Summary
ATBF1 is a large nuclear protein that contains multiple zinc-finger motifs and four homeodomains. In mammals, ATBF1 regulates differentiation, and its mutation and/or downregulation is involved in tumorigenesis in several organs. To gain more insight into the physiological functions of ATBF1, we generated and validated a conditional allele of mouse Atbf1 in which exons 7 and 8 were flanked by loxP sites (Atbf1flox). Germline deletion of a single Atbf1 allele was achieved by breeding to EIIa-cre transgenic mice, and Atbf1 heterozygous mice displayed reduced body weight, preweaning mortality, increased cell proliferation, and attenuated cytokeratin 18 (CK18) expression, indicating haploinsufficiency of Atbf1. Floxed Atbf1 mice will help us understand such biological processes as neuronal differentiation and tumorigenesis.
Keywords: Atbf1, Cre-loxP, conditional knockout, heterozygous, preweaning mortality
ATBF1 (AT-motif binding factor 1), also named ZFHX3 for zinc finger homeobox 3, belongs to a small family that is structurally characterized by multiple zinc-finger motifs and three or four homeodomains. In human and mouse, the ATBF1 gene contains 11 exons, and exon 10 is the largest (5454 bps). Using distinct promoters, ATBF1 produces two splicing variants, ATBF1-A and -B, which share exons 4 to 11 but differ in 5′ exons. Both isoforms encode large proteins. ATBF1-A uses a start codon in exon 3 to generate a protein of 404-kD. ATBF1-B uses a start codon in exon 4 to produce a protein of 300-kD (Miura et al., 1995a; Morinaga et al., 1991). Although both isoforms were reported, ATBF1-A is the predominant one with an expression level much higher than ATBF1-B. In mouse, only Atbf1-A was detectable at the mRNA and protein levels (Ishii et al., 2003; Miura et al., 1995b).
ATBF1 functions in multiple biological processes including cell differentiation and tumorigenesis. It could regulate region-specific central nervous system differentiation (Ishii et al., 2003); its expression decreases at postnatal stage but remains detectable in mature neurons (Jung et al., 2005; Watanabe et al., 1996); and its expression is up-regulated in retinoic acid-treated p19 cells, an in vitro model of neuronal differentiation (Ido et al., 1994; Jung et al., 2005; Miura et al., 1995a). ATBF1 may also function in myogenic differentiation (Berry et al., 2001). ATBF1 regulates several differentiation genes including alpha-fetoprotein (AFP), aminopeptidase N and neurod1 (Jung et al., 2005; Kataoka et al., 2000; Morinaga et al., 1991; Yasuda et al., 1994). ATBF1 may affect atrial fibrillation, since genetic variations in introns of ATBF1 are associated with atrial fibrillation (Benjamin et al., 2009; Gudbjartsson et al., 2009; Li et al., 2011).
ATBF1 has also been suggested to be a tumor suppressor in cancers of the breast, prostate, stomach and liver. In humans, ATBF1 is located at 16q22, frequently deleted in prostate and breast cancers (Dong, 2001). ATBF1 undergoes frequent somatic mutations in human prostate cancer, some of which are protein-truncating (Sun et al., 2005). A germline variant of ATBF1 was linked to prostate cancer risk (Xu et al., 2006), and ATBF1 mutation was detected in gastric cancers (Cho et al., 2007). In highly malignant AFP-producing gastric cancers, ATBF1 expression is absent, and its role in transcriptional repression of the AFP oncogene has been demonstrated (Kataoka et al., 2001; Yasuda et al., 1994). Re-establishing the expression of ATBF1 in human prostate and breast cancer cell lines inhibits cell growth (Dong et al., 2010; Sun et al., 2005; Sun et al., 2007), and changes in subcellular localization alter its function in cancer gene regulation (Mabuchi et al., 2010; Mori et al., 2007) (our unpublished data). ATBF1 may therefore be a tumor suppressor in human tissues, though this has not been examined in vivo.
For functional studies of ATBF1 in vivo, we developed a mouse strain in which exons 7 and 8 of Atbf1 were flanked by loxP sites so they can be deleted upon spatiotemporally controlled expression of Cre recombinase. Removal of exons 7 and 8 leads to a frame shift in Atbf1 translation, resulting in the removal of approximately 70% of the Atbf1-A protein sequence including most of the zinc finger motifs and all four homeodomains. The targeting vector pFlexible was used, containing the positive/negative selectable marker PGK-puroΔtk and loxP and Frt sequences (van der Weyden et al., 2005).
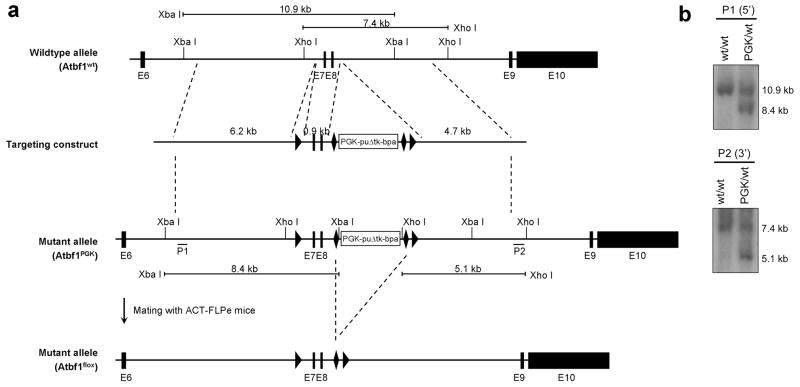
Three Atbf1 genomic fragments were amplified by PCR from genomic DNA of TL-1 embryonic stem cells (derived from 129S6/SvEvTac mice) and subcloned into the pFlexible vector (Fig. 1a). The linearized targeting construct was then electroporated into TL1 embryonic stem cells. After selection in puromycin-containing medium, ES cell clones were screened for homologous recombination by Southern blot analysis. Two clones showed the expected 10.9-kb wildtype and 8.4-kb targeted fragments using the 5′ probe (P1) (Fig. 1b). One had the expected 5.1-kb targeted fragment for the 3′ probe (P2), indicating that this clone contained a correctly targeted allele in which exons 7 and 8 are flanked by loxP sites and the PGK-puroΔtk selection marker cassette by Frt sites (Fig. 1b). The mutant allele was termed Atbf1PGK.
Figure 1. Generation of the floxed Atbf1 allele (Atbf1flox) in mice.
a. Design of cloning vectors and targeting strategies. Part of the Atbf1 gene (from exon 6 to exon 10) is shown at top (wildtype allele). LoxP and Frt sequences are marked by triangle and diamond-shaped boxes respectively. The targeting construct was generated by inserting three Atbf1 fragments (6.2 kb, 0.9 kb and 4.7 kb) into pFlexible. Two hybridization probes (P1 and P2) used in Southern blot analyses are indicated in the mutant allele. The PGK-puroΔtk selection cassette was deleted by mating Atbf1PGK mice with ACT-FLPe mice to generate the Atbf1flox allele. Digestion of targeted genomic DNA by Xba I yields a 10.9 kb fragment for the wildtype allele and an 8.4 kb fragment for the mutant allele when the 5′ probe (P1) is used, while digestion by Xho I yields 7.4 kb wildtype and 5.1 kb mutant bands when the 3′ probe (P2) is used. b. Southern blot analyses of wildtype (wt) and targeted (PGK) alleles in ES cells, showing expected mutant fragments for both 5′ and 3′ probes.
The ES cell clone with correct recombinations was injected into blastocysts and implanted into recipient female mice. Nine chimeric male mice were obtained. Of the six male mice bred with C57BL/6J female mice, three had germline transmission of the targeted allele. Genotypes of the Atbf1PGK mice were confirmed by Southern blot analysis using the same strategy as for ES cells. All subsequent genotyping was performed by a PCR strategy (Fig. 1a, 2a).
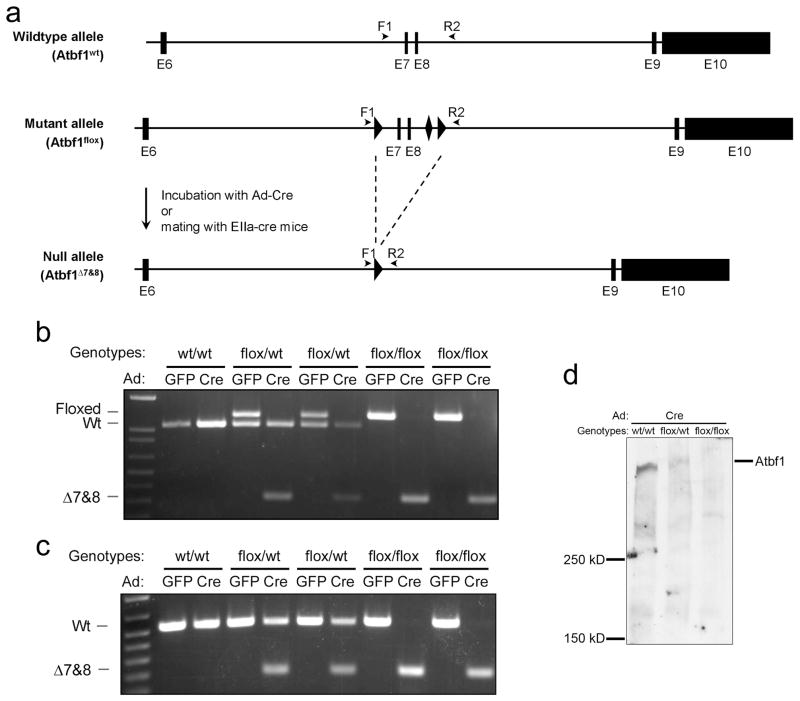
Figure 2. Excision of exons 7 and 8 of Atbf1 by the Cre recombinase.
a. Schematic representation of Cre-mediated deletion of exons 7 and 8. LoxP and Frt sequences are marked by triangle and diamond-shaped boxes respectively. Primers F1 and R2 are indicated by arrow heads. The Atbf1Δ7&8 deletion allele was induced by Cre from adenoviruses or EIIa-cre mice. b. Five MEF clones incubated with adenoviruses expressing GFP or Cre were genotyped by a PCR using primers F1 and R2, identifying three Atbf1 alleles: Atbf1flox (1248 bp), Atbf1wt (1071 bp) and Atbf1Δ7&8 (289 bp). c. Detection of the wildtype (532 bp) and truncated (197 bp) Atbf1 mRNA by RT-PCR in MEFs with different status of Atbf1 deletion. d. Detection of Atbf1 protein in MEFs with different Atbf1 deletion status by immunoblotting. Molecular masses of protein standards are shown to the left of the gel.
The Atbf1PGK mice were then bred with ACT-FLPe transgenic mice, which universally express the FLP enzyme, and the PGK-puroΔtk cassette was removed from the genome (Fig. 1a). Mice were then bred to C57BL/6J mice to obtain floxed Atbf1 allele without Flp, which was termed Atbf1flox/wt. Mice with single (Atbf1flox/wt) and double (Atbf1flox/flox) floxed alleles were viable and fertile, and showed no gross abnormality. The allele with the deletion of exons 7 and 8 was termed Atbf1Δ7&8.
Using a PCR-based genotyping strategy (Fig. 2a), three Atbf1 genotypes could be detected with one pair of primers: Atbf1flox, Atbf1wt and Atbf1Δ7&8. To verify the excision of exons 7 and 8 by Cre, Atbf1flox/wt mice were inbred and mouse embryonic fibroblasts (MEFs) were prepared from embryos at day 13.5 of gestation. Cultured MEFs were then infected with adenoviruses bearing either green fluorescent protein (Ad-GFP) or Cre (Ad-Cre). After Ad-Cre infection, the Atbf1flox allele was abolished and the Atbf1Δ7&8 allele appeared, indicating the excision of exons 7 and 8 by Cre (Fig. 2b). Deletion of exons 7 and 8 generated a truncated mRNA, which was also detected by a PCR approach (Fig. 2c). The Atbf1 protein was significantly reduced in Atbf1flox/wt MEFs and was not detectable in Atbf1flox/flox MEFs after the infection of Ad-Cre (Fig. 2d). Because the antibody was produced by a peptide sequence within the deleted region, it is unknown whether the deleted Atbf1 allele is expressed as a truncated protein.
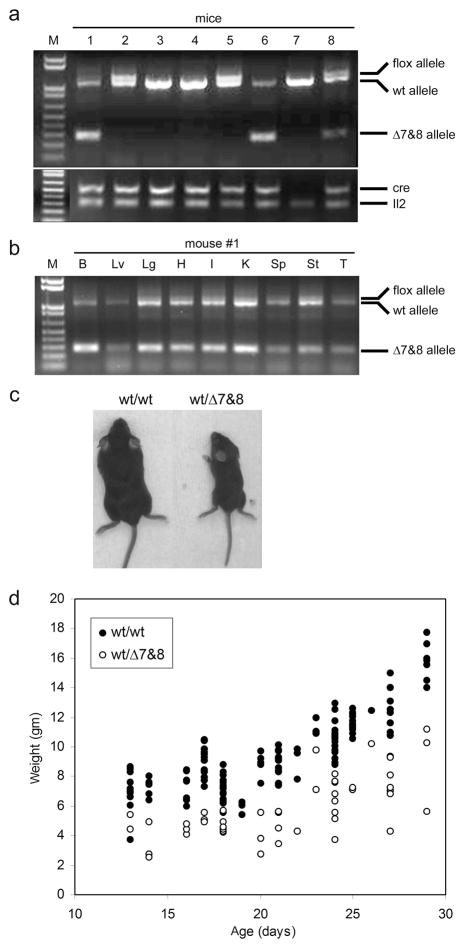
To examine the effect of whole-body deletion of Atbf1, we crossed Atbf1flox/wt mice with heterozygous EIIa-cre transgenic mice, which express Cre in early stage of embryogenesis under the control of the adenoviral EIIa promoter and thus cause the deletion of loxP-flanked sequences in most if not all tissues including germ cells (Lakso et al., 1996). Some F1 mice were further bred with C57BL/6J mice to achieve germline transmission of the deletion. All F1 and F2 offspring were genotyped with tail DNA and the PCR method (Fig. 3a), which detected different status of Atbf1 deletion in the Cre+/Atbf1flox/wt bigenic mice: no band for the Atbf1Δ7&8 allele (no deletion, mice 2 and 5 in Fig. 3a); complete excision of Atbf1 in all cells (mouse 6); and incomplete excision (3 bands, mice 1 and 8), reflecting that the Atbf1 deletion only occurred in some but not all cells due to heterogeneous expression of Cre post the zygote stage. In individual mice, a mosaic genotype was present at a similar extent in all major organs tested (Fig. 3b). Unexpectedly, only 55% (51 of 92) of the Cre+/Atbf1flox/wt bigenic mice survived weaning, suggesting a preweaning mortality phenotype in these mice (Table 1). Most of the dead mice had complete excision of Atbf1. Breeding Cre+/Atbf1flox/wt mice with C57BL/6J mice demonstrated germline transmission of the Atbf1Δ7&8 allele and more severe preweaning mortality in the heterozygous mice. Among a total of 70 mice with heterozygous Atbf1wt/Δ7&8, 29 (41%) died within the first 48 hours after birth, additional and 20 (29%) died within 4 weeks after birth (Table 1). Some of the remaining Atbf1wt/Δ7&8 mice died after weaning. The mortality was sex-independent.
Figure 3. Reduced body size and weight in mice with deletion of one Atbf1 allele.
a. Representative genotyping PCR. Eight mice were examined for Atbf1, Cre, and Il2 (an internal control) genotypes. Three Atbf1 alleles (Atbf1flox, Atbf1wt and Atbf1Δ7&8) were detected in a single PCR. Two mice with the Cre+/Atbf1flox/wt genotype (#1 and #8) showed a mosaic genotype. M, 1 Kb plus DNA standard (Invitrogen). b. Mosaic genotypes in different organs. Nine organs from mouse #1 in panel 3a were examined for the Atbf1 genotype. B, brain; Lv, liver; Lg, lung; H, heart; I, intestine; K, kidney; Sp, spleen; St, stomach; T, tail. c. Representative images of 21-day old mice with the wildtype (wt/wt) or heterozygous deletion (wt/Δ7&8) of Atbf1. d. Weights of wildtype mice (wt/wt, filled circles) and heterozygous mice (wt/Δ7&8, open circles) aged at 13 to 29 days. Atbf1 heterozygous mice were significantly lighter than their wildtype littermates.
Table 1.
Numbers of live mice obtained for different genotypes.
| Atbf1 genotypes | Cre+or− wt/wt |
Cre+
|
Cre− wt/Δ7&8 | |||
|---|---|---|---|---|---|---|
| flox/wt | flox/wt/Δ7&8* | wt/Δ7&8§ | total | |||
| At birth | 442 | 7 | 25 | 60 | 92 | 70 |
| 48 hrs after birth | 434 | 7 | 23 | 41 | 71 | 41¶ |
| Weaning (4 weeks) | 431 | 6 | 21 | 24 | 51 | 21¶ |
, deletion of exons 7 and 8 occurred only in some but not all cells in Cre+/Atbf1flox/wt bigenic mice, showing a mosaic genotype.
excision of exons 7 and 8 was complete in all cells in Cre+/Atbf1flox/wt mice.
P values for comparisons between Cre−/Atbf1wt/Δ7&8 and Atbf1wt/wt at different time points were all smaller than 0.0001.
Except for the early postnatal mortality, Atbf1wt/Δ7&8 mice were not distinguishable from their wildtype littermates at birth. However, whereas Atbf1wt/Δ7&8 mice kept growing and gaining weight until they died, it became obvious at day 10 after birth that they were much smaller, and their body weights were significantly lighter (p<0.0001) than the wildtype littermates (Fig. 3c, 3d). These mice were also hypoactive and usually required extended weaning.
Mating mice with heterozygous deletion (Atbf1wt/Δ7&8) with wildtype C57BL/6J mice (Atbf1wt/wt) produced 83 Atbf1wt/wt and 33 Atbf1wt/Δ7&8 pups, which was different from the expected ratio of 1:1 wildtype to heterozygous pups according to Mendelian distribution. A reduction of heterozygous pups by 60.2% suggests that deletion of one Atbf1 allele also causes partial prenatal lethality in mice. When Atbf1wt/Δ7&8 mice were inbred, conception time was prolonged and only two pregnancies were observed so far. No pups were delivered, further suggesting that defects in embryonic development had occurred. Due to the limited number of sexually mature Atbf1wt/Δ7&8 mice and a low pregnancy rate in these mice, we were unable to fully characterize Atbf1 null embryos or mice. In addition, preweaning mortality and growth retardation were not observed when Atbf1 was knocked out in a tissue-specific manner (e.g., the prostate and mammary gland, unpublished data).
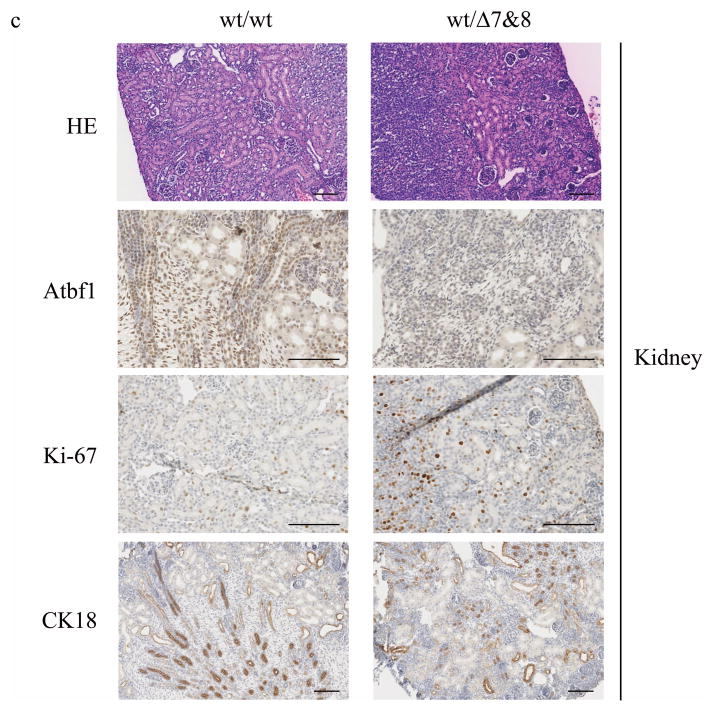
Several mechanisms could be responsible for the postnatal mortality in heterozygous mice, including haploinsufficiency, monoallelic expression, imprinting and dominant negative property of the mutant allele. As the first step in addressing this question, we measured Atbf1 expression at mRNA and protein levels in 8 major organs including the brain, liver, lung, heart, intestine, kidney, spleen and stomach. Tissues were collected from both the wildtype (Atbf1wt/wt) and heterozygous (Atbf1wt/Δ7&8) mice at postnatal day 1 and day 21. Real-time RT-PCR with primers from exon 6 and exon 8 of mouse Atbf1 was performed to measure the Atbf1 mRNA expression. At postnatal day 1, Atbf1 mRNA level was the highest in the brain, whereas at postnatal day 21, the expression level was relatively higher in the liver, lung, heart and kidney and lower in the brain, intestine, spleen and stomach (Fig. 4a). At both time points, Atbf1 mRNA level in the heterozygous (Atbf1wt/Δ7&8) mice was about half of that in the wildtype mice in most organs examined (Fig. 4a). Immunohistochemistry (IHC) for Atbf1 on tissue sections confirmed that deletion of one Atbf1 allele reduced Atbf1 protein expression (Fig. 4c). These results excluded the possibility of monoallelic expression or imprinting, while suggesting that haploinsufficiency of Atbf1 causes preweaning mortality in the Atbf1 heterozygous mice. RT-PCR with a pair of primers from exons 4 and 5 of Atbf1, which amplifies both the wildtype (wt) and the expected truncated (mt) Atbf1 mRNA showed similar mRNA expression in the majority of the tissues at both postnatal day 1 and day 21 (Fig. 4b). Taken together with the results from the primers from exons 6 and 8, these results indicate the existence of a truncated Atbf1 mRNA in these organs of the Atbf1 heterozygous mice. Therefore, we can not exclude the possibility that the Atbf1Δ7&8 allele may produce a truncated protein that interferes with the wildtype Atbf1 function.
Figure 4. Reduced Atbf1 expression in mice with Atbf1 deletion.
a. Knockout of Atbf1 reduced the expression of the wildtype Atbf1 mRNA in most organs of mice at postnatal day 1 (left) and day 21 (right), as detected by real-time RT-PCR. The ratio of Atbf1 to β-actin mRNA levels for wildtype mouse brain was set to 1, and that for other organs were adjusted accordingly. The reduction by about half is obvious in the brain (B), liver (Lv), lung (Lg), heart (H), kidney (K) and stomach (St) but not in the intestine (I) and spleen (Sp) in mice at 21-day-old (right). b. Detection of the truncated Atbf1 mRNA by real-time RT-PCR in the brain, liver, heart, intestine, kidney and spleen but not in the lung and stomach of the Atbf1 heterozygous mice. c. Molecular and histological analyses of mouse kidneys from both Atbf1 wildtype (wt/wt) and heterozygous (wt/Δ7&8) mice. HE, representative images from Hematoxylin and Eosen stained sections. IHC staining confirmed reduced Atbf1 expression, increased Ki-67 expression and attenuated CK18 expression in the Atbf1 heterozygous mice (wt/Δ7&8) when compared to wildtype mice (wt/wt). All scale bars are 100 μm.
Histologically, we did not detect any abnormalities in the organs by comparative pathologic analysis and blood cell counting (Fig. 4c and data not shown). The preweaning mortality and growth retardation were not due to milk sucking or lactation defects, because milk was visible in the stomach of the Atbf1 heterozygous pups. Human ATBF1 could modulate cell proliferation and differentiation, so we measured the expression of Ki-67, a proliferation marker, and cytokeratin 18 (CK18), a differentiation marker, in the Atbf1wt/Δ7&8 mice. In the kidney, loss of one Atbf1 allele caused significant increase in Ki-67 expression while attenuating CK18 expression (Fig. 4c). However, it is unknown whether these changes caused preweaning mortality.
In summary, we developed a mouse strain in which the Atbf1 gene is flanked by loxP sequences (Atbf1flox/wt) so the gene can be conditionally deleted by controlled expression of Cre. Atbf1 is haploinsufficient, and knockout of one allele induced prenatal and preweaning mortality in mice. Availability of mice with a floxed Atbf1 allele could facilitate functional examination of Atbf1 under biological conditions. For example, we specifically knocked out prostate Atbf1 and observed prostatic intraepithelial neoplasia, supporting a tumor suppressor role of Atbf1 (manuscript in preparation).
Methods
Generation of the targeting construct
Genomic DNA of TL-1 mouse embryonic stem cells was used as the template for generating mouse Atbf1 genomic fragments. The 5′ homologous arm was a 6.2-kb fragment from intron 6 of Atbf1, and the 3′ homologous arm was 4.7-kb from intron 8. The targeting sequence was a 0.9-kb fragment spanning part of intron 6, exon 7, intron 7, exon 8 and part of intron 8. All three fragments were amplified by PCR using the LA Taq polymerase (TaKaRa, Berkeley, CA) and were cloned into the T-vector (Promega, Madison, WI). After confirmation by DNA sequencing, the 5′ homologous arm, the targeting sequence, and the 3′ homologous arm were sequentially inserted into the EcoR I, Hind III, and Not I sites of the pFlexible vector (van der Weyden et al., 2005). The gene-targeting construct was then confirmed by DNA sequencing.
Generation of Atbf1flox and Atbf1Δ7&8 mice
After linearization by digestion with the Sfi I restriction enzyme, the gene-targeting construct was electroporated into TL1 embryonic stem cells, which were then cultured in puromycin-containing medium to kill cells without gene integration. Puromycin-resistant clones were picked, expanded sequentially in 96-well and 24-well plates, and screened for homologous recombination by Southern blotting. PCR primers for generating hybridization probes were: 5′-TGGCTGGAGGCGGACTGAGA-3′ and 5′-ATGTTGTGGAGCCTCTGAAAATGG-3′ for the P1 probe and 5′-CATATGCTGGGGTCCTCCTGTGA-3′ and 5′-TGAGGCATGGCAGCACAGTCT-3′ for the P2 probe. The ES clone with correct recombination was then expanded, injected into blastocysts, and implanted into pseudopregnant recipient female mice. Chimeric mice were bred with C57BL/6J mice to obtain mice with germline transmission. The mutant allele was termed Atbf1PGK. Mice with an Atbf1PGK allele were bred with ACT-FLPe transgenic mice (Jackson Laboratory, Bar Harbor, ME) to remove the Frt-flanked PGK-puroΔtk cassette, resulting in the floxed Atbf1 allele Atbf1flox. Heterozygous Atbf1flox/wt mice were further bred to EIIa-Cre transgenic mice (kindly provided by Dr. Tongzhong Ju, Emory University) to delete exons 7 and 8 to produce mice with the deleted Atbf1 allele (Atb1Δ7&8). All alleles were maintained in the 129S6/C57BL/6J background. The generation and usage of the mice were approved by the Emory University Institutional Animal Care and Use Committee.
Genotyping by PCR
PCR was applied to genomic DNA from tail biopsies and MEFs to genotype various alleles of Atbf1 and other genes. PCR products of the Atbf1PGK allele (528 bp) were distinguished from that of the Atbf1wt allele (457 bp) when using primers F1 (5′-GGCCCTTGACTGCATTTCTTTCCTGT-3′) and R1 (5′-CCCAAGACCCAGTGCCCCAAAA-3′). Similarly, alleles of Atbf1flox (1248 bp), Atbf1wt (1071 bp) and Atbf1Δ7&8 (289 bp) were detected in a single PCR by using primers F1 and R2 (5′-ATTCGTTAATGGGAAGGTGTCAGA-3′). Primers used for genotyping other genes included: 5′-CGGTCGATGCAACGAGTGAT-3′ and 5′-CCACCGTCAGTACGTGAGAT-3′ for Cre (529 bp), 5′-CTAGGCCACAGAATTG AAAGATCT-3′ and 5′-GTAGGTGGAAATTCTAGCATCATCC-3′ for Il2 (324 bp), and 5′-CACTGATATTGTAAGTAGTTTGC-3′ and 5′-CTAGTGCGAAGTAGTGATCAGG-3′ for Flp (725 bp).
RT-PCR and real-time RT-PCR
Total RNA was extracted from MEFs or mouse tissues by Trizol reagent (Invitrogen, Carlsbad, CA) and cDNA was synthesized from 1 μg of total RNA using the iScript reverse transcription kit (Bio-Rad, Hercules, CA). PCR primers used for amplifying fragments transcribed from all three Atbf1 alleles (Atbf1flox, 532 bp; Atbf1wt, 532 bp; and Atbf1Δ7&8, 197 bp) were 5′-GGCCAGATCTTCACCATCC-3′ and 5′-CAGGGAGGAACATGCTACTAGG-3′, with the former from exon 5 and the latter from exon 9 of mouse Atbf1. Atbf1 mRNA levels were measured in mouse organs by real-time RT-PCR using 1) primers 5′-AGAGCAAGAGGGCAGCGTCATC-3′ and 5′-CGGTTCACGTCAGCGTTGCTATAC-3′, which span exons 6 to 8 and only amplify the wildtype Atbf1 cDNA; 2) primers 5′-AAGCTGCGGCTGCACACAGTCAAC-3′ and 5′-TCACTGCGCTGGTGCTTCATGG-3′, which span exons 4 and 5 and amplify both the wildtype and the truncated Atbf1 cDNA. Mouse β-actin mRNA, which was used as the internal control, was detected with primers 5′-GATCTGGCACCACACCTTCT-3′ and 5′-GGGGTGTTGAAGGTCTCAAA-3′.
Immunoblotting and immunohistochemistry (IHC)
Atbf1 protein in MEFs was analyzed by immunoblotting. Briefly, MEFs were infected with Cre-expressing or control adenoviruses at an MOI of 200. Cell lysates were then prepared in the RIPA buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 10% glycerol, 20 mM β-glycerophosphate) supplemented with 1% protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). A total of 50 μg protein lysates were separated by 4% SDS-polyacrylamide gel electrophoresis (PAGE) and electrophoretically transferred to nitrocellulose membranes. After incubation with an anti-Atbf1 antibody (kindly provide by Dr. Masakiyo Sasahara of Toyama Medical and Pharmaceutical University, Toyama, Japan) (Ishii et al., 2003), blots were then incubated with horseradish peroxidase-conjugated anti-IgG secondary antibodies (Sigma-Aldrich, St. Louis, MO) and visualized with an enhanced chemiluminescence system (Thermo Scientific Pierce Protein Research Products, Rockford, IL).
Atbf1 protein was analyzed by immunohistochemistry (IHC) in mouse tissues according to standard procedures. Briefly, tissue sections were deparaffinized, rehydrated, and washed in PBS. Antigen retrieval was done by heating slides in a microwave oven for 15 min in a citrated buffer (pH 6.0, 10 mM trisodium citrate). After blocking with 5% normal goat serum in Tris-buffered saline with 0.1% Tween-20 (TBST), tissue sections were incubated with primary antibodies at 4°C overnight, followed by an incubation with EnVision Polymer-HPR secondary antibodies (Dako, Glostrup, Denmark) at room temperature for 40 min. After the application of DAB-chromogen, tissue sections were mounted and visualized under microscopes. Primary antibodies used in this study included anti-Atbf1 (Ishii et al., 2003), anti-Ki-67 (Lab Vision of Thermo Scientific, Kalamazoo, MI), and anti-CK18 (GeneTex, San Antonio, TX) antibodies.
Body weight measurement
Body weights were measured for the Atbf1 heterozygous mice and their wildtype littermates aged at 13 to 29 days. Only a few litters were followed at multiple time points because of the frequent mortality. Body weights of the two groups (Atbf1wt/wt vs. Atbf1wt/Δ7&8) at different ages were graphically plotted against ages, and a general linear model (GLM) was used to test whether there was a significant difference in mouse body weight between the two groups. A P value of 0.05 was considered significant. The SAS statistical package V9.2 (SAS Institute Inc., Cary, NC) was used for data management and analysis.
Acknowledgments
Contract grant sponsor: NIH, Grant number: CA121459 (JT Dong).
We thank Dr. Tamas Nagy of University of Georgia College of Veterinary Medicine for comparative pathologic examination and Dr. Mohammad S. Hossain of Emory University School of Medicine for blood analyses. We thank Dr. Lily Yang of Emory University School of Medicine for providing the Ad-Cre virus.
References
- Benjamin EJ, Rice KM, Arking DE, Pfeufer A, van Noord C, Smith AV, Schnabel RB, Bis JC, Boerwinkle E, Sinner MF, Dehghan A, Lubitz SA, D’Agostino RB, Sr, Lumley T, Ehret GB, Heeringa J, Aspelund T, Newton-Cheh C, Larson MG, Marciante KD, Soliman EZ, Rivadeneira F, Wang TJ, Eiriksdottir G, Levy D, Psaty BM, Li M, Chamberlain AM, Hofman A, Vasan RS, Harris TB, Rotter JI, Kao WH, Agarwal SK, Stricker BH, Wang K, Launer LJ, Smith NL, Chakravarti A, Uitterlinden AG, Wolf PA, Sotoodehnia N, Kottgen A, van Duijn CM, Meitinger T, Mueller M, Perz S, Steinbeck G, Wichmann HE, Lunetta KL, Heckbert SR, Gudnason V, Alonso A, Kaab S, Ellinor PT, Witteman JC. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet. 2009;41:879–881. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry FB, Miura Y, Mihara K, Kaspar P, Sakata N, Hashimoto-Tamaoki T, Tamaoki T. Positive and negative regulation of myogenic differentiation of C2C12 cells by isoforms of the multiple homeodomain zinc finger transcription factor ATBF1. J Biol Chem. 2001;276:25057–25065. doi: 10.1074/jbc.M010378200. [DOI] [PubMed] [Google Scholar]
- Cho YG, Song JH, Kim CJ, Lee YS, Kim SY, Nam SW, Lee JY, Park WS. Genetic alterations of the ATBF1 gene in gastric cancer. Clin Cancer Res. 2007;13:4355–4359. doi: 10.1158/1078-0432.CCR-07-0619. [DOI] [PubMed] [Google Scholar]
- Dong JT. Chromosomal deletions and tumor suppressor genes in prostate cancer. Cancer Metastasis Rev. 2001;20:173–193. doi: 10.1023/a:1015575125780. [DOI] [PubMed] [Google Scholar]
- Dong XY, Sun X, Guo P, Li Q, Sasahara M, Ishii Y, Dong JT. ATBF1 inhibits estrogen receptor (ER) function by selectively competing with AIB1 for binding to the ER in ER-positive breast cancer cells. J Biol Chem. 2010;285:32801–32809. doi: 10.1074/jbc.M110.128330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, Gulcher J, Mathiesen EB, Njolstad I, Nyrnes A, Wilsgaard T, Hald EM, Hveem K, Stoltenberg C, Kucera G, Stubblefield T, Carter S, Roden D, Ng MC, Baum L, So WY, Wong KS, Chan JC, Gieger C, Wichmann HE, Gschwendtner A, Dichgans M, Kuhlenbaumer G, Berger K, Ringelstein EB, Bevan S, Markus HS, Kostulas K, Hillert J, Sveinbjornsdottir S, Valdimarsson EM, Lochen ML, Ma RC, Darbar D, Kong A, Arnar DO, Thorsteinsdottir U, Stefansson K. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ido A, Miura Y, Tamaoki T. Activation of ATBF1, a multiple-homeodomain zinc-finger gene, during neuronal differentiation of murine embryonal carcinoma cells. Dev Biol. 1994;163:184–187. doi: 10.1006/dbio.1994.1134. [DOI] [PubMed] [Google Scholar]
- Ishii Y, Kawaguchi M, Takagawa K, Oya T, Nogami S, Tamura A, Miura Y, Ido A, Sakata N, Hashimoto-Tamaoki T, Kimura T, Saito T, Tamaoki T, Sasahara M. ATBF1-A protein, but not ATBF1-B, is preferentially expressed in developing rat brain. J Comp Neurol. 2003;465:57–71. doi: 10.1002/cne.10807. [DOI] [PubMed] [Google Scholar]
- Jung CG, Kim HJ, Kawaguchi M, Khanna KK, Hida H, Asai K, Nishino H, Miura Y. Homeotic factor ATBF1 induces the cell cycle arrest associated with neuronal differentiation. Development. 2005;132:5137–5145. doi: 10.1242/dev.02098. [DOI] [PubMed] [Google Scholar]
- Kataoka H, Joh T, Miura Y, Tamaoki T, Senoo K, Ohara H, Nomura T, Tada T, Asai K, Kato T, Itoh M. AT motif binding factor 1-A (ATBF1-A) negatively regulates transcription of the aminopeptidase N gene in the crypt-villus axis of small intestine. Biochem Biophys Res Commun. 2000;267:91–95. doi: 10.1006/bbrc.1999.1911. [DOI] [PubMed] [Google Scholar]
- Kataoka H, Miura Y, Joh T, Seno K, Tada T, Tamaoki T, Nakabayashi H, Kawaguchi M, Asai K, Kato T, Itoh M. Alpha-fetoprotein producing gastric cancer lacks transcription factor ATBF1. Oncogene. 2001;20:869–873. doi: 10.1038/sj.onc.1204160. [DOI] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wang F, Yang Y, Fu F, Xu C, Shi L, Li S, Xia Y, Wu G, Cheng X, Liu H, Wang C, Wang P, Hao J, Ke Y, Zhao Y, Liu M, Zhang R, Gao L, Yu B, Zeng Q, Liao Y, Yang B, Tu X, Wang QK. Significant association of SNP rs2106261 in the ZFHX3 gene with atrial fibrillation in a Chinese Han GeneID population. Hum Genet. 2011;129:239–246. doi: 10.1007/s00439-010-0912-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuchi M, Kataoka H, Miura Y, Kim TS, Kawaguchi M, Ebi M, Tanaka M, Mori Y, Kubota E, Mizushima T, Shimura T, Mizoshita T, Tanida S, Kamiya T, Asai K, Joh T. Tumor suppressor, AT motif binding factor 1 (ATBF1), translocates to the nucleus with runt domain transcription factor 3 (RUNX3) in response to TGF-beta signal transduction. Biochem Biophys Res Commun. 2010;398:321–325. doi: 10.1016/j.bbrc.2010.06.090. [DOI] [PubMed] [Google Scholar]
- Miura Y, Tam T, Ido A, Morinaga T, Miki T, Hashimoto T, Tamaoki T. Cloning and characterization of an ATBF1 isoform that expresses in a neuronal differentiation-dependent manner. J Biol Chem. 1995a;270:26840–26848. doi: 10.1074/jbc.270.45.26840. [DOI] [PubMed] [Google Scholar]
- Miura Y, Tam T, Ido A, Morinaga T, Miki T, Hashimoto T, Tamaoki T. Cloning and characterization of an ATBF1 isoform that expresses in a neuronal differentiationdependent manner. J Biol Chem. 1995b;270:26840–26848. doi: 10.1074/jbc.270.45.26840. [DOI] [PubMed] [Google Scholar]
- Mori Y, Kataoka H, Miura Y, Kawaguchi M, Kubota E, Ogasawara N, Oshima T, Tanida S, Sasaki M, Ohara H, Mizoshita T, Tatematsu M, Asai K, Joh T. Subcellular localization of ATBF1 regulates MUC5AC transcription in gastric cancer. Int J Cancer. 2007;121:241–247. doi: 10.1002/ijc.22654. [DOI] [PubMed] [Google Scholar]
- Morinaga T, Yasuda H, Hashimoto T, Higashio K, Tamaoki T. A human alpha-fetoprotein enhancer-binding protein, ATBF1, contains four homeodomains and seventeen zinc fingers. Mol Cell Biol. 1991;11:6041–6049. doi: 10.1128/mcb.11.12.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Frierson HF, Chen C, Li C, Ran Q, Otto KB, Cantarel BL, Vessella RL, Gao AC, Petros J, Miura Y, Simons JW, Dong JT. Frequent somatic mutations of the transcription factor ATBF1 in human prostate cancer. Nat Genet. 2005;37:407–412. doi: 10.1038/ng1528. [DOI] [PubMed] [Google Scholar]
- Sun X, Zhou Y, Otto KB, Wang M, Chen C, Zhou W, Subramanian K, Vertino PM, Dong JT. Infrequent mutation of ATBF1 in human breast cancer. J Cancer Res Clin Oncol. 2007;133:103–105. doi: 10.1007/s00432-006-0148-y. [DOI] [PubMed] [Google Scholar]
- van der Weyden L, Adams DJ, Harris LW, Tannahill D, Arends MJ, Bradley A. Null and conditional semaphorin 3B alleles using a flexible puroDeltatk loxP/FRT vector. Genesis. 2005;41:171–178. doi: 10.1002/gene.20111. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Miura Y, Ido A, Sakai M, Nishi S, Inoue Y, Hashimoto T, Tamaoki T. Developmental changes in expression of the ATBF1 transcription factor gene. Brain Res Mol Brain Res. 1996;42:344–349. doi: 10.1016/s0169-328x(96)00204-5. [DOI] [PubMed] [Google Scholar]
- Xu J, Sauvageot J, Ewing CM, Sun J, Liu W, Isaacs SD, Wiley KE, Diaz L, Zheng SL, Walsh PC, Isaacs WB. Germline ATBF1 mutations and prostate cancer risk. Prostate. 2006;66:1082–1085. doi: 10.1002/pros.20430. [DOI] [PubMed] [Google Scholar]
- Yasuda H, Mizuno A, Tamaoki T, Morinaga T. ATBF1, a multiple-homeodomain zinc finger protein, selectively down-regulates AT-rich elements of the human alphafetoprotein gene. Mol Cell Biol. 1994;14:1395–1401. doi: 10.1128/mcb.14.2.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]