Abstract
Two major difficulties encountered in implementing Magnetic Resonance Spectroscopic Imaging (MRSI) in a clinical setting are limited coverage and difficulty in prescription. The goal of this project was to completely automate the process of 3D PRESS MRSI prescription, including placement of the selection box, saturation bands and shim volume, while maximizing the coverage of the brain. The automated prescription technique included acquisition of an anatomical MRI image, optimization of the oblique selection box parameters, optimization of the placement of OVS saturation bands, and loading of the calculated parameters into a customized 3D MRSI pulse sequence. To validate the technique and compare its performance with existing protocols, 3D MRSI data were acquired from 6 exams from 3 healthy volunteers. To assess the performance of the automated 3D MRSI prescription for patients with brain tumors, the data were collected from 16 exams from 8 subjects with gliomas. This technique demonstrated robust coverage of the tumor, high consistency of prescription and very good data quality within the T2 lesion.
Keywords: 3D 1H MRSI, automated prescription, brain, outer volume suppression
Introduction
Proton Magnetic Resonance Spectroscopic Imaging (MRSI) is a valuable modality for diagnosis and evaluation of brain diseases. It allows the measurement of relative concentrations of brain metabolites and hence evaluation of functional behavior of the tissue. MRSI has been successfully used for tumor classification, evaluating the extent of infiltrative gliomas, evaluating tumor malignancy, planning radiation and surgical treatments and predicting response to therapy and disease progression (1). It has also been used in evaluating patients with neurodegenerative diseases such as multiple sclerosis (MS) and Alzheimer’s disease (2).
Two major difficulties with implementing MRSI in a clinical setting are limited coverage and difficulty in prescription. Clinical MRSI protocols typically keep the excited area (PRESS box) small to avoid exciting subcutaneous lipids and sinuses that could result in signal contamination in voxels far from the skull due to the “ringing” artifact and off-resonance artifacts due to magnetic susceptibility effects. Outer-volume suppression (OVS) with saturation bands, such as the previously developed Very Selective Suppression (VSS) bands (3) is typically used to suppress the signal from those areas.
In order to evaluate disease progression, the area covered by the MRSI data needs to match in subsequent exams from the same patient, which can be a challenge if the coverage volume is small and if there are differences in head position. Small coverage presents a problem when imaging multifocal and highly diffuse lesions, such as the ones commonly seen in patients receiving anti-angiogenic treatment. Improving coverage is also important for imaging patients with MS, since lesions often occur in multiple regions of the brain.
Several MRSI acquisition methods that did not use PRESS selection have been proposed to increase the brain coverage. All of them required additional steps to minimize the lipid contamination. Henning et al. (4) presented the spin-echo MRSI localized by OVS (SELOVS) technique that used two cycles of OVS (10 suppression pulses in each cycle) with numerically optimized flip angles to minimize the residual lipid signal in the regions where OVS bands crossed. The technique was applied to slice selected 2D MRSI at 3T and later to free-induction decay MRSI at 7T (5). Similarly, Posse et al. described their proton echo-planar spectroscopic imaging (PEPSI) sequence with a nine-band OVS module and 32x32 spatial matrix for slice-selected 2D MRSI within clinically acceptable acquisition times (6). Application of PEPSI to 3D MRSI (7) applied filtering of data in spatial domain to reduce the lipid ringing. Maudsley et al. used a combination of lipid inversion nulling, increased in-plane resolution (50x50) and iterative k-space extrapolation to reduce ringing artifacts from subcutaneous lipids (8).
All of these approaches have benefits and trade-offs that must considered in the context of the requirements of a clinical protocol. While effective in lipid suppression, inversion recovery negatively affects the SNR of other metabolites and suppresses the lipid peak within the lesion, which is a valuable biomarker of tumor malignancy (9). Spatial filtering can reduce ringing artifacts, but leads to loss of spatial resolution. Using a large phase encoding matrix results in longer acquisition times that may be unacceptable for a clinical exam. Over-prescribed PRESS selection together with effective OVS scheme can help avoid the major downsides of PRESS (chemical shift artifacts, rectangular shape), while still preventing the excitation of a large part of the subcutaneous lipids.
The delicate balance between coverage volume and quality of data is difficult to achieve for any operator. An oblique PRESS box should usually cover the tumor and as much of the healthy tissue as possible, while avoiding the skull and sinuses. A number of saturation bands require manual adjustment. The use of high-order shims (10) significantly improves the quality of the data, but usually requires an operator to outline a region of interest over which the shim would be optimized. Finally, the operator usually does not receive immediate feedback on the quality of prescription, since the data are often reconstructed offline. All of this makes acquiring 3D MRSI data a challenge in the clinical setting.
Earlier Ozhinsky et al. developed a technique for optimization of the OVS saturation band placement for 3D MRSI (11). While allowing the acquisition of data from a larger volume of interest, this technique still requires manual adjustment of the PRESS box size and position. The PRESS box was kept in the axial orientation, making it possible to cover only the regions of the brain above the level of the orbits. In their recent paper Martinez-Ramon et al. proposed using a manually prescribed para-axial slab with automatically placed saturation bands (12).
Automated prescription has been used in brain MRI previously to ensure the consistency of image orientation. Among the proposed approaches was calculation of mid-sagittal planes (13,14), surface matching (15), active shapes (13,16), aligning to earlier images from the same subject (17) and to an atlas(18). Yung et al. proposed transforming an atlas-based manual prescription of the selected slice and OVS bands for MRSI (7).
The goal of this project was to completely automate the process of 3D PRESS MRSI prescription, including placement of the selection box, saturation bands and shim volume, while maximizing the coverage of the brain. Unlike MR imaging prescriptions, where the whole brain can be easily covered, defining the MRSI prescription requires calculation of the optimal size and position of both the PRESS volume and OVS saturation bands, while taking into account the constraints imposed by the anatomy. This makes robust automated prescription crucial for acquiring good quality MRSI data from a large volume of the brain.
Methods
Overview
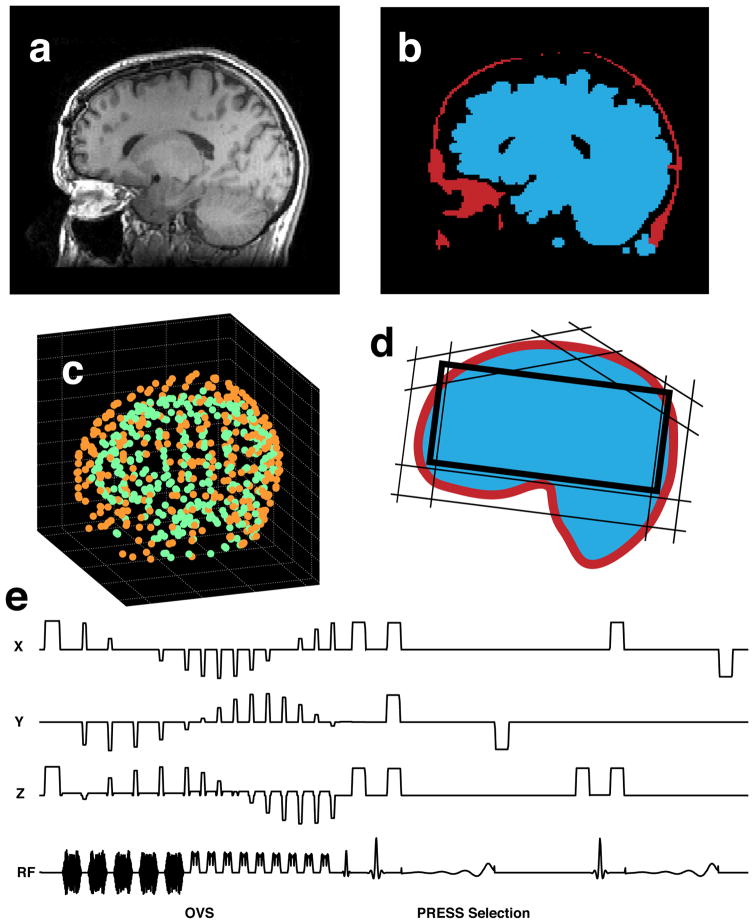
The automated prescription technique used in this study included acquisition of an anatomical MRI image (fig. 1a), optimization of the oblique PRESS box geometry, optimization of the placement of OVS saturation bands (fig. 1d, fig. 2), and loading the calculated parameters into a customized 3D MRSI pulse sequence (fig. 1e).
Fig. 1.
Technique overview: (a) anatomical MRI image, (b) segmented lipid and brain tissue masks, (c) sets of points, defining the brain and lipid surfaces (d) calculated OVS saturation band and PRESS box configuration, (e) saturation band and PRESS box parameters loaded into the MRSI pulse sequence.
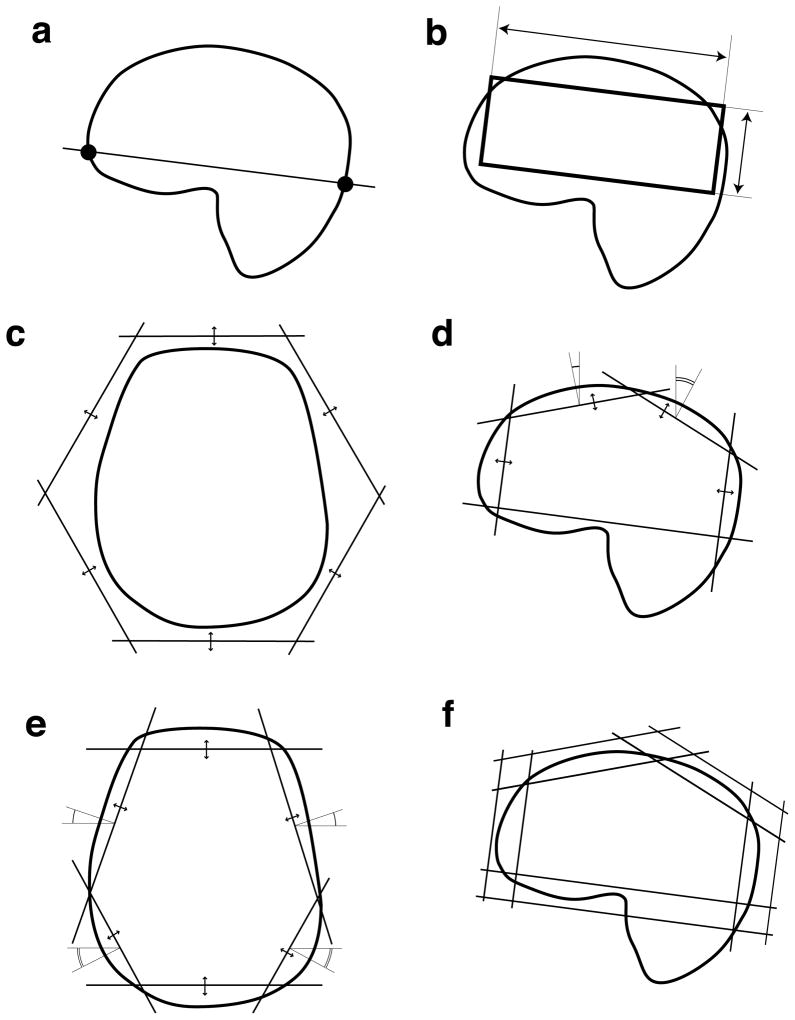
Fig. 2.
The stages of prescription optimization: (a) the lower oblique plane was defined using anatomical landmarks, (b) optimization of dimension of the PRESS box, (c) optimization of distances to individual saturation bands, (d) fine optimization of saturation bands in the sagittal plane, (e) fine optimization of saturation bands in the axial plane, (f) defining the inner and outer surfaces of the saturation bands.
To validate the technique and compare its performance with existing protocols, 3D MRSI data were acquired from 6 exams of 3 healthy volunteers. To assess the performance of the automated 3D MRSI prescription for patients with brain tumors, the data were collected from 16 exams of 8 subjects with gliomas.
Image Acquisition and Processing
A series of axial T1-weighted 3D IR-SPGR anatomic images without lipid suppression (fig. 1a) was acquired as part of the clinical imaging protocol (256x256x120 matrix, 240x240x180 mm FOV, 15 flip angle, TE = 2.5 ms, TR=7.8 ms) using a 3T MR scanner (GE Healthcare, Waukesha, WI). Image processing and automated prescription were performed by a custom software package, that was implemented in Matlab (Mathworks, Natick, MA), compiled into executable code and installed on the scanner console. The images were segmented into 3D lipid and brain tissue masks using the k-means clustering algorithm (19) with k=3 (fig. 1b). Morphological opening (brain mask) and closing (lipid mask) were performed to remove segmentation artifacts. The opening operation uses erosion, followed by dilation (20) to remove thin and small structures, while preserving the size of large objects. The closing operation (dilation followed by erosion) fills in the holes and gaps in the images. A number of inferior slices were removed by finding a cutoff level where the area of the brain mask was less than 10% of the total area of the image.
Two sets of 3D points, approximating the lipid and brain surfaces, (fig. 1c) were generated by casting rays onto the brain and lipid tissue masks from the superior, left, right, anterior and posterior directions with a step of 10 pixels. The coordinate system was defined as X -right-left, Y -anterior-posterior and Z -inferior-superior with the center of mass of the brain surface as the origin.
Optimization of PRESS Box Placement
An oblique plane was defined using anterior-inferior and posterior-inferior landmarks on the brain tissue mask (fig. 2a). The anterior landmark was defined as the most inferior point of the brain mask, in which the Y coordinate lay within 5 mm of the minimum Y-value among all the points of the mask. Similarly, the posterior landmark was defined as the most inferior point, in which the Y-coordinate lay within 5 mm of the maximum Y-value. These two landmarks defined the inferior face of the PRESS box.
To find the optimum PRESS box placement, the algorithm iteratively searched for parameters that minimized the following cost function (fig. 2b):
| [1] |
where:
Si – faces of the PRESS box
o – origin (center of mass of the brain surface)
Vin – volume inside the PRESS box
d(p,Si) – distance from point p on the lipid surface to face Si
wdist, wpen – weights for distance and penalty terms
Intuitively, the first term of the cost function approximates the volume of the press box, and if the volume were larger, the cost function would be lower. The second term balances the effect of the first one by adding a penalty, proportional to the square distance to any points on the lipid surface within the PRESS box. This discourages the PRESS box from growing outside the skull. The adjustable parameters were the distances from origin to the faces of the PRESS box. They were initialized with the standard deviations of X, Y and Z coordinates of points on the brain surface. The weights were fine-tuned manually to achieve convergence of optimization to an acceptable configuration. The active-set optimization algorithm, implemented by Matlab fmincon function (21) was used to find the parameters that minimized the cost function. The dimensions of the PRESS box were constrained by the maximum absolute values of X, Y and Z coordinates of the points of the lipid surface.
OVS Saturation Band Placement Optimization
The algorithm that optimized the saturation bands searched for a configuration where the bands closely approximated the skull surface. Placement of each band was defined by α -angle around inferior-superior axis, β -angle between the saturation band normal and the inferior-superior axis and d -distance from origin to the plane of the saturation band. The inferior band was placed obliquely along the inferior face of the PRESS box. The other eight bands were initially placed with six bands, forming a hexagon in the axial-oblique plane (fig. 2c) and two bands placed superior and tilted in the anterior and posterior direction by 30°.
The OVS saturation band configuration was optimized in 3 stages by minimizing the following cost function with different constraints:
| [2] |
where:
Si – planes of sat bands
d(p,Si) – distance from point p on the lipid surface to saturation band i
o – origin (center of mass of the brain surface)
Vout,i – outer volume of band Si
wdist, wpen – weights for distance and penalty terms
Intuitively term 1 of the cost function would be minimal when the planes lie closest to the lipid surface. The distance term (term 2) added regularization when the planes were far from the surface. The penalty term (term 3) insured that the planes remained on the outer side of the lipid surface during optimization.
At the initial stage, only the distances to the bands were allowed to vary (fig. 2c). In the second stage, the distances to the bands, orthogonal to the sagittal plane along with the tilt angles of the superior planes were optimized (fig. 2d) In the third stage the placement of bands, orthogonal to the axial-oblique plane was optimized (fig. 2e). Additional constraints ensuring that the configuration was symmetrical in right-left direction were imposed throughout the optimization. At the end of the optimization, the saturation bands ended up approximately tangential to the lipid surface. The inner plane of each saturation band was then set 15 mm inward and the outer plane −30 mm outward from the tangent plane (fig. 2f). This enclosed the lipid surface between the polyhedrons, defined by the inner and outer planes of the saturation bands.
The calculated PRESS box and saturation bands were overlaid on the anatomical MR images and displayed using an interactive 3D visualization tool Orthogonal Slicer (L. Balkay, Univ. of Debrecen, Hungary). If required, the user had an option to override the automated prescription and place PRESS box and OVS saturation bands manually.
Shimming
The vendor-supplied high-order shimming routine (10) was modified to automatically define an ROI based on the calculated MRSI PRESS box. This was defined as an ellipsoid fitting within the PRESS box. The shim currents were calculated to minimize the B0 field inhomogeneity within the ROI and downloaded to the hardware before the MRSI acquisition.
Pulse Sequence
The MRSI pulse sequence was modified to allow PRESS box parameters to be loaded from a file: center (L, P and S), size (X, Y and Z) and rotation angles (α, β, γ). These parameters were used to calculate the gradient amplitude and RF frequency offsets for PRESS excitation.
The sequence also allowed the parameters of up to 16 saturation bands to be loaded, as described in (11). Octagonal selection volume was implemented within the MRSI pulse sequence by using 5 cosine-modulated saturation pulses (22) to produce 10 saturation bands: 6 along the faces of the PRESS box and 4 over the corners of the PRESS box in the axial-oblique plane. A protocol could utilize any combination of these OVS methods.
MRSI acquisition
Data were acquired using a commercially available 8-channel head coil and an EPSI flyback sequence (23) (TE = 144 ms, TR = 1100 ms, 1 cm3 isotropic voxels, NEX=1 ). J-difference lactate editing with dual basing pulses was used to provide summed data for Cho, Cr, NAA, and lipids and difference data for lactate as required by the clinical protocol. The PRESS volume was over-prescribed (OverPRESS) by the factor of 1.5 to minimize the chemical shift artifact of PRESS excitation
Raw data were reconstructed offline using an approach that has been described previously (24,25). Data from each channel were filtered with 4 Hz apodization, Fourier-transformed in spatial and frequency domains. Phase and frequency corrections, residual water and baseline removal were performed. Data from different channels were combined using the weights, obtained from a low-resolution proton-density calibration image.
MRSI Protocols
The data obtained with automated prescription were compared with two existing protocols (table 1):
Table 1.
3D MRSI acquisition protocols that were considered in this study: MANUAL = MRSI with manual prescription, AUTOSAT = MRSI with automatically generated saturation band placement, AUTO MRSI = automated oblique PRESS box and saturation band placement.
| MANUAL | AUTOSAT | AUTO MRSI | |
|---|---|---|---|
| Scan Plane | axial | axial | axial-oblique |
| Automated PRESS box placement | ✓ | ||
| Fixed ROI-edge saturation bands | ✓ | ||
| Manually prescribed saturation bands | ✓ | ||
| Automatically prescribed saturation bands | ✓ | ✓ | |
| Octagonal selection | ✓ | ✓ | |
| FOV (cm) | 16×16×16 | 18×18×16 | 18×18×16 |
| TR (ms) | 1104 | 1500 | 1140 |
| Tacq (min) | 9 | 13 | 13 |
Manually Prescribed MRSI (MANUAL)
This protocol used a manually prescribed PRESS box (superior-inferior dimension: 4 cm) with 6 fixed saturation bands along the edges of the PRESS box and 6 additional saturation bands that were manually placed by operators to cover regions of lipid.
MRSI with Automatically Generated Saturation Band Placement (AUTOSAT)
This protocol used the automatically prescribed saturation bands together with octagonal selection (11). The PRESS box (SI dimension: 6 cm) was prescribed manually by the operator in the axial plane. Since the algorithm placed the lower saturation band to cover the nasal cavities and orbits, the PRESS box was placed above the eye-level.
Automatically Prescribed MRSI (AUTO MRSI)
This protocol used the automatically prescribed oblique PRESS box, together with an improved implementation of automated OVS (9 bands). The optimal X, Y and Z dimensions of the PRESS volume were determined automatically as described above. Octagonal selection (10 bands, 5 OVS pulses) was used for additional suppression in the corners of the selected volume.
Subjects
The data was collected from three healthy volunteers. Each volunteer received two MRSI exams on different days with different operators within the period of two weeks. Both exams included MRSI acquisitions with all the protocols listed above.
The automated MRSI protocol has been implemented in a clinical research protocol for patients with brain tumors and was performed on 54 subjects during the study timeframe (June 2010 -January 2011). In total, the technique was used in 62 MRSI exams and was successful in 52 cases. The automated prescription failed in 2 cases. In 5 cases the tumor was not covered by AUTO MRSI due to its inferior location and MANUAL protocol had to be used instead. In 3 cases interpretable data could not be acquired due to scanner hardware or software failures.
The 8 patients who received two MRSI examinations with the automated protocols during the study timeframe were selected for this study. The exams were performed after the patients had undergone surgery and were 3–4 months apart from each other. The patients were undergoing a course of treatment that included radiotherapy, temozolomide and other chemotherapy agents. Since the patients were not part of any clinical study, their detailed medical history was not available. All subjects provided informed consent as approved by the Institutional Review Board.
Data analysis
Parameters that were used to evaluate the data included the coverage volume, consistency of prescription and the quality of spectral data. The coverage volume of the MRSI acquisition was calculated by generating masks from the parameters, obtained from the headers of raw data files, produced by the MRSI acquisition. The coverage volume was defined as the number of spectroscopy voxels within the PRESS box excluding those covered by a saturation band by more than 50%. Coverage volumes between the STANDARD, AUTOSAT and AUTO MRSI protocols were compared with 1-tailed t-test with unequal variances.
To estimate the consistency of the prescription, the volume of brain tissue that was covered by two MRSI exams from the same subject was calculated as the volume of the intersection of the aligned coverage masks. For this purpose, the exams were aligned to the same coordinate system based on the anatomic images using VTK CISG Registration Toolkit (Computational Imaging Science Group, King’s College London). Bland-Altman repeatability coefficient RC = 1.96 σΔV and mean ΔV were calculated for covered volumes for exams from the same subjects (26). Assuming the differences between measurements are approximately normally distributed, the absolute difference between them is expected to differ by no more than the RC for 95% of the time.
To compare data quality, the spectroscopic parameters, such as peak SNR, metabolite ratios and peak linewidth were calculated within the whole covered volume and within the intersection of all volumes covered by different protocols using software developed in our laboratory (24,25). Spectral quality within the tumor (for patients with brain tumors) was assessed by analyzing the same parameters within the T2 lesions as defined by manually outlined ROIs. The peak heights (real and magnitude) were measured based on predefined peak regions in spectral data that had baseline, phase and frequency corrections applied, as described by Nelson et al. (24). The SNR of individual peaks was calculated using the tail end of the spectra for noise estimation. The SNR values were corrected for the difference in acquisition times and repetition times (SNR efficiency) using the relaxation constants for individual metabolites (27,28). Bland-Altman repeatability coefficients and mean differences (26) were calculated for changes in metabolite ratios within the whole brain (volunteers) and T2 lesions (patients) using the same AUTO MRSI protocol (test-retest experiment) and for comparing the values obtained with AUTO MRSI and MANUAL protocols (measurement technique validation).
Results
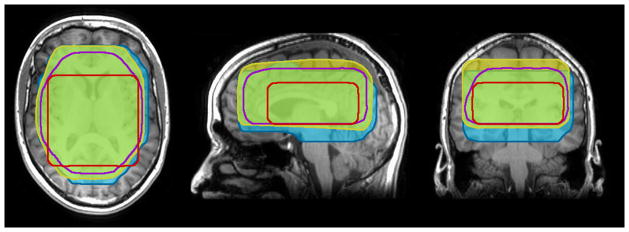
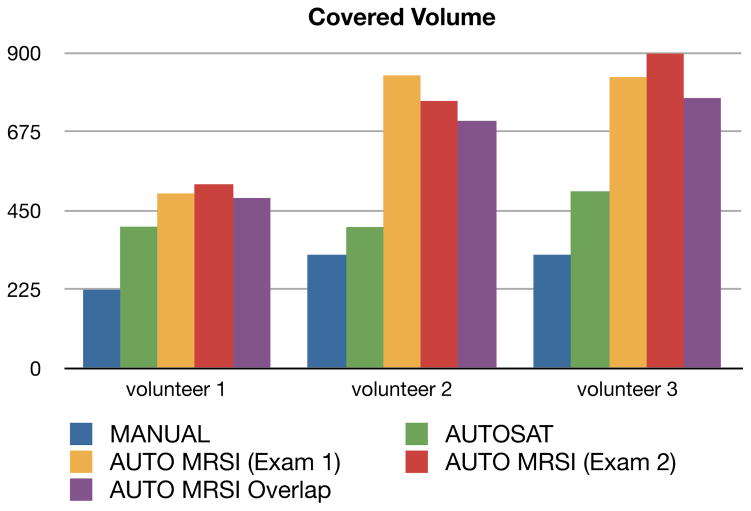
All three protocols showed good quality data for the six exams from the three normal volunteers. Figure 3 shows an example of the covered volumes from different protocols, overlaid over an anatomical image. Figure 4 shows the calculated coverage volumes of those protocols in healthy volunteers. The mean fraction of the total brain volume (including the cerebellum), covered by MRSI was 21±2% for MANUAL, 32±3% for AUTOSAT and 53±7% for AUTO MRSI. Two of the previously used protocols (MANUAL and AUTOSAT) had PRESS volume dimensions determined by the operators with default SI dimensions of 4 and 6 cm, respectively. These values were considered optimal by the operators performing routine clinical-research scans with these protocols and could be changed on a case-by-case basis. The mean coverage volumes for these protocols in patients were 227.67 cm3 for MANUAL and 621.8 cm3 for AUTOSAT, as published previously (11). The resulting covered volumes in these protocols, however, were likely suboptimal and biased by the operators’ fear of compromising the quality of data and limited time to prescribe a scan. In contrast, the AUTO MRSI protocol always tailored the prescription to the subject’s anatomy. While it was not possible to compare the automated prescription to an ideal manual prescription, our results show that by avoiding the biases, resulting from the pre-defined protocol parameters and manual prescription, AUTO MRSI achieved significantly larger covered volume than the MANUAL protocol with manual prescription (p<0.0005) and the semi-automated AUTOSAT protocol (p<0.005).
Fig. 3.
Example of the covered volumes acquired from a healthy volunteer using the following protocols: red -MANUAL, purple -AUTOSAT, yellow and blue -AUTO MRSI (from different exams).
Fig. 4.
Covered volume of the three volunteers using the three protocols that were compared. The volunteers received two AUTO MRSI acquisitions on different days (yellow and red). Purple column shows the volume that was covered by both of those acquisitions.
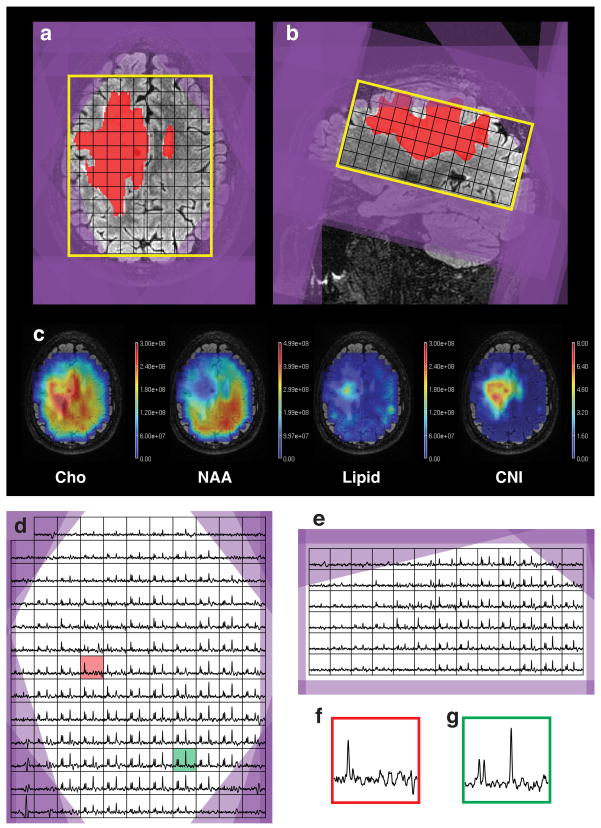
Figure 5 shows an example of a dataset, acquired from a patient using the AUTO MRSI protocol. The automated prescription was able to cover most of the supratentorial brain, including almost all of the T2 lesion. The oblique PRESS box covered inferior areas of the brain posteriorly, while avoiding the orbits and sinuses in the anterior areas of the brain. Figure 5c shows one slice of the metabolite maps for Cho, NAA, Lipid and Choline to NAA Index (CNI)(29). Figures 5d-g show axial and sagittal slices through the spectral data as well as sample voxels from the tumor and healthy tissue. The Cho and NAA metabolite maps show decreased metabolite concentrations throughout the anterior-right side of the brain, beyond the region of elevated choline, which could be attributed to the effect of the radiation therapy. The lipid map shows that only a few voxels on the periphery of the brain had noticeable lipid contamination. There is a region of elevated lipid within the T2 lesion, which appears to be coming from the necrotic core of the tumor.
Fig. 5.
(a, b) Automatically generated MRSI prescription (yellow -PRESS box, purple -saturation bands), overlaid on top of T2 FLAIR images and T2 lesion mask (red); (c) metabolite maps; (d, e) axial and sagittal slices through the spectral grid; (f, g) sample spectral voxels from T2 lesion (red, location shown on Fig. 6d) and healthy tissue (green).
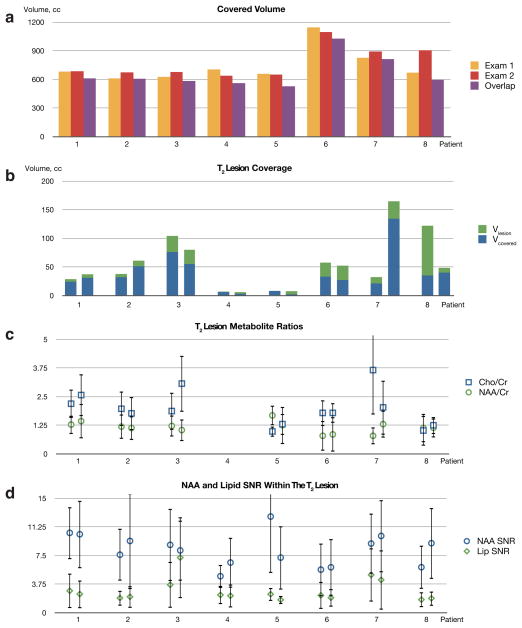
The algorithm was able to achieve high consistency of prescription despite differences in subject positioning. The purple column on figure 4 shows that in the exams of the same healthy volunteers, the covered volume was largely the same (average overlap: 91%). In patients, the technique also demonstrated very good consistency of prescription (average overlap: 88%). Figure 6a shows the covered volume for two exams from each patient as well as the volume of the overlap area between the two of them. Mean fraction of the total brain volume (including the cerebellum) that was covered by MRSI was 56±10%.
Fig. 6.
(a) Covered volume (cm3) for the two exams of the patients with brain tumors (yellow and red) and the volume of the overlap area between those exams (purple); (b) The volume in cm3 of the T2 lesion (green) and T2 lesion covered by MRSI acquisition (blue); (c) Mean Cho/Cr (blue) and NAA/Cr (green) metabolite ratios and their standard deviations, calculated within the T2 lesion; (d) Mean magnitude NAA (blue) and lipid (green) SNR within the T2 lesion and their standard deviations.
The volumes of the T2 lesions and their coverage by MRSI are presented on figure 6b. In almost all exams the T2 lesion was well covered by the MRSI volume of interest. There was only one exam (patient 8, exam 1) where a large part of the lesion could not be covered because most of it was in the inferior regions of the brain. Changes in the T2 lesion volumes between exams could be attributed to response to therapy or disease progression.
The mean metabolite ratios, calculated within the T2 lesion areas are shown on fig. 6c. In most cases they show very consistent results between exams of the same patient. Unlike the healthy volunteers, most patients showed choline that was elevated relative to NAA. For patient 4 it was not possible to reliably quantify the ratios due to the T2 lesion being very small (less than 7 cm3). For patient 7 choline became less elevated in the second exam. This could be attributed to averaging over a much larger T2 lesion. Bland-Altman reproducibility coefficients and mean differences for coverage volumes and metabolite ratios (test-retest for AUTO MRSI and comparison to MANUAL) from volunteers and patients are shown in table 3. All parameters show good reproducibility and small bias for measurement in the same subjects, compared to standard deviations of parameters within subjects with an exception of Cho/Cr ratio in patients. This is to be expected, since choline is a major metabolite that changes due to treatment and disease progression.
Table 3.
Mean values across all subjects, Bland-Altman reproducibility coefficients and mean differences between measurements of the same subject for coverage volumes and metabolite ratios
| Mean | RC | Mean Δ | ||
|---|---|---|---|---|
| V, cm3 | volunteers | 725.33 | 141.07 | 6.67 |
| patients | 758.5 | 184.51 | 37 | |
|
| ||||
| Cho/Cr | volunteers | 1.24 | 0.045 | 0.031 |
| volunteers (AUTO MRSI vs. MANUAL) | 0.041 | 0.059 | ||
| patients | 1.77 | 1.683 | 0.043 | |
|
| ||||
| NAA/Cr | volunteers | 2.23 | 0.018 | 0.002 |
| volunteers (AUTO MRSI vs. MANUAL) | 0.027 | 0.13 | ||
| patients | 1.20 | 0.57 | 0.009 | |
The mean NAA SNR efficiency for the whole covered volume in all exams was 0.92±0.63, and for the T2 lesions it was 0.58±0.31. These numbers were comparable to those of the healthy volunteers (table 2) and were expected to be lower in the patients due to the effects of the disease and therapy. Mean NAA linewidth was 12.01±2.81 Hz throughout the whole volume and 11.43±2.87 Hz within the T2 lesion, which are similar to the linewidths, observed in volunteers with existing protocols.
Table 2.
Mean SNR efficiency and linewidth and their standard deviations of the NAA peak in the datasets, acquired from healthy volunteers with the protocols that were compared.
| MANUAL | AUTOSAT | AUTO MRSI | |
|---|---|---|---|
| NAA SNReff | 1.25±0.54 | 1.38±0.53 | 1.34±0.48 |
| NAA linewidth (Hz) | 10.53±2.1 | 10.67±1.94 | 10.92±1.89 |
Figure 6d shows the mean SNR of the magnitude NAA and lipid peaks within the T2 lesions in all datasets and their standard deviations. It indicates that the lipid signal was much smaller than the signal from NAA and did not interfere with NAA quantification, which is crucial for interpretation of brain MRSI data.
Discussion
Realizing the true clinical value of brain MRSI requires that data are acquired from the whole brain as easily as conventional imaging data. The technique developed in this project came closer to this goal than any of the previous approaches (7,11,12). The data from patients and volunteers showed that automated prescription was able to achieve much larger coverage volumes while allowing the acquisition of high-quality data from both the lesions and the healthy tissue.
The approach taken in this study was different from that of Yung et al., which included affine transformation of a manually placed MRSI slice and OVS saturation bands from an atlas coordinate space to that of the patient exam (7). Our technique used a multistep iterative algorithm to achieve a configuration optimal for the anatomy of each individual subject. The cost functions were designed for faster convergence using a gradient-based optimization method. Constraints were imposed on the parameters, which were subdivided into groups and optimized in multiple steps to minimize the dimensionality of the parameter space and to avoid converging on a local minimum. That allowed us to achieve a much shorter calculation time (about 30 sec) than the previous approaches and made it possible to run the automated prescription software directly on the scanner console. The optimization of the PRESS box size enabled acquisition of data from the largest possible volume for a given brain. Finally, in the present technique, the pulse sequence was customized to read all the prescription parameters from files, automating the entire scan prescription.
Fully automated prescription of MRSI exams has resulted in more comprehensive and consistent MRSI datasets and reduced the need for time-consuming operator training. A large coverage volume reduced the risk that some of the diagnostically relevant tissues would be left outside the volume of interest, while the oblique orientation of the PRESS box ensured better coverage of the inferior regions of the brain. This is especially critical for diseases such as multi-focal gliomas, where previously a single acquisition was not enough to cover all parts of the tumor. Including a large number of healthy voxels in the volume of interest also improved statistical quantification of metabolic abnormality parameters, such as Choline to NAA Index (CNI, (29)), which estimates how different a given spectrum is from that of a “normal” brain.
Automated prescription also improved the consistency of prescription. This is especially important for serial scans of the same patient. The tissue, that is unremarkable in an earlier scan might become of interest in a later one. A large overlap between the covered volumes in these scans will improve the detection of metabolic changes as disease progresses.
Manual prescription of MRSI exams is a difficult and time-consuming task. It took an experienced operator about 5 minutes to set up a regular MRSI acquisition, compared to calculation time for automated prescription of about 30 seconds. The use of automated prescription software will ensure that in most cases the operators could acquire good quality data by following a simple protocol. This will make adding MRSI to clinical protocols much easier and will solve many problems in organizing multi-center clinical trials that employ MRSI.
The proposed technique gave rise to a number of new challenges. Since the volume of interest is much larger than in conventional methods, B0 variations over the volume of interest can cause peak broadening and loss of spectral resolution. This was addressed by automated high-order shimming over an ellipsoidal shim volume that was based on the oblique PRESS box and produced excellent results in most cases.
The algorithm could fail to achieve a good prescription in some cases. This could be due to failures of segmentation logic or failure of the optimizations to converge. These cases were rare and often corresponded to the subjects with unusual brain structure, such as large portions of the brain tissue missing due to prior surgery. The automatically prescribed volume of interest may fail to cover the lesion, or the lesion may be obscured by OVS saturation bands. That could happen, for instance when the lesion is located in the extremely superior or inferior regions of the brain. In such cases, the protocol can fall back to manual prescription. While no automated image analysis algorithms succeed 100% of the time, effective quality assurance, together with the option of the manual alternative, ensured that data could be obtained in all cases, as required in the clinical setting.
In some datasets, as with conventional methods, lipid contamination was detectable within the brain, usually in the voxels adjacent to the skull. Usually the lipid peak was not big enough to affect the NAA peak. This could be minimized by making saturation band placement more aggressive. There is an inherent tradeoff, however, between the increase of the lipid coverage and the decrease of the area from which MRSI data can be acquired. In this work we aimed to achieve balance between these competing effects by making the usable volume as large as possible, as long as the level of the lipid contamination was low enough for data interpretation.
In conclusion, the automated prescription technique described here provided larger coverage volumes than previous manual protocols while maintaining high quality of data. It also yielded consistent prescription quality, not dependent on operator skill, and made possible to include 3D MRSI into the clinical research protocols without the need for extensive operator training.
Acknowledgments
The authors would like to thank Adam Elkhaled, Trey Jalbert, Wei Bian, Angela Jakary and Chris Williams for help with data acquisition and processing, and Jason Crane and Beck Olson for help with research software.
This work was supported by NIH grants R01CA127612and P01CA118816.
References
- 1.Horska A, Barker PB. Imaging of brain tumors: MR spectroscopy and metabolic imaging. Neuroimaging clinics of North America. 2010;20(3):293–310. doi: 10.1016/j.nic.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonavita S, Di Salle F, Tedeschi G. Proton MRS in neurological disorders. Eur J Radiol. 1999;30(2):125–131. doi: 10.1016/s0720-048x(99)00051-0. [DOI] [PubMed] [Google Scholar]
- 3.Tran TK, Vigneron DB, Sailasuta N, Tropp J, Le Roux P, Kurhanewicz J, Nelson S, Hurd R. Very selective suppression pulses for clinical MRSI studies of brain and prostate cancer. Magn Reson Med. 2000;43(1):23–33. doi: 10.1002/(sici)1522-2594(200001)43:1<23::aid-mrm4>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 4.Henning A, Schar M, Schulte RF, Wilm B, Pruessmann KP, Boesiger P. SELOVS: brain MRSI localization based on highly selective T1-and B1-insensitive outer-volume suppression at 3T. Magn Reson Med. 2008;59(1):40–51. doi: 10.1002/mrm.21374. [DOI] [PubMed] [Google Scholar]
- 5.Henning A, Fuchs A, Murdoch JB, Boesiger P. Slice-selective FID acquisition, localized by outer volume suppression (FIDLOVS) for (1)H-MRSI of the human brain at 7 T with minimal signal loss. NMR Biomed. 2009;22(7):683–696. doi: 10.1002/nbm.1366. [DOI] [PubMed] [Google Scholar]
- 6.Posse S, Otazo R, Caprihan A, Bustillo J, Chen H, Henry PG, Marjanska M, Gasparovic C, Zuo C, Magnotta V, Mueller B, Mullins P, Renshaw P, Ugurbil K, Lim KO, Alger JR. Proton echo-planar spectroscopic imaging of J-coupled resonances in human brain at 3 and 4 Tesla. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2007;58(2):236–244. doi: 10.1002/mrm.21287. [DOI] [PubMed] [Google Scholar]
- 7.Yung KT, Zheng W, Zhao C, Martinez-Ramon M, van der Kouwe A, Posse S. Atlas-based automated positioning of outer volume suppression slices in short-echo time 3D MR spectroscopic imaging of the human brain. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2011;66(4):911–922. doi: 10.1002/mrm.22887. [DOI] [PubMed] [Google Scholar]
- 8.Maudsley AA, Domenig C, Sheriff S. Reproducibility of serial whole-brain MR spectroscopic imaging. NMR in biomedicine. 2010;23(3):251–256. doi: 10.1002/nbm.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford FW, Khayal IS, McGue C, Saraswathy S, Pirzkall A, Cha S, Lamborn KR, Chang SM, Berger MS, Nelson SJ. Relationship of pre-surgery metabolic and physiological MR imaging parameters to survival for patients with untreated GBM. J Neurooncol. 2009;91(3):337–351. doi: 10.1007/s11060-008-9719-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim DH, Adalsteinsson E, Glover GH, Spielman DM. Regularized higher-order in vivo shimming. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2002;48(4):715–722. doi: 10.1002/mrm.10267. [DOI] [PubMed] [Google Scholar]
- 11.Ozhinsky E, Vigneron DB, Nelson SJ. Improved spatial coverage for brain 3D PRESS MRSI by automatic placement of outer-volume suppression saturation bands. Journal of magnetic resonance imaging : JMRI. 2011;33(4):792–802. doi: 10.1002/jmri.22507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez-Ramon M, Gallardo-Antolin A, Cid-Sueiro J, Heileman GL, Yung KT, Zheng W, Zhao C, Posse S. Automatic placement of outer volume suppression slices in MR spectroscopic imaging of the human brain. Magn Reson Med. 2010;63(3):592–600. doi: 10.1002/mrm.22275. [DOI] [PubMed] [Google Scholar]
- 13.Young S, Bystrov D, Netsch T, Bergmans R, van Muiswinkel A, Visser F, Sprigorum R, Gieseke G. Automated planning of MRI neuro scans. Proc SPIE. 2006;6144:61441M. [Google Scholar]
- 14.Tao X, Gupta S. Automatic Scan Plane Planning Method for Brain MRI. Proceedings 17th Scientific Meeting, International Society for Magnetic Resonance in Medicine; 2009 April; Honolulu. p. 2903. [Google Scholar]
- 15.Itti L, Chang L, Ernst T. Automatic scan prescription for brain MRI. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine/Society of Magnetic Resonance in Medicine. 2001;45(3):486–494. doi: 10.1002/1522-2594(200103)45:3<486::aid-mrm1064>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 16.Nelles M, Gieseke J, Urbach H, Semrau R, Bystrov D, Schild HH, Willinek WA. Pre-and postoperative MR brain imaging with automatic planning and scanning software in tumor patients: an intraindividual comparative study at 3 Tesla. Journal of magnetic resonance imaging : JMRI. 2009;30(3):672–677. doi: 10.1002/jmri.21888. [DOI] [PubMed] [Google Scholar]
- 17.Gedat E, Braun J, Sack I, Bernarding J. Prospective registration of human head magnetic resonance images for reproducible slice positioning using localizer images. Journal of magnetic resonance imaging : JMRI. 2004;20(4):581–587. doi: 10.1002/jmri.20153. [DOI] [PubMed] [Google Scholar]
- 18.van der Kouwe AJ, Benner T, Fischl B, Schmitt F, Salat DH, Harder M, Sorensen AG, Dale AM. On-line automatic slice positioning for brain MR imaging. NeuroImage. 2005;27(1):222–230. doi: 10.1016/j.neuroimage.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 19.Seber GAF. Multivariate observations. New York: Wiley; 1984. p. 686. [Google Scholar]
- 20.van den Boomgaard R, van Balen R. Methods for fast morphological image transforms using bitmapped binary images. CVGIP: Graphical Models and Image Processing. 1992;54(3):252–258. [Google Scholar]
- 21.Byrd RH, Gilbert JC, Nocedal J. A Trust Region Method Based on Interior Point Techniques for Nonlinear Programming Mathematical Programming. 2000;89(1):149–185. [Google Scholar]
- 22.Osorio JA, Xu D, Cunningham CH, Chen A, Kerr AB, Pauly JM, Vigneron DB, Nelson SJ. Design of cosine modulated very selective suppression pulses for MR spectroscopic imaging at 3T. Magn Reson Med. 2009;61(3):533–540. doi: 10.1002/mrm.21842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cunningham CH, Vigneron DB, Chen AP, Xu D, Nelson SJ, Hurd RE, Kelley DA, Pauly JM. Design of flyback echo-planar readout gradients for magnetic resonance spectroscopic imaging. Magn Reson Med. 2005;54(5):1286–1289. doi: 10.1002/mrm.20663. [DOI] [PubMed] [Google Scholar]
- 24.Nelson SJ. Analysis of volume MRI and MR spectroscopic imaging data for the evaluation of patients with brain tumors. Magn Reson Med. 2001;46(2):228–239. doi: 10.1002/mrm.1183. [DOI] [PubMed] [Google Scholar]
- 25.Osorio JA, Ozturk-Isik E, Xu D, Cha S, Chang S, Berger MS, Vigneron DB, Nelson SJ. 3D 1H MRSI of brain tumors at 3. 0 Tesla using aneight-channel phased-array head coil. J Magn Reson Imaging. 2007;26(1):23–30. doi: 10.1002/jmri.20970. [DOI] [PubMed] [Google Scholar]
- 26.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1(8476):307–310. [PubMed] [Google Scholar]
- 27.Li Y, Srinivasan R, Ratiney H, Lu Y, Chang SM, Nelson SJ. Comparison of T(1) and T(2) metabolite relaxation times in glioma and normal brain at 3T. J Magn Reson Imaging. 2008;28(2):342–350. doi: 10.1002/jmri.21453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han E, Gold G, Stainsby J, Wright G, Beaulieu C, Brittain J. In-Vivo T1 and T2 Measurements of Muskuloskeletal Tissue at 3T and 1.5T. Proceedings 11th Scientific Meeting, International Society for Magnetic Resonance in Medicine; 2003 July; Toronto. p. 450. [Google Scholar]
- 29.McKnight TR, Noworolski SM, Vigneron DB, Nelson SJ. An automated technique for the quantitative assessment of 3D-MRSI data from patients with glioma. Journal of magnetic resonance imaging : JMRI. 2001;13(2):167–177. doi: 10.1002/1522-2586(200102)13:2<167::aid-jmri1026>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]