total peptides. Pooling the data led to identification of 2064 GG peptides (1786 GG sites in 921 proteins, supplemental Table S1) with 637 GG peptides shared by both strategies (Fig. 4B). When examining the peptides identified in every SCX fraction, most of GG peptides were found in the fraction containing at least triply charged peptides (fraction 17 and above, Fig. 4C). In addition, we did observe enrichment of GG peptides in several SCX fractions (e.g. 6-fold more enrichment in fraction 17 than fraction 16). However, some additional modifications (e.g. N-terminal acetylation before or after first Met removal, and phosphorylation) decrease the solution charge state of GG peptides and should be detected in early SCX fractions (Fig. 3B). Indeed, we performed dual modification search and found 37 N-terminal acetylated GG peptides, largely in the fractions with doubly charged peptides (fraction 5–15, Fig. 5A and 5B). Only one phosphorylated GG peptide was identified and was matched to integral membrane protein 2C (Fig. 5C). The reliability of this modification was supported by the detection of the same GG modification site (Lys20 residue) in a corresponding unphosphorylated counterpart (Table S1). Moreover, according to N-end rule for protein degradation47, some proteins are deaminated at the N-terminus during enzymatic modification. In our analysis, we identified three deaminated GG peptides (Fig. 5D). In summary, both the approaches with or without SCX enabled the identification of a large number of GG peptides in the brain tissue, and SCX fractionation improved GG peptide identification by approximately 2-fold.
Fig. 3. Strategies for GG peptide enrichment with or without SCX fractionation.
(A). The scheme of the strategies.
(B). The distribution of possible GG peptides in SCX fractions based on solution charge states. Under low pH condition (pH =3), three residues (H. K and R) and N-terminal amine groups are positive charged, while phosphorylation introduces a negative charged group, and acetylation neutralizes the amine groups.
Fig. 4. Rat cortical GG peptides were analyzed by immunoaffinity purification with or without SCX fractionation.
(A). The identified total and GG peptides by multiple cycle of Ab enrichment.
(B). The identified total and GG peptides by SCX chromatography and Ab enrichment. The data was pooled for the final dataset.
(C). A small portion of every SCX fraction (with shown elution profile) was analyzed to determine solution charge distribution, and the remaining sample was subjected to Ab pulldown and LC-MS/MS analysis.
Fig. 5. Identification of GG peptides with additional modifications.
(A). An MS/MS scan assigned to an acetylated GG peptide.
(B). The SCX fractions containing identified N-terminal acetylated GG peptides.
(C). An MS/MS scan assigned to a phosphorylated GG peptide.
(D) An MS/MS scan assigned to a deamidated GG peptide.
Analysis of GG peptides modified on C-terminal Lys residues
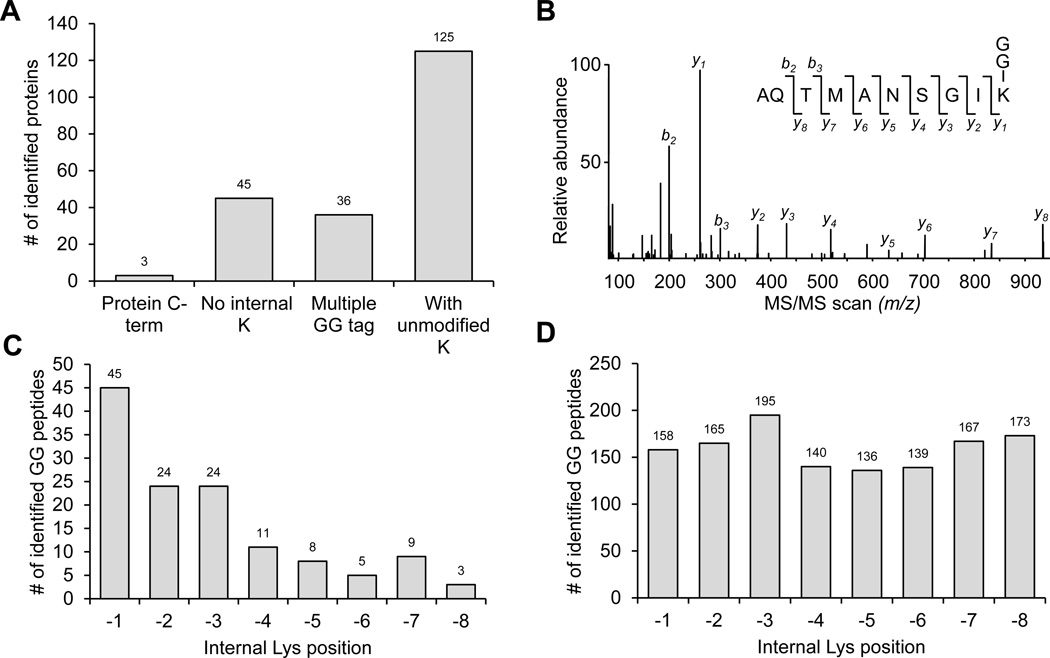
As GG modification of lysine residues leads to trypsin miscleavage5,48, GG peptides with C-terminal Lys modification (C-term GG peptides) derived from database search are considered as false positives38. In this large scale analysis, we sought to examine these 209 C-term GG peptides in detail and classified these peptides into different categories (Fig. 6A). First, 3 peptides were directly derived from proteins that end with Lys at C-termini, which showed well-matched product ions during manual interpretation and were considered to be true positives (Fig. 6B). Then, we tested whether an internal K-GG modification was misassigned to a C-terminal Lys residue by database searching algorithms. 45 GG peptides without internal Lys were removed, and 36 peptides containing multiple GG tags were considered as false positives, as we noticed that the frequency of multiple GG modification is rare (0.4%, 9/2064) in our final dataset. Finally, we found that in the remaining 125 C-term GG peptides, unmodified internal Lys residues were positioned in the C-terminal region with much higher frequency (Fig. 6C). For example, 45, 24, and 24 peptides displayed a Lys residue right next to the C-terminus on −1, −2, and −3 positions, respectively, whereas only 7±3 peptides had Lys on other positions (−4 to −8). This C-terminal bias of internal lysine distribution was not observed in our large GG peptide dataset (Fig. 6D). This result strongly suggested that such GG sites were misassigned to C-terminal Lys instead of the adjacent Lys at −1, −2 or −3 position. Therefore we manually repositioned the GG sites for the 92 (n = 45+24+24) peptides. In addition, we noticed that 67% of the repositioned GG peptides were already identified by other MS/MS scans in the dataset, providing further evidence for this correction of misassigned K-GG sites.
Fig. 6. The evaluation of C-terminal modified GG peptides identified in the brain proteome.
(A). The classification of these C-term GG peptides.
(B). An MS/MS scan assigned to a GG peptide derived from the C-terminus of a protein.
(C). The internal Lys residue distribution in identified C-term GG peptides.
(D) The internal Lys residue distribution in the accepted GG peptide dataset (Table S1)
The vast majority of GG peptides were produced from Ub modification
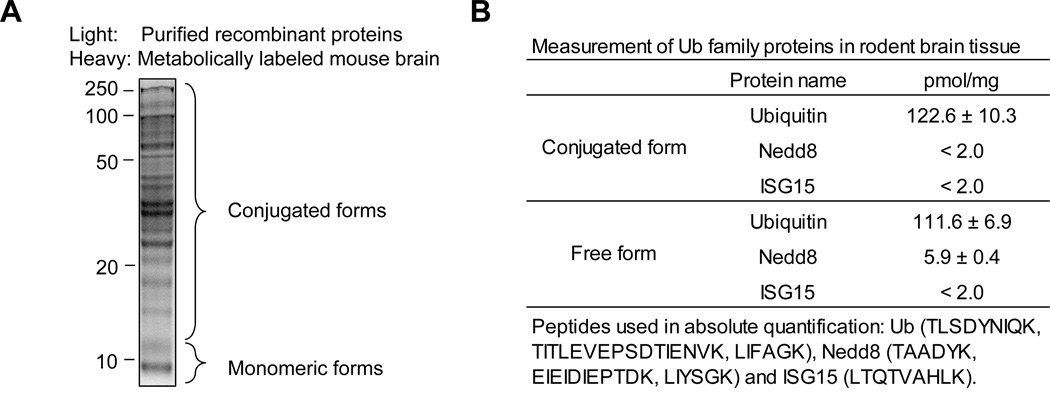
The K-GG Ab enrichment analysis may be confounded because GG tags can be generated from Ub, Nedd8 or ISG15 modification. Therefore, we quantified the level of free and conjugated forms of the three proteins in rodent brain to evaluate their contribution to the pool of GG peptides by a targeted LC-MS approach. Highly purified recombinant proteins were used as internal standard to measure the level of these Ub family proteins in metabolically labeled mouse brain lysate (Fig. 7A). Based on the analysis of multiple peptides, we detected 111.6±6.9 picomoles of free Ub and 122.6±10.3 picomoles of conjugated Ub per mg total protein (Fig. 7B). Although we also detected 5.9±0.4 picomoles/mg of free Nedd8, the levels of conjugated Nedd8 or ISG15 were below the detection limit (~2.0 picomoles/mg), consistent with previous estimations38,49. Therefore, the level of conjugated Nedd8 or ISG15 in the brain was at least 60-fold lower than that of ubiquitin, indicating that ubiquitination is credited for the vast majority of GG peptides.
Fig. 7. The measurement of Ub, Nedd8 and ISG15 in metabolically labeled mouse brain.
(A). Light recombinant proteins were mixed with monomeric (less than 12 KDa) or conjugated (>12 KDa) metabolically labeled mouse cerebral cortex (heavy Lys, +6.0201 Da). These mixed proteins were digested with trypsin and analyzed on LC-MS to quantitate Ub and Nedd8 levels.
(B). The quantified protein level listed.
Comparison the rat brain analysis with previously reported large datasets of ubquitome
We compared our rat brain dataset (2,064 GG peptides) with three recent reported large datasets from human cell lines (totaling 27,318 GG peptides)38,39,50. Only 30.5% (631/2,064) of the brain peptides were overlapped with the reported studies, and 14.9% (308/2,064) of the rat peptides were homologous to the human counterparts (supplemental Table S2), showing conservation of ubiqutination sites in these two species. Importantly, more than half of our dataset (54.5%, 1,125/2,064) are novel, revealing ubiquitination events that may be specific to brain tissue.
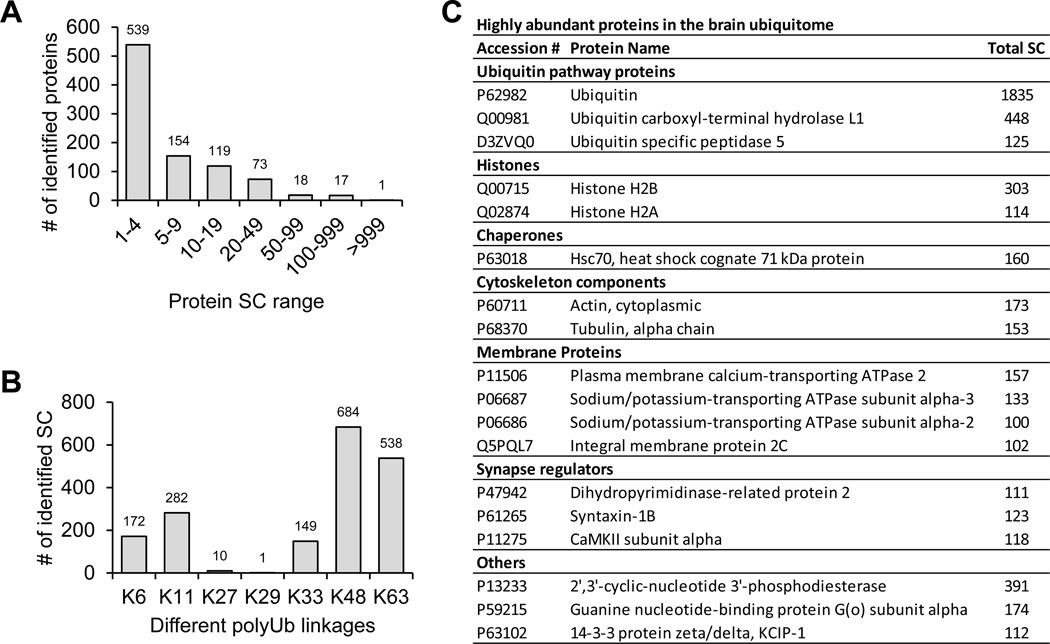
The brain ubiquitome is dominated by a small group of highly modified Ub-conjugates
We analyzed the abundance of individual proteins in the ubiquitome by spectral counting, the total number of MS/MS spectra matched a designated protein, which is commonly used as a semi-quantitative index during MS analysis51. While 539 (59%) out of 921 identified proteins had only 1–4 spectra counts, 18 (2%) proteins had spectra counts of more than 100, suggesting that the ubiquitome displayed a large dynamic range in protein concentration (Fig. 8A, 8B). The levels of seven polyUb linkages were also compared and ranked in a descending order of K48, K63, K11, K6, K33, K27 and K29 (Fig. 8C). This result is in good agreement with our previous measurement in mammalian cells based on stable isotope labeled GG peptides41, except that the K29 GG peptide was greatly underrepresented. The discrepancy in the K29 linkage levels was due to the loss of this hydrophilic K29 GG peptide (AK(GG)IQDK) during desalting and LC runs.
Fig. 8. The abundance of modified proteins in the Ub/Nedd8-modified proteome.
(A). Distribution of GG tagged proteins based on total spectral counts.
(B) The list of ubiquitinated proteins with spectral count of at least 100.
(C) Relative level of polyUb linkages in the brain.
Analysis of synaptic proteins and Ub/Nedd8 pathway proteins in the dataset
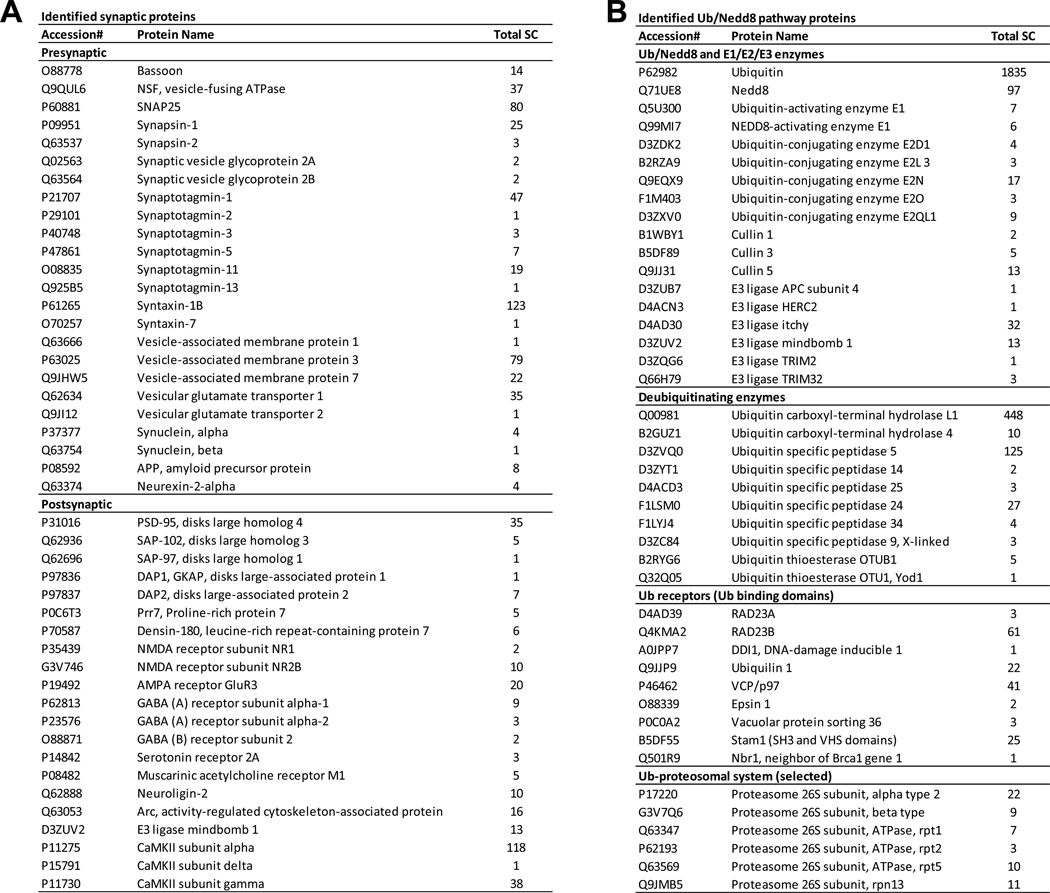
To evaluate the depth of this ubiquitome analysis, it is important to examine if we were able to identify previously known key ubiquitin substrates in the brain20–23. We identified 45 synaptic substrates (Fig. 9A), including 24 presynaptic proteins (e.g. Basson, NSF, SNAP25, Synapsin, Sv2, Synaptotagmin, Syntaxin, VAMP, and synuclein) and 23 postsynaptic components (e.g. PSD-95, GKAP, CaMKII, and receptors for NMDA, AMPA, GABA, serotonin, and acetylcholine). The consequence of these ubiquitination events may be linked to proteasomal degradation and membrane sorting. For instance, ubiquitination of AMPA receptor is proposed to regulate its degradation and trafficking52,53. In addition, as active Ub pathway proteins are often self-modified by ubiquitination, we collected these Ub pathway proteins in our dataset, including Ub/Nedd8 and their corresponding E1s, 5 E2s, 9 E3s, 10 DUBs, 9 Ub-binding proteins and multiple proteasome subunits (Fig. 9B),. Although this list is small compared to all potential Ub pathway proteins (up to ~1,000) in the predicted proteome, it overlapped significantly with known Ub enzymes functioning in the neuronal system (e.g. APC, Mindbomb, TRIM, FAM, UCH-L1, and USP14, see the introduction), suggesting these detected proteins are highly active in the brain tissue. A number of proteins related to neurodegenerative diseases were also identified, such as APP, alpha-synuclein, UCH-L, USP14, and VCP/p9723. However, the role of most of the discovered Ub events is not known. Thus this proteomics analysis provides important targets for future investigation.
Fig. 9. Identified proteins involved in synaptic function or Ub/Nedd8 modifications.
(A). Synaptic proteins classified by the location.
(B) Ub/Nedd8 pathway proteins classified by the function.
DISCUSSION
Prior to the development of K-GG monoclonal antibodies, ubiquitome studies had been solely based on affinity purification of Ub-conjugates, leading to global identification of as many as 5756 putative Ub substrates in mammalian cells36. The validation of these substrates relies on individual immunoblotting, large-scale virtual western blotting48, identification of GG modification sites5., and quantitative comparison upon perturbation of Ub system6,32. The analysis of GG modification sites is, however, hampered by the “undersampling” issue of the LC-MS/MS system when K-GG peptides compete with much higher levels of non-GG peptides in the same sample for ionization and selection for MS/MS. The development of the K-GG monoclonal Abs largely alleviates this problem and permits selective enrichment of K-GG peptides37–39, increasing the identified GG sites up to 19,000. In one single analysis with four rounds of immunoisolation, the Gygi group identified ~4000 GG peptides from a cell line treated with proteasome inhibitor38. Using similar reagents, procedures and instrument settings, we identified 932 GG peptides from the brain sample. The difference may be largely due to the ubiquitome abundance variation in a treated cell line versus the brain, as the bulk of GG peptides were isolated during the first round of our experiment. Indeed, previous reports indicated that fast growing cells have a much higher level of ubiquitination than the cells in the brain tissue41,54.
Comparing the direct Ab enrichment with the SCX-Ab method, the latter method provides only ~2-fold increase in GG peptide identification. The effectiveness of SCX implementation appears to be lower than that for the analysis of total proteome or phosphoproteome. This may be also affected by the low abundance of ubiquitome and sample loss during fractionation. During the preparation of this manuscript, the Carr group also performed similar comparison and found 3–4 fold improvement with a minimal SCX separation (i.e. 4 fractions), consistent with our observation50. To fully realize the benefits of SCX chromatography, more starting materials and/or higher sensitive LC-MS/MS methods will be needed.
In summary, we showed that the K-GG Ab enrichment method differentiated K-GG peptides from linear GG peptides or pseudo-GG peptides derived from iodoacetamide, and we addressed several important issues for GG peptide analysis, such as the reliability of the target-decoy strategy, and the analysis of C-terminal K-GG peptides. The SCX-Ab method can separate peptides by their charge state and lead to improved GG peptide identification. More importantly, we showed that the current method is sensitive and able to detect these critical regulatory components during brain development and function, demonstrating the feasibility of this approach for the tissue analysis. Combining this method with quantitative approaches will allow the analysis of the ubiquitome dynamics in the brain.
Supplementary Material
ACKNOWLEDGEMENTS
This work was partially supported by National Institutes of Health grant RR025822 and American Cancer Society grant RSG-09-181, and ALSAC (American Lebanese Syrian Associated Charities). We thank Dr. John Rush for reagents, Drs. Woong Kim and Steven Gygi for discussion and data analysis.
ABBREVIATIONS
- LC-MS/MS
liquid chromatography with tandem mass spectrometry
REFERENCES
- 1.Hershko A, Ciechanover A. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Varshavsky A. Trends Biochem. Sci. 2005;30:283–286. doi: 10.1016/j.tibs.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Ciechanover A, Ben-Saadon R. Trends Cell. Biol. 2004;14:103–106. doi: 10.1016/j.tcb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K. EMBO J. 2006;25:4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. Nat. Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 6.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cadwell K, Coscoy L. Science (New York, N.Y. 2005;309:127–130. doi: 10.1126/science.1110340. [DOI] [PubMed] [Google Scholar]
- 8.Shimizu Y, Okuda-Shimizu Y, Hendershot LM. Mol. Cell. 2010;40:917–926. doi: 10.1016/j.molcel.2010.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Herr RA, Hansen TH. Traffic. 2012;13:19–24. doi: 10.1111/j.1600-0854.2011.01269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Annu. Rev. Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weake VM, Workman JL. Mol. Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 12.Harper JW, Elledge SJ. Mol. Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 13.Mukhopadhyay D, Riezman H. Science (New York, N.Y. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 14.Kirkin V, McEwan DG, Novak I, Dikic I. Mol. Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 15.Jiang X, Chen ZJ. Nat. Rev. Immunol. 2012;12:35–48. doi: 10.1038/nri3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Semple CA. Genome Res. 2003;13:1389–1394. doi: 10.1101/gr.980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, Chanda SK, Batalov S, Joazeiro CA. PLoS ONE. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hochstrasser M. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darwin KH. Nat. Rev. Microbiol. 2009;7:485–491. doi: 10.1038/nrmicro2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hegde AN, DiAntonio A. Nat. Rev. Neurosci. 2002;3:854–861. doi: 10.1038/nrn961. [DOI] [PubMed] [Google Scholar]
- 21.Tai HC, Schuman EM. Nat. Rev. Neurosci. 2008;9:826–838. doi: 10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]
- 22.Bingol B, Sheng M. Neuron. 2011;69:22–32. doi: 10.1016/j.neuron.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Mabb AM, Ehlers MD. Annu. Rev. Cell Dev. Biol. 2010;26:179–210. doi: 10.1146/annurev-cellbio-100109-104129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, Jiang H, Hirano M, Rampersaud E, Jansen GH, Donkervoort S, Bigio EH, Brooks BR, Ajroud K, Sufit RL, Haines JL, Mugnaini E, Pericak-Vance MA, Siddique T. Nature. 2011;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciechanover A, Brundin P. Neuron. 2003;40:427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 26.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 27.Peng J, Gygi SP. J. Mass Spectrom. 2001;36:1083–1091. doi: 10.1002/jms.229. [DOI] [PubMed] [Google Scholar]
- 28.Marotti LA, Jr, Newitt R, Wang Y, Aebersold R, Dohlman HG. Biochemistry. 2002;41:5067–5074. doi: 10.1021/bi015940q. [DOI] [PubMed] [Google Scholar]
- 29.Kirkpatrick DS, Denison C, Gygi SP. Nat. Cell Biol. 2005;7:750–757. doi: 10.1038/ncb0805-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu P, Peng J. Biochimica et biophysica acta. 2006;1764:1940–1947. doi: 10.1016/j.bbapap.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeram SM, Srikumar T, Pedrioli PG, Raught B. Proteomics. 2009;9:922–934. doi: 10.1002/pmic.200800666. [DOI] [PubMed] [Google Scholar]
- 32.Meierhofer D, Wang X, Huang L, Kaiser P. J. Proteome Res. 2008;7:4566–4576. doi: 10.1021/pr800468j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franco M, Seyfried NT, Brand AH, Peng J, Mayor U. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.002188. M110 002188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Starita LM, Lo RS, Eng JK, von Haller PD, Fields S. Proteomics. 2012;12:236–240. doi: 10.1002/pmic.201100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi Y, Chan DW, Jung SY, Malovannaya A, Wang Y, Qin J. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.002089. M110 002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Danielsen JM, Sylvestersen KB, Bekker-Jensen S, Szklarczyk D, Poulsen JW, Horn H, Jensen LJ, Mailand N, Nielsen ML. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.003590. M110 003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu G, Paige JS, Jaffrey SR. Nat. Biotechnol. 2010;28:868–873. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M111.013284. M111 013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen ML, Vermeulen M, Bonaldi T, Cox J, Moroder L, Mann M. Nat. Methods. 2008;5:459–460. doi: 10.1038/nmeth0608-459. [DOI] [PubMed] [Google Scholar]
- 41.Dammer EB, Na CH, Xu P, Seyfried NT, Duong DM, Cheng D, Gearing M, Rees H, Lah JJ, Levey AI, Rush J, Peng J. J. Biol. Chem. 2011;286:10457–10465. doi: 10.1074/jbc.M110.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. J. Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 43.Elias JE, Gygi SP. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 44.Kruger M, Moser M, Ussar S, Thievessen I, Luber CA, Forner F, Schmidt S, Zanivan S, Fassler R, Mann M. Cell. 2008;134:353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 45.Xu P, Duong DM, Peng J. J. Proteome Res. 2009;8:3944–3950. doi: 10.1021/pr900251d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi Y, Xu P, Qin J. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.R110.006882. R110 006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tasaki T, Sriram SM, Park KS, Kwon YT. Annu. Rev. Biochem. 2012;81:261–289. doi: 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seyfried NT, Xu P, Duong DM, Cheng D, Hanfelt J, Peng J. Anal. Chem. 2008;80:4161–4169. doi: 10.1021/ac702516a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skaug B, Chen ZJ. Cell. 2010;143:187–190. doi: 10.1016/j.cell.2010.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Udeshi ND, Mani DR, Eisenhaure T, Mertins P, Jaffe JD, Clauser KR, Hacohen N, Carr SA. Mol. Cell. Proteomics. 2012;11:148–159. doi: 10.1074/mcp.M111.016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu H, Sadygov RG, Yates JR., 3rd Anal. Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 52.Lin A, Hou Q, Jarzylo L, Amato S, Gilbert J, Shang F, Man HY. J. Neurochem. 2011;119:27–39. doi: 10.1111/j.1471-4159.2011.07221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fu AK, Hung KW, Fu WY, Shen C, Chen Y, Xia J, Lai KO, Ip NY. Nat. Neurosci. 2011;14:181–189. doi: 10.1038/nn.2715. [DOI] [PubMed] [Google Scholar]
- 54.Kaiser SE, Riley BE, Shaler TA, Trevino RS, Becker CH, Schulman H, Kopito RR. Nat. Methods. 2011;8:691–696. doi: 10.1038/nmeth.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.